Abstract
Tiamulin is a pleuromutilin antibiotic that is used in veterinary medicine. The recently published crystal structure of a tiamulin-50S ribosomal subunit complex provides detailed information about how this drug targets the peptidyl transferase center of the ribosome. To promote rational design of pleuromutilin-based drugs, the binding of the antibiotic pleuromutilin and three semisynthetic derivatives with different side chain extensions has been investigated using chemical footprinting. The nucleotides A2058, A2059, G2505, and U2506 are affected in all of the footprints, suggesting that the drugs are similarly anchored in the binding pocket by the common tricyclic mutilin core. However, varying effects are observed at U2584 and U2585, indicating that the side chain extensions adopt distinct conformations within the cavity and thereby affect the rRNA conformation differently. An Escherichia coli L3 mutant strain is resistant to tiamulin and pleuromutilin, but not valnemulin, implying that valnemulin is better able to withstand an altered rRNA binding surface around the mutilin core. This is likely due to additional interactions made between the valnemulin side chain extension and the rRNA binding site. The data suggest that pleuromutilin drugs with enhanced antimicrobial activity may be obtained by maximizing the number of interactions between the side chain moiety and the peptidyl transferase cavity.
Tiamulin and valnemulin are used in veterinary medicine to treat enteric diseases in pigs and enzootic pneumonia in pigs and poultry. Valnemulin has also been used in human medicine to treat isolated cases of immunocompromised patients with resistant Mycoplasma infections (6). Timaulin-resistant isolates of Brachyspira hyodysenteriae and Brachyspira pilosicoli, the causative agents of swine dysentery and porcine intestinal spirochetosis, respectively, have been reported recently in various parts of Europe (4, 9, 10). However, there is only limited knowledge on mechanisms of resistance to the pleuromutilin antibiotics. The development of tiamulin resistance occurs in a slow, stepwise fashion in vitro (1, 3, 8). An investigation of tiamulin resistance in Escherichia coli showed that mutation of ribosomal protein L3 at position 149 proximal to the peptidyl transferase center leads to a tiamulin resistance phenotype in a single mutant (2). More recently, mutations in ribosomal protein L3 and 23S rRNA have been associated with reduced susceptibility to tiamulin in Brachyspira isolates (13). Mutations were found at two amino acid positions of L3 and six nucleotides of 23S rRNA in two groups of laboratory-selected mutants. Each mutant contained a unique combination of mutations, implying that a single mutation was not sufficient to cause the highest levels of resistance. A set of British veterinary B. hyodysenteriae field isolates contained a single mutation in ribosomal protein L3 at position 148. These results emphasize the need for the development of improved pleuromutilin derivatives. The mutated positions are consistent with the X-ray structure of tiamulin bound in the 50S subunit in that all mutations are clustered around nucleotide U2504, which forms part of the wall of the tiamulin binding cavity (13, 15). A combination of structural data from the X-ray structure of a tiamulin-50S complex and biochemical information from chemical footprinting can improve our understanding of drug binding and thus pave the way for rational design of new pleuromutilin derivatives. Precedence for the improvement of ribosomal antibiotics through the development of semisynthetic derivatives that make additional contacts with the ribosome is illustrated in the case of the ketolides that are semisynthetic derivatives of erythromycin.
In this work, chemical footprinting was used to investigate the binding of four pleuromutilin derivatives at the peptidyl transferase center. In addition, the susceptibility of an E. coli L3 mutant strain to these pleuromutilin derivatives was examined, as well as their binding to L3 mutant ribosomes. We conclude that the thioether and acyl carbamate side chain extensions of pleuromutilin derivatives can adopt distinct conformations within the peptidyl transferase cavity and that additional interactions between the side chain extension of valnemulin and the cavity are responsible for the lack of valnemulin cross-resistance in an L3 mutant strain.
MATERIALS AND METHODS
E. coli strains.
Strain MRE600 was used to prepare ribosomes for the chemical footprinting experiments. The wild-type parent strain CN2476 and L3 mutant strain JB5 (2) were used for susceptibility testing and to prepare ribosomes for the chemical footprinting experiments.
