Abstract
The rmpA2 gene, which encodes an activator for capsular polysaccharide (CPS) synthesis, was isolated from a 200-kb virulence plasmid of Klebsiella pneumoniae CG43. Based on the sequence homology with LuxR at the carboxyl-terminal DNA-binding motif, we hypothesized that RmpA2 exerts its effect by activating the expression of cps genes that are responsible for CPS biosynthesis. Two luxAB transcriptional fusions, each containing a putative promoter region of the K. pneumoniae K2 cps genes, were constructed and were found to be activated in the presence of multicopy rmpA2. The activation is likely due to direct binding of RmpA2 to the cps gene promoter through its C-terminal DNA binding motif. Moreover, the loss of colony mucoidy in a K. pneumoniae strain deficient in RcsB, a regulator for cps gene expression, could be recovered by complementing the strain with a multicopy plasmid carrying rmpA2. The CPS production in Lon protease-deficient K. pneumoniae significantly increased, and the effect was accompanied by an increase of RmpA2 stability. The expression of the rmpA2 gene was negatively autoregulated and could be activated when the organism was grown in M9 minimal medium. An IS3 element located upstream of the rmpA2 was required for the full activation of the rmpA2 promoter. In summary, our results suggest that the enhancement of K2 CPS synthesis in K. pneumoniae CG43 by RmpA2 can be attributed to its transcriptional activation of K2 cps genes, and the expression level of rmpA2 is autoregulated and under the control of Lon protease.
Klebsiella pneumoniae, an important nosocomial pathogen, causes suppurative infection, pneumonia, urinary tract infection, and septicemia in humans. Clinically isolated K. pneumoniae usually produces large amounts of capsular polysaccharides (CPS) as reflected by the formation of glistening mucoid colonies with viscid consistency (25). The degree of mucoidy has been shown to positively correlate with the establishment of infection (19, 20). Among at least 77 K serotypes distinguished (18), K. pneumoniae strains belonging to serotypes K1 and K2 are the most virulent to mice (17). It has been well documented that CPS protects K. pneumoniae from complement-mediated serum killing and phagocytosis (6, 11, 30, 37).
Bacterial CPS can be classified into two groups by chemical and physical criteria. Group I polysaccharides contain uronic acid as the acidic component, have high molecular mass, and are coexpressed with specific O polysaccharides. Group II polysaccharides contain a large variety of acidic components and have a relatively low molecular mass (12). Klebsiella K2 CPS has been determined as [→)4-Glc-(1→3)-α-Glc-(1→4)-β-Man-(3←1)-α-GlcA)-(1→]n (34), which is made from a biosynthetic pathway similar to that of the group I CPS in Escherichia coli (36). The genomic organization of the chromosomal cps (CPS synthesis) region that is responsible for K2 CPS biosynthesis in K. pneumoniae Chedid has been determined, and a total of 15 open reading frames organized into two transcriptional units have been identified as indispensable (2) (Fig. 1A).
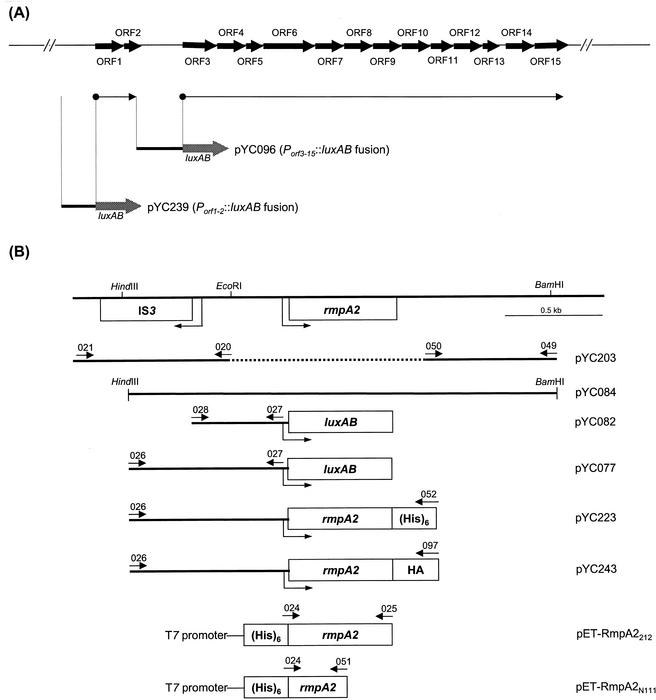
FIG. 1.
(A) Organization of the K. pneumoniae K2 cps gene cluster. The horizontal arrows that begin with a solid circle represent the putative transcriptional units. The putative promoter regions that were cloned into pYC017 as luxAB transcriptional fusions are indicated. (B) Physical map of the rmpA2 gene. The positions of the IS3 element, the primers used for PCR amplification, and the extents of subclones used in this study are indicated.
The regulatory strategy of colanic acid biosynthesis employed by E. coli usually serves as a model for that of group I CPS synthesis. It is built around the RcsC/RcsB two-component pair, where RcsC is the transmembrane sensor and RcsB is the response regulator. The active transcriptional regulator that binds upstream of the cps gene cluster (9) consists of activated RcsB and an accessory positive regulator, RcsA. The availability of RcsA is normally limited because the RcsA protein is rapidly degraded by the ATP-dependent Lon protease; therefore, mutations at lon have been found to lead to the accumulation of colanic acid (9). In E. coli K-12, the presence of RcsB is indispensable for CPS biosynthesis, whereas an rcsA-negative phenotype can be suppressed by multicopy rcsB (4).
K. pneumoniae CG43, a clinical isolate of K2 serotype, showed strong virulence in a mouse peritonitis model with a 50% lethal dose (LD50) as low as 10 CFU (5). A large plasmid of approximately 200-kb in size was noted to be present in this strain as well as in most, if not all, bacteremic isolates of K. pneumoniae. The cure of the 200-kb plasmid from K. pneumoniae CG43 resulted in a loss of colony mucoidy and a 1,000-fold decrease in virulence. This plasmid was therefore designated as pLVPK (for large virulence plasmid in Klebsiella). Sequence analysis of a pathogenicity island carried by pLVPK has revealed a locus named rmpA2, which has been reported to enhance the colony mucoidy of various serotypes of K. pneumoniae (34). Introduction of multicopy rmpA2 could confer on E. coli HB101 the ability to produce Klebsiella K2 capsule in the presence of K2 cps gene cluster, and the transcription of a long-strand mRNA from K2 cps operon was simultaneously increased (1, 33). These findings indicated that RmpA2 functions as a trans-acting activator for the CPS biosynthesis. Due to the essential role of CPS in K. pneumoniae pathogenesis, it would be important to understand how RmpA2 exerts its activation to the K2 CPS biosynthesis. We report here our results on characterizing RmpA2 as a transcriptional activator for the Klebsiella K2 cps genes.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli, K. pneumoniae CG43 (5, 22), and its derivatives were propagated at 37°C in Luria-Bertani (LB) broth. M9 minimal medium was prepared as described previously (26). The density of the bacterial culture was determined by measuring the absorbance of adequate dilutions at an optical density at 600 nm (OD600) with a Shimadzu UV-1201 spectrophotometer and expressed as OD600 × dilution factor.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| K. pneumoniae | ||
| CG43 | Clinical isolates | 22 |
| CG43-17 | CG43 with chromosome Tn5 insertion, galU Kmr | 5 |
| CG43S3 | CG43 Smr | This study |
| CG43S3-R2035 | CG43S3 ΔrmpA2 Smr | This study |
| CG43S3-B2202 | CG43S3 ΔrcsB Smr | This study |
| CG43S3-L2117 | CG43S3 Δlon Smr | This study |
| CG43S3-RB01 | CG43S3 ΔrmpA2 ΔrcsB Smr | This study |
| CG43S3-RL01 | CG43S3 ΔrmpA2 Δlon Smr | This study |
| E. coli | ||
| S17-1 | hsdR recA pro RP4-2 (Tc::Mu; Km::Tn7) | 28 |
| S17-1λpir | hsdR recA pro RP4-2 (Tc::Mu; Km::Tn7)(λ pir) | 28 |
| NovaBlue(DE3) | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB laclqZΔM15Tn10] (DE3) | Novagen |
| Plasmids | ||
| pET30a | His-tagged protein expression vector, Kmr | Novagen |
| pKAS46 | Suicide vector, rpsL Apr Kmr | 28 |
| pRK415 | Shuttle vector, mob+ Tcr | 14 |
| pYC017 | Promoter selection vector, GalU+ LuxAB+ Cmr | This study |
| pYC077 | Fragment containing the 0.9-kb region upstream of rmpA2 cloned into pYC017 | This study |
| pYC082 | Fragment containing the 0.5-kb region upstream of rmpA2 cloned into pYC017 | This study |
| pYC084 | 2.1-kb HindIII/BamHI fragment containing the entire rmpA2 locus cloned into pRK415 | This study |
| pYC096 | 1.0-kb fragment containing the promoter region of K2 CPS orf3 through orf15 cloned into pYC017 | This study |
| pYC203 | 2.0-kb fragment containing a 530-bp deletion in rmpA2 locus cloned into pKAS46 | This study |
| pYC211 | 1.9-kb fragment containing a 1.3-kb deletion in lon locus cloned into pKAS46 | This study |
| pYC220 | 2.0-kb fragment containing a 763-bp deletion in rcsB locus cloned into pKAS46 | This study |
| pYC223 | 1.5-kb fragment containing a C-terminally fused His tag with the rmpA2-coding region cloned into pRK415 | This study |
| pYC239 | 0.7-kb fragment containing the promoter region of K2 CPS orf1-orf2 cloned into pYC017 | This study |
| pYC243 | 1.5-kb fragment containing a C-terminally fused HA tag with the rmpA2-coding region cloned into pRK415 | This study |
| pET-RmpA2212 | 636-bp fragment containing full-length RmpA2 cloned into pET30a | This study |
| pET-RmpA2N111 | 333-bp fragment containing the N-half of RmpA2 cloned into pET30a | This study |
Construction of gene-specific mutants.
