Abstract
Microcins are ribosomally synthesized peptide antibiotics that are produced by enterobacterial strains. Although the first studies concentrated on plasmid-encoded activities, in the last years three chromosome-encoded microcins have been described: H47, E492, and M. Here, a new microcin, I47, is presented as a fourth member of this group. Common features exhibited by chromosome-encoded microcins were searched for. The comparison of the genetic clusters responsible for microcin production revealed a preserved general scheme. The clusters essentially comprise a pair of activity-immunity genes which determine antibiotic specificity and a set of microcin maturation and secretion genes which are invariably present and whose protein products are highly homologous among the different producing strains. A strict functional relationship between the maturation and secretion pathways of microcins H47, I47, and E492 was demonstrated through genetic analyses, which included heterologous complementation assays. The peptide precursors of these microcins share a maturation process which implies the addition of a catecholate siderophore of the salmochelin type. Microcins thus acquire the ability to enter gram-negative cells through the catechol receptors. In addition, they employ a common mode of secretion to reach the external milieu by means of a type I export apparatus. The results presented herein lead us to propose that chromosome-encoded microcins constitute a defined subgroup of peptide antibiotics which are strictly related by their modes of synthesis, secretion, and uptake.
Many enterobacterial strains produce peptide antibiotics called microcins. They are synthesized as gene-encoded peptides that, thereafter, may undergo posttranslational modifications to be converted into mature molecules. Although early descriptions indicated that microcins were plasmid-encoded antibacterial activities (4), a few examples of chromosome-encoded microcins appeared later (9, 13, 14, 17, 25).
The first description of a chromosome-encoded activity of this type was microcin H47 (MccH47) produced by Escherichia coli H47 (14). Its genetic determinants are clustered, forming a genetic system which ensures microcin synthesis and secretion and the specific immunity against its antibiotic action (Fig. 1). MccH47 is synthesized as a 75-residue peptide precursor which is encoded by the activity gene mchB and then undergoes a maturation process carried out by the products of the mchA, mchC, and mchD genes. Maturation consists of the addition of a salmochelin to the peptide. Salmochelins are mono- or diglucosylated derivatives of the catecholate siderophore enterobactin (Ent). Thus, the cell must provide enterobactin, which is converted into salmochelins, and these are employed for MccH47 synthesis (2, 7). After being synthesized, MccH47 is secreted through a type I apparatus that is formed by the products of the mchE and mchF genes and by the outer membrane protein TolC. MccH47 would be processed in its 15 N-terminal residues during its export (3, 20). MccH47 is then composed of two moieties, peptide and salmochelin. The peptide possesses the deleterious activity, and the salmochelin confers the uptake properties to the molecule (2, 20). Thus, the antibiotic is taken up by E. coli K-12 cells through the catechol pathway, involving the receptor Cir, Fiu, or FepA, and the TonB complex (24). This mode of synthesis, which endows the peptide antibiotic with the ability to enter gram-negative cells through the catechol uptake pathway, has been called the “catechol strategy” (2).
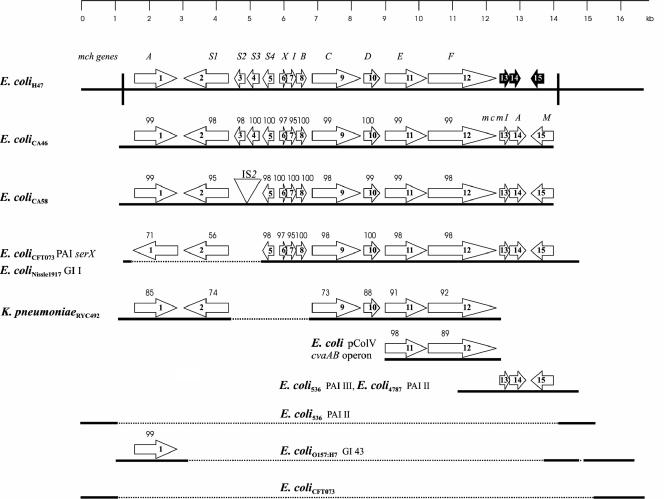
FIG. 1.
Comparison of the DNA region containing the MccH47 genetic system of E. coli strain H47. The sequenced DNA region and the genetic structure of the MccH47 system are shown above. Extensive DNA segments exhibiting high levels of identity (>80%) with the compared sequence are depicted as full lines, which are connected by dotted lines when they belong to the same genomic region of an organism. PAI, pathogenicity island; GI, genomic island. Vertical lines correspond to the direct repeat flanking the microcin cluster in E. coli H47. Arrows represent the mch genes of E. coli H47 and homologues from other organisms, whose correspondence is shown by the numbers in the arrows. Above the genes, numbers denote the percentage of identity of their protein products compared to the proteins of E. coli H47. Truncated genes related to MccM (mcmI, mcmA, and mcmM) are shown as black arrows.