Footprinting experiments.
Ribosome isolation, chemical modification with dimethyl sulfate (DMS) and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT), and primer extension procedures were carried out essentially as described previously (12). Kethoxal modifications were performed by incubating 5 pmol of the antibiotic-70S complexes for 30 min at 37°C in 50 μl modification buffer (70 mM HEPES-KOH, pH 7.8, 10 mM MgCl2, 270 mM KCl), followed by modification with 5 μl kethoxal (40 mg/ml in 20% ethanol) for 20 min at 37°C. The reactions were stopped by adding 5 μl SB buffer (0.3 M sodium acetate, 0.25 M H3BO3), followed by precipitation with ethanol. The washed pellet was redissolved in 100 μl SB buffer, extracted with phenol, (1:1) phenol-chloroform, and chloroform, followed by precipitation with 3 volumes of ethanol. The pellet was then dissolved in 7 μl TEB buffer (10 mM Tris-Cl, pH 7.8, 0.1 mM EDTA, 50 mM potassium borate) and used in primer extension reactions.
The following DNA oligonucleotide primers were used in the extension reactions: 5′-TCCGGTCCTCTCGTACT-3′, complementary to nucleotides 2654 to 2670 of 23S rRNA; 5′-CCATGCAGACTGGCGTC-3′, complementary to nucleotides 2141 to 2157 of 23S rRNA; and 5′-GAACAGCCATACCCTTG-3′, complementary to nucleotides 2540 to 2556 of 23S rRNA. The cDNA products of the primer extension reactions were separated on 6 or 8% polyacrylamide-7 M urea sequencing gels. The intensities of the modification bands were quantified using a phosphorimager.
Antimicrobial susceptibility testing.
For MIC determination, overnight liquid cultures were diluted and 102 to 103 cells were spread onto antibiotic-containing NZY agar plates. MICs are the results of at least two to three independent experiments and are expressed as ranges, where the lower value represents the highest antibiotic concentration at which there are easily visible single colonies after 24 h and the higher value represents the lowest tested concentration where there are no easily visible colonies after 24 h. The highest antibiotic concentration tested was 400 μg/ml.
RESULTS AND DISCUSSION
Chemical footprinting of pleuromutilin derivatives.
The interaction of pleuromutilin antibiotics with the peptidyl transferase cavity was examined using a set of derivatives with different side chain extensions. Chemical footprinting was used to compare ribosomal binding of the fungal natural product pleuromutilin and the semisynthetic derivatives tiamulin, valnemulin, and SB-264128 (Fig. 1). Tiamulin and valnemulin have thioether side chains substituted at the hydroxyl group of the ester moiety of the parent compound pleuromutilin. SB-264128 is an acyl carbamate derivative with improved metabolic stability and bioavailability that was synthesized as part of an effort to develop a pleuromutilin analog for human use (7).
FIG. 1.
Chemical structures of the pleuromutilin derivatives used in this study.
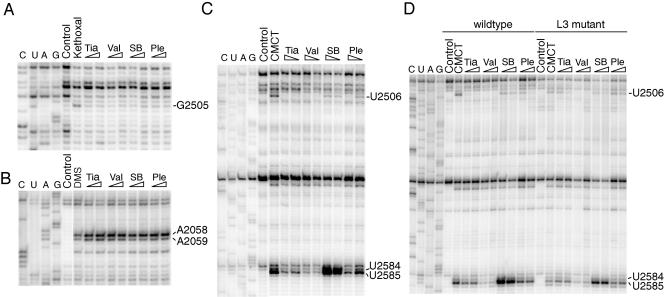
The drug binding sites on the ribosome were studied by chemical probing, where accessible rRNA nucleotides are modified by chemical reagents. After formation of pleuromutilin derivative-70S ribosome complexes, the antibiotic-70S complexes and control samples of free ribosomes were treated with DMS to probe the accessibility of the N1 position of adenine and N3 of cytosine, with CMCT to probe N3 of uracil and N1 of guanine, and with kethoxal to probe N1 and N2 of guanine. Primer extension with reverse transcriptase was then used to identify alterations induced by drug binding. Autoradiograms showing the affected nucleotides in the central part of domain V of 23S rRNA are presented in Fig. 2A to C. The observed protection and enhancement effects are tabulated in Table 1 and indicated on the secondary structure of domain V of Escherichia coli 23S rRNA (Fig. 3A).
