Abstract
Candida glabrata, a yeast with intrinsically low susceptibility to azoles, frequently develops increased azole resistance during prolonged treatment. Transposon mutagenesis revealed that disruption of CgPDR1 resulted in an 8- to 16-fold increase in fluconazole susceptibility of C. glabrata. CgPDR1 is a homolog of Saccharomyces cerevisiae PDR1, which encodes a transcriptional regulator of multidrug transporters. Northern blot analyses indicated that CgPDR1 regulated both constitutive and drug-induced expression of CgCDR1, a multidrug transporter gene. In agreement with the Northern analysis, the Cgpdr1 mutant had increased rhodamine accumulation, in contrast to the decreased accumulation in the CgPDR1-overexpressing strain. Northern analyses also indicated the importance of CgPDR1 in fluconazole resistance arising during therapy. Two clinically resistant isolates had higher expression of CgPDR1 and CgCDR1 compared to their paired susceptible isolates. Integrative transformation of CgPDR1 from the two resistant isolates converted the Cgpdr1 mutant into azole-resistant strains with upregulated CgPDR1 expression. Two different amino acid substitutions, W297S in one isolate and F575L in the other, accounted for the upregulated CgPDR1 expression and the resistance. Finally, CgPDR1 was shown to be required for the azole resistance due to mitochondrial deficiency. Thus, CgPDR1 encodes a transcriptional regulator of a pleiotropic drug resistance network and contributes to the azole resistance of clinical isolates and petite mutants.
Candida glabrata has emerged as a common cause of bloodstream and mucosal infections in many countries, and it exhibits intrinsically low susceptibility to fluconazole (29, 35). This species is naturally about eightfold more resistant to fluconazole than C. albicans and easily develops further fluconazole resistance following prolonged therapy of patients with fluconazole (2, 30).
Azole antifungals target cytochrome P-450-dependent C14 lanosterol demethylase encoded by ERG11 and interfere with ergosterol biosynthesis (17, 23, 43). Inhibition of ergosterol biosynthesis leads to the depletion of ergosterol, the major component of the fungal plasma membrane, and possibly accumulation of toxic sterol intermediates. The major mechanisms of azole resistance described to date have been primarily based on studies done in C. albicans and Saccharomyces cerevisiae and include overexpression of C14 lanosterol demethylase (13), alteration of the azole-binding site of C14 lanosterol demethylase (19, 22, 37), mutation in other ergosterol biosynthetic genes (18), and increased drug efflux. Increased drug efflux in Candida species is mainly due to increased expression of ATP-binding cassette (ABC) and major facilitator superfamily transporters (25, 28, 31, 34, 40). In addition to studies of the transporter and ergosterol biosynthetic genes at a transcriptional level, there also have been studies of posttranscriptional events in the genes of C. glabrata. Fluconazole treatment was shown to increase the presence of CgErg11p and CgCdr1p in the plasma membrane, and activity of CgCdr1p was modulated by phosphorylation (27, 39).
There are two lines of evidences which suggest that CgPDR1 is a transcriptional regulator of ABC transporters and plays a central role in fluconazole resistance acquired by C. glabrata during azole therapy or through mitochondrial dysfunction. High-frequency acquired resistance (HFAR) to fluconazole has been reported in C. glabrata and S. cerevisiae in the laboratory setting (4, 5, 7, 16, 33, 44), and overexpression of fluconazole-effluxing ABC transporters in the petite mutants has been documented in both S. cerevisiae and C. glabrata (33, 44). Two transcriptional factors, Pdr1p and Pdr3p, are known to regulate the pleiotropic drug resistance (PDR) network in S. cerevisiae (1, 9, 15). To date, PDR3 but not PDR1 has been shown to be essential for HFAR in S. cerevisiae (12, 44). However, the homolog of PDR1 but not PDR3 was annotated in the recently published C. glabrata genome sequences from the Pasteur Institute (11). The other line of evidence comes from Vermitsky and Edlind (38), who reported that expression of CgPDR1 was increased in one of seven C. glabrata mutants selected in vitro for fluconazole resistance. The strain with increased CgPDR1 expression differed from the parental Pdr1p by a single deduced amino acid, P927L, in the activation domain, raising the question of whether this mutation had caused the resistance.
We report here the role of CgPDR1 in fluconazole resistance using several approaches. We studied Cgpdr1 mutants obtained by transposon mutagenesis (36) and by targeted gene disruption, including Cgpdr1 mutants in a petite background. We also sequenced CgPDR1 and its 5′ flanking region in clinical isolates obtained before and after the appearance of fluconazole resistance (2). Sequence analysis and gene substitution experiments found that azole resistance and CgPDR1 expression were determined by single amino acid differences in the CgPDR1 open reading frame (ORF). The results indicate the central role of CgPDR1 in fluconazole resistance arising both during clinical use and through mitochondrial dysfunction.
MATERIALS AND METHODS
Strains and culture conditions.
Plasmids were maintained in Escherichia coli XL1-Blue (Stratagene, La Jolla, CA), Top 10 (Invitrogen, Carlsbad, CA), or Top 10F′ (Invitrogen) host cells grown in 50 μg/ml ampicillin or 12.5 μg/ml chloramphenicol. EC100D pir+ and EC100D pir-116 (Epicenter, Madison, WI) were used as hosts for the EZ::TN transposon (Tn) (Epicenter) which relies on the R6K origin for replication.
C. glabrata strains (Table 1) were cultured on either YPD, containing 1% Bacto yeast extract (Difco Laboratories, Detroit, MI), 2% Bacto peptone (Difco Laboratories), and 2% glucose (Sigma, St. Louis, MO), or MIN, containing 0.67% yeast nitrogen base plus 2% glucose without amino acids (Difco Laboratories). YEPG agar was used for drug sensitivity assay and petite growth phenotype, containing 1% Bacto yeast extract (Difco Laboratories), 2% Bacto peptone (Difco Laboratories), 3% glycerol (Invitrogen), 1% ethanol (Warner-Graham Inc., Cockeysville, MD), and 2% agar (Difco Laboratories).
TABLE 1.
