Abstract
A mutant strain of Escherichia coli was created by inserting a cassette encoding sucrose sensitivity and neomycin resistance (sacB-neo) into the small-subunit rRNA-encoding gene rrs in the rrnB operon. During growth in a complex medium, the cassette was lost from the population, and a complete rrs gene was restored at a rate of 5 × 10−9 per cell division. Repair of this lesion required flanking regions of DNA that were similar to the six remaining intact rRNA operons and reestablished the full complement of seven rRNA operons. The relative fitness of strains with restored rrnB operons was 1 to 2% higher than that of the mutant strain. The rrnB operon normally contains a spacer region between the 16S and 23S rRNA-encoding genes that is similar in length and tRNA gene content to the spacer in rrnC, -E, and -G. In 2 of the 14 strains in which rrnB was restored, the spacer region had the same length as the spacer region in rrnA, -D, and -H. The requirement for flanking regions of nearly identical DNA and the replication of the spacer region from other rRNA operons during the repair of rrnB suggest that the restoration was accomplished via gene conversion. The rate of gene conversion was 10-fold less than the fixation of point mutations in the same region of the chromosome but was apparently sufficient to homogenize the sequences of rRNA genes in E. coli. These findings are discussed in the context of a conceptual model describing the presence of sequence heterogeneity in coevolving rRNA genes.
Unlike the majority of genes in Bacteria and Archaea that are present in single copies, there are as many as 15 copies of each of the rRNA-encoding genes (25). When multiple copies of rRNA genes are present in an organism, they are nearly (≥99%) identical in most cases, suggesting that there must be some form of homogenization of the gene sequences to remove neutral mutations that would otherwise accumulate. Apparent exceptions to this phenomenon of concerted evolution include the rRNA genes from Thermomonospora chromogena, Thermobispora bispora, and Haloarcula marismortui, in which the 16S and 23S rRNA gene sequences differ by up to 10% (11, 34, 45, 46). It has been suggested that lateral gene transfer is responsible for the presence of divergent rRNA genes in an organism, but co-occurrences of divergent rRNA genes in microbes appear to be exceptions to the rule.
The extensive regions of sequence similarity provided by repeated rRNA genes present opportunities for recombination that can lead to inversions, duplications, deletions, and transpositions of the chromosome. Recombinational events involving rRNA operons have been observed in Escherichia coli at frequencies of 10−3 to 10−5 in overnight liquid cultures (3, 16, 18-21). This high recombination frequency suggests the potential for dynamic reorganization of the E. coli chromosome, yet the overall arrangement of rRNA operons in the chromosome is remarkably stable. Comparative genomic analysis of E. coli (strain K-12) and Salmonella enterica serovar Typhimurium (strain LT2) offers evidence for this stability because the number, chromosomal location, and orientation of rRNA operons have been preserved in these strains even though their genomes differ by roughly 1 million base substitutions (3, 22). Conservation of rRNA operon organization and location on the chromosome, despite the potential for extensive recombination, suggests strong selection for maintenance of these characteristics.
This study focuses on factors that influence the concerted evolution of rRNA gene sequences in bacteria, using E. coli as an experimental system. The rate of recombination involving rrnB and the six other rRNA operons in E. coli was measured to estimate the homogenizing force of recombination, while the rate at which point mutations occur was determined to provide an estimate for the divergence rate of gene sequences. The fitness effects of interoperon recombination were also measured to estimate the selective pressures affecting the eventual fixation or loss of homogenized operons from a population. These results were then used to develop a conceptual model of rRNA operon evolution, focusing on an explanation for the presence of minor sequence heterogeneity between rRNA genes.
MATERIALS AND METHODS
Strains and genotype verification.
Insertional inactivation of the rrnB operon was produced by linear transformation of a recD mutant strain of E. coli (D308) with a DNA fragment containing an rrnB operon with a sacB-neo cassette (5) inserted in the BglII site of rrsB. Generalized transduction (P1vir) was used to move the rrnB operon disrupted by the sacB-neo cassette from D308 into a strain of E. coli B that had been propagated in the laboratory for 10,000 generations (27). The resulting strain, BSV1, contains an rrnB operon with intact promoters and regions of DNA both upstream and downstream from the inserted cassette that are identical to the other rrn operons (Fig. 1). A second strain (BSV2) was constructed in which the sacB-neo cassette replaced the promoters (P1 and P2), rrsB, and the 5′ end of rrlB, leaving a promoterless rrnB operon with no homology upstream of the sacB-neo cassette to any of the other rRNA operons (Fig. 1).