FIG. 2.
(A to C) Gel autoradiograms showing the antibiotic footprints on MRE600 ribosomes modified with (A) kethoxal, (B) DMS, and (C) CMCT. (D) Comparison of CMCT footprints on CN2476 (wild-type) and JB5 (L3 mutant) ribosomes. Nucleotides exhibiting altered reactivities in the presence of the pleuromutilin derivatives are indicated. Dideoxy sequencing lanes are designated G, A, U, and C. Lanes are labeled to indicate reactions with chemically unmodified 70S ribosomes in the absence of drugs (control) and 70S ribosomes modified in the absence of drugs (kethoxal, DMS, and CMCT). Ribosomes modified in the presence of tiamulin (Tia), valnemulin (Val), SB-264128 (SB), or pleuromutilin (Ple) are marked, where wedges are used to indicate a low (2 μM) or high (20 μM) drug concentration.
TABLE 1.
Summary of chemical footprinting data with pleuromutilin derivativesa
| 23S rRNA position | No drug | Tiamulin | Valnemulin | SB-264128 | Pleuromutilin |
|---|---|---|---|---|---|
| A2058 | ++ | ++++ E | ++++ E | +++ E | +++ E |
| A2059 | + | +++ E | +++ E | ++ E | + |
| G2505 | + | − P | − P | − P | − P |
| U2506 | ++ | − P | − P | + P | (+) P |
| U2584 | ++ | + P | + P | +++ E | ++ |
| U2585 | +++ | + P | (+) P | ++++ E | ++ P |
E, enhancement; P, protection. The extent of modification with DMS, CMCT, or kethoxal is indicated with + symbols, with (+) and − indicating very weak and no modification, respectively.
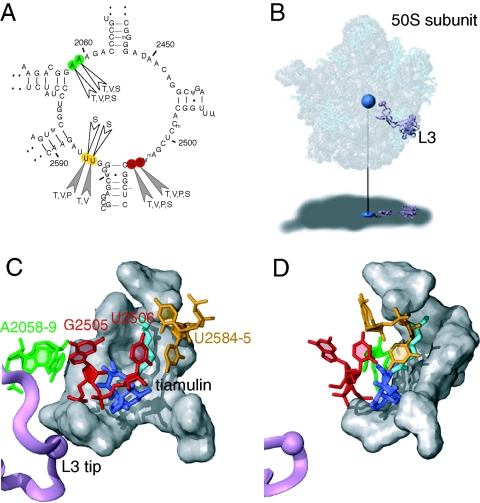
FIG. 3.
Ribosomal binding site of the pleuromutilin antibiotics. (A) Secondary structure of domain V of E. coli 23S rRNA, showing the chemical footprints of the four pleuromutilin antibiotics in color (A2058 and A2059 in green, G2505 and U2506 in red, and U2584 and U2585 in yellow). The nucleotide positions exhibiting altered reactivities in the presence of each drug (T, tiamulin; V, valnemulin; P, pleuromutilin; and S, SB-264128) are indicated. Protection effects are shown as filled arrowheads and enhancement effects as open arrowheads. (B) The structure of the 50S subunit from Deinococcus radiodurans complexed with tiamulin (15) (PDB accession no. 1XBP), where tiamulin is represented as a blue sphere and ribosomal protein L3 as a purple ribbon. RNA is represented as gray spheres, and proteins are shown as light blue ribbons. The subunit is rendered transparently to show the internal positions of tiamulin and L3, which are projected onto the shadow below. (C) An expanded view of the tiamulin binding site. Tiamulin is shown in stick representation, where the mutilin core is in dark blue and the side chain extension is in cyan. The nucleotides in the footprints and L3 are colored as described for panels A and B. Other nucleotides involved in hydrophobic interactions with tiamulin (15) are shown in gray. (D) As in panel C, but rotated 60 degrees around the y axis.