Strains used in this study
| Strain | Parental strain | Genotype and/or description | Reference or source |
|---|---|---|---|
| Candida glabrata | |||
| 38a | Clinical isolate | 25 | |
| NCCLS84 | Wild type | ATCC 90030 | |
| 84u | NCCLS84 | ura3 | 14 |
| 84870 | 84u | Cgcdr1::URA3 pdh1Δ::ura3 | 14 |
| Cg12581 | Clinical isolate, pair 1 | 2 | |
| Cg13928 | Clinical resistant isolate, pair1 | 2 | |
| Cg1660 | Clinical isolate, pair 4 | 2 | |
| Cg4672 | Clinical resistant isolate, pair 4 | 2 | |
| Cg1660u | Cg1660 | ura3 | 36 |
| CgTn173S | Cg1660u | ura3 Cgpdr1::Tn5<Cm URA3> | This study |
| CgB4 | 84u | ura3 Cgpdr1::Tn5<Cm URA3> | This study |
| CgB4u | CgB4 | ura3 Cgpdr1::Tn5<Cm ura3> | This study |
| Cg173Cu | CgB4 | ura3CgPDR1-13928a | This study |
| Cg173C | Cg173Cu | URA3 CgPDR1-13928 URA3 revertant of Cg173Cu | This study |
| CgB4u/pCgACU | CgB4u | ura3 Cgpdr1::Tn5<Cm ura3> URA3 | This study |
| CgB4u/pCgPDR1 | CgB4u | ura3 Cgpdr1::Tn5<Cm ura3> URA3 ADH1p-CgPDR138a-ADH1T | This study |
| CgB4Ca12581 | CgB4 | ura3 CgPDR1-12581a | This study |
| CgB4Cb12581 | CgB4 | ura3 CgPDR1-12581a | This study |
| CgB4Ca13928 | CgB4 | ura3 CgPDR1-13928a | This study |
| CgB4Cb13928 | CgB4 | ura3 CgPDR1-13928a | This study |
| CgB4Ca1660 | CgB4 | ura3 CgPDR1-1660a | This study |
| CgB4Cb1660 | CgB4 | ura3 CgPDR1-1660a | This study |
| CgB4Ca4672 | CgB4 | ura3 CgPDR1-4672a | This study |
| CgB4Cb4672 | CgB4 | ura3 CgPDR1-4672a | This study |
| 84p1 | NCCLS84 | Mitochondrion-deficient mutant | This study |
| 84p2 | NCCLS84 | Mitochondrion-deficient mutant | This study |
| 84p3 | NCCLS84 | Mitochondrion-deficient mutant | This study |
| CgB4p3 | CgB4 | Mitochondrion-deficient mutant | This study |
| CgB4p4 | CgB4 | Mitochondrion-deficient mutant | This study |
| CgB4p5 | CgB4 | Mitochondrion-deficient mutant | This study |
| Saccharomyces cerevisiae | |||
| BY4741 | MATa his3Δ1 leu2Δ0 metΔ0 ura3Δ0 | 3 | |
| 4381 | BY4741 | MATa his3Δ1 leu2Δ0 metΔ0 ura3Δ0 pdr1 | 41 |
| 4381/pH 392 | 4381 | MATa his3Δ1 leu2Δ0 metΔ0 ura3Δ0 pdr1 URA3 | This study |
| 4381/pHCgPDR1 | 4381 | MATa his3Δ1 leu2Δ0 metΔ0 ura3Δ0 pdr1 URA3 ADH1p-CgPDR138a-ADH1T | This study |
Complementation by integrative transformation.
The two pairs of clinical isolates were selected because each pair came from a patient receiving fluconazole; the strains acquired increased fluconazole resistance during therapy, while the karyotype remained unchanged (2).
The Cgpdr1 mutant, CgTn173S, was generated by transposon mutagenesis using the custom transposon Tn5<Cm URA3>, as described by Tsai et al. (36). The ura3 mutants were selected on a MIN plus uracil agar plate containing 0.1% 5-fluoroorotic acid (FOA) (Lancaster, Pelham, NH).
Candida glabrata petite mutants were obtained from cultures grown on a YPD agar plate containing 40 μg/ml ethidium bromide for 6 days at 30°C. The petite phenotype was confirmed by inability to grow on YEPG agar.
A haploid S. cerevisiae pdr1 mutant, 4381, and its parental strain, BY4741, were obtained from Open Biosystems (Huntsville, AL) (41).
Identification of the Tn-inserted gene.
The plasmid pTn173S (Table 2) was rescued from the Cgpdr1 mutant, CgTn173S. Briefly, the Tn-inserted genomic DNA of CgTn173S was digested with BglII, self ligated, and transformed into E. coli EC100D pir+. The rescued plasmid pTn173S was sequenced using the Tn primers SqFP and SqRP (Epicenter) to obtain the sequence information of the Tn insertion site. Nucleotide sequences obtained were then used for BlastX searches against the protein database to identify homologs. The plasmid pTn173S was found to contain sequences homologous to S. cerevisiae PDR1. The full nucleotide sequence of the C. glabrata PDR1 homolog (CgPDR1) was obtained from the Pasteur Institute genomic database (Génolevures Consortium; http://cbi.labri.fr/Genolevures/). We also sequenced CgPDR1 from two pairs of clinical isolates.
TABLE 2.
Plasmids used in this study
| Strain | Genotype and/or description | Reference or source |
|---|---|---|
| pCC1FOS | Copy control fosmid vector | Epicentre |
| pCC1URA3 | Copy control fosmid vector | 36 |
| pCgACU | C. glabrata centromere vector | 20 |
| pH 392 | S. cerevisiae centromere overexpression vector | H. Edskesa |
| pTn173S | Plasmid rescued from CgTn173S Cgpdr1::Tn5<Cm URA3> | This study |
| P16C3 | Cg13928 CgPDR1 genomic fosmid clone | This study |
| P2F5 | Cg4672 CgPDR1 genomic fosmid clone | This study |
| pCgPDR138a | 38a CgPDR1 genomic clone in pBluscript SK− | This study |
| pCgPDR1 | ADH1p-CgPDR138a-ADH1T in pCgACUb | This study |
| pHCgPDR1 | ADH1p-CgPDR138a-ADH1T in pH 392b | This study |
| pCgPDR112581 | CgPDR1 ORF of Cg12581 in pCR-Blunt II-TOPO | This study |
| pCgPDR113928 | Cg13928 CgPDR1 gene in pCgACU | This study |
| pCgPDR11660 | CgPDR1 ORF of Cg1660 in pCR-Blunt II-TOPO | This study |
| pCgPDR14672 | Cg4672 CgPDR1 gene in pCgACU | This study |
A kind gift of Herman Edskes, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD.
P, promoter; T, terminator.
CgPDR1 disruption.
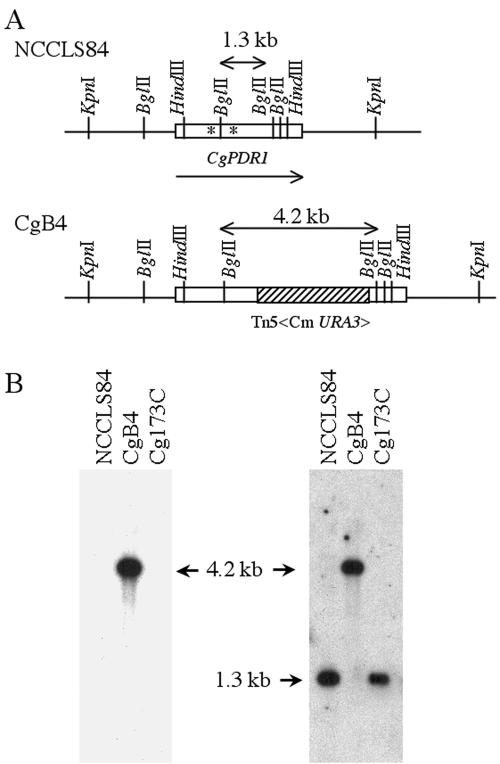
To confirm the association of Cgpdr1 mutations and increases in fluconazole sensitivity, the rescued plasmid, pTn173S, was linearized with BglII and transformed into 84u (ura3 mutant of the wild-type strain NCCLS84) to construct a Cgpdr1 disruptant via a double-crossover homologous recombination. The disruption of CgPDR1 was confirmed by Southern blot analysis of the BglII-digested genomic DNA. The purified 2.9-kb Tn probe detected a single 4.2-kb signal in the putative Cgpdr1 mutant CgB4, while there was no signal in the parental strain NCCLS84 (Fig. 1B, left panel). The 1.3-kb CgPDR1 probe detected the 1.3-kb signal in NCCLS84 but only hybridized to the 4.2-kb DNA in CgB4 (Fig. 1B, right panel). Cg173C, a CgPDR1-complemented strain, was obtained by introducing the 8-kb KpnI genomic DNA containing the CgPDR1 gene of Cg13928 into the Cgpdr1 locus of the mutant, CgB4, via a targeted gene replacement (Fig. 1).
FIG. 1.