The MccH47 maturation genes, mchA, mchC, and mchD, together with a fourth gene, mchS1, whose participation in MccH47 antibiosis is as yet uncertain, appear to be characteristic of the genetic systems of chromosome-encoded microcins (Fig. 1). Two of these, mchA and mchS1, are homologous to the iroB and iroD genes described for Salmonella enterica and several pathogenic E. coli strains (2). In these organisms, they integrate the salmochelin genetic system iroA, composed of five genes (iroBCDEN), which is responsible for iron uptake mediated by the siderophores salmochelins. The iroB gene product is the glucosyltransferase which performs enterobactin modification, and iroD encodes an enterobactin esterase (5, 7, 15). In our case, the mchA-encoded glucosyltransferase would contribute to MccH47 synthesis by glucosylating enterobactin to produce salmochelins, a step that is absolutely required for antibiotic synthesis. The mchS1-encoded enterobactin esterase is not essential for MccH47 synthesis since mutations affecting this gene decrease but do not abolish microcin production. The other two gene products required for MccH47 maturation, MchC and MchD, could be in charge of joining the two MccH47 moieties, peptide and salmochelin (2, 8).
Other E. coli strains have been reported to contain the MccH47 genetic determinants, and in two cases, their chromosomal location was confirmed: the uropathogenic E. coli CFT073 strain and the probiotic E. coli Nissle1917 strain (9, 25). In all of the strains, the MccH47 system includes determinants for microcin M (MccM), a recently described antibacterial activity whose uptake also depends on the catechol receptors (17).
Microcin E492 (MccE492), which is produced by Klebsiella pneumoniae RYC492, is another example of a chromosome-encoded microcin. It differs from MccH47 in its final mode of action: MccE492 acts upon the membrane as an ionophore, while MccH47 acts on the proton channel of ATP synthase (13, 19). MccE492 carries a posttranslational modification consisting of the linkage of a linear monoglucosylated salmochelin to the C terminus of the peptide (23) and, as is the case for microcins H47 and M, its uptake depends on the catechol pathway (17). In addition, MccE492 also reaches the extracellular medium by means of a type I secretion system which is very similar to that of MccH47 (13).
In this work, we perform a comparative analysis of chromosome-encoded microcins and propose that they constitute a defined subgroup with the following characteristics: their genetic clusters exhibit a preserved general scheme, they are synthesized following the catechol strategy and exported by type I secretion systems, and their uptake by gram-negative bacteria is performed through the catechol receptors in a TonB-dependent manner. The versatility of these microcin maturation and export pathways is assessed in heterologous genetic constructions, and their suitability to adopt incoming microcin activity and immunity determinants is discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli H47 and K. pneumoniae RYC492 are natural producers of MccH47 and MccE492, respectively (13, 14). E. coli K-12 strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB)-rich medium and M63 minimal medium supplemented with glucose were used (16). To create iron deprivation conditions, plates were supplemented with 200 μM 2,2′-dipyridyl. The following antibiotics were added to the media at the indicated final concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 60 μg/ml; and tetracycline (Tc), 12 μg/ml.
TABLE 1.
E. coli K12 bacterial strains and plasmids
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| RYC1000 | araD gyrA lacU169 rbs relA rpsL thiA | Laboratory collection |
| KL14 | PO68 relA | Laboratory collection |
| MC4100 | araD lacU169 relA rpsL thiA | Laboratory collection |
| FGB099 | MC4100 atpB::Tn5 99 | 19 |
| FGB11 | MC4100 tonB::Tn5 | 24 |
| FGB1876 | MC4100 rpsL+cir fepA::Tn10 fiu::Mud1X | This work |
| FGB1870 | MC4100 rpsL+cir fepA::Tn10 | This work |
| FGB1728 | MC4100 rpsL+cir fiu::Mud1X | This work |
| FGB1594 | MC4100 rpsL+fiu::Mud1X | This work |
| FGB102 | MC4100 fepA::Tn10 | 2 |
| FGB103 | MC4100 Δ(ompT-fepC)267 | 2 |
| Plasmids | ||
| pAB492 | pBR322 carrying mceAB from the MccE492 system (Apr) | This work |
| pB492 | pUC13 carrying mceB from the MccE492 system (Apr) | This work |
| pMVDS3 | pACYC184 carrying mchS3 from the MccH47 system (Cmr) | This work |
| pMVD41 | pACYC184 carrying mchEF from the MccH47 system (Cmr) | This work |
| pUY69 | pUC13 carrying mchI from the MccH47 system (Apr) | 18 |
| pEX100 | pACYC184 carrying the MccH47 system (Cmr) | 8 |
| pEX100::Tnlac7.2 | pEX100 mchA::Tnlac | 8 |
| pEX100::Tnlac5.4 | pEX100 mchS1::Tnlac | 8 |
| pEX100::Tn576 | pEX100 mchB::Tn5 | 20 |
| pEX100::Tnlac7.7 | pEX100 mchC::Tnlac | 8 |
| pEX100::Tnlac3.5 | pEX100 mchD::Tnlac | 8 |
| pEX100::Tnlac7.1 | pEX100 mchE::Tnlac | 3 |
| pEX4 | pUC13 carrying the MccH47 system (Apr) | 14 |
| pEX4::Tn538 | pEX4 mchS2::Tn5 | 8 |
Strain construction.