The footprinting data show that all four derivatives protect G2505 from kethoxal modification (Fig. 2A). The tiamulin and valnemulin footprints include enhancements at A2058 and A2059 (Fig. 2B) and protections at U2506, U2584, and U2585 (Fig. 2C) and are consistent with the results of a previous study using DMS and CMCT (12). Attenuated effects are observed at A2058, U2506, and U2585 in the presence of pleuromutilin, whose footprint is generally weaker than those of tiamulin and valnemulin (Fig. 2B and C). In addition, there are no significant alterations in reactivity observed at U2584 in the presence of pleuromutilin. In the SB-264128 footprint, strong enhancements are observed at U2584 and U2585, as well as weak enhancements at A2058 and A2059 and a weak protection at U2506 (Fig. 2B and C). The differences in the footprints of the four drugs are primarily at nucleotides U2506, U2584, and U2585. The degree of protection at U2506 upon drug binding increases from SB-264128 to pleuromutilin to tiamulin to valnemulin, with relative protections of 35, 80, 95, and 100%, respectively, at a 20 μM drug concentration. The strength of the protection effects at U2584 and U2585 is correlated with increasing side chain length from pleuromutilin to tiamulin to valnemulin. However, the strong enhancements observed with SB-264128 at U2584 and U2585 show that these nucleotides are more, rather than less, accessible to CMCT modification upon SB-264128 binding compared to ribosomes with no drug bound. Thus, the footprinting data show that the side chain extensions of the derivatives are able to interact with neighboring rRNA nucleotides and influence their conformation.
E. coli L3 mutant strain susceptibility to pleuromutilins and drug binding to L3 mutant ribosomes.
A previous investigation showed that a point mutation resulting in an Asn-to-Asp alteration at position 149 of ribosomal protein L3 causes reduced susceptibility to tiamulin in E. coli (2). In order to test whether this phenotype applies to other pleuromutilin derivatives, the susceptibilities of the parent strain to the four pleuromutilins were determined and compared with those of the L3 mutant strain. The results are summarized as MIC ranges in Table 2. The L3 mutant strain has a decreased susceptibility to tiamulin, pleuromutilin, and SB-264128 relative to the parent strain. In contrast, no difference in susceptibility to valnemulin is observed between the parent and L3 mutant strains. Thus, the L3 mutation does not confer cross-resistance to valnemulin. This is important, as mutations at corresponding positions of ribosomal protein L3 occur in both laboratory-selected Brachyspira mutants and B. hyodysenteriae field isolates with decreased susceptibility to tiamulin (13). As the mutated position is approximately 10 to 12 Å from the drug binding pocket, the mutation apparently functions indirectly through perturbation of the drug binding cavity. The mutation presumably induces an altered conformation of the binding cavity, perturbing the drug-rRNA interactions at the mutilin core. For tiamulin, pleuromutilin, and SB-264128, this leads to decreased susceptibility of the L3 mutant strain and reduced binding to L3 mutant ribosomes. The lack of valnemulin cross-resistance in the L3 mutant strain suggests that valnemulin makes additional interactions with the binding cavity through its side chain extension and remains bound despite an altered binding surface around the mutilin core. The footprinting technique is limited to obtaining information on the subset of nucleotide bases that are accessible to chemical probes and the portions of these bases involved in the modification reactions. Therefore, additional interactions may be present without necessarily being detectable via chemical footprinting.
TABLE 2.