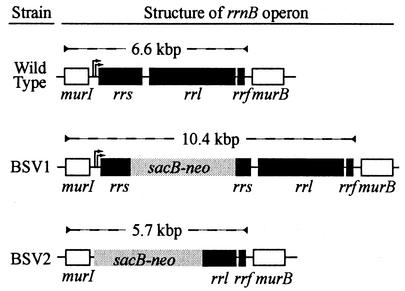
FIG. 1.
Structure of rrnB operon regions in wild-type, BSV1, and BSV2 strains. rRNA genes (black), the sacB-neo cassette (3,825 bp; grey), and the adjacent genes involved in murein synthesis (murI and murB; white) are shown along with the rrnB promoters (arrows). Dashed lines indicate the region bound by the PCR primers upB and dnB along with the resulting PCR product sizes. BSV1 has the sacB-neo cassette inserted into the BglII site of rrsB (after position 704) and upstream sequence similarity to the other rrn operons, and the promoters P1 and P2 are preserved. BSV2 has the sacB-neo cassette replacing sequence upstream of rrnB, rrsB, the ITS region, and the 5′ end of rrl, leaving no promoters or upstream sequence similarity to the other rrn operons.
BSV1 and BSV2 genotypes were verified by PCR and Southern hybridization. Following denaturation of genomic DNA at 94°C for 3 min, PCR amplification of rrnB was carried out using the upB forward (5′-CCGAATTACATATGACCGTGCTGGTGTTTGAC-3′) and dnB reverse (5′-CTACTCTAGACCTGATGCAAAAACGAGGCTAGTTTA-3′) primers and rTth DNA polymerase XL (PE Applied Biosystems; Foster City, Calif.). The amplification consisted of 30 cycles (94°C for 0.5 min, 60°C for 1 min, and 72°C for 10 min) followed by a 10-min extension at 72°C. The rRNA operons were visualized by Southern hybridization of genomic DNA digested with PvuII (New England Biolabs, Beverly, Mass.) and probed with a digoxigenin-dUTP-labeled DNA probe for a conserved region of rrs (positions 8 to 536) (24).
Growth conditions and media.
Cultures were typically grown in Luria-Bertani liquid medium (LB), with 1.5% (wt/vol) Bacto agar (Difco Laboratories; Detroit, Mich) added for solid medium. When selecting for sucrose resistance, sucrose was added at a final concentration of 6% (wt/vol) to LB with NaCl omitted (LBsuc) (5). Where noted, kanamycin was added to LBsuc at a final concentration of 50 mg/liter (LBsuc-kan). Overnight cultures consisted of 2 ml of LB in culture tubes (16 by 125 mm), inoculated from freezer stocks. Competition experiments, conditioning cultures, and cultures for rate measurements were done in either 10 ml of medium in a 50-ml Erlenmeyer flask or 50 ml of medium in a 250-ml Erlenmeyer flask with shaking at 225 rpm at 37°C. The conditioning cultures and competition experiments used to measure relative fitness were grown in Davis-Mingoli broth (DM) supplemented with 2.0 mg of thiamine hydrochloride and 25 mg of glucose (DM + 25) per liter (27). Tetrazolium arabinose (TA) indicator plates were used to differentiate strains with an arabinose (Ara) marker; Ara+ colonies appear white, and Ara− colonies appear red (28).
Recombination and mutation rate measurements.
The recombination rate and sacB mutation rate measurements were carried out as outlined by Crane et al. (9) and are a modification of the fluctuation analysis of Luria and Delbruck (31). Eleven flasks with 10 ml of LB were inoculated with approximately 100 cells from an overnight LB culture. The cultures were incubated at 37°C with shaking at 225 rpm for 15 h. The stationary-phase cultures were sampled by spreading 75 μl of the culture on four LBsuc plates and four LBsuc-kan plates, and diluted and spread on four LB plates. After overnight incubation at 30°C, the number of colonies on each plate was counted. For each replicate culture, the number of colonies on the LB plates represented the total population size, while the population density of the recombinant strain was calculated by using the number of colonies on LBsuc minus the number of colonies on LBsuc-kan. Colonies appearing on LBsuc-kan represented individuals that retained the cassette, including kanamycin resistance, but lost sensitivity to sucrose through a mutation in sacB.
To determine the rate of recombination and sacB mutation, the median value for each of the populations was determined from 11 replicate cultures. The following median estimator of mutation rates was used to determine rates of mutation and recombination (9, 23):
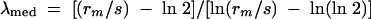 |
(1) |
where rm is the median number of recombinants or mutants per culture, s is the dilution factor, and λmed is the median number of recombination events or mutation events per culture. The recombination or mutation rate was then determined by dividing λmed by the median number of cells per culture.