K. pneumoniae CG43 mutants disrupted specifically at rmpA2, rcsB, or lon genes were constructed by the allelic exchange strategy. The primer sets used for PCR amplification of the DNA fragments that flank the regions to be deleted are listed in Table 2. For homologous recombination, the generated DNA fragments were cloned into pKAS46, and the resulting plasmids were then mobilized to K. pneumoniae CG43S3 through conjugation from E. coli S17-1λpir. Plasmid pKAS46 (a generous gift of K. Skorupski, University of New Hampshire) is a suicide vector containing the rpsL gene, which allows positive selection with streptomycin for the loss of the vector (28). One of the kanamycin-resistant transconjugants was picked, grown overnight, and then spread onto an LB plate supplemented with streptomycin (500 μg/ml). After the occurrence of double crossover, the streptomycin-resistant and kanamycin-sensitive colonies were selected, and the deletions of rmpA2, rcsB, or lon were verified by PCR and by Southern blot analysis with a gene-specific probe. Three gene-specific mutant strains—K. pneumoniae R2035 (ΔrmpA2), B2202 (ΔrcsB), and L2117 (Δlon)—were obtained, and K. pneumoniae R2035 was used further to generate double-mutant strains K. pneumoniae RB01 (ΔrmpA2 ΔrcsB) and RL01 (ΔrmpA2 Δlon) (Table 1).
TABLE 2.
Primers used in this study
| Primer no. | Sequence | Enzyme cleaved | Complementary position |
|---|---|---|---|
| 020 | 5′-CTGTGTGATTAAGAATTCATA-3′ | 5′-EcoRI | −223 relative to the rmpA2 start codon |
| 021 | 5′-ATCCTCTAGAGTCGACGCGTT-3′ | 5′-XbaI | −1392 relative to the rmpA2 start codon |
| 024 | 5′-CATGCCATGGGAAAATATATTTAC-3′ | 5′-NcoI | +1 of the rmpA2 coding region |
| 025 | 5′-ATATCTCGAGTTATCTAGGTATTTGATG-3′ | 5′-XhoI | +636 of the rmpA2 coding region |
| 026 | 5′-ACGCGGATCCTAGCTCCACAGGTAAAGT-3′ | 5′-BamHI | −812 relative to the rmpA2 start codon |
| 027 | 5′-ACGCGGATCCGTCCAGTTAACTGCTTTA-3′ | 5′-BamHI | +96 relative to the rmpA2 start codon |
| 028 | 5′-ATATGGATCCCACTTAGTCCTGTGTCCA-3′ | 5′-BamHI | −390 relative to the rmpA2 start codon |
| 040 | 5′-ACTGGATCAGGCCTGGTAATCGCCATT-3′ | 5′-BamHI | −891 relative to the orf3 start codon |
| 041 | 5′-ACTGGATCCCGCTGTCGTATCTCAATG-3′ | 5′-BamHI | +60 relative to the orf3 start codon |
| 049 | 5′-GCCGAGCTCTGGACATAGACAGC-3′ | 5′-SacI | +639 relative to the rmpA2 stop codon |
| 050 | 5′-CCGGAATTCCTAAAGGGTGTGATTATG-3′ | 5′-EcoRI | +295 relative to the rmpA2 start codon |
| 051 | 5′-ATATCTCGAGTTAAATTTCCTTGCATGTTGAC-3′ | 5′-HindIII | +333 of the rmpA2 coding region |
| 052 | 5′-CCCAAGCTTCAGTGGTGGTGGTGGTGGTGGGTATTTGATGTGC ACCA-3′ | 5′-HindIII | +636 of the rmpA2 coding region |
| 053 | 5′-TGCTCTAGAAGCGACTCATCGATCAG-3′ | 5′-XbaI | +999 relative to the lon stop codon |
| 054 | 5′-CCCAAGCTTGCGCTGCAGAACGCC-3′ | 5-HindIII | +61 relative to the lon stop codon |
| 055 | 5′-CCCAAGCTTCACGCGCTCCAGGCCATA-3′ | 5-HindIII | +993 relative to the lon start codon |
| 056 | 5′-AGAGAGCTCTATGAATCCTGAGCG-3′ | 5′-SacI | +10 relative to the lon start codon |
| 061 | 5′-AGAGAGCTCTGCAGCTGCTCATCAACA-3′ | 5′-SacI | −983 relative to the rcsB start codon |
| 062 | 5′-CCCAAGCTTGCGCATCCTTTTCGCGA-3′ | 5′-HindIII | −10 relative to the rcsB start codon |
| 063 | 5′-CCCAAGCTTATTCCCGCCCTTTACGCA-3′ | 5-HindIII | +47 relative to the rcsB stop codon |
| 064 | 5′-TGCTCTAGAGGGGATCCCGGCGAAA-3′ | 5′-XbaI | +1018 relative to the rcsB stop codon |
| 074 | 5′-ACTGGATCCACGATCATGGATAAGAT-3′ | 5′-BamHI | −723 relative to the orf1 start codon |
| 075 | 5′-ACTGGATCCTGCGACCGGAATAACC-3′ | 5′-BamHI | +42 relative to the orf1 start codon |
| 097 | 5′-CCCAAGCTTCTAAGCGTAGTCTGGGACGTCGTATGGGTAGGTATTT GATGTGCACCAA-3′ | 5′-HindIII | +636 of the rmpA2 coding region |
Construction of luxAB transcriptional fusions.