Southern hybridization analysis confirmed the disruption and targeted replacement of PDR1 in C. glabrata. (A) Restriction enzyme map. (B) Southern blot analysis. Total DNA was digested with BglII. The membrane was hybridized with the 2.9-kb Tn5<Cm URA3> (left panel) or 1.3-kb BglII CgPDR1 (right panel) probe. Lanes: 1, strain NCCLS84, C. glabrata laboratory wild-type strain; 2, strain CgB4, Cgpdr1 mutant; 3, strain Cg173C, CgB4 complemented with CgPDR1 of clinical fluconazole-resistant isolate Cg13928. An asterisk indicates the codon mutations in the clinical fluconazole-resistant isolates, W297S in Cg4672 and F575L in Cg13928.
Drug sensitivity assay.
Susceptibilities to fluconazole and voriconazole (both courtesy of Pfizer, Sandwich, United Kingdom) and itraconazole (Janssen Pharmaceuticals, Titusville, NJ) were tested using the NCCLS microtiter test using 80% growth reduction (MIC80) as the MIC and modified by addition of 2% glucose to the RPMI media and incubation at 37°C for 48 h (26). Disk diffusion susceptibility to cerulenin (Sigma), chloramphenicol (Sigma), cycloheximide (Sigma), and rhodamine 6G (R6G; Sigma) was tested by adding 10 μl of drug solution to 6-mm paper disks (Becton Dickinson, Sparks, MD), and we placed the disks on the agar plate seeded with 2 × 105 cells. The plates were photographed after 2 days of incubation at 30°C.
Complementation using CgPDR1 from different clinical isolates.
A fosmid genomic library of Cg13928, a fluconazole-resistant clinical isolate, was constructed in copy control fosmid vector pCC1URA3 as described in Tsai et al. (36). The average insert size of the genomic library was about 40 kb. The Cg4672 fosmid genomic library was constructed similarly, except it was in a different copy control fosmid vector, pCC1FOS (Epicenter). For functional complementation, the fosmid genomic clones P16C3 (Cg13928) and P2F5 (Cg4672), carrying the full-length CgPDR1 gene, were identified via PCR screening of the fosmid genomic library using the primer set CgPDR2S (5′-TATCCTAAGTATGGACAACG-3′) and CgPDR4AS (5′-GATTCCTTAAGCCCGATAAG-3′) with the following parameters: 95°C for 2 min; 35 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 2 min; with extension on the last cycle at 72°C for 10 min.
To complement the Tn-disrupted Cgpdr1 in CgB4, P16C3 was digested with KpnI, and the 8-kb KpnI fragment containing the CgPDR1 ORF, 2.5 kb of the 5′ upstream region, and 1.9 kb of the 3′ downstream region was used to transform CgB4. Transformants were plated on MIN plus uracil agar plates containing 100 μg/ml fluconazole and incubated at 30°C for 3 days; the transformants were then replica plated onto MIN plus uracil agar plates containing 0.1% FOA. FOA-resistant transformants were obtained and analyzed by PCR using primers CgPDR2S and CgPDR4AS with the above conditions. Southern hybridization and DNA sequence analysis confirmed that the Tn-inserted Cgpdr1 was replaced with the intact CgPDR1 gene of P16C3, including the 5′ upstream region via a targeted gene replacement in the complemented strain, Cg173C (Fig. 1). Absence of a tandem repeat was also confirmed by Southern blot analysis (data not shown).
The CgPDR1 ORF of Cg12581 and Cg1660 was obtained by PCR using Pfu Ultra DNA polymerase (Stratagene) with primers CgPDR8S (5′-GGTGGAGCTCTTTAGCTACGTTATTGAG) and CgPDR5AS (5′-GGTTACACCACTACTAGTTG). The 3.6-kb PCR product was cloned into pCR-BluntII-TOPO (Invitrogen) to yield the plasmids pCgPDR112581 and pCgPDR11660, which were sequenced and confirmed. The plasmid pCgPDR113928, containing the CgPDR1 gene of Cg13928, was obtained by subcloning the 8-kb KpnI DNA fragment from the fosmid clone P16C3 into pCgACU (20). Similarly, the plasmid pCgPDR14672, containing the CgPDR1 gene of Cg4672, was obtained by subcloning the 8-kb KpnI DNA fragment from the fosmid clone P2F5 into pCgACU. The 2.9-kb HindIII fragments of the susceptible (Cg12581 and Cg1660) and resistant (Cg13928 and Cg4672) clinical isolates were purified from HindIII-digested pCgPDR112581, pCgPDR11660, pCgPDR113928, and pCgPDR14672; the 2.9-kb HindIII fragments containing the partial CgPDR1 ORF were then used to transform the Cgpdr1 mutant CgB4, which is highly susceptible to fluconazole. Putative transformants were obtained based on the restoration of wild-type fluconazole susceptibility at 50 μg/ml and loss of URA3. Briefly, the transformants were plated on a MIN plus uracil agar plate containing 50 μg/ml fluconazole and incubated at 30°C for 3 days; they were then replica plated onto a MIN plus uracil agar plate containing 0.1% FOA. Restoration of CgPDR1 could only result from the targeted integration of the partial CgPDR1 into the mutated Cgpdr1 locus. An ectopic integration of the partial CgPDR1 could not result in an intact CgPDR1. Two independent PCR amplifications using Pfu Ultra DNA polymerase and DNA sequencing confirmed the targeted replacement of CgPDR1 ORF in the transformants.
CgPDR1 overexpression.
The promoter of S. cerevisiae ADH1 was used for CgPDR1 overexpression in both S. cerevisiae and C. glabrata. The CgPDR1 ORF, along with the 24-bp 5′ upstream region and 335-bp 3′ downstream region, was obtained by PCR using Pfu Turbo DNA polymerase with primers CgPDR5AS (5′-GGTTACACCACTACTAGTTG) and CgPDR8S (5′-GGTGGAGCTCTTTAGCTACGTTATTGAG). The underlined SacI site was created for cloning purposes. The 3.7-kb PCR product was cloned into pCRScript Amp (Stratagene). DNA sequencing confirmed the sequence accuracy of the 0.5-kb insert at both ends. The 3.7-kb CgPDR1 insert was excised with NotI (blunt-ended with T4 DNA polymerase)-SacI and was subcloned into the HindIII (also blunt-ended)-SacI site of the yeast centromere overexpression vector pH392. To avoid mutations introduced by PCR in the nonsequenced coding region, the 2.9-kb HindIII coding region was replaced with the 2.9-kb HindIII fragment from the CgPDR1 genomic clone pCgPDR138a to give the construct pHCgPDR1. The plasmid pCgPDR138a was obtained from a custom genomic library of C. glabrata isolate 38a (Stratagene) (25) by hybridization screening using the CgPDR1-specific probe. For overexpression in C. glabrata, the overexpression construct pCgPDR1 was obtained by excising the CgPDR1 overexpression cassette from pHCgPDR1 with BglI plus SpeI and subcloning into the SmaI site of the C. glabrata centromere vector pCgACU.
Rhodamine 6G accumulation analysis.
Accumulation of rhodamine 6G (R6G) (Sigma) was measured by flow cytometry in a FACSCalibur fluorescence-activated cell cytometer (Becton Dickinson, San Jose, CA) (14, 24). Overnight cultures grown at 30°C in MIN were diluted to an optical density at 600 nm of 1 with MIN and shaken an additional 2 h at 30°C. Rhodamine 6G (R6G) was added to give a final concentration of 0.2 μg/ml and was shaken an additional 4 h at 30°C. After incubation, 10 μl of the culture was transferred to 0.9 ml of ice-cold phosphate-buffered saline (PBS) at pH 7.0 and incubated on ice for 5 min before fluorescent-activated cell sorter (FACS) analysis. A total of 20,000 cells were scanned using the 488-nm laser and FL-2 filter. Cultures without R6G were also analyzed and served as unstained controls. Data were analyzed with CellQuest (Becton Dickinson) and FlowJo programs (Tree Star Inc., San Carols, CA). Fluorescence was expressed as geometric mean values.