Several strains with mutations in genes (fepA, fiu, and cir) encoding catechol receptors were constructed by conjugation on the basis of strains H1876 (MC4100 aroB cir fepA::Tn10 fiu::Mud1X), H1728 (MC4100 aroB cir fiu::Mud1X), and H1594 (MC4100 aroB fiu::Mud1X) (11). Since the aroB mutation implies that these strains have multiple requirements for their growth in minimal media, Hfr KL14 was mated with each of them to construct aroB+-derivative strains following standard procedures (16). Recombinants FGB1876 and FGB1870 were obtained in minimal M63 medium with tetracycline in the KL14 X H1876 experiment; FGB1594 results from the crossing with H1594, and FGB1728 results from the crossing with H1728, in both cases being selected in minimal plates with ampicillin. When necessary, the presence of the fiu::Mud1X allele was detected by the ampicillin resistance encoded by Mud1X; the cir-defective allele was corroborated by the conferred resistance to colicin V. Besides carrying mutations for the catechol receptors and being aroB+, all the strains constructed were rpsL+.
Plasmid construction.
pMVDS3 is a pACYC184-derivative plasmid with an SphI-SphI DNA segment from pEX4::Tn535 (8). The insert contains the mchS3 gene and part of a Tn5 inserted in the mchS4 gene. For plasmid pAB492 construction, a DNA segment containing the MccE492 activity and immunity genes mceAB was amplified by PCR with oligonucleotides A (5′-CCTCCCCATGAATTTTGTAGA-3′) and B (5′-ACATACGTAACTACAGGCTTTG-3′). The DNA segment was cloned into a polylinker-containing intermediate vector to finally insert it between the HindIII and AvaI sites in pBR322. Plasmid pB492 is a pUC13 derivative carrying a PCR-generated DNA segment containing the MccE492 immunity gene mceB inserted in the HincII site of the vector. The oligonucleotides used for this construction were A and C (5′-TAATCCGCCAGGAGCGCCAAGA-3′). Plasmid pMVD41 is a pACYC184 derivative with an insert containing the MccH47 mchEF export genes in the HindIII site of the vector. The cloned fragment extends from a HindIII site in a Tn5 inserted in mchD (pEX4::Tn5B41) (8) to the HindIII site at the end of the MccH47-sequenced DNA (accession no. AJ009631).
Microcin production and sensitivity assays.
Microcin production was assayed by patch test on minimal M63 glucose plates as previously described (20). To analyze the maturation and secretion requirements for microcin production, strains carrying a collection of pEX100-derivative plasmids with mutations in the following MccH47 genes were employed: mchA (pEX100::Tnlac7.2), mchS1 (pEX100::Tnlac5.4), mchB (pEX100::Tn576), mchC (pEX100::Tnlac7.7), mchD (pEX100::Tnlac3.5), and mchE (pEX100::Tnlac7.1) (8). Considering that many of the strains assayed produced two microcins, one of them MccH47, several isogenic indicator strains were employed: (i) MC4100, sensitive to all microcins, (ii) FGB099, an atp mutant resistant to MccH47 (19), (iii) MC4100(pB492), resistant (immune) to MccE492, and (iv) FGB099(pB492), resistant to both MccH47 and MccE492. A patch test on minimal M63 glucose plates was also employed to assay the microcin sensitivity of different strains mutated for the catechol uptake pathway: FGB102 (fepA), FGB1594 (fiu), FGB1728 (cir and fiu), FGB1870 (cir and fepA), and FGB1876 (cir, fiu, and fepA).
DNA sequencing.
DNA sequencing was performed at the “DNA Sequencing Core Laboratory Service” of the University of Florida.
Nucleotide sequence accession number.
Sequencing data have been deposited in the EMBL database under accession no. AJ009631.
RESULTS
Comparative analysis of the MccH47 genetic system.