For complementation of the rmpA2 deletion, a 2.1-kb HindIII/BamHI fragment that comprises the entire rmpA2 locus (Fig. 1B) was cloned into a shuttle vector pRK415 (14) to generate pYC084. To construct Pcps::luxAB fusion plasmids compatible with pYC084, the primer sets (Table 2) were designed based upon the published sequence of K2 cps genes (GenBank accession no. D21242) and used for PCR amplification of the putative promoter regions. Primer sets used to amplify the upstream region of rmpA2 gene are also listed in Table 2. These generated promoter fragments were then cloned into pYC017, a derivative of pYC016 (15), which contains a copy of promoterless luxAB of Vibrio fischeri as a reporter and a control cassette from pKK232-8 that ensures transcriptional fusion and prevents interference from a fortuitous plasmid promoter. The fragments that encode a full-length RmpA2 protein with a C-terminally fused six-His tag and a hemagglutinin (HA) tag driven by its own promoter were also amplified by PCR with respective primer sets 026/052 and 026/097 (Table 2), cloned into pRK415 to generate pYC223 or pYC243. All these constructs were transferred into K. pneumoniae CG43S3, and its mutant derivatives through conjugation from E. coli S17-1.
Luciferase activity assay.
The expression of different promoter::luxAB fusions was assessed by measuring the luciferase activity as follows. Overnight bacterial cultures were recovered with 1:10 dilution in M9 medium at 25°C for 4.5 h. Five hundred microliters of the recovered culture was mixed with 500 μl of 0.1% (vol/vol) n-decyl aldehyde (Sigma-Aldrich, Milwaukee, Wis.). The mixture was made to stand at room temperature for 60 s and then read in full integral, auto ranging mode of a 30 s integration time with a luminometer (TD-20/20; Turner Designs). The data shown was normalized and expressed as relative light units/OD600.
Mouse lethality assay.
Female BALB/c mice with an average weight of 25 g were obtained from the animal center of National Taiwan University and were acclimatized in an animal house for 3 days. The tested bacterial strains were cultured in LB medium at 37°C for overnight. Five mice of a group were injected intraperitoneally with bacteria resuspended in 0.2 ml of saline in 10-fold steps graded doses. The LD50s, based on the number of survivors after one week, were calculated by the method of Reed and Muench (24) and expressed as CFU.
Resistance to serum killing.
One hundred microliters of bacterial suspension in saline was mixed with 100 μl of pooled serum from healthy volunteers, and the mixture was incubated at 37°C for 30 min. The number of viable bacteria in the mixture was then determined by plating.
Extraction and quantification of CPS.
CPS was extracted by the method described previously (7). Five hundred microliters of bacterial culture was mixed with 100 μl of 1% Zwittergent 3-14 detergent (Sigma-Aldrich) in 100 mM citric acid (pH 2.0), and then the mixture was incubated at 50°C for 20 min. After centrifugation, 250 μl of the supernatant was transferred to a new tube, and CPS was precipitated with 1 ml of absolute ethanol. The pellet was then dissolved in 200 μl of distilled water, and a 1,200-μl volume of 12.5 mM borax (Sigma-Aldrich) in H2SO4 was added. The mixture was vigorously vortexed, boiled for 5 min, and cooled, and then 20 μl of 0.15% 3-hydroxydiphenol (Sigma-Aldrich) was added and the absorbance at 520 nm was measured. The uronic acid content was determined from a standard curve of glucuronic acid (Sigma-Aldrich) and expressed as micrograms per 109 CFU (3).
RNA dot blotting analysis.
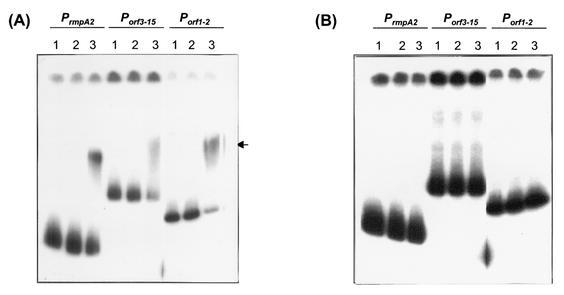
Total RNA was isolated from mid-log-phase K. pneumoniae cells (OD600 = 0.6 to 0.8) by extraction with the TRI reagent (Molecular Research Center, Cincinnati, Ohio). Contaminating DNA was eliminated from the RNA samples with RQ1 RNase-free DNase (Promega, Madison, Wis.). Probes used in hybridization assay were labeled with fluorescein-11-dUTP by random priming with the Gene Images kit (Amersham-Pharmacia, Piscataway, N.J.). Five and ten micrograms of total RNA was transferred onto a Hybond-N+ membrane (Amersham-Pharmacia) by dot blotting, prehybridized for 1 h at 65°C, hybridized overnight at the same temperature, washed, and detected with the CDP-Star reagent (Amersham-Pharmacia).
Expression and purification of recombinant RmpA2.
The coding region of rmpA2 was amplified with primers 024 and 025 (Table 2), and cloned as an NcoI/XhoI fragment into pET30a (Novagen, Madison, Wis.). The resulting plasmid pET-RmpA2212 allowed in-frame fusion of the full-length rmpA2 coding region to six histidine codons at the N terminus and transcription from a T7 promoter. A C-terminal truncated form of RmpA2, comprising codons 1 to 111 of rmpA2, was amplified with primers 024 and 051 (Table 2), cloned into pET30a, and resulted in plasmid pET-RmpA2N111. The overexpressed His-RmpA2 proteins were then purified from the soluble fraction of total cell lysate by affinity chromatography with His-Bind resin (Novagen). The purified His-RmpA2212 and His-RmpA2N111 were then concentrated and dialyzed against 1× storage buffer (100 mM KCl, 20 mM MgCl2, 10 mM Na2HPO4 [pH 7.4], 1.8 mM KH2PO4 [pH 7.4], and 10% glycerol), and the purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
DNA electrophoretic mobility shift assay (EMSA).
DNA fragments comprising the testing promoter regions were obtained by PCR amplification with primer sets 027-028, 040-041, and 074-075 (Table 2), respectively, followed by end labeling with [γ-32P]ATP. The purified His-RmpA2 proteins, ranging from 50 ng to 1 μg, were mixed with DNA probes (0.1 ng) in 50-μl reaction mixtures containing 12 mM HEPES (pH 7.4), 100 mM KCl, 20 mM MgCl2, 0.6 mM dithiothreitol, and 5% glycerol. The mixtures were incubated at room temperature for 25 min, mixed with 0.1 volume of DNA loading dye, and then loaded onto 5% nondenaturing polyacrylamide gels containing 5% glycerol in 0.5× TBE buffer (45 mM Tris-HCl [pH 8.0], 45 mM boric acid, 1.0 mM EDTA). Gels were electrophoresed with a 20-mA current at 4°C and dried under a vacuum, and the results were detected by autoradiography.
Determination of turnover of RmpA2 protein.
Pulse-chase and immunoprecipitation experiments were performed as described previously (32). K. pneumoniae cells were grown at 37°C to an OD600 of 0.6 and labeled for 2 min with 10 μl of l-[35S]methionine (1,000 Ci/mmol; New England Nuclear) per ml in M63 medium (27). The labeled cells were chased with M63 medium containing 0.5% l-methionine, and 1.5 ml of the samples was collected at the time indicated. The cell precipitates were resuspended in 30 μl of 1× lysis buffer (1% SDS, 1 mM EDTA, 10 mM Tris-HCl [pH 7.4]), boiled for 5 min, and diluted 30-fold with 1× immunoprecipitation buffer (1 mM EDTA, 10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 0.5% NP-40, and Complete protease inhibitor [Roche Molecular Biochemicals, Mannheim, Germany]). Five microliters of anti-His monoclonal antibody (MAb) (Novagen) or anti-HA MAb (Roche Molecular Biochemicals) was added, and the incubation was continued at 4°C overnight. The immunoprecipitates were adsorbed onto protein A-Sepharose (Amersham-Pharmacia) and were washed three times before being resuspended in SDS-PAGE loading buffer and electrophoresed. The amount of RmpA2 protein on the gel was determined with a densitometer (Molecular Dynamics).
RESULTS
Isolation and characterization of an rmpA2 deletion mutant.