Effect of drugs on CgCDR1 expression.
A C. glabrata overnight culture was refreshed in MIN broth and incubated for 3 h in a 30°C shaker; it was subsequently incubated for an additional 2 h after addition of 1 μg/ml of oligomycin (Sigma), 1 μg/ml of cycloheximide, or 100 μg/ml of rhodamaine 6G prior to extraction of RNA. C. glabrata RNA was isolated from log-phase cultures with a FastRNA Pro-Red kit (QBIOgene, Carlsbad, CA).
Techniques and reagents.
C. glabrata genomic DNA was isolated from overnight cultures using the MasterPure Yeast Purification kit (Epicenter). Purified DNA fragments were recovered using a Geneclean Spin kit (QBIOgene). Hybond-N nylon membranes (Amersham, Arlington Heights, IL) were used for Southern and Northern blot analyses. DNA probes were labeled with [α-32P]dCTP or [α-32P]dATP (Amersham) using the Prime-It II kit. DNA cloning and hybridization analyses were done according to the standard protocols (32). DNA sequencing was done using a DNA sequencing kit with a dRhodamine terminator (Applied Biosystems, Foster City, CA) and an ABI automatic DNA Sequencing system (Perkin-Elmer, Foster City, CA). For sequencing of PCR products, Pfu Ultra DNA polymerase (Stratagene) was used for PCR amplification to minimize the rate of PCR-introduced mutation. The PCR products were cleaned with a Strataprep PCR purification kit (Stratagene) and used as templates for DNA sequencing.
Nucleotide sequence accession numbers. The sequence data determined in the course of this work have been submitted to the GenBank under accession numbers DQ174090 (Cg12581), DQ174091 (Cg13928), DQ174092 (Cg1660), and DQ174093 (Cg4672).
RESULTS
CgPDR1 affected azole susceptibility and complemented the S. cerevisiae pdr1 mutant.
The Cgpdr1 mutant CgTn173S obtained via transposon (Tn) mutagenesis showed increased sensitivity to fluconazole compared to its parental clinical isolate, Cg1660 (Table 3). Southern blot analysis using the transposon probe showed that there was a single transposon insertion in CgTn173S (data not shown). The inserted Tn along with the flanking genomic DNA from CgTn173S was recovered and sequenced to characterize the gene disrupted by the transposon. BlastX search of GenBank with the obtained sequences indicated that the sequences had the greatest homology with S. cerevisiae PDR1, which encodes a zinc finger transcriptional factor of 1,068 amino acids. Protein sequence comparison using the GCG Bestfit program showed that the putative protein had 40% identity and 52% similarity with Pdr1p versus 35% identity and 47% similarity with Pdr3p. Therefore, the gene was designated CgPDR1, which encodes a putative protein of 1,107 amino acids. The transposon had inserted at the codon of amino acid 748.
TABLE 3.
Azole susceptibility of C. glabrata strains
| Strain | MIC80 (μg/ml) of:
|
||
|---|---|---|---|
| Fluconazole | Voriconazole | Itraconazole | |
| 38a | 32 | NDb | ND |
| Cg12581 | 16 | ND | ND |
| Cg13928 | 128 | ND | ND |
| Cg1660 | 32-64 | ND | ND |
| Cg4672 | 256 | ND | ND |
| CgTn173S | 4 | 0.0625 | 0.5 |
| NCCLS84 | 64 | 1 | 4-8 |
| CgB4 | 4-8 | 0.125 | 0.5 |
| CgB4u/pCgACU | 4-8 | 0.0625 | 0.5 |
| CgB4u/pCgPDR1 | 512 | 8 | >32 |
| Cg173C | 512 | 4-8 | >32 |
| NCCLS84 | 64a | ND | ND |
| CgB4 | 16a | ND | ND |
| CgB4Ca12581 | 32-64a | ND | ND |
| CgB4Cb12581 | 32-64a | ND | ND |
| CgB4Ca13928 | 256a | ND | ND |
| CgB4Cb13928 | 256-512a | ND | ND |
| CgB4Ca1660 | 64a | ND | ND |
| CgB4Cb1660 | 32-64a | ND | ND |
| CgB4Ca4672 | 512a | ND | ND |
| CgB4Cb4672 | >512a | ND | ND |
RPMI medium supplemented with 8 μg/ml uracil.
ND, not determined.
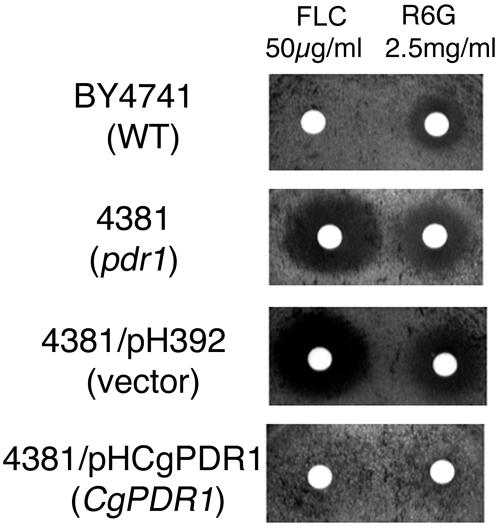
CgPDR1 was disrupted in a laboratory strain to further confirm the association of Cgpdr1 mutations and increased fluconazole susceptibility. The Cgpdr1 disruptant CgB4 had 8- to 16-fold increased susceptibility to the azoles fluconazole, voriconazole, and itraconazole compared to its parental strain, NCCLS84 (Fig. 1 and Table 3). Complementation of the Cgpdr1 disruption by overexpressing CgPDR1 with the S. cerevisiae ADH1 promoter conferred drug resistance. CgB4u, the ura3 mutant of CgB4, transformed with pCgACU, the shuttle vector, was used as a negative control and did not reduce azole susceptibility. In contrast, CgB4u/pCgPDR1, the complemented strain, showed an 8- to 16-fold increased resistance to the three azoles compared to NCCLS84 (Table 3). Finally, CgPDR1 was shown to complement a commercially obtained S. cerevisiae pdr1 mutant (Fig. 2). Disk diffusion assays showed that S. cerevisiae pdr1 mutant 4381 had increased sensitivities to fluconazole and rhodamine 6G compared to its wild-type strain, BY4741 (Fig. 2). Importantly, the CgPDR1 overexpression construct, pHCgPDR1, reduced the sensitivities of pdr1 mutant 4381 to fluconazole and rhodamine 6G (Fig. 2).
FIG. 2.
Overexpression of CgPDR1 in the pdr1 mutant of S. cerevisiae reduced its sensitivities to fluconazole and rhodamine 6G. A disk diffusion assay was used to determine the drug sensitivities of S. cerevisiae strains. Strains: BY4741, wild type; 4381, pdr1 mutant; 4381/pH392, pdr1 mutant carrying the vector pH392; 4381/pHCgPDR1, pdr1 mutant carrying the CgPDR1 overexpression cassette ADH1p-CgPDR1-ADH1T. FLC, fluconazole; R6G, rhodamine 6G. Plates were photographed after 2 days of incubation at 30°C. WT, wild type.
CgPDR1 regulated the gene expression of ABC transporters and efflux of rhodamine 6G.