The sequence of a 16,823-bp DNA segment containing the MccH47 genetic system, proceeding from the chromosome of E. coli H47, was elucidated and compared with sequences in databanks using the BLASTN program (1). High levels of identity were found with genomic sequences from enterobacterial strains (Fig. 1). The most significant matches were with microcin H47, M, and E492 determinants from E. coli strains CA46 (EMBL accession no. AJ515251), CA58 (AJ515252), Nissle 1917 (AJ586887), and CFT073 (AE014075) and K. pneumoniae strain RYC492 (AF063590). In addition, plasmid determinants devoted to colicin V secretion were also highly similar. On the other hand, the flanking DNA regions of the MccH47 cluster presented high scores with pathogenicity islands from the enterohemorrhagic E. coli strain O157:H7 (BA000007), the uropathogenic E. coli strains CFT073 (AE014075) and 536 (AF301153 and AJ494981), and the septicemic E. coli strain 4787 (AY560914).
The overall G+C content of the sequenced DNA segment was 39.9%, which is significantly below the genome content of E. coli K-12. At the left and right ends of the MccH47 genetic system, there was an imperfect 148-bp direct repeat (Fig. 1). This remarkable feature appears to indicate the ends of a DNA segment dedicated to microcin functions, which presents an extension of 12,927 bp.
Open reading frames adequately preceded by a possible ribosome binding site were assigned to the genes of the MccH47 system previously described by genetic approaches. The deduced amino acid sequences for the mch genes were compared with sequences in databanks using the program BLASTP 2.2.5 (1). The comparison revealed that other strains encoding microcins H47, M, or E492 contain homologues to the E. coli H47 mch genes (Fig. 1). All these strains invariably contain microcin determinants that are homologous to the E. coli H47 mchA, mchS1, mchC, mchD, mchE, and mchF genes. In addition, each antibiotic genetic system includes at least a couple of microcin activity-immunity determinants which differ depending on the encoded microcin. Other genes may be present in the microcin clusters; some of them that were detected in the present study, e.g., mchS2, mchS3, and mchS4, had not been previously described in strains other than E. coli H47 (Fig. 1). Finally, it must be noticed that microcin determinants exhibit variations in their arrangement. For example, in the case of strains H47, CA46, CA58, and RYC492, all microcin genes are clustered in a single locus, while in strains CFT073 and Nissle 1917, which are identical to each other in the region under consideration, the iroBD genes are separated from the remaining microcin genes and integrate the locus iroA.
At the right end of the MccH47 system, there are three open reading frames clearly related to the described MccM gene cluster from other strains (17). They are truncated homologues of the mcmI, mcmA, and mcmM genes.
A second microcin is encoded by the MccH47 system in E. coli H47.
The ability of strains Nissle 1917, CA46, and CA58 to produce two microcins, H47 and M, has been related to the presence of two pairs of activity-immunity determinants and of a single set of maturation and export genes in their microcin clusters, suggesting that both antibiotics would share the same production pathways (17). If the maturation and export proteins were able to act upon either of the chromosome-encoded microcins, then they should recognize specific common features in these peptides. In fact, a double-glycine signal peptide, dedicated to microcin secretion, is clearly recognizable at the N terminus of the microcin H47, E492, and M precursors (12, 13, 20). In addition, it has been noticed that the C-terminal region of these microcin can be aligned in a particularly serine-rich segment (17). This could well be the motif that is recognized by the microcin maturation proteins, which would agree with the fact that the catechol addition to MccE492 is linked to the C terminus of the peptide (23).
In the light of these considerations, it was observed that the mchS2 gene coded for a 77-residue peptide with a possible double-glycine signal sequence and with a serine-rich C end (Fig. 2). It was then suspected that the mchS2 and mchS3 genes could be a new pair of microcin activity-immunity determinants. However, no additional antibiotic activity had been detected to be produced by strains carrying the MccH47 genetic system. Thus, this putative second activity was searched for by assaying these strains on the MccH47-resistant strain FGB099 under different conditions. It was found that growth inhibition halos appeared in low-iron medium: they were very turbid after 24 h but progressively gained clearness upon further incubation (72 h). A strain bearing the MccH47 system with a mutation in the mchS2 gene, RYC1000(pEX4::Tn538), did not produce this second antibiotic activity, indicating that mchS2 was indeed involved in its production. In addition, the mchS3 gene, carried by plasmid pMVDS3, conferred resistance to the new microcin, as expected of an immunity determinant. At this point, the absence of cross-immunity with MccH47 was confirmed: a strain producing only MccH47, RYC1000(pEX4::Tn538), generated halos on a lawn of mchS3-bearing cells and, conversely, a strain producing only the second activity, RYC1000(pEX100::Tn576), inhibited the growth of a strain containing the MccH47 immunity gene mchI carried by plasmid pUY69. Thus, the second activity was clearly differentiated from MccH47 and named MccI47 (data not shown).
FIG. 2.
Alignment of the amino acid sequences of microcins H47, E492, and M peptide precursors and of MchS2. The alignment was performed with Multalin (version 5.4.1) (6). Identical or conserved residues throughout the four sequences are in bold capital letters. The arrow points to the putative site of processing of the double-glycine signal peptide during secretion.