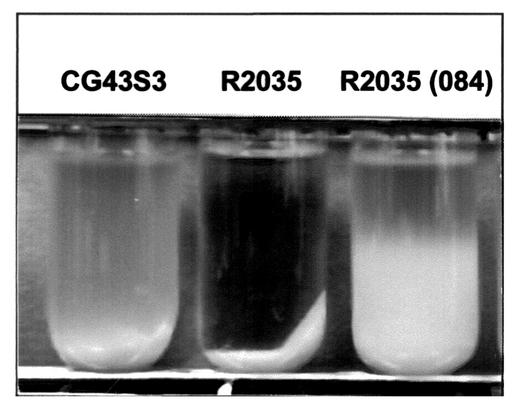
To assess the role of RmpA2 in the regulation of K2 CPS biosynthesis, a K. pneumoniae rmpA2 deletion mutant designated R2035 was first generated. The mutant strain displayed a large, glistening colony morphology on LB agar that was indistinguishable from that of wild-type strains. However, the colony mucoidy of R2035 appeared to reduce, as determined by the inability of the colony to form a string using a toothpick. The reduction of mucoidy was also evident when the bacterial cultures were subjected to low-speed centrifugation. As shown in Fig. 2, R2035 cells precipitated much faster than its parental strain CG43S3. Introduction of pYC084 (Fig. 1B) that contains a functional rmpA2 gene into R2035 restored the colony mucoidy as reflected by both the string formation test and the slower precipitation (Fig. 2). The amount of CPS produced in R2035 was further quantified by measuring the glucuronic acid content, which serves as an indicator of Klebsiella K2 CPS (21). As shown in Table 3, R2035 synthesized less K2 CPS than the wild-type strain, and the effect was even more pronounced when the cells were grown in M9 medium with 0.4% glucose. The complementation strain R2035[pYC084] produced twice as much CPS as that produced by R2035 (Table 3). Finally, in a mouse peritonitis model, the deletion of rmpA2 resulted in an increase of LD50 by 25-fold, and the reduction in virulence could be restored and even enhanced in strain R2035 complemented with pYC084 (Table 4). However, the rmpA2 mutant R2035 was as resistant as its parental strain to the human serum killing effect (Table 4).
FIG. 2.
Comparison of precipitation speeds of K. pneumoniae CG43S3, K. pneumoniae R2035 (ΔrmpA2), and K. pneumoniae R2035 [pYC084]. The strains tested were cultured overnight in LB broth at 37°C and subjected to centrifugation at 1,000 ×g for 5 min.
TABLE 3.
Effects of RmpA2 on CPS synthesis in K. pneumoniae CG43 strains with different genetic backgrounds
| Strains | CPS synthesized when grown as indicatedd
|
Mucoid phenotypec | |||||
|---|---|---|---|---|---|---|---|
| LB incubation
|
Minimal medium (0.4% glucose)
|
Minimal medium (1% glycerol)
|
|||||
| Mean quantity ± SDa | Foldb | Mean quantity ± SD | Fold | Mean quantity ± SD | Fold | ||
| CG43S3 | 15.4 ± 0.7 | 110.4 ± 3.9 | 51.6 ± 6.3 | + | |||
| R2035 | 13.5 ± 0.5 | 79.1 ± 3.6 | 48.4 ± 5.1 | − | |||
| B2202 | 12.9 ± 0.8 | 72.2 ± 2.9 | 33.3 ± 2.8 | − | |||
| L2117 | 97.4 ± 2.8 | 600.6 ± 32.4 | 232.6 ± 9.7 | +++ | |||
| RB01 | 10.4 ± 1.2 | 62.3 ± 4.7 | 27.3 ± 5.6 | − | |||
| RL01 | 10.9 ± 1.4 | 60.7 ± 5.3 | 35.1 ± 6.2 | − | |||
| S3[pYC084] | 29.2 ± 2.5 | 1.89 | 166.1 ± 6.2 | 1.50 | 65.3 ± 6.4 | 1.27 | ++ |
| R2035[pYC084] | 30.9 ± 1.8 | 2.29 | 235.3 ± 13.4 | 2.97 | 87.4 ± 8.9 | 1.81 | ++ |
| B2202[pYC084] | 31.4 ± 2.7 | 2.43 | 158.5 ± 8.4 | 2.20 | 61.2 ± 7.7 | 1.84 | ++ |
| L2117[pYC084] | 111.8 ± 6.7 | 1.15 | 814.3 ± 31.5 | 1.36 | 417.3 ± 9.6 | 1.79 | +++ |
| RB01[pYC084] | 30.3 ± 3.2 | 2.91 | 188.5 ± 5.8 | 3.03 | 72.0 ± 3.5 | 2.64 | + |
| RL01[pYC084] | 26.1 ± 2.9 | 2.39 | 268.2 ± 7.5 | 4.42 | 67.8 ± 6.4 | 1.93 | ++ |
Values are the averages of triplicate samples and are given as micrograms of uronic acid per 109 CFU.
Compared with each parental strain without pYC084.
Assessed by string formation test after 48 h of growth on M9-glucose medium. Symbols: −, negative; +, positive; ++, strong; +++, very strong.
The production of polysaccharides on the cell surface of each strain was ascertained as K2 CPS by the Quellung test.
TABLE 4.
Virulence properties of K. pneumoniae rmpA2 mutant
| Strain | LD50 (CFU) | Survival rate in human serum (%)a |
|---|---|---|
| CG43S3 | 4 × 103 | >90 |
| R2035 | 1 × 105 | >90 |
| R2035[pYC084] | 8 × 102 | >90 |
| CG43-17 (control)b | 1 × 106 | 0 |
Percent survival rate in human serum is expressed as 100 × (the number of viable bacteria after treatment/the number of viable bacteria before treatment).
CG43-17, a galU mutant strain with defective CPS and lipopolysaccharide (5).
Enhancement of K2 cps gene expression by rmpA2.
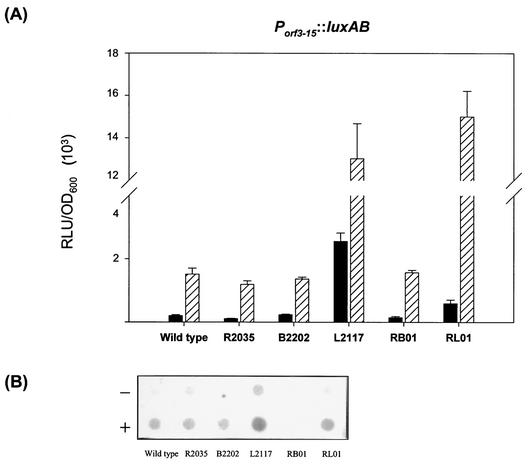
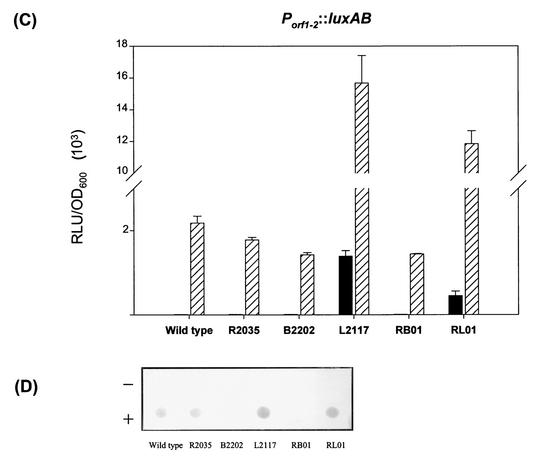
In E. coli K-12 strains, the amount of colanic acid produced has been reported to be correlated with the transcriptional level of cps genes (9). We reasoned that the production of CPS enhanced by rmpA2 might also have resulted from an elevated transcriptional level of Klebsiella K2 cps genes. To test the possibility, two luxAB reporter fusions, pYC096 (Porf3-15::luxAB), which carries the putative promoter region responsible for initiating transcription of the operon containing orf3 to orf15, and pYC239 (Porf1-2::luxAB), which comprises the region controlling the expression of orf1 and orf2 (Fig. 1A), were generated. The luxAB fusion plasmids were then individually transformed into K. pneumoniae strains CG43S3 and R2035 in the presence or absence of pYC084. As shown in Fig. 3A and C, the activity of Porf3-15 and Porf1-2 in the wild-type strain CG43S3 was barely detectable and indistinguishable from that in the rmpA2 deletion strain R2035. However, in the presence of pYC084, expression of the Porf3-15::luxAB fusion increased 7-fold in CG43S3 and 11-fold in R2035 (Fig. 3A). A similar enhancement by pYC084 was also observed for Porf1-2, in which RmpA2 performed as a strong activator (Fig. 3C).