Northern blot analysis with the CgPDR1 probe showed that the CgPDR1 transcript was estimated to be 3.5 kb in size and was present at a low level in the wild-type strain NCCLS84 (Fig. 3). In contrast, the native CgPDR1 transcript was absent in the Cgpdr1 mutant CgB4, which had slightly increased expression of a truncated Cgpdr1 transcript 2.2 kb in size. As PDR1 regulates the gene expression of ABC transporters in S. cerevisiae, the effect of CgPDR1 on the gene expression of the two ABC transporters, CgCdr1p and Pdh1p, was analyzed. The expression of CgCDR1 in CgB4 was reduced compared to that in NCCLS84, while the PDH1 transcript was not detected in both NCCLS84 and CgB4 due to its low basal expression. In contrast, overexpression of CgPDR1 in CgB4u (strain CgB4u/pCgPDR1) resulted not only in abundant expression of CgPDR1 but also a dramatic increase in the expression of both ABC transporter genes, CgCDR1 and PDH1. A putative pleiotropic drug resistance responsive element (PDRE), TTCCGTGGAA, is present in the upstream region of both CgCDR1 (position −1201 to −1192 from the translation start site ATG) and PDH1 (position −560 to −551 from the translation start site ATG). Rhodamine 6G, which is effluxed by these two ABC transporters, was used to assess the effect of regulation by CgPDR1. FACS analyses showed that the strain CgB4u/pCgACU (vector only) had greater rhodamine 6G accumulation than NCCLS84, and rhodamine 6G accumulation was greatly reduced in the strain CgB4u/pCgPDR1 (Table 4). Therefore, we conclude that CgPDR1 affected the expression of ABC transporter genes as well as rhodamine 6G efflux.
FIG. 3.

Disruption or overexpression of CgPDR1 altered the expression of CgCDR1 and PDH1. Northern hybridization analyses were used to determine the expression of CgPDR1, CgCDR1, and PDH1. Ten micrograms of total RNA was used for Northern blot analysis. The membrane was hybridized with the following probe: 2.3-kb BglII CgPDR1 DNA (top panel), 1.1-kb NotI-BamHI CgCDR1 DNA from pCRScript-CDR1 (14) (middle panel), or 3.6-kb PDH1 DNA from pClaI (25) (bottom panel). Sizes of putative transcripts are indicated in kilobases on the right. The rRNA stained with ethidium bromide was used as the loading control. Lanes: 1, strain NCCLS84, wild type; 2, strain CgB4, Cgpdr1 mutant; 3, strain CgB4/pCgACU, Cgpdr1 mutant carrying the vector pCgACU; 4, strain CgB4/pCgPDR1, Cgpdr1 mutant overexpressing CgPDR1. WT, wild type.
TABLE 4.
Rhodamine 6G accumulation in C. glabrata strains
| Strain | Geometric mean fluorescence (in arbitrary units)
|
|
|---|---|---|
| Without R6G | With R6G | |
| NCCLS84 | 2.6 | 9.6 |
| CgB4 | 2.2 | 23.1 |
| CgB4u/pCgACU | 2.0 | 25.4 |
| CgB4u/pCgPDR1 | 2.0 | 3.1 |
| Cg173C | 1.9 | 2.7 |
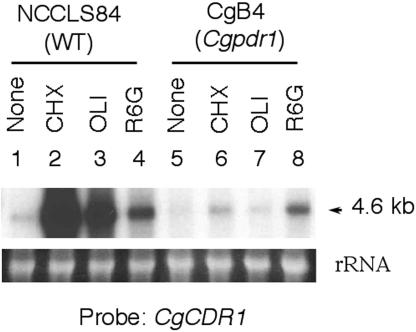
CgPDR1 mediated the drug-induced expression of CgCDR1.
The expression of CgCDR1 can be induced by drug treatment. Northern blot analyses showed that CgCDR1 expression was induced moderately by rhodamine 6G, and disruption of Cgpdr1 slightly reduced the rhodamine 6G-induced expression (Fig. 4). Fluconazole, on the other hand, is a weak inducer based on our Northern blot analysis (data not shown). In contrast, the CgCDR1 expression was induced dramatically by cycloheximide and oligomycin in NCCLS84. Northern blot analyses showed that disruption of CgPDR1 abolished the induced expression of CgCDR1 by cycloheximide and oligomycin in CgB4. Thus, CgPDR1 is the major regulator of the cycloheximide- and oligomycin-induced CgCDR1 expression.
FIG. 4.
Cycloheximide- and oligomycin-induced CgCDR1 expression depended on CgPDR1. Ten micrograms of total RNA was used for the Northern hybridization analysis to determine the level of drug-induced CgCDR1 expression. The membrane was hybridized with the 1.1-kb NotI-BamHI CgCDR1 DNA probe. The sizes of putative transcripts are indicated in kilobases. The rRNA stained with ethidium bromide was used as the loading control. Lanes: 1 to 4, NCCLS84 (wild type); 5 to 8, CgB4 (Cgpdr1 mutant). CHX, cycloheximide; OLI, oligomycin; R6G, rhodamine 6G. WT, wild type.
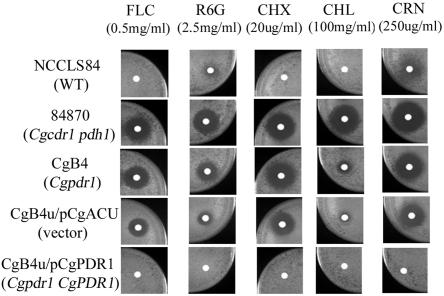
CgPDR1 affected drug resistance.
The Cgpdr1 mutant CgB4 exhibited increased sensitivity to rhodamine 6G, cycloheximide, chloramphenicol, and cerulenin (Fig. 5). Disk diffusion assays showed that NCCLS84 was resistant to 0.5 mg/ml of fluconazole, 2.5 mg/ml of rhodamine 6G (R6G), 20 μg/ml of cycloheximide, and 100 mg/ml of chloramphenicol, with only borderline inhibition by 250 μg/ml cerulenin (Fig. 5). In contrast to NCCLS84, in which no clear growth inhibition zones were observed, clear growth inhibition zones were observed with CgB4, indicating its increased drug sensitivities. The plasmid pCgACU alone (CgB4u/pCgACU) seemed to reduce the growth inhibition zone of CgB4u slightly in general. However, overexpression of CgPDR1 in CgB4u (CgB4u/pCgPDR1) completely eliminated the growth inhibition zone, which indicated restoration of drug resistance by CgPDR1 overexpression in CgB4u. As CgPDR1 regulated the expression of ABC transporter genes CgPDR1 and PDH1, the drug sensitivity of a transporter disruptant was also analyzed. The Cgcdr1 and pdh1 double disruptant, 84870, had greater sensitivity to rhodamine 6G and chloramphenicol than CgB4. It is possible that basal expression of these transporters was not completely absent when transcriptional regulation by CgPDR1 was lost.
FIG. 5.
Altered drug sensitivity in C. glabrata caused by the disruption or overexpression of CgPDR1. Disk diffusion assay was used to determine the drug sensitivity of C. glabrata. Strains: NCCLS84, wild type; 84870, Cgcdr1 and pdh1 double disruptant; CgB4, Cgpdr1 mutant; CgB4u/pCgACU, Cgpdr1 carrying the vector pCgACU; CgB4u/pCgPDR1, Cgpdr1 mutant overexpressing CgPDR1. FLC, fluconazole; R6G, rhodamine 6G; CHX, cycloheximide; CHL, chloramphenicol; CRN, cerulenin. Plates were photographed after 2 days of incubation at 30°C. WT, wild type.