Taking into account that the mchS2S3 genes are immersed in the MccH47 system, MccI47 could eventually be modified and secreted by the same proteins operating on MccH47. To identify the mch genes that are necessary to produce MccI47, strains carrying the MccH47 system with mutations in different mch genes were assayed by patch test, as described in Materials and Methods. The strains that were affected in the maturation genes mchA, mchC, or mchD or in the secretion gene mchE did not produce the antibiotic, while those impaired for mchS1 or the MccH47 activity gene mchB still produced MccI47. In sum, results indicated that MccI47 was produced following the same pathways of maturation and secretion as MccH47. Then, MccI47 was assessed for its activity on the catechol receptor mutant strains as described in Materials and Methods. For this purpose, a strain producing only MccI47, RYC1000(pEX100::Tn576), was employed. All the single- and double-receptor mutants were sensitive to MccI47 and only the triple cir, fiu, and fepA mutant was resistant (data not shown). Therefore, MccI47 fulfilled the requirements of synthesis and uptake of the catechol strategy.
Heterologous complementation for MccE492 production by MccH47 maturation and secretion genes.
The sequence comparison analyses obviously raised the question of whether the remaining chromosome-encoded microcins, E492 and M, also employed the catechol strategy for their synthesis. The fact that genes devoted to MccH47 maturation had clear counterparts in the other microcin clusters strongly supported this view. To analyze this issue, we focused on MccE492 by performing heterologous complementation assays: MccH47 maturation genes were analyzed for their ability to modify the MccE492 precursor peptide MceA. MccH47 secretion genes were included in the experimental design with the expectation that they would complement for MccE492 export.
The experiments consisted of introducing into E. coli K-12 cells the MccE492 activity and immunity genes, mceA and mceB, respectively, in the presence of the MccH47 genetic system in order to assay MccE492 production. Considering that mutations affecting any of the maturation or export genes from the MccE492 system completely abolish microcin production (13), the single presence of the mceAB genes should not determine MccE492 production. On this basis, the possible ability of MccH47 gene products for MccE492 maturation and secretion would be amenable to assay. Since most of the strains under study would have both MccE492 and MccH47 activity genes, microcin production was assayed by patch test on a set of indicator strains so as to discern between MccH47 and MccE492-generated halos, as described in Materials and Methods.
Plasmid pAB492, containing only the mceAB genes, was introduced in several genetic contexts. As expected, its single presence did not determine antibiotic production. The same result was observed when cells carried the MccH47 secretion genes mchE and mchF (pMVD41). On the other hand, a strain bearing the entire MccH47 genetic system (pEX100) produced antibiosis halos on all of the indicator strains except for the doubly resistant one (Fig. 3 and data not shown). Thus, a heterologous complementation for MccE492 maturation and secretion had taken place. The strain produced both microcins and, judging by the size of the halos, it produced more MccE492 than MccH47.
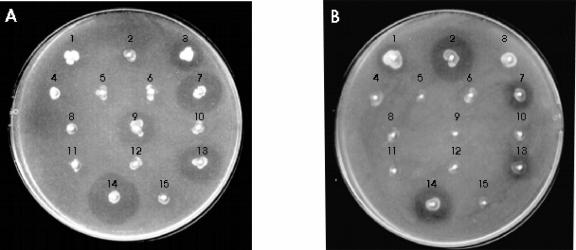
FIG. 3.
Heterologous complementation for MccE492 production. A patch test was performed on the indicator strains (A) FGB099, to detect heterologous MccE492 production, and (B) MC4100(pB492), to detect MccH47 production. The stabs correspond to the following strains: 1, H47; 2, RYC1000(pEX100); 3, RYC492; 4, RYC1000; 5, RYC1000(pAB492); 6, RYC1000(pAB492, pMVD41); 7, RYC1000(pAB492, pEX100); 8, RYC1000(pAB492, pEX100 mchA::Tnlac7.2); 9, RYC1000(pAB492, pEX100 mchB::Tn576); 10, RYC1000(pAB492, pEX100 mchC::Tnlac7.7); 11, RYC1000(pAB492, pEX100 mchD::Tnlac3.5); 12, RYC1000(pAB492, pEX100 mchE::Tnlac7.1); 13, RYC1000(pAB492, pEX100 mchS1::Tnlac5.4); 14, MC4100(pAB492, pEX100) (Ent+); and 15, FGB103(pAB492, pEX100) (Ent−).