FIG. 3.
Expression of K2 cps genes in various genetic backgrounds. The luciferase activity of K2 Pcps::luxAB transcriptional fusions in strains CG43S3 (wild type), R2035 (ΔrmpA2), B2202 (ΔrcsB), L2117 (Δlon), RB01 (ΔrmpA2 ΔrcsB), and RL01 (ΔrmpA2 Δlon) carrying either the Porf3-15::luxAB fusion (A) or the Porf1-2::luxAB fusion (C) in the presence (striped bars) or absence (solid bars) of pYC084 was determined (error bars, standard deviations). Ten micrograms of total RNA extracted from different K. pneumoniae cells was spotted on a Hybond-N+ membrane and hybridized with probes specific to either orf3 (B) or orf1 (D). + and −, presence and absence of pYC084, respectively. RLU, relative light units.
The activation was further verified by RNA dot blotting analysis with fluorescein-labeled DNA fragments containing the coding region of orf3 and orf1, respectively, as a probe. As shown in Fig. 3B and D, the amount of both orf3- and orf1-containing transcripts in the testing strains was increased in the presence of pYC084. Therefore, it can be concluded that the transcriptional level of K2 cps genes could be elevated by the multicopy rmpA2 in a trans-acting way.
Requirement of the C-terminal half of RmpA2 for its trans-activating properties.
The rmpA2 gene contains a poly(G) tract located at position +276 relative to the A residue of the first in frame start codon. The length of the poly(G) tract in the rmpA2 genes has been investigated in a number of clinical isolates of K. pneumoniae and was found to be variable, ranging from 9 to 12. Only the 11-G tract allowed rmpA2 remain in frame to encode a full-length RmpA2 protein. Other G tracts would incur a truncated RmpA2 protein by an amber stop codon in the reading frames. Since truncated forms of RmpA2 are frequently observed in clinical strains, it would be of interest to know whether these gene products retain its trans-activating function. Expression constructs for the full-length RmpA2 and a truncated RmpA2 with a tract of 10 G residues were generated and designated pET-RmpA2212 and pET-RmpA2N111, respectively. These two plasmids were then cotransformed individually with one of the two Pcps::luxAB transcriptional fusions into E. coli NovaBlue(DE3). After IPTG (isopropyl-β-d-thiogalactopyranoside) induction, the luciferase activity of each luxAB fusion was measured. Consistent with the effects of pYC084, pET-RmpA2212 activated the expression of Porf1-2::luxAB and Porf3-15::luxAB (Table 5). However, pET-RmpA2N111, which produced a truncated form of RmpA2, did not display similar trans-activating properties.
TABLE 5.
Effect of RmpA2 on expression of luxAB transcriptional fusions in E. coli NovaBlue(DE3)
| Strain | Mean LuxAB activity ± SD (RLU/OD600)a
|
||
|---|---|---|---|
| Porf3-15::luxAB | Porf1-2::luxAB | PrmpA2::luxAB | |
| NovaBlue(DE3) | 215 ± 13 | 5 ± 0.74 | 12,638 ± 2,168 |
| NovaBlue(DE3)[pET-RmpN111] | 328 ± 52 | 9 ± 1 | 11,237 ± 1,045 |
| NovaBlue(DE3)[pET-Rmp212] | 1,236 ± 83 | 104 ± 2 | 1,813 ± 112 |
Luciferase activity was measured after 8 h of induction with 1 mM IPTG at 25°C. Values are the averages of triplicate samples. RLU, relative light units.
Binding of RmpA2 to its target promoters.
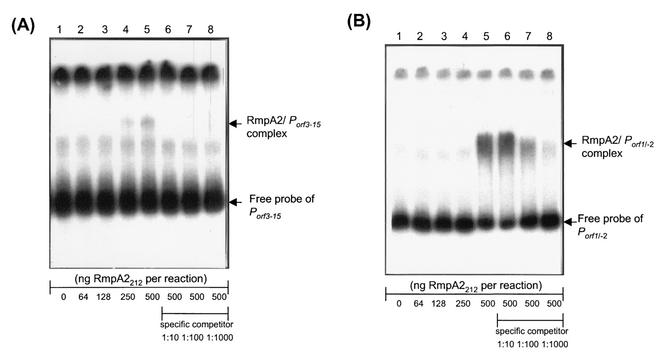
To validate RmpA2 as a transcriptional regulator that interacts with its target promoter directly, DNA EMSA was performed with the purified RmpA2 protein and each of the DNA fragments carrying the promoter region of K2 cps genes. As shown in Fig. 4, RmpA2 is capable of binding to the DNA fragments containing Porf3-15 and Porf1-2, whereas no DNA-protein complexes could be observed with the C-terminal truncated RmpA2. The DNA-protein interaction is specific, and the formation of RmpA2-promoter complex could be inhibited in the presence of specific competitor DNA (Fig. 5A and B).
FIG. 4.
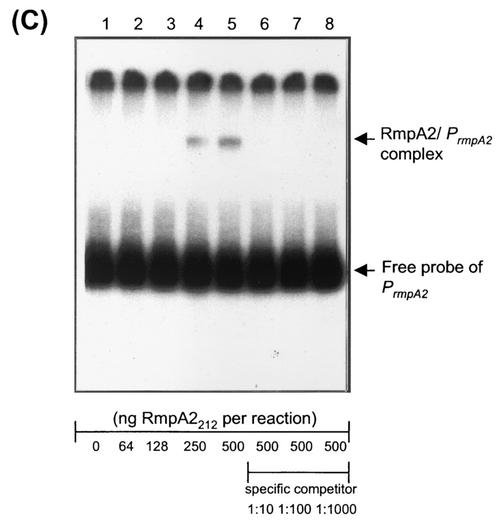
DNA EMSA of RmpA2 and its target promoters. The 32P-labeled DNA probes of PrmpA2, Porf3-15, or Porf1-2 were incubated with the full-length His-RmpA2212 (A) or the C-terminally truncated His-RmpA2N111 (B). Lanes 1 to 3 contain RmpA2 amounts of 0, 50 ng, and 500 ng, respectively. The arrow indicates the protein-DNA complex.
FIG. 5.
Binding specificity of RmpA2 to its target promoters. The 32P-labeled DNA probes of Porf3-15 (A), Porf1-2 (B), and PrmpA2 (C) were incubated with increasing amounts of His-RmpA2212 as indicated. Lanes: 1, no protein; 2, 64 ng of protein; 3, 128 ng of protein; 4, 250 ng of protein; 5 to 8, 500 ng of protein. Specific inhibition of binding was investigated by adding the indicated amounts of unlabeled DNA fragment of a target promoter: lane 6, 1 ng; lane 7, 10 ng; lane 8, 100 ng.
Effects of rmpA2 on K2 CPS biosynthesis in rcsB mutant K. pneumoniae cells.
Analysis of predicted protein sequence of RmpA2 has shown that it belongs to the UhpA-LuxR family, which also includes RcsA and RcsB (29). To further investigate the relationship between RmpA2 and the Rcs system in CPS biosynthesis, an rcsB deletion mutant, B2202 and a double mutant, RB01 (ΔrmpA2 ΔrcsB), were generated in K. pneumoniae CG43S3 (Table 1). Similar to the rmpA2 deletion mutant R2035, strain B2202 lost the colony mucoid characteristic and displayed reduced K2 CPS synthesis (Table 3). In the double-mutant strain RB01, the reduction of K2 CPS was more pronounced (Table 3). The deletion of rcsB, however, did not affect the activation of Porf1-2 and Porf3-15 by RmpA2 (Fig. 3). Complementation of B2202 and RB01with pYC084 not only restored but also enhanced colony mucoidy and CPS production in these strains (Table 3).