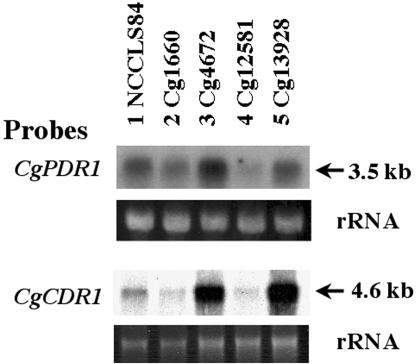
Increased CgPDR1 and CgCDR1 expression in clinical isolates.
As overexpression of CgPDR1 in C. glabrata contributed to drug resistance in the laboratory setting, it would be of interest to determine whether it also contributes to drug resistance developed in a clinical setting. Northern blot analyses were done with two pairs of clinical azole-sensitive and -resistant isolates, which had four- to eightfold differences in fluconazole MIC80s. Both resistant isolates Cg4672 and Cg13928 had higher expression of CgPDR1 and CgCDR1 than their paired fluconazole-sensitive strains, Cg1660 and Cg12581 (Fig. 6).
FIG. 6.
Clinical fluconazole-resistant isolates overexpressed CgPDR1 and CgCDR1. Two pairs of clinical isolates, Cg1660 versus Cg4672 and Cg12581 versus Cg13928, were analyzed by Northern blot hybridization along with NCCLS84. Probes used for each Northern blot are labeled on the left, and the estimated sizes of hybridized transcripts in kilobases are labeled on the right. The rRNA stained with ethidium bromide was used as the loading control. Probes: top, 2.3-kb BglII CgPDR1 DNA; bottom, 1.1-kb NotI-BamHI CgCDR1 DNA.
Upregulated PDR gene expression and increased fluconazole resistance in Cg13928 linked to its CgPDR1 locus.
CgPDR1 of Cg13928, along with its promoter region, was introduced into the disrupted Cgpdr1 locus of CgB4 via an integrative transformation to complement the Cgpdr1 mutation. The complemented strain, Cg173C, showed an 8- to 16-fold increase in the resistance to three azoles compared to that of NCCLS84 (Table 3 and Fig. 1). Northern blot analysis showed that the expression of CgPDR1 and CgCDR1 was also upregulated in Cg173C (data not shown). In agreement, FACS analysis showed that the rhodamine 6G accumulation was greatly reduced in Cg173C compared to NCCLS84 and CgB4 (Table 4). Together, our data indicated that CgPDR1 and its promoter region were responsible for the fluconazole resistance in the clinical fluconazole-resistant isolate Cg13928.
F575L and W297S codon substitutions in CgPDR1 increased CgPDR1 expression and fluconazole resistance of clinical isolates.
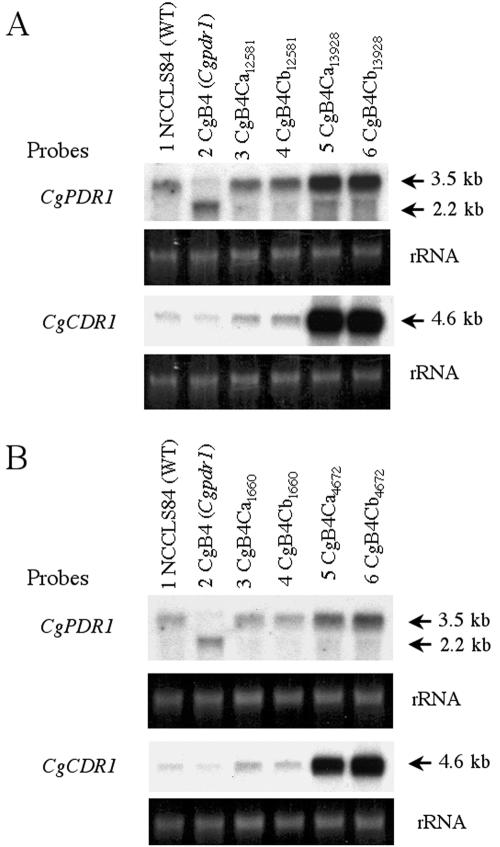
It was unclear whether the azole resistance and increased CgPDR1 expression in the resistant isolates of each pair was due to the promoter or regulatory elements in the flanking regions or to mutations in the CgPDR1 ORF. Therefore, CgPDR1 from the two pairs of clinical isolates was sequenced. Analyses of the CgPDR1 coding region as well as the 2.5-kb upstream regions revealed no differences in the flanking regions, and the CgPDR1 ORF differed between the susceptible and resistant isolates at a single codon in each pair.
The CgPDR1 sequence of Cg13928 contained a point mutation at codon 575 (TTC to CTC) compared to the CgPDR1 sequences of Cg12581. F575L is located in the fungus-specific transcription factor domain of CgPdr1p, according to the homology of CgPdr1p with S. cerevisiae Pdr1p. The CgPDR1 sequence of Cg4672 contained a single mutation at codon 297 (TGG to TCG) compared to the CgPDR1 sequences of Cg1660. W297S is located in the inhibition domain of CgPdr1p in Cg4672, according to the homology with S. cerevisiae Pdr1p. The data indicated that even though the two clinical fluconazole-resistant isolates had upregulated CgPDR1 expression, they harbored different mutations.
Direct correlation of the mutations with upregulated CgPDR1 expression as well as increased fluconazole resistance was further investigated by gene substitution and Northern blot analyses. Both mutated codons, W297S and P595L, were within the 2.9-kb HindIII region of the CgPDR1 ORF. The 2.9-kb HindIII fragments containing the partial CgPDR1 ORF (amino acids 30 to 974) from Cg12581, Cg13928, Cg1660, and Cg4672 were introduced into the mutated Cgpdr1 locus of CgB4 independently by integrative transformation. Two transformants, labeled “CgB4Ca” and “CgB4Cb,” were selected from each integrative transformation for sequence analysis and fluconazole susceptibility assay. The presence of the correct nucleotide sequences in the transformants was verified by DNA sequencing. In Table 3, the fluconazole susceptibilities of the transformants can be seen to correspond to those of the clinical isolates from which the CgPDR1 gene was obtained. In agreement, Northern blot analyses showed that the transformants carrying the Cg13928 or Cg4672 CgPDR1 had increased expression of CgPDR1 and CgCDR1 compared to the transformants carrying the Cg12581 or Cg1660 CgPDR1 as well as the wild type (Fig. 7). Thus, we concluded that the W297S and F575L single-amino-acid substitutions were the cause of upregulated CgPDR1 and CgCDR1 expression as well as the increased fluconazole resistance in the clinical fluconazole-resistant isolates Cg4672 and Cg13928, respectively.
FIG. 7.
W297S or F595L single-amino-acid substitution of CgPdr1p led to upregulated expression of CgPDR1 and CgCDR1. The expression of CgPDR1 and CgCDR1 was analyzed by Northern blot analysis. The membrane was hybridized with the 2.9-kb HindIII CgPDR1 DNA or 1.1-kb NotI-BamHI CgCDR1 DNA probe. The rRNA stained with ethidium bromide was used as the loading control. All complementations were done with integrative transformation. (A) Lanes: 1, strain NCCLS84, wild type; 2, strain CgB4, Cgpdr1 mutant; 3 and 4, strain CgB4Ca12581 and strain CgB4Cb12581, CgB4 complemented by CgPDR1 from Cg12581; 5 and 6, strain CgB4Ca13928 and strain CgB4Cb13928, CgB4 complemented by CgPDR1 from Cg13928. (B) Lanes: 1, strain NCCLS84, wild type; 2, strain CgB4, Cgpdr1 mutant; 3 and 4, strain CgB4Ca1660 and strain CgB4Cb1660, CgB4 complemented by CgPDR1 from Cg1660; 5 and 6, strain CgB4Ca4672 and strain CgB4Cb4672, CgB4 complemented by CgPDR1 from Cg4672. WT, wild type.