For the rate of double mutation in sacB and neo in BSV2 (recombinant phenotype, sucrose resistant, kanamycin sensitive), a different equation was used because many of the replicate cultures produced no colonies with the recombinant phenotype. The following equation is more suited for results with many “mutant-free” cultures (9, 23):
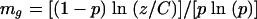 |
(2) |
where p is the dilution factor, z is the number of cultures devoid of mutants, C is the total number of cultures, and mg is the number of mutation events per culture.
Relative fitness assay.
Overnight LB cultures for each strain to be used in a competition experiment were inoculated from freezer stocks. The following day, conditioning cultures, consisting of 10 ml of DM + 25 in 50-ml Erlenmeyer flasks, were inoculated with a final dilution of 1:10,000 from the overnight cultures. Competition cultures were initiated 24 h after inoculating the conditioning cultures by adding 0.1 ml of a 1:1 mix of the two competitors to five 50-ml flasks containing 10 ml of DM + 25. BSV1 and the BSV1 mutants and recombinants carry the neutral arabinose utilization marker (Ara+), while the control strain carries the opposite marker (Ara−), allowing relative fitness to be measured for each group against the control strain (27). Cultures were transferred at 24-h intervals for 4 days and monitored daily by plating a 5 × 10−6 dilution of the each replicate culture on three TA plates.
Relative fitness measurements were calculated by using the following equations:
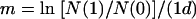 |
(3) |
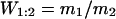 |
(4) |
where N(0) and N(1) are the population sizes of the strains at 0 and 1 day (d), respectively, and m is the Malthusian parameter for that strain. The relative fitness (W) of the Ara+ strain compared to the control strain (Ara−) was calculated by using equation 4, where m1 is the Malthusian parameter of the Ara+ strain and m2 is the Malthusian parameter of the Ara− strain (27, 41). Statistical analysis of the relative fitness measured for BSV1, the BSV1 recombinant strains, and the BSV1 sacB strains included analysis of variance and pairwise comparisons with the Bonferroni multiple-comparisons procedure.
rRNA operon sequence alignments.
Sequences of small- and large-subunit rRNA genes were obtained from the Institute for Genomic Research Microbial Genome Database (www.tigr.org/tdb/mdb/mdbcomplete.html) or the National Center for Biotechnology Information Microbial Genome Database (www.ncbi.nlm.nih.gov/PMGifs/Genomes/bact.html) and analyzed with the Genetics Computer Group software package (version 10; Genetics Computer Group, Madison, Wis.). The following microbial genomes containing multiple rRNA operons were used in the analysis and represent all available sequenced genomes with multiple rRNA operons at the time of analysis: Aquifex aeolicus VF5, Bacillus subtilis ATCC 23857, Campylobacter jejuni ATCC 700819, Escherichia coli ATCC 10798, Haemophilus influenzae ATCC 51907, Helicobacter pylori 26695, Methanococcus jannaschii DSMZ 2661, Methanobacterium thermoautotrophicum ATCC 29096, Neisseria meningitidis MC58, Synechocystis sp. strain PCC6803, Treponema pallidum ATCC 25870, Ureaplasma urealyticum serovar 3, Vibrio cholerae ATCC 39315, and Xylella fastidiosa 9a5c. Pairwise differences were calculated for the rrs and rrl genes of each organism. Distances between rRNA operons were calculated based on the published locations of the operons (4, 6, 10, 13, 14, 17, 26, 35, 36, 38, 40, 42, 43).
RESULTS
Rates of mutation and recombination.
The presence of the sacB-neo cassette in the rrnB operon of E. coli conferred sensitivity to sucrose and resistance to kanamycin. The cassette was either inserted into rrs (strain BSV1) or engineered to replace the promoter region of rrnB as well as rrs and a portion of rrl (strain BSV2). The frequency at which sucrose sensitivity, kanamycin resistance, or both of these phenotypes was lost in overnight cultures was enumerated on selective medium (Table 1).
TABLE 1.
Occurrence of derived phenotypes used to estimate rates of recombination and point mutationsa
| Primary strain (Sucs, Kanr) | Implied change | Resulting phenotype | Expt | rm | Dilution factor (s) | λmed | Population size |
|---|---|---|---|---|---|---|---|
| BSV1 | Recombination | Sucr, Kans | a | 41 | 0.030 | 180 | 3.76 × 1010 |
| Point mutation | Sucr, Kanr | a | 523 | 0.030 | 1,720 | 3.76 × 1010 | |
| Recombination | Sucr, Kans | b | 20 | 0.004 | 562 | 1.20 × 1011 | |
| Point mutation | Sucr, Kanr | b | 424 | 0.004 | 8,879 | 1.20 × 1011 | |
| BSV2 | Point mutation | Sucr, Kanr | a | 447 | 0.030 | 1,493 | 2.91 × 1010 |
Primary strain phenotype (sucrose sensitive, Sucs; kanamycin resistant, Kanr), recombination phenotype (sucrose resistant, Sucr; kanamycin sensitive, Kans), or point mutation phenotype (sucrose resistant, Sucr; kanamycin resistant, Kanr). Measurements of the rate of recombination and point mutations at different loci were conducted in two independent, replicate experiments, a and b. rm, median number of recombinants or mutants per culture. λmed, median number of recombination events or mutation events per culture.