Effects of rmpA2 on K2 CPS biosynthesis in lon mutant K. pneumoniae cells.
To investigate the interplay between RmpA2 and Lon protease, a lon deletion mutant L2117 and a double-deletion mutant RL01 (ΔrmpA2 Δlon) were generated. Similar to the E. coli lon mutant strains, K. pneumoniae L2117 displayed extremely mucoid colony morphology and produced at least four times as much K2 CPS as the wild-type strain (Table 3). The transcription level and promoter activity of K2 cps genes were also found to increase in L2117; however, when the rmpA2 and lon genes were simultaneously deleted, the enhancement of CPS production following lon deletion was diminished (Fig. 3 and Table 3). The result suggested that the K2 CPS biosynthesis was negatively regulated by Lon protease and this action was mediated by RmpA2.
Comparison of RmpA2 stability in wild-type and lon mutant K. pneumoniae strains.
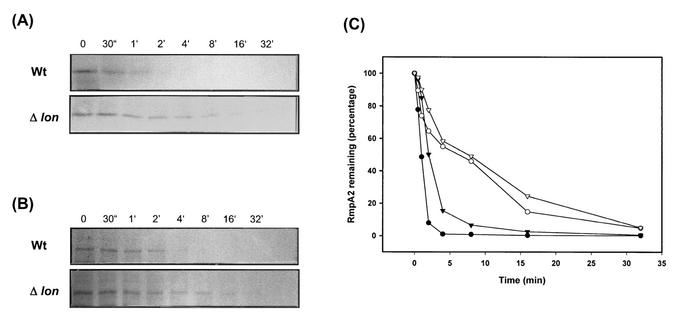
In E. coli K-12 strains, the increase of CPS production in lon mutant cells can be explained by the stabilization of RcsA protein (29). Whether the accumulation of CPS in Lon protease-deficient K. pneumoniae L2117 is due to stabilization of RmpA2 was investigated. The stability of RmpA2 protein was determined by a pulse-chase analysis with [35S]methionine as a label. We have found that the concentration of RmpA2 in K. pneumoniae was rather low and was virtually undetectable even in the lon deletion strain. To increase the concentration of RmpA2, the low-copy-number plasmid pYC223 or pYC243, which encodes an RmpA2 protein with a C-terminally fused His tag or HA tag, were constructed and subsequently transferred into CG43S3 and L2117. After pulse-chase treatment, the RmpA2 protein was precipitated with anti-His or anti-HA MAb. As shown in Fig. 6, the half-life of either His-RmpA2 or HA-RmpA2 was found to increase from 1 to 2 min in the wild-type K. pneumoniae CG43S3 to approximately 10 min in the lon mutant strain.
FIG. 6.
Stability of RmpA2 protein in K. pneumoniae. The bacterial cells were pulse-labeled with [35S]methionine and chased at the indicated time points. Tag-fused RmpA2 protein was immunoprecipitated with anti-His MAb or with anti-HA MAb and then subjected to SDS-PAGE. (A) Turnover of His-RmpA2 protein. (B) Turnover of HA-RmpA2 protein in the wild-type strain K. pneumoniae CG43S3 (Wt) or in the lon mutant strain L2117 (Δlon). (C) Quantification of the autoradiogram shown in panel A of wild-type (solid circles) or lon mutant (open circles) cells and of that in panel B of wild-type (solid triangles) or lon mutant (open triangles) cells. The quantity of labeled protein at time zero was set at 100%.
Autoregulation of rmpA2.
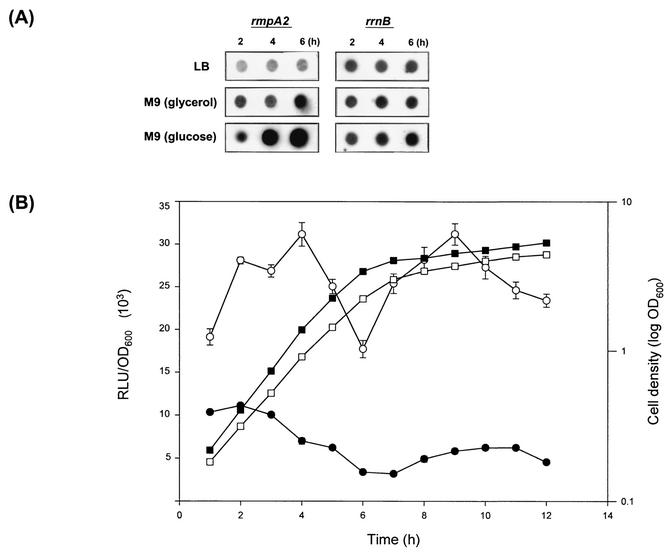
As shown in Table 3, the growth of K. pneumoniae strains in M9 minimal medium increased the production of CPS. The enhancement might have resulted from an activation of rmpA2 gene expression. To test the possibility, total RNA was extracted from R2035 [pYC084] grown in LB or under CPS-inducing conditions, with M9 minimal medium supplemented with glycerol or glucose as the sole carbon source. As shown in Fig. 7A, the CPS-inducing conditions increased the amount of rmpA2 transcripts. Consistent with the result shown in Table 3, the growth in M9 medium-0.4% glucose that induced the highest level of expression of the rmpA2 gene also led to the highest yield of K2 CPS. To quantitatively analyze the activity of the rmpA2 promoter, the plasmid pYC082, which comprises the putative control region of rmpA2, was generated as a luxAB fusion construct (Fig. 1B). At different time points, the expression of PrmpA2::luxAB was found to be at least twofold more when grown under the CPS-inducing conditions than when grown in LB broth.
FIG. 7.
(A) Dot blot analysis of rmpA2 transcripts in cells grown under different nutritional conditions. Three different time points—2, 4, and 6 h—were investigated. The rrnB gene was an internal control. (B) Time course analysis of PrmpA2::luxAB expression. K. pneumoniae R2035[pYC082] was grown in LB (solid symbols) or M9-glycerol (open symbols). Luciferase activity (circles) and the bacterial cell density (squares) were measured every hour and are represented as the average of triplicate results (error bars, standard deviations). RLU, relative light units.
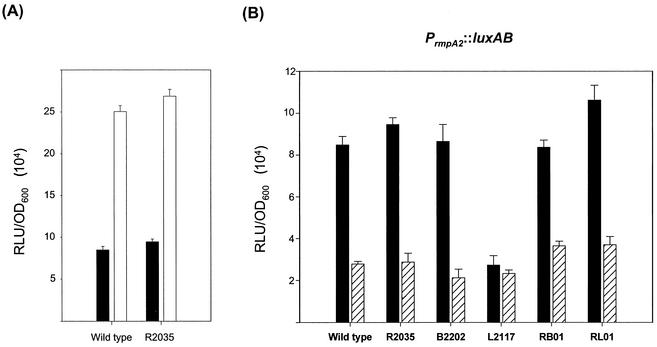
An IS3 element was noted to be located upstream of the rmpA2 locus and be transcribed in the opposite direction (Fig. 1B). To examine whether the IS3 element affects the gene expression of rmpA2, the plasmid pYC077, which contains the upstream region of rmpA2, including a 0.4-kb fragment of IS3, was generated as a luxAB fusion construct (Fig. 1B). As shown in Fig. 8A, the luciferase activity of K. pneumoniae CG43S3 [pYC077] was approximately threefold higher than that of CG43S3 [pYC082], in which IS3 was omitted.
FIG. 8.
(A) Comparison of luciferase activity of pYC082 (solid bars) and pYC077 (open bars) in K. pneumoniae CG43S3 (wild type) and R2035 (ΔrmpA2). (B) Luciferase activity of pYC082 in strains CG43S3, R2035, B2202 (ΔrcsB), L2117 (Δlon), RB01 (ΔrmpA2 ΔrcsB), and RL01 (ΔrmpA2 Δlon) was measured in the presence (striped bars) or absence (solid bars) of pYC084. Error bars, standard deviations; RLU, relative light units.