To identify the MccH47 genes necessary to produce MccE492, pAB492 was put in the presence of pEX100-derivative plasmids with mutations in different mch genes and the resulting strains were assayed for microcin production (Fig. 3). Once again, strains that were defective for MccH47 maturation or secretion did not produce either antibiotic. Cells that were impaired for MchS1 still produced both microcins, and those affected in the MccH47 activity gene mchB produced only MccE492. The requirement of enterobactin synthesis for heterologous MccE492 production was also confirmed: two isogenic strains, Ent+ and Ent−, proved to be proficient and deficient for microcin production, respectively (Fig. 3).
A strain producing only MccE492 activity was analyzed on the set of indicator strains carrying different mutations in the genes for catechol receptors (Fig. 4). The pattern of sensitivity of single-, double-, and triple-receptor mutants clearly indicated that MccE492 produced in the heterologous context presented the same spectrum of activity as did microcins from the reference strains RYC492 and H47: halos appeared on every lawn tested with the exception of that corresponding to the cir, fiu, and fepA triple mutant. In addition, the assay was performed on a tonB-null mutant (FGB11), which was also completely resistant. In sum, MccE492 was heterologously produced following the catechol strategy and was taken up by E. coli K-12 cells through the catechol uptake pathway.
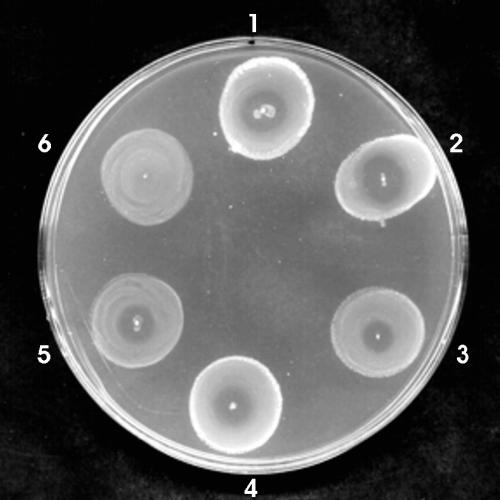
FIG. 4.
Sensitivity of catechol receptor mutants to heterologously produced MccE492. A patch test was performed on small medal-like lawns of the following indicator strains: 1, MC4100 (WT); 2, FGB1594 (fiu); 3, FGB102 (fepA); 4, FGB1728 (fiu cir); 5, FGB1870 (fiu fepA); and 6, FGB1876 (cir fiu fepA). Patches correspond to the strain RYC1000(pAB492, pEX100 mchB::Tn576), which produces only MccE492 activity.
Screening for antibiotic activities employing the catechol receptors Cir, Fiu, and FepA.
A report concerning the distribution of mch-specific sequences among E. coli strains from different sources revealed their overwhelming presence in uropathogenic E. coli isolates, while they seldom appeared in intestinal (pathogenic or nonpathogenic) or in other extraintestinal E. coli isolates (9). Taking this information into account, 160 gram-negative clinical isolates from urine were analyzed for their ability to produce antibiotic activities that depended on the presence of any of the catechol receptors and of the TonB pathway for their action on E. coli K-12 cells. The idea was that such activities could be chromosome-encoded microcins produced following the catechol strategy.
A total of 49 isolates (48 E. coli and 1 K. pneumoniae) inhibited the growth of an E. coli K-12 indicator strain. Of these, 35 (71%) did not generate inhibition halos on a tonB-deficient mutant (FGB11), indicating that the TonB pathway would be involved in their uptake. When tested on the collection of catechol receptor mutant strains, 18 (37%) of all the antibiotic-producing isolates exhibited the same pattern as microcins H47 and E492: only the triple mutant was resistant. These 18 activities also belonged to the TonB-dependent subgroup.
To assess whether any of these 18 antibiotic activities produced MccH47 or MccE492, they were analyzed on indicator strains that were immune to these microcins. One E. coli isolate did not generate halos on the MccH47 immune strain, indicating that it produced MccH47. Another isolate produced MccE492 since it could not inhibit the growth of MccE492 immune cells. Interestingly, this latter isolate corresponded to the single K. pneumoniae strain producing an antibacterial activity. The remaining isolates that were assayed originated inhibition halos on all the indicator strains, and therefore, they produced antibiotic activities that were different from microcins H47 and E492.
DISCUSSION
In this work, we attempted to identify common features presented by chromosome-encoded microcins, a group now integrated by microcins H47, I47, E492, and M. In organisms whose genome has been extensively sequenced, microcin gene clusters were found to integrate genomic islands: the serX pathogenicity island of E. coli CFT073 and the genomic island I of E. coli Nissle1917 (9, 25). Accordingly, the E. coli H47-sequenced DNA segment containing the MccH47 system presented strong similarities with sequences belonging to genomic islands of other enterobacteria, most of them involved in pathogenicity. The extensive direct repeat located at the boundaries of the E. coli H47 mch gene cluster recalls similar structures that often flank genomic islands (10). This repeat, presumably generated during the mch integration event into the chromosome, suggests that the MccH47 cluster might be an islet, which could behave as a unit for transfer. Considering its surrounding sequences, it appears as an islet inside an island.