Loss of sucrose sensitivity with retention of kanamycin resistance was attributed to mutations in sacB. In every one of the 18 sacB mutants (sucrose resistant, kanamycin resistant) examined by either PCR (Fig. 2) or Southern hybridization (Fig. 3), the sacB-neo cassette was present in the rrnB operon, confirming that sucrose resistance was due to a mutation in sacB rather than loss of the sacB-neo cassette. The average sacB mutation rate was 5.7 × 10−8 per cell division in both strains BSV1 and BSV2 (Table 2).
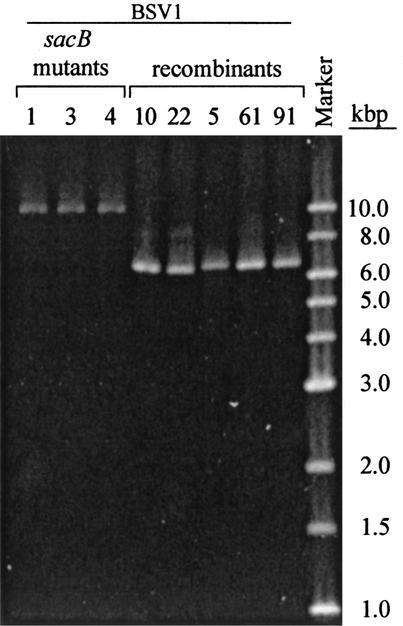
FIG. 2.
PCR amplification of the rrnB region from strains used in the relative fitness assay. Amplification of the rrnB operon from BSV1 sacB mutant strains, which contain the sacB-neo cassette, results in a product of approximately 10 kbp. Amplification of the recombinant or wild-type rrnB operon results in a product of approximately 6.6 kbp.
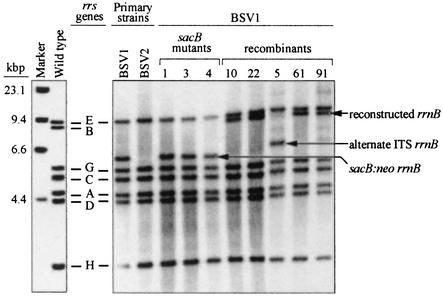
FIG. 3.
Southern hybridization of PvuII-digested genomic DNA from “wild-type” E. coli B, strains BSV1 and BSV2, sacB mutants, and recombinant strains used in the relative fitness assay. The locations of hybridized bands representing native rrs genes are indicated for the wild-type and BSV1 strains. Fragments that correspond to rrnB operons that contain the sacB-neo cassette or have been reconstructed with a native or alternative ITS region are also labeled. Operons containing the alternative ITS region result in a smaller hybridized fragment due to the introduction of an additional PvuII site.
TABLE 2.
Estimated rates (per cell division) at which recombination and point mutations were fixed in populations of E. colia
| Strain | Expt | Recombination rate | Mutation rate |
|---|---|---|---|
| BSV1 | a | 4.8 × 10−9 | 4.6 × 10−8 |
| b | 4.7 × 10−9 | 7.4 × 10−8 | |
| BSV2 | a | 0 | 5.1 × 10−8 |
See Table 1, footnote a.
Loss of both sucrose sensitivity and kanamycin resistance in strain BSV1 was attributed to recombination. Two independent fluctuation analysis experiments with BSV1 revealed an average rate of recombination of 4.8 × 10−9 per cell division (Table 2). Fourteen independent BSV1 recombinant strains (each originating from a different culture) were screened by PCR and Southern hybridization for the presence of the sacB-neo cassette in rrsB. The amplification product from the rrnB operon with the sacB-neo cassette in BSV1 was approximately 10 kbp, while the amplification product from the rrnB operon without the cassette (either wild type or recombinant) was 6.6 kbp (Fig. 2). Southern hybridization of genomic DNA digested with PvuII and probed for rrs produced an 8.4-kbp fragment in wild-type strains and recombinant strains of E. coli and a 6.1-kbp fragment in BSV1 (Fig. 3). The restriction fragment length polymorphism pattern for each of the recombinants was indicative of a restored rrnB operon (Fig. 3). Two of the 14 recombinants contained an rrnB operon whose size was indicative of recombination with an rRNA operon containing the tRNAIle1-tRNAAla1B internally transcribed spacer (ITS) (rrnA, rrnD, or rrnH; Fig. 4).