Susceptibilities of E. coli parent and L3 mutant strains to pleuromutilins
| Strain | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Tiamulin | Valnemulin | SB264128 | Pleuromutilin | |
| CN2476 (parent) | 100-125 | 15-20 | 50-75 | 200-250 |
| JB5 (L3 mutant) | 350-400 | 15-20 | 75-100 | >400 |
Chemical footprinting was used to compare binding of the four pleuromutilin drugs on wild-type and L3 mutant ribosomes. Ribosome-drug complexes were modified with CMCT and isolated rRNA analyzed by primer extension to monitor the protection effect at position U2506 at antibiotic concentrations of 2 and 20 μM. In the wild-type ribosomes, complete protection at U2506 is observed at 2 μM tiamulin and 20 μM pleuromutilin (Fig. 2D, lanes 5 to 8, 13, and 14). In contrast, a nearly complete protection of U2506 in L3 mutant ribosomes is observed only at 20 μM tiamulin, and only partial protection is observed at 20 μM pleuromutilin (Fig. 2D, lanes 15 to 18, 23, and 24). The U2506 protection in the presence of 2 or 20 μM SB-264128 is around 55% for wild-type ribosomes, while there is no significant protection of U2506 at 2 μM and around 30% protection at 20 μM SB-264128 in the case of L3 mutant ribosomes, showing that SB-264128 binds less efficiently to L3 mutant than to wild-type ribosomes. No significant difference in the U2506 protection with valnemulin is detected between wild-type and L3 mutant ribosomes in the concentration range of 0.02 to 20 μM (Fig. 2D, lanes 9, 10, 19, and 20, and data not shown). The results show that drug binding to L3 mutant ribosomes is markedly reduced except in the case of valnemulin, consistent with the susceptibility data and lack of valnemulin cross-resistance of the L3 mutant strain.
Correlations to the pleuromutilin binding site at the peptidyl transferase center.
It is possible to directly relate the positions of nucleotides in the footprints to the bound drug by using the X-ray structure of the Deinococcus radiodurans 50S subunit bound to tiamulin (15) (Fig. 3B). The positions of the nucleotides affected in the pleuromutilin footprints relative to bound tiamulin are illustrated in Fig. 3C and D. The tricyclic core of tiamulin is positioned in a cavity defined in part by nucleotides G2505 and U2506 (15). The protections at G2505 and U2506 can be rationalized in that these nucleotides are within 3 to 4 Å from the mutilin core and close enough to contact the drugs directly through hydrogen bonding and hydrophobic interactions. The protection effects at U2584 and U2585 can be assigned to the side chain extensions off of the mutilin core. The strength of the U2585 protection is correlated with the side chain length and increases from pleuromutilin to tiamulin to valnemulin. This is consistent with the fact that the distance from U2585 to the tiamulin side chain extension is about 4 Å. The data further suggest that the side chain extension of SB-264128 induces a change in rRNA conformation within the cavity, where U2584 and U2585 are more accessible to modification by CMCT than without drug bound. Nucleotides A2058 and A2059 are located in the peptide exit tunnel and are 7 to 8 Å from the bound mutilin core, suggesting that the footprints here are indirect effects induced by drug binding. Similar enhancement effects at these positions have also been observed with streptogramin A drugs (11, 14), whose binding site at the peptidyl transferase center overlaps extensively with that of tiamulin (5, 15).
Concluding remarks.
The pleuromutilin antibiotics examined in this work all affect the reactivities of nucleotides A2058, A2059, G2505, and U2506, reflecting the binding of the common tricyclic mutilin core of pleuromutilins to the peptidyl transferase center. However, the side chain extensions of the derivatives adopt different conformations within the cavity as evidenced by the diverse effects observed at positions U2584 and U2585, including both protections and enhancements. The susceptibility of the L3 mutant to pleuromutilins suggests that resistance caused by mutation of ribosomal protein L3 can be overcome by increasing the number of side chain interactions with the binding cavity. There is thus a potential for drug improvement by lengthening and derivitization of the pleuromutilin side chain according to the information on tiamulin binding to ribosomes derived from X-ray crystallography.