All the mch genes devoted to microcin maturation and secretion present in the H47 strain have clear homologues in the other enterobacterial strains producing chromosome-encoded microcins, irrespective of the kind of microcin being produced. This statement would also apply to E. coli CFT073, although its ability to produce antibiotic has not been reported. The MccH47 activity and immunity genes form a tightly linked pair, the immunity determinant being upstream of the activity one, an arrangement invariably found for all the cognate activity-immunity genes in the microcin systems under consideration. The analyzed MccH47 genetic system shows a compacted arrangement of the mch genes, as do the microcin systems in E. coli CA46, E. coli CA58, and K. pneumoniae RYC492. On the contrary, a different gene disposition is found in E. coli Nissle1917 and E. coli CFT073, where the iroBD genes, related to the salmochelin system, are not included in the microcin cluster but integrate the separate iroA locus. From a functional point of view, this would not be a complete obstacle for microcin production but could be the reason for the “residual” levels of microcin activity reported for strain Nissle 1917 (17). This latter arrangement of genes that are involved in microcin production could correspond to an earlier evolutionary stage in relation to the compacted systems of other strains. The recruitment of the iroBD genes into a single microcin cluster probably implies their integration into the regulatory circuits of gene expression devoted to microcin production.
In this study, chromosome-encoded microcins are correlated with a specific mode of synthesis that determines their uptake through the catechol receptors. MccI47, a new microcin encoded by the E. coli H47 MccH47 system, was identified and shown to follow the catechol strategy in all its terms. Its production is related to a pair of activity-immunity genes, mchS2S3, plus the participation of the MccH47 maturation and export pathways, which are shared by microcins I47 and H47. The mchS2S3 genes were searched for in the MccH47 systems from other strains, being found quasi-identical in E. coli CA46, mutated in E. coli CA58, and absent in E. coli Nissle1917 and E. coli CFT073. MccM-specific determinants could be in a similar situation, as they are present in all of the mentioned E. coli strains except for E. coli H47, which contains only truncated remnants. Apparently, these antibiotic systems would possess great genetic plasticity, acquiring and losing information to produce different microcins. Presumably, when strains produce more than one chromosome-encoded microcin, these activities would sum up their effects since they are targeted to the same bacterial population.
The fact that the E. coli H47 MccH47 system codes for two microcins and that these are produced from different precursor peptides, which are then processed through a common route of maturation and secretion, indicates that their antibiotic specificity resides in their peptide portion and that maturation and export proteins would have a relaxed substrate specificity. Then, these latter proteins could eventually function with any microcin peptide precursor able to follow the catechol strategy. This was confirmed for MccE492 through heterologous complementation assays: the maturation and export genes from the MccH47 system, put in the presence of the MccE492 activity-immunity genes, proved to efficiently complement for MccE492 production. Although it cannot be discarded that the MccE492 molecule thus produced might not be exactly the same as the naturally produced antibiotic, no indication in this sense was detected. In fact, the results showed that the heterologous MccE492 suffered posttranslational modifications following the catechol strategy, was secreted through the type I apparatus, and inhibited the growth of E. coli K-12 cells depending on the presence of any of the catechol receptors.
The heterologous secretion of MccE492 was indeed not surprising: the protein components of the type I exporters for different microcins, either plasmid or chromosome encoded, are extremely similar in their sequences. In this sense, the reported precedent of MccH47 secretion by the colicin V exporter was auspicious (3). On the contrary, MccE492 heterologous maturation was not obvious since proteins involved in microcins H47 and E492 maturation pathways, although being clearly homologous, presented enough differences in their sequence to eventually account for more specific interactions with their substrates. In addition, it should be stressed that MccE492 genes have never been found together with MccH47 genes and that these two microcins are naturally produced by different enterobacterial species.
If the maturation proteins were able to proceed with any of the chromosome-encoded microcin precursors, then they should recognize a specific common feature in these peptides. The serine-rich C-terminal region of microcins H47, I47, E492, and M could well be the motif recognized by the microcin modification proteins. On the basis of these considerations, it is tempting to think that the versatility of these microcins' maturation and export proteins could allow the functional incorporation of new pairs of activity-immunity determinants proceeding from lateral transfer events, provided that the encoded antibiotic peptide precursors possessed the required double-glycine and serine-rich signal motifs. The new incorporations would then have the opportunity to be assayed so that they could be maintained or discarded depending on the advantages conferred to the cell.