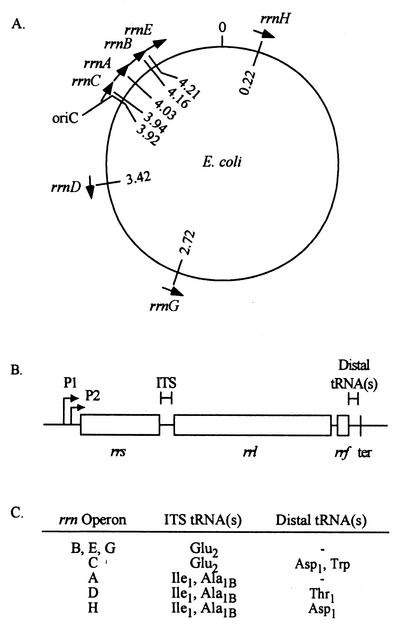
FIG. 4.
Location and organization of rRNA operons in E. coli (determined for strain K-12). (A) Chromosomal map with location (megabase pairs) and orientation of rRNA operons. (B) Organization of rRNA operons, including tandem promoters P1 and P2, the rRNA-encoding genes (rrs, rrl, and rrf), the ITS, and the rho-independent termination site (ter). (C) ITS and distal tRNA species associated with each rRNA operon.
In cultures of strain BSV2, the recombinant phenotype (sucrose resistant, kanamycin sensitive) appeared at a rate of 1.4 × 10−10 per cell division, a rate that is within the range of values expected for the spontaneous inactivation of both phenotypes of the sacB-neo cassette by point mutations (5). Seven independent isolates of BSV2 with the recombinant phenotype (sucrose resistant, kanamycin sensitive) were screened by PCR or Southern hybridization and all maintained the 3.8-kbp cassette in rrsB. The presence of the cassette revealed that the loss of both phenotypes encoded by the sacB-neo cassette was a result of mutations rather than removal of the cassette through recombination (data not shown).
Fitness effects of recombination between rRNA operons.
There was no significant difference between the fitness of BSV1 and that of the BSV1 sacB mutant strains (P = 0.2823). However, the BSV1 recombinant populations had a 1 to 2% greater relative fitness (Table 3) than either the parental BSV1 strain or the BSV1 sacB mutant strains (P = 0.0089 and P = 0.0034, respectively). The relative fitness of one of the recombinant strains with an alternate ITS region was indistinguishable from that of the other recombinants, and it is included in the BSV1 recombinant group.
TABLE 3.
Relative fitness of derived strains compared to the parental strain
| Derived strains | sacB-neo cassette | Phenotype | No. of isolatesa | Relative fitnessb | Variance |
|---|---|---|---|---|---|
| BSV1 | + | Sacs Neor | 1 | 0.99 | 3.8 × 10−5 |
| BSV1 sacB | + | Sacr Neor | 3 | 0.98 | 1.4 × 10−5 |
| BSV1 recombinant | − | Sacr Neos | 4 | 1.00c | 2.5 × 10−5 |
Number of independent isolates used in experiments to determine relative fitness.
Relative fitness values are the arithmetic averages of five replicate measurements for each isolate.
The fitness of BSV1 recombinants was significantly different from that of BSV1 (P = 0.0089) and BSV1 sacB (P = 0.034) based on analysis of variance.
Comparison of rRNA gene sequences and relative chromosomal position.
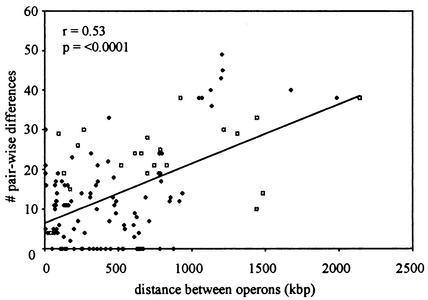
The result of pairwise comparisons of rrs and rrl sequences from microbial genomes with multiple rRNA operons are summarized in Fig. 5. There was a significant positive correlation between the total pairwise differences present in the rrs and rrl genes and the distance between them, suggesting a link between the proximity of genes and the rates of gene conversion.
FIG. 5.
Correlation of pairwise rrs and rrl sequence differences with distance between operons for all available bacterial and archaeal genome sequences containing multiple rRNA operons. E. coli comparisons are represented as open diamonds, and the other organism comparisons are represented as solid diamonds.