In addition to regulating the target genes, it is not uncommon for a bacterial transcription factor to also control its own expression. As shown in Fig. 8B, in almost all strains tested, the gene expression driven by the rmpA2 promoter was reduced at least 50% by the presence of multicopy rmpA2. In the lon mutant strain L2117, the PrmpA2::luxAB fusion exhibited threefold-lower activity than that in the wild-type strain, presumably due to the increased availability of RmpA2 protein in the lon mutant cells. The direct interaction of RmpA2 protein with its own promoter was further verified by EMSA and a specific binding was demonstrated in Fig. 5C.
Differential regulation of RmpA2-responsive promoters.
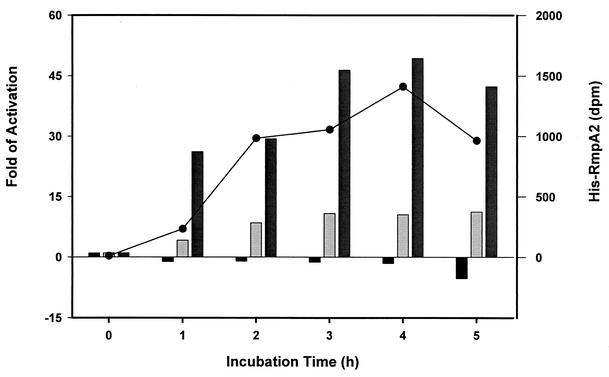
As demonstrated above, RmpA2 plays dual functional roles: it activates the K2 cps promoters while repressing its own gene expression. To understand how these two types of promoter respond to different concentrations of RmpA2 in cells, their activity was measured over time in an IPTG-inducible RmpA2 expression system in E. coli NovaBlue(DE3). As shown in Fig. 9, a relatively small amount of RmpA2 was detected 1 h after the addition of IPTG. At this time, an approximately 20-fold increase in Porf1-2 activity was observed, in contrast to nearly no reduction in PrmpA2 activity. A more pronounced repressive effect on rmpA2 promoter was seen 5 h after IPTG induction, presumably due to an accumulation of RmpA2 protein.
FIG. 9.
Response of RmpA2 target promoters. The luxAB fusions were cotransformed individually with the RmpA2 expression vector pET-RmpA2212 into E. coli NovaBlue(DE3). Upon 1 mM IPTG induction, the luciferase activity of PrmpA2::luxAB (black bars), Porf3-15::luxAB (light gray bars), or Porf1-2::luxAB (gray bars) was measured every hour and represented as the activation relative to that at time zero. The amount of His-RmpA2 synthesized in E. coli NovaBlue(DE3) after IPTG induction was determined by using a His tag-specific MAb followed by counting with a densitometer, and the result is shown on the right-hand axis (closed circles).
DISCUSSION
The gene rmpA2 was originally identified as a gene that encodes the ability to enhance CPS synthesis in K. pneumoniae (34). Nevertheless, the molecular mechanism underlying the activation exerted by rmpA2 has been elusive. In this study, we demonstrated that RmpA2 functions as a transcriptional regulator that activates the K2 cps gene expression and that the concentration of RmpA2 in K. pneumoniae cells is governed by autoregulation at transcriptional level as well as by Lon protease posttranslationally.
Despite the fact that deletion of rmpA2 only results in a slight reduction in CPS production in K. pneumoniae, it is apparent that the colony mucoidy is lost in the mutant strain. It has been reported that the colony mucoidy of K. pneumoniae is proportional to the complexity of the fibrils surrounding the capsule (34). The loss of mucoidy in the rmpA2 mutant might be due to a reduction in branching degrees of CPS. Comparative analysis with the group I cps region of E. coli K30 reveals that the orf7-15 of the K. pneumoniae K2 cps operon encodes enzymes responsible for synthesizing the repeating units of the K2 antigen (23). Among them, orf14 encodes a glycosyltransferase (2) that assembles the K2-specific tetrasaccharide units onto the preformed CPS and is likely the key enzyme in controlling the CPS branching. Therefore, when the expression of orf14 was elevated in the presence of RmpA2, the enhanced activity of the glycosyltransferase would increase the branching degree of CPS and hence result in a mucoid colony.
Sequence analysis of RmpA2 suggests that it belongs to the UhpA-LuxR family of transcription factors, which also includes RcsA and RcsB of E. coli (29). A conserved RcsAB box (35) with a sequence of TAAGATTATTCTCA could be identified in the region 168 to 181 nucleotides upstream of the K2 orf1 gene. It is not known, however, whether this RcsAB box is critical for RmpA2-mediated gene activation. No obvious similarity could be observed among the other RmpA2-responsive promoters tested here. When the cellular concentration of RmpA2 reached high levels, either by the introduction of a multicopy plasmid or by the increased stability in lon mutant cells, the K2 cps gene transcription could be activated even in an rcsB mutant genetic background. Despite the results which suggest that RmpA2 is capable of exerting the transactivation function independent of RcsB, the possibility that RmpA2 interacts with an additional protein factor such as RcsB to achieve a stronger activation could not be ruled out. Wacharotayankun et al. (34) reported that the central domain of RmpA2 protein displayed on average a 16.5% similarity to that of the NtrC, which activates transcription by the σ54-holoenzyme. Close examination of the sequence has revealed that the region does not include the major conserved segments that are shared by members of the σ54 activator family (16) Thus, RmpA2 is likely to exert the effects in a σ54-independent pathway. Therefore, the detailed mechanisms by which RmpA2 binds and activates its responsive promoters remain to be demonstrated.
It has been reported that E. coli K30 strains defective at rcsA and rcsB remain capable of synthesizing group I CPS (13). We also demonstrated that the basal expression levels of K2 cps genes as well as the amount of CPS produced in K. pneumoniae were not affected in rmpA2 mutant and in rmpA2 and rcsB double mutants. The results suggest that neither RmpA2 nor RcsB is essential for basal expression of K2 cps genes. Rather, these two factors are required to maintain high expression levels of cps genes and hence the production of a thick capsule, which is advantageous for K. pneumoniae during infection in humans.
Phase variation due to slip strand DNA synthesis is one of the means of controlling the bacterial gene expression (10). The presence of a poly(G) tract of various lengths in rmpA2 of different K. pneumoniae strains indicates that the bacterium might employ this strategy to regulate CPS production. Despite a full-length RmpA2, which could increase the virulence of K. pneumoniae CG43 in mice, it is probably not essential for the bacterium to infect immunocompromised patients, since some of the bacteremic isolates of our laboratory collection produce the truncated version of RmpA2. The functional switching on RmpA2 might be useful for the opportunistic K. pneumoniae to adjust its metabolic carbon flow upon facing different environments, such as during free-living or parasitic stages.
It has been demonstrated that the lon mutations in E. coli could elevate the transcription levels of genes responsible for CPS biosynthesis (31). The phenomenon can be explained by the enhanced stability of RcsA, which is a positive regulator of cps genes and acts as a substrate for Lon protease. A similar result was observed in the K. pneumoniae lon mutant strain L2117, in which the half-life of RmpA2 is increased and is accompanied by an accumulation of K2 CPS. However, other Lon-dependent positive regulators, such as RcsA, may also play a role, since the expression of K2 Pcps::luxAB fusions was still higher in the double-mutant strain RL01 (ΔrmpA2 Δlon) than that in the wild-type strain. Despite the fact that RmpA2 behaves like RcsA as a Lon-limited regulator, the way they control the expression of their own genes is different. A 100-fold increase in expression of a rcsA::lacZ transcriptional fusion has been demonstrated in E. coli strains with high levels of RcsA protein (8). While RcsA activates its own expression, RmpA2 was found to down-regulate the expression of PrmpA2::luxAB fusion by acting as a repressor to its own promoter.