The screening performed on clinical urine isolates revealed that 11% produced antibiotic activities which needed the presence of any of the three catechol receptors as well as the TonB pathway for their action on E. coli K-12 cells. We presume that these antibiotics that are employing the catechol uptake pathway are most probably related to microcins H47, I47, E492, and M in their genetics, synthesis, secretion, and overall structure. Their significant prevalence among urine isolates agrees with results from other authors relating MccH47 with uropathogenicity. Sequences from the MccH47 system were found almost exclusively in uropathogenic E. coli strains (9). Moreover, it was reported that iro and mch genes from the uropathogenic strain E. coli CFT073 are up-regulated during urinary tract infections as well as during growth in human urine (22). In more general terms, the participation of iro genes in the synthesis of chromosome-encoded microcins directly relates the production of these antibacterial activities with that of the siderophore salmochelins, which are considered virulence factors of extraintestinal (mainly uropathogenic) E. coli strains (9). Therefore, chromosome-encoded microcins could be aimed at enhancing the fitness of uropathogenic enterobacteria at some stage of their lives in the urinary tract. Their strict observance of a synthesis scheme determining their uptake through the catechol receptors indicates the importance of this way of entry into competing cells. It seems obvious that chromosome-encoded microcins should be more effective in low-iron conditions, where the production of catechol receptors is induced, and thus susceptible bacteria become more sensitive to microcin action. These conditions would be met by the urinary tract, where it has been reported that Fur-regulated genes would enhance their expression (21, 22). Therefore, the production of chromosome-encoded microcins might be a valuable resource among enterobacteria reaching the urinary tract and, as such, these microcins could be seen as virulence factors, a point that deserves further study.
Acknowledgments
This work was supported by Comisión Sectorial de Investigación Científica, by Programa de Desarrollo Tecnológico grant 141, and by Programa de Desarrollo de las Ciencias Básicas, Uruguay.
We are indebted to Graciela Borthagaray, who kindly supplied the clinical isolates from urine cultures from the Servicio de Microbiología, Hospital Central de las Fuerzas Armadas, Uruguay, and to Eliana Rodríguez for critical reading of the manuscript. We are also grateful to María Parente for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., W. T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azpiroz, M. F., and M. Laviña. 2004. Involvement of enterobactin synthesis pathway in production of microcin H47. Antimicrob. Agents Chemother. 48:1235-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azpiroz, M. F., E. Rodríguez, and M. Laviña. 2001. The structure, function, and origin of the microcin H47 ATP-binding cassette exporter indicate its relatedness to that of colicin V. Antimicrob. Agents Chemother. 45:969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baquero, F., and F. Moreno. 1984. The microcins. FEMS Microbiol. Lett. 23:117-124. [Google Scholar]
- 5.Bäumler, A. J., T. L. Norris, T. Lasco, W. Voigt, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischbach, M. A., L. Hening, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. USA 102:571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaggero, C., F. Moreno, and M. Laviña. 1993. Genetic analysis of microcin H47 antibiotic system. J. Bacteriol. 175:5420-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 11.Hantke, K. 1990. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol. Lett. 67:5-8. [DOI] [PubMed] [Google Scholar]
- 12.Havarstein, L. S., H. Holo, and I. F. Nes. 1994. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by gram-positive bacteria. Microbiology 140:2383-2389. [DOI] [PubMed] [Google Scholar]
- 13.Lagos, R., M. Baeza, G. Corsini, C. Hetz, E. Strahsburger, J. A. Castillo, C. Vergara, and O. Monasterio. 2001. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42:229-243. [DOI] [PubMed] [Google Scholar]
- 14.Laviña, M., C. Gaggero, and F. Moreno. 1990. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J. Bacteriol. 172:6585-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127:11075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Patzer, S. I., M. R. Baquero, D. Bravo, F. Moreno, and K. Hantke. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149:2557-2570. [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez, E., and M. Laviña. 1998. Genetic analysis of microcin H47 immunity. Can. J. Microbiol. 44:692-697. [DOI] [PubMed] [Google Scholar]
- 19.Rodríguez, E., and M. Laviña. 2003. The proton channel is the minimal structure of ATP synthase necessary and sufficient for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 47:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez, E., C. Gaggero, and M. Laviña. 1999. The structural gene for microcin H47 encodes a peptide precursor with antibiotic activity. Antimicrob. Agents Chemother. 43:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russo, T. A., U. B. Carlino, A. Mong, and S. T. Jodush. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 67:5306-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. T. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas, X., D. Destoumieux-Garzon, J. Peduzzi, C. Afonso, A. Blond, N. Birlirakis, C. Goulard, L. Dubost, R. Thai, J. C. Tabet, and S. Rebuffat. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279:28233-28242. [DOI] [PubMed] [Google Scholar]
- 24.Trujillo, M., E. Rodríguez, and M. Laviña. 2001. ATP synthase is necessary for microcin H47 antibiotic action. Antimicrob. Agents Chemother. 45:3128-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]