DISCUSSION
The objective of this study was to explore the coevolution of rRNA genes in bacteria by studying the rate and fitness effects of recombination among rRNA operons in E. coli. As in many prokaryotes, the genes encoding the 16S rRNA (rrs), 23S rRNA (rrl), and 5S rRNA (rrf) in E. coli are organized into an operon that also contains tRNA-encoding genes in the ITS region between rrs and rrl or distal to rrf (Fig. 4). There are seven rRNA operons in E. coli, positioned asymmetrically about the chromosome (Fig. 4) (3). When multiple copies of rRNA genes are present in Bacteria and Archaea, they are nearly (≥98%) identical, suggesting that the gene sequences are homogenized. Otherwise, neutral mutations would be expected to accumulate in individual rRNA genes at a rate similar to that at which they accumulate between species (29).
Gene conversion, a form of recombination resulting in the nonreciprocal transfer of genetic information, is typically invoked to explain the homogenization of replicate genes in eukaryotes. A mechanism for gene conversion involving a nontraditional resolution of Holliday structures has been proposed based on studies in the ascomycete fungi, in which all products of meiosis are readily examined (32, 33). Gene conversion results in the conversion of one allele to another and is thought to involve recombination of relatively short (ca. 500-bp) segments of DNA (15). The nature of the recombination events described in the current study suggests that gene conversion drives the homogenization of rRNA genes in bacteria.
The involvement of recombination in genetic alterations is typically shown by the requirement of one or a combination of genes whose products are involved in recombination (e.g., recA, recB, and recF). Although we did not test directly for the requirement of any of these genes, two lines of evidence clearly indicate that recombination was involved in the restoration of rrnB in E. coli strain BSV1. First, loss of the 3.8-kbp sacB-neo insertion was observed only when flanking regions of homology to the other rRNA operons were present, a common requirement for recombination. Second, on at least two occasions the restoration of rrnB included replacement of the ITS region between rrs and rrl. The presence of the alternative ITS region in rrnB could only occur through a recombinational event between rrnB and an operon with the tRNAIle1-tRNAAla1B ITS region (rrnA, rrnD, or rrnH). This result clearly implicates gene conversion in the restoration of rrnB, since reciprocal recombination would have simply moved the sacB-neo cassette to one of the other rRNA operons.
The possibility that a portion of the genome was duplicated in any of the lineages used in the study was also considered. Duplication of portions of the chromosome could have led to the amplification of one or more of the rRNA operons, but differences in the abundance of any of the operons would have been readily observed in the results from Southern hybridization. There was a consistent level of probe binding for each of the rRNA operons (Fig. 3), indicating that each operon was present in equal abundance per chromosome.
The recombination rate measured for the restoration of rrnB (5 × 10−9 per cell division) is within the range of values reported for the recombination in the tuf genes of S. enterica serovar Typhimurium (3 × 10−9 to 2 × 10−8) (1). Unfortunately, mutation and recombination events are commonly reported as frequencies in a population, which is a measure that includes both the rate of recombination and the subsequent growth of the recombinant. This makes direct comparison of rates and frequencies problematic, because the growth of recombinants can vary considerably. Improved growth rates of the recombinant is the likely explanation for the discrepancy in the rates of recombination reported here (per cell division) and the reported frequency (5 × 10−5) of recombination involving the tRNAGlu2 spacer regions in another strain of E. coli (16).
The restoration of rrnB via nonreciprocal recombination (gene conversion) with one of the other six intact rRNA operons resulted in a 1 to 2% increase in relative fitness. It has been reported previously that interruptions of rrnA, rrnB, or rrnD with antibiotic cassettes resulted in no significant change in growth rate in various types of medium and at different temperatures (7, 8). Results reported here do not necessarily contradict these earlier studies, because the comparisons of relative fitness made here were measured in head-to-head competition experiments, where it is possible to detect small differences in the duration of lag phase and the rate of exponential growth.
The relative fitness of the BSV1 strains that retained the sacB-neo cassette but contained a mutation in sacB was indistinguishable from that of its primary strain and significantly lower than that of strains with a restored rrnB, suggesting that a functional sacB gene has little impact on relative fitness in the absence of sucrose. The negative effect on relative fitness of the mutation in strain BSV1 is therefore most likely attributable to inactivation of the rrnB operon by the insertion of the sacB-neo cassette.
The relative fitness of the recombinant strain with an alternative ITS region was similar to that of the other recombinant strains, indicating that duplication of one of the alternative ITS regions had little effect on relative fitness. However, the conservation of three ITS regions encoding tRNAIle and tRNAAla and four ITS regions encoding tRNAGlu in strains from the Escherichia coli reference collection (2) suggests a selective advantage for maintaining the balance of tRNAs in the spacer region and that there is selective pressure under natural conditions that maintains the four plus three dosage of ITS tRNAs in E. coli.