CgPDR1 is required for fluconazole resistance in petite mutants.
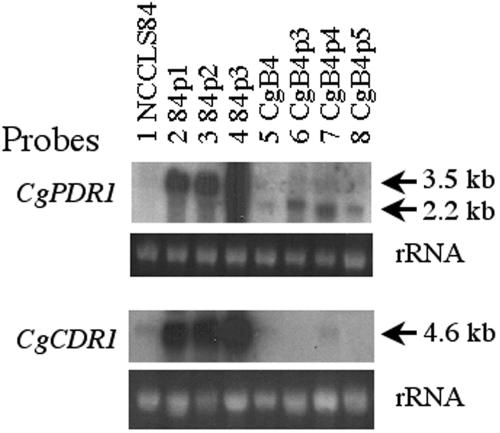
Similar to S. cerevisiae, petite mutants of C. glabrata also exhibited increased fluconazole resistance. PDR3, not PDR1, was overexpressed in the S. cerevisiae petite mutants and was shown to be essential for the upregulated expression of PDR5 in the petite mutants. To date, only the PDR1 homologue has been annotated in the genome database of C. glabrata. We postulated that CgPDR1 might be the transcriptional factor responsible for azole resistance in the C. glabrata petite mutants. Therefore, three petite mutants from both the wild-type strain, NCCLS84, as well as the Cgpdr1 mutant, CgB4, were analyzed. MIC80 assays showed that the petite mutants of NCCLS84 exhibited 8-fold or greater resistance to fluconazole, 16-fold or greater resistance to voriconazole, and 4-fold or greater resistance to itraconazole compared to its parental strain NCCLS84 (Table 5). In addition, Northern blot analyses showed the increased expression of CgPDR1, the transcriptional factor, and its transcriptional target, CgCDR1, the transporter gene in the three petite mutants of NCCLS84 (Fig. 8). In contrast, three petite mutants of CgB4 exhibited unaltered susceptibility to fluconazole, one- to fourfold increased resistance to voriconazole, and two- to fourfold increased resistance to itraconazole compared to its parental strain, CgB4 (Table 5). Northern blot analysis showed that the expression of the truncated Cgpdr1 gene was slightly increased in the three petite mutants of CgB4 compared to their parental strain, CgB4, but there was no obvious change in the CgCDR1 expression of the three CgB4 petite mutants. We conclude that the functioning CgPDR1 was essential for mediating fluconazole resistance in the petite mutants of C. glabrata.
TABLE 5.
Azole susceptibility of C. glabrata and petite mutants
| Strain | MIC80 (μg/ml) of:
|
||
|---|---|---|---|
| Fluconazole | Voriconazole | Itraconazole | |
| NCCLS84 | 64 | 1 | 4 |
| 84p1 | >512 | 16 | >32 |
| 84p2 | 512 | >32 | >32 |
| 84p3 | 512 | 16 | 16 |
| CgB4 | 8 | 0.125 | 0.5 |
| CgB4p3 | 8 | 0.125 | 1-2 |
| CgB4p4 | 8 | 0.5 | 2 |
| CgB4p5 | 8 | 0.5 | 2 |
FIG. 8.
CgPDR1 is required for upregulated expression of PDR genes caused by mitochondrial dysfunction. For Northern blot analysis, the membrane was hybridized with the 2.3-kb BglII CgPDR1 DNA (top) or 2.3-kb BamHI-NotI CgCDR1 (bottom) probe. The rRNA stained with ethidium bromide was used as the loading control. Lanes: 1, strain NCCLS84, wild type; 2 and 3, petite mutant strains derived from NCCLS84; 5, strain CgB4, Cgpdr1 mutant; 6 to 8, petite mutant strains derived from CgB4.
DISCUSSION
Increased expression of multidrug transporter genes has been shown in both laboratory and clinical azole-resistant isolates of Candida species (25, 28, 31, 34, 40). In S. cerevisiae, gain-of-function mutations in either PDR1 or PDR3 led to increased expression of the ABC transporter PDR5. However, CgPDR1 mutation and upregulation has only been identified in one laboratory azole-resistant C. glabrata isolate to date, with no evidence that the mutation caused the upregulation (38). Here, for the first time, we showed that upregulation of the CgPDR1 expression indeed occurred in the clinical azole-resistant isolates and petite mutants, which contributed to the drug resistance by upregulating the expression of CgCDR1.
We identified that only a single-amino-acid substitution in CgPdr1p, W297S in Cg4672 and F575L in Cg13928, could lead to increased CgPDR1 expression and azole resistance. The putative functional domains that W297S and F575L mutations located were determined based on the homology of CgPdr1p with S. cerevisiae Pdr1p. Amino acid 297 is located near the region corresponding to the inhibitory domain of S. cerevisiae Pdr1p. In S. cerevisiae, several gain-of-function mutations have been observed in the domain, e.g., K302Q (pdr1-6) and M308I (pdr1-2) (6). In contrast, the altered F595L codon is located in the region corresponding to the fungus-specific transcription factor domain, which has not been reported to associate with fluconazole resistance. In S. cerevisiae, gain-of-function mutations in PDR1 are also located in the activation domain (amino acids 879 to 1036) (21, 42). The amino acid substitution identified by Vermitsky and Edlind as possibly increasing transcriptional activity, P927L, was located in the carboxy-terminal domain of CgPdr1p, not the fungus-specific domain (38). It would be of interest to further investigate the importance of the codon 575 in the function of CgPdr1p.
Both W297S and F575L mutations have resulted in the upregulated expression of CgPDR1. One explanation is that the mutations altered CgPdr1p conformation and CgPdr1p became hyperactive, which may directly or indirectly affect the expression of CgPDR1. The possibility of autoregulation will be investigated as a putative PDRE, “TTCCGTGGAA,” is observed at the 5′ upstream region of CgPDR1 and located at position −558 to −549 (upstream from the translation start site ATG). The other possibility is that the mutations increase the CgPDR1 mRNA half-life and thus resulted in the accumulation of CgPDR1 mRNA. However, as the two mutations are located at different functional domains, the upregulation of CgPDR1 expression likely involved different mechanisms. As shown in Fig. 8, mitochondrial dysfunction increased the expression of CgPDR1. In addition, Northern blot analysis showed that cycloheximide also induced the expression of CgPDR1 (data not shown). These facts suggested the possibility that multiple pathways regulate CgPDR1 expression.
Our studies showed that CgPDR1 is essential in the azole resistance caused by mitochondrial dysfunction in C. glabrata petite mutants. In contrast, PDR3 is the transcriptional factor essential for drug resistance in the petite mutants and PDR1 is not involved in the resistance due to mitochondrial dysfunction S. cerevisiae (12, 44). In that species, PDR3 was shown to be under positive autoregulation by Pdr3p (8), and retrograde signaling pathways have been shown to upregulate PDR3 expression in the petite mutants of S. cerevisiae (10, 12). Our observation of dramatic increases of CgPDR1 expression in the NCCLS84 petite mutants but not in the CgB4 petite mutants suggests that CgPDR1 expression also may be regulated by itself in responding to mitochondrial dysfunction. However, the precise interplay mechanisms of PDR transcription factors and mitochondrial dysfunction require further exploration in both S. cerevisiae and C. glabrata.
Disruption of Cgpdr1 led to slightly decreased rhodamine 6G-induced expression of CgCDR1. In contrast, the dramatic induction of CgCDR1 expression by cycloheximide and oligomycin was abolished by disruption of CgPDR1. These data indicated that CgPDR1 is the major regulator of cycloheximide- and oligomycin-induced CgCDR1 expression but not rhodamine 6G-induced expression.