Acknowledgments
We thank Novartis for furnishing tiamulin and valnemulin and GlaxoSmithKline for pleuromutilin. We thank Scott Champney for discussions and for providing the SB-264128 derivative as part of a collaboration. We thank Jacob Poehlsgaard for assistance with Fig. 3.
B.V. was supported by the Nucleic Acid Center funded by the Danish National Research Council. K.S.L. was supported by a grant from the European Commission's 5th Framework Program (grant QLK2-CT-2002-00892).
REFERENCES
- 1.Böck, A., F. Turnowsky, and G. Högenauer. 1982. Tiamulin resistance mutations in Escherichia coli. J. Bacteriol. 151:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bøsling, J., S. M. Poulsen, B. Vester, and K. S. Long. 2003. Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein L3. Antimicrob. Agents Chemother. 47:2892-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drews, J., A. Georgopoulos, G. Laber, E. Schutze, and J. Unger. 1975. Antimicrobial activities of 81.723 hfu, a new pleuromutilin derivative. Antimicrob. Agents Chemother. 7:507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fossi, M., T. Saranpää, and E. Rautiainen. 1999. In vitro sensitivity of the swine Brachyspira species to tiamulin in Finland 1995-97. Acta Vet. Scand. 40:355-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harms, J. M., F. Schlünzen, P. Fucini, H. Bartels, and A. Yonath. 2004. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heilmann, C., L. Jensen, J. S. Jensen, K. Lundstrom, D. Windsor, H. Windsor, and D. Webster. 2001. Treatment of resistant mycoplasma infection in immunocompromised patients with a new pleuromutilin antibiotic. J. Infect. 43:234-238. [DOI] [PubMed] [Google Scholar]
- 7.Hunt, E. 2000. Pleuromutilin antibiotics. Drugs Future 25:1163-1168. [Google Scholar]
- 8.Karlsson, M., A. Gunnarsson, and A. Franklin. 2001. Susceptibility to pleuromutilins in Brachyspira (Serpulina) hyodysenteriae. Anim. Health Res. Rev. 2:59-65. [PubMed] [Google Scholar]
- 9.Karlsson, M., A. Aspan, A. Landen, and A. Franklin. 2004. Further characterization of porcine Brachyspira hyodysenteriae isolates with decreased susceptibility to tiamulin. J. Med. Microbiol. 53:281-285. [DOI] [PubMed] [Google Scholar]
- 10.Lobova, D., J. Smola, and A. Cizek. 2004. Decreased susceptibility to tiamulin and valnemulin among Czech isolates of Brachyspira hyodysenteriae. J. Med. Microbiol. 53:287-291. [DOI] [PubMed] [Google Scholar]
- 11.Porse, B. T., and R. A. Garrett. 1999. Sites of interaction of streptogramin A and B antibiotics in the peptidyl transferase loop of 23S rRNA and the synergism of their inhibitory mechanisms. J. Mol. Biol. 286:375-387. [DOI] [PubMed] [Google Scholar]
- 12.Poulsen, S. M., M. Karlsson, L. B. Johansson, and B. Vester. 2001. The pleuromutilin drugs tiamulin and valnemulin bind to the RNA at the peptidyl transferase centre on the ribosome. Mol. Microbiol. 41:1091-1099. [DOI] [PubMed] [Google Scholar]
- 13.Pringle, M., J. Poehlsgaard, B. Vester, and K. S. Long. 2004. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 54:1295-1306. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Fonseca, C., R. Amils, and R. A. Garrett. 1995. Fine structure of the peptidyl transferase centre on 23S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J. Mol. Biol. 247:224-235. [DOI] [PubMed] [Google Scholar]
- 15.Schlünzen, F., E. Pyetan, P. Fucini, A. Yonath, and J. M. Harms. 2004. Inhibition of peptide bond formation by pleuromutilins: the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol. Microbiol. 54:1287-1294. [DOI] [PubMed] [Google Scholar]