If the reestablishment of a functional rRNA operon results in a 1% fitness increase, fixation of recombination events between rRNA operons would occur only rarely. In an environment similar to the conditions of the relative fitness assay, the probability of fixation of a recombinant with a fitness advantage of 1% over the rest of the population is 4.4 × 10−3 and would occur on average after approximately 2,500 generations (27). Furthermore, the 1 to 2% increase in the relative fitness of the recombinant strains may not be solely attributable to the reestablishment of the rrnB operon but could also be due to loss of the sacB-neo cassette. Thus, even though homogenization of the rRNA operons may be occurring at a considerable rate, the fixation of the homogenized operons may be somewhat slower and could explain the presence of some heterogeneity in rRNA operons. In light of these observations, it is possible to explain the maintenance of minor sequence heterogeneity in the rRNA operons of E. coli through a model that balances the homogenizing effect of recombination between rRNA operons and the heterogeneity arising from point mutations (44).
Comparisons of the relative positions and sequences of rRNA operons from bacterial and archaeal genomes add an additional dimension to this model. For prokaryotes with two or more rRNA operons, there is a positive correlation between the number of pairwise sequence differences (of 16S and 23S genes within an organism) and the distance between the operons on the chromosome. In other words, the further apart two rRNA operons are, the more likely they are to contain sequence heterogeneity. Thus, by being distant from other rRNA operons, an operon may escape homogenization and begin to diverge.
The dependence of recombination rate on the distance between the regions participating in the exchange has been shown for plasmid- and chromosome-based systems in phylogenetically diverse organisms (12, 30, 37, 39). In E. coli, recombination rates were reduced by up to 100-fold when homologous regions were separated by 7 kbp (30). In S. enterica serovar Typhimurium, recombination between 5-kbp regions is reduced by over 10-fold when the distance separating the homologies is increased from 2 to 17 min (37). In Eukarya, the pattern of decreasing recombination rate with increasing distance between regions has been observed in the nontranscribed spacers of rRNA genes (12). The pattern of heterogeneity in the nontranscribed spacer regions of wheat, rye, maize, and mice was better fit by a model that assumes an increased probability of sequence exchange for closely located regions (12). Thus, the observed positive correlation of sequence difference and distance has empirical support based on recombination rates between homologous sequences separated by various distances and may be a widespread phenomenon impacting the concerted evolution of replicated genes in all domains of life.
Conclusion.
The degree of sequence similarity among the rRNAs expressed from the seven rRNA operons in E. coli reveals that the rRNA-encoding genes are being homogenized, and data presented here suggest that gene conversion is the mechanism responsible for the homogenization. Through independent fluctuation experiments, we have provided direct measurements of the rate of gene conversion between rrnB and the other rRNA operons of E. coli (5 × 10−9 per cell division). The fitness effect associated with restoration of the rrnB operon was approximately 1 to 2%. This relatively small fitness benefit provides a steep barrier for fixation of recombinant operons. Thus, relatively few homogenizing events would be expected to rise to fixation in a population, but the few that do become fixed result in the overall similarity seen in the rRNA operons. The presence of minor heterogeneity in ribosomal DNA sequences can be explained by the introduction of de novo mutations balanced by the homogenizing force of gene conversion, which is dependent upon the proximity of rRNA operons. Selection for variant rRNAs would have a dramatic influence on the maintenance of heterogeneous rRNA genes in an organism, but there are as yet no documented examples of an advantage conferred by the maintenance of heterogeneous rRNA-encoding genes.
Acknowledgments
This work was supported by a grant from the National Science Foundation (IBN 9875254), awarded to T.M.S.
We thank Ann Stafford for expert assistance in conducting the competition experiments.