Though the Cgcdr1 and pdh1 double disruptant (84870) and the Cgpdr1 disruptant (CgB4) both had increased sensitivities to various drugs, their sensitivity profiles are not identical. The mutant 84870 had greater sensitivity to rhodamine 6G and chloramphenicol than CgB4. Even though rhodamine 6G can be effluxed by CgCdr1p, rhodamine 6G and chloramphenicol were reported to be the preferred substrates of Pdh1p (14); the difference of 84870 and CgB4 in rhodamine 6G and chloramphenicol sensitivity may be mainly due to PDH1 expression. It is possible that disruption of CgPDR1 has not completely abolished the expression of one or both of these transporters.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIAID.
REFERENCES
- 1.Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux, and A. Goffeau. 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262:16871-16879. [PubMed] [Google Scholar]
- 2.Bennett, J. E., K. Izumikawa, and K. A. Marr. 2004. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob. Agents Chemother. 48:1773-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 4.Brun, S., C. Aubry, O. Lima, R. Filmon, T. Berges, D. Chabasse, and J. P. Bouchara. 2003. Relationships between respiration and susceptibility to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 47:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brun, S., T. Berges, P. Poupard, C. Vauzelle-Moreau, G. Renier, D. Chabasse, and J. P. Bouchara. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48:1788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvajal, E., H. B. van den Hazel, A. Cybularz-Kolaczkowska, E. Balzi, and A. Goffeau. 1997. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256:406-415. [DOI] [PubMed] [Google Scholar]
- 7.Defontaine, A., J. P. Bouchara, P. Declerk, C. Planchenault, D. Chabasse, and J. N. Hallet. 1999. In-vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J. Med. Microbiol. 48:663-670. [DOI] [PubMed] [Google Scholar]
- 8.Delahodde, A., T. Delaveau, and C. Jacq. 1995. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol. Cell. Biol. 15:4043-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delaveau, T., A. Delahodde, E. Carvajal, J. Subik, and C. Jacq. 1994. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet. 244:501-511. [DOI] [PubMed] [Google Scholar]
- 10.Devaux, F., E. Carvajal, S. Moye-Rowley, and C. Jacq. 2002. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 515:25-28. [DOI] [PubMed] [Google Scholar]
- 11.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44.15229592 [Google Scholar]
- 12.Hallstrom, T. C., and W. S. Moye-Rowley. 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:37347-37356. [DOI] [PubMed] [Google Scholar]
- 13.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izumikawa, K., H. Kakeya, H. F. Tsai, B. Grimberg, and J. E. Bennett. 2003. Function of Candida glabrata ABC transporter gene, PDH1. Yeast 20:249-261. [DOI] [PubMed] [Google Scholar]
- 15.Katzmann, D. J., P. E. Burnett, J. Golin, Y. Mahe, and W. S. Moye-Rowley. 1994. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 14:4653-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur, R., I. Castano, and B. P. Cormack. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 48:1600-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly, S. L., A. Arnoldi, and D. E. Kelly. 1993. Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans. 21:1034-1038. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, S. L., D. C. Lamb, J. Loeffler, H. Einsele, and D. E. Kelly. 1999. The G464S amino acid substitution in Candida albicans sterol 14alpha-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262:174-179. [DOI] [PubMed] [Google Scholar]
- 20.Kitada, K., E. Yamaguchi, and M. Arisawa. 1996. Isolation of a Candida glabrata centromere and its use in construction of plasmid vectors. Gene 175:105-108. [DOI] [PubMed] [Google Scholar]
- 21.Kolaczkowska, A., M. Kolaczkowski, A. Delahodde, and A. Goffeau. 2002. Functional dissection of Pdr1p, a regulator of multidrug resistance in Saccharomyces cerevisiae. Mol. Genet. Genomics 267:96-106. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, D. C., D. E. Kelly, W. H. Schunck, A. Z. Shyadehi, M. Akhtar, D. J. Lowe, B. C. Baldwin, and S. L. Kelly. 1997. The mutation T315A in Candida albicans sterol 14alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J. Biol. Chem. 272:5682-5688. [DOI] [PubMed] [Google Scholar]
- 23.Lees, N. D., M. Bard, and D. R. Kirsch. 1999. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 34:33-47. [PubMed] [Google Scholar]
- 24.Maesaki, S., P. Marichal, H. Vanden Bossche, D. Sanglard, and S. Kohno. 1999. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 44:27-31. [DOI] [PubMed] [Google Scholar]
- 25.Miyazaki, H., Y. Miyazaki, A. Geber, T. Parkinson, C. Hitchcock, D. J. Falconer, D. J. Ward, K. Marsden, and J. E. Bennett. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCCLS. 1995. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.Niimi, M., Y. Nagai, K. Niimi, S. Wada, R. D. Cannon, Y. Uehara, and B. C. Monk. 2002. Identification of two proteins induced by exposure of the pathogenic fungus Candida glabrata to fluconazole. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 782:245-252. [DOI] [PubMed] [Google Scholar]
- 28.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller, M. A., R. N. Jones, G. V. Doern, A. C. Fluit, J. Verhoef, H. S. Sader, S. A. Messer, A. Houston, S. Coffman, R. J. Hollis, and SENTRY Participant Group (Europe). 1999. International surveillance of blood stream infections due to Candida species in the European SENTRY Program: species distribution and antifungal susceptibility including the investigational triazole and echinocandin agents. Diagn. Microbiol. Infect. Dis. 35:19-25. [DOI] [PubMed] [Google Scholar]
- 30.Pfaller, M. A., S. A. Messer, R. J. Hollis, R. N. Jones, and D. J. Diekema. 2002. In vitro activities of ravuconazole and voriconazole compared with those of four approved systemic antifungal agents against 6,970 clinical isolates of Candida spp. Antimicrob. Agents Chemother. 46:1723-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad, R., P. De Wergifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627-630. [DOI] [PubMed] [Google Scholar]
- 36.Tsai, H. F., M. Bard, K. Izumikawa, A. A. Krol, A. M. Sturm, N. T. Culbertson, C. A. Pierson, and J. E. Bennett. 2004. Candida glabrata erg1 mutant with increased sensitivity to azoles and to low oxygen tension. Antimicrob. Agents Chemother. 48:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanden Bossche, H., P. Marichal, J. Gorrens, D. Bellens, H. Moereels, and P. A. Janssen. 1990. Mutation in cytochrome P-450-dependent 14 alpha-demethylase results in decreased affinity for azole antifungals. Biochem. Soc. Trans. 18:56-59. [DOI] [PubMed] [Google Scholar]
- 38.Vermitsky, J. P., and T. D. Edlind. 2004. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 48:3773-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada, S., K. Tanabe, A. Yamazaki, M. Niimi, Y. Uehara, K. Niimi, E. Lamping, R. D. Cannon, and B. C. Monk. 2005. Phosphorylation of Candida glabrata ATP-binding cassette transporter Cdr1p regulates drug efflux activity and ATPase stability. J. Biol. Chem. 280:94-103. [DOI] [PubMed] [Google Scholar]
- 40.White, T. C., S. Holleman, F. Dy, L. F. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 42.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2001. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 152:375-389. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida, Y., and Y. Aoyama. 1987. Interaction of azole antifungal agents with cytochrome P-45014DM purified from Saccharomyces cerevisiae microsomes. Biochem. Pharmacol. 36:229-235. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, X., and W. S. Moye-Rowley. 2001. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the F(0) component of the mitochondrial ATPase. J. Biol. Chem. 276:47844-47852. [DOI] [PubMed] [Google Scholar]