If RmpA2 negatively regulates its own gene expression, why is the transcription of K2 cps genes increased in lon mutant cells, in which more RmpA2 is available? As demonstrated in Fig. 9, before RmpA2 inhibits its own expression, the K2 cps promoters could be activated by a relatively small amount of RmpA2. The higher responsiveness of K2 cps promoter to RmpA2 is presumably due to a better DNA binding affinity, which is evident by comparing the EMSA results in Fig. 5B with those in Fig. 5C. Therefore, in lon mutant K. pneumoniae cells, though the amount of stabilized RmpA2 protein increases slightly, it is enough to activate K2 cps gene expression; whereas, the repressive effect on rmpA2 promoter would be seen afterwards when cells accumulate more RmpA2 protein.
Based on these results, the overall regulation scheme exerted by RmpA2 on CPS biosynthesis is proposed as follows. Under normal growth conditions, K. pneumoniae synthesizes only a small quantity of RmpA2 protein, which is rapidly eliminated by the Lon protease. Upon encountering certain environmental signals, such as those found in M9 minimal medium, the expression of rmpA2 gene is activated. In some strains, the rmpA2 expression could be further enhanced by an upstream IS3 element, which is likely to be acquired through an in vivo selection for the enhancement of virulence. The increased availability of RmpA2 proteins then binds to its target promoters, including those of cps genes, leading to the activation of K2 cps gene expression and eventually the accumulation and increase in the mucoidy of K2 capsule. When the RmpA2 protein increases to a threshold level, it negatively autoregulates its own expression to prevent an overwhelming effect on the capsule production.
Acknowledgments
This work was supported by the National Science Council of the Republic of China (NSC-90-2320-B-007-004 and NSC-90-2321-B-007-001 to H.-Y.C.) and VTY Joint Research Program, Tsou's Foundation (VTY90-p4-28 to H.-Y.C.).
We are grateful to J. Vatsyayan for critical reading of the manuscript.
REFERENCES
- 1.Arakawa, Y., M. Ohta, R. Wacharotayankun, M. Mori, N. Kido, H. Ito, T. Komatsu, T. Sugiyama, and N. Kato. 1991. Biosynthesis of Klebsiella K2 capsular polysaccharide in Escherichia coli HB101 requires the functions of rmpA and the chromosomal cps gene cluster of the virulent strain Klebsiella pneumoniae Chedid (O1:K2). Infect. Immun. 59:2043-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, Y., R. Wacharotayankun, T. Nagatsuka, H. Ito, N. Kato, and M. Ohta. 1995. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J. Bacteriol. 177:1788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumenkrantz, N., and G. Asboe-Hansen. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484-489. [DOI] [PubMed] [Google Scholar]
- 4.Brill, J. A., C. Quinlan-Walshe, and S. Gottesman. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170:2599-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, H. Y., J. H. Lee, W. L. Deng, T. F. Fu, and H. L. Peng. 1996. Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb. Pathog. 20:255-261. [DOI] [PubMed] [Google Scholar]
- 6.Cryz, S. J., Jr., F. Furer, and R. Germanier. 1984. Experimental Klebsiella pneumoniae burn wound sepsis: role of capsular polysaccharide. Infect. Immun. 43:440-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domenico, P., S. Schwartz, and B. A. Cunha. 1989. Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect. Immun. 57:3778-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebel, W., and J. E. Trempy. 1999. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J. Bacteriol. 181:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman, S., and V. Stout. 1991. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5:1599-1606. [DOI] [PubMed] [Google Scholar]
- 10.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 11.Highsmith, A. K., and W. R. Jarvis. 1985. Klebsiella pneumoniae: selected virulence factors that contribute to pathogenicity. Infect. Control 6:75-77. [DOI] [PubMed] [Google Scholar]
- 12.Jann, A., and K. Jann. 1990. Structure and Biosynthesis of the capsular antigens of Escherichia coli. Curr. Top. Microbiol. Immunol. 150:19-42. [DOI] [PubMed] [Google Scholar]
- 13.Jayaratne, P., W. J. Keenleyside, P. R. MacLachlan, C. Dodgson, and C. Whitfield. 1993. Characterization of rcsB and rcsC from Escherichia coli O9:K30:H12 and examination of the role of the rcs regulatory system in expression of group I capsular polysaccharides. J. Bacteriol. 175:5384-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 15.Lai, Y. C., H. L. Peng, and H. Y. Chang. 2001. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, J., L. Passaglia, I. Rombel, D. Yan, and S. Kustu. 1999. Mutations affecting motifs of unknown function in the central domain of nitrogen regulatory protein C. J. Bacteriol. 181:5443-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuta, K., M. Ohta, M. Mori, T. Hasegawa, I. Nakashima, and N. Kato. 1983. Virulence for mice of Klebsiella strains belonging to the O1 group: relationship to their capsular (K) types. Infect. Immun. 40:56-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori, M., M. Ohta, N. Agata, N. Kido, Y. Arakawa, H. Ito, T. Komatsu, and N. Kato. 1989. Identification of species and capsular types of Klebsiella clinical isolates, with special reference to Klebsiella planticola. Microbiol. Immunol. 33:887-895. [DOI] [PubMed] [Google Scholar]
- 19.Nassif, X., N. Honore, T. Vasselon, S. T. Cole, and P. J. Sansonetti. 1989. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol. Microbiol. 3:1349-1359. [DOI] [PubMed] [Google Scholar]
- 20.Nassif, X., and P. J. Sansonetti. 1986. Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, S. H., J. Eriksen, and S. D. Henriksen. 1967. Structure of the capsular polysaccharide of Klebsiella pneumoniae type 2 (B). Acta Pathol. Microbiol. Scand. 69:431-436. [Google Scholar]
- 22.Peng, H. L., P. Y. Wang, J. L. Wu, C. T. Chiu, and H. Y. Chang. 1991. Molecular epidemiology of Klebsiella pneumoniae. Clin. J. Microbiol. Immunol. (Taipei) 4:264-271. [PubMed] [Google Scholar]
- 23.Rahn, A., J. Drummelsmith, and C. Whitfield. 1999. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 181:2307-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed, L. J., and H. Muench. 1938. A simple method of estimating the fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 25.Richards, H., and N. Datta. 1982. Plasmids and transposons acquired by Salmonella typhi in man. Plasmid 8:9-14. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor laboratory, Cold Spring Harbor, N.Y.
- 27.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 29.Stout, V., A. Torres-Cabassa, M. R. Maurizi, D. Gutnick, and S. Gottesman. 1991. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173:1738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suerbaum, S., S. Friedrich, H. Leying, and W. Opferkuch. 1994. Expression of capsular polysaccharide determines serum resistance in Escherichia coli K92. Zentbl. Bakteriol. 281:146-157. [DOI] [PubMed] [Google Scholar]
- 31.Trisler, P., and S. Gottesman. 1984. lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J. Bacteriol. 160:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trun, N. J., and T. J. Silhavy. 1989. PrlC, a suppressor of signal sequence mutations in Escherichia coli, can direct the insertion of the signal sequence into the membrane. J. Mol. Biol. 205:665-676. [DOI] [PubMed] [Google Scholar]
- 33.Wacharotayankun, R., Y. Arakawa, M. Ohta, T. Hasegawa, M. Mori, T. Horii, and N. Kato. 1992. Involvement of rcsB in Klebsiella K2 capsule synthesis in Escherichia coli K-12. J. Bacteriol. 174:1063-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wacharotayankun, R., Y. Arakawa, M. Ohta, K. Tanaka, T. Akashi, M. Mori, and N. Kato. 1993. Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 61:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wehland, M., and F. Bernhard. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 275:7013-7020. [DOI] [PubMed] [Google Scholar]
- 36.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 37.Williams, P., P. A. Lambert, M. R. Brown, and R. J. Jones. 1983. The role of the O and K antigens in determining the resistance of Klebsiella aerogenes to serum killing and phagocytosis. J. Gen. Microbiol. 129:2181-2191. [DOI] [PubMed] [Google Scholar]