REFERENCES
- 1.Abdulkarim, F., and D. Hughes. 1996. Homologous recombination between the tuf genes of Salmonella typhimurium. J. Mol. Biol. 260:506-522. [DOI] [PubMed] [Google Scholar]
- 2.Anton, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1998. Sequence diversity in the 16S-23S intergenic spacer region (ISR) of the rRNA operons in representatives of the Escherichia coli ECOR collection. J. Mol. Evol. 47:62-72. [DOI] [PubMed] [Google Scholar]
- 3.Bachellier, S., E. Gilson, M. Hofnung, and C. W. Hill. 1996. Repeated sequences, p. 2012-2040. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 4.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 6.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. M. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 7.Condon, C., S. French, C. Squires, and C. L. Squires. 1993. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. EMBO J. 12:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condon, C., D. Liveris, C. Squires, I. Schwartz, and C. L. Squires. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J. Bacteriol. 177:4152-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crane, G. J., S. M. Thomas, and M. E. Jones. 1996. A modified Luria-Delbruck fluctuation assay for estimating and comparing mutation rates. Mutat. Res. 354:171-182. [DOI] [PubMed] [Google Scholar]
- 10.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, P. P., S. Ziesche, and S. Mylvaganam. 1998. Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 180:4804-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dvorak, J., D. Jue, and M. Lassner. 1987. Homogenization of tandemly repeated nucleotide sequences by distance-dependent nucleotide sequence conversion. Genetics 116:487-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. Mckenney, G. Sutton, W. Fitzhugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. Mcdonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, C. M., S. J. Norris, C. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, C. Fujii, S. Garland, B. Hatch, K. Horst, K. Roberts, M. Sandusky, J. Weidman, H. O. Smith, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 15.Frisse, L., R. R. Hudson, A. Bartoszewicz, J. D. Wall, J. Donfack, and A. Di Rienzo. 2001. Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am. J. Hum. Genet. 69:831-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey, S., and C. W. Hill. 1990. Exchange of spacer regions between rRNA operons in Escherichia coli. Genetics 125:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, C. W., R. H. Grafstrom, B. W. Harnish, and B. S. Hillman. 1977. Tandem duplications resulting from recombination between ribosomal RNA genes in Escherichia coli. J. Mol. Biol. 116:407-428. [DOI] [PubMed] [Google Scholar]
- 19.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, C. W., and B. W. Harnish. 1982. Transposition of a chromosomal segment bounded by redundant rRNA genes into other rRNA genes in Escherichia coli. J. Bacteriol. 149:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, C. W., S. Harvey, and J. A. Gray. 1990. Recombination between rRNA genes in Escherichia coli and Salmonella typhimurium, p. 335-340. In K. Drlica and M. Riley (ed.), The bacterial chromosome. American Society for Microbiology, Washington, D.C.
- 22.Hughes, D. 1999. Impact of homologous recombination on genome organization and stability, p. 109-128. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 23.Jones, M. E., S. M. Thomas, and A. Rogers. 1994. Luria-Delbruck fluctuation experiments: design and analysis. Genetics 136:1209-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S. Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M. F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. KlaerrBlanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S. M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. OReilly, K. Ogawa, A. Ogiwara, B. Oudega, S. H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Lenski, R. E., M. R. Rose, S. C. Simpson, and S. C. Tadler. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138:1315-1341. [Google Scholar]
- 28.Levin, B. R., F. M. Stewart, and L. Chao. 1977. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111:3-24. [Google Scholar]
- 29.Liao, D. 2000. Gene conversion drives within genic sequences: Concerted evolution of ribosomal RNA genes in Bacteria and Archaea. J. Mol. Evol. 51:305-317. [DOI] [PubMed] [Google Scholar]
- 30.Lovett, S. T., T. J. Gluckman, P. J. Simon, V. A. Sutera, Jr., and P. T. Drapkin. 1994. Recombination between repeats in Escherichia coli by a recA-independent, proximity-sensitive mechanism. Mol. Gen. Genet. 245:294-300. [DOI] [PubMed] [Google Scholar]
- 31.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, M. B. 1955. Aberrant recombination of pyridooxine mutants of Neurospora. Proc. Natl. Acad. Sci. USA 41:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, M. B. 1955. Further evidence of aberrant recombination in Neurospora. Proc. Natl. Acad. Sci. USA 41:935-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mylvaganam, S., and P. P. Dennis. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura, Y., T. Kaneko, M. Hirosawa, N. Miyajima, and S. Tabata. 1998. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 26:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 37.Segall, A. M., and J. R. Roth. 1994. Approaches to half-tetrad analysis in bacteria: recombination between repeated, inverse-order chromosomal sequences. Genetics 136:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvestri, M. L., W. J. Siqueira, A. A. de Souza, A. P. de Souza, M. F. Terenzi, D. Truffi, S. M. Tsai, M. H. Tsuhako, H. Vallada, M. A. Van Sluys, S. Verjovski-Almeida, A. L. Vettore, M. A. Zago, M. Zatz, J. Meidanis, and J. C. Setubai. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157.10910347 [Google Scholar]
- 39.Singer, B. S. 1988. On the role of homologous sequences in chromosomal rearrangements. Genes Dev. 2:1800-1811. [DOI] [PubMed] [Google Scholar]
- 40.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicaire, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, J. N. Reeve, et al. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson, B. S. 2000. Life history implications of ribosomal RNA gene copy number in Escherichia coli. Doctoral dissertation. Michigan State University, East Lansing, Mich.
- 42.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 43.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. X. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weldman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, J. M. Weidman, C. Fujii, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 44.Ueda, K., T. Seki, T. Kudo, T. Yoshida, and M. Kataoka. 1999. Two distinct mechanisms cause heterogeneity of 16S rRNA. J. Bacteriol. 181:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Y., Z. Zhang, and N. Ramanan. 1997. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 179:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap, W. H., Z. Zhang, and Y. Wang. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]