Abstract
A cluster containing five similarly oriented genes involved in the metabolism of galactose via the Leloir pathway in Lactococcus lactis subsp. cremoris MG1363 was cloned and characterized. The order of the genes is galPMKTE, and these genes encode a galactose permease (GalP), an aldose 1-epimerase (GalM), a galactokinase (GalK), a hexose-1-phosphate uridylyltransferase (GalT), and a UDP-glucose 4-epimerase (GalE), respectively. This genetic organization reflects the order of the metabolic conversions during galactose utilization via the Leloir pathway. The functionality of the galP, galK, galT, and galE genes was shown by complementation studies performed with both Escherichia coli and L. lactis mutants. The GalP permease is a new member of the galactoside-pentose-hexuronide family of transporters. The capacity of GalP to transport galactose was demonstrated by using galP disruption mutant strains of L. lactis MG1363. A galK deletion was constructed by replacement recombination, and the mutant strain was not able to ferment galactose. Disruption of the galE gene resulted in a deficiency in cell separation along with the appearance of a long-chain phenotype when cells were grown on glucose as the sole carbon source. Recovery of the wild-type phenotype for the galE mutant was obtained either by genetic complementation or by addition of galactose to the growth medium.
Understanding the mechanisms involved in carbohydrate metabolism and its regulation in lactic acid bacteria (LAB) is a prerequisite for improving the industrial properties of these organisms by metabolic engineering (16, 29). The primary function of LAB in industrial dairy fermentations is conversion of lactose to lactic acid. Utilization of this disaccharide can occur in LAB via several mechanisms (18, 47, 48). The glucose moiety resulting from lactose hydrolysis is metabolized via glycolysis, whereas the galactose moiety follows different pathways depending on the LAB. While some thermophilic strains of LAB are known to release the galactose moiety of lactose into the medium (16, 30, 37), other LAB metabolize this disaccharide via two possible pathways depending on the mode of transport of the sugar (18, 48). When lactose or galactose is transported via a phosphoenolpyruvate-dependent phosphotransferase system (PTS), the resulting galactose 6-phosphate is metabolized by the enzymes of the tagatose 6-phosphate pathway (3, 51). In the case of galactose or lactose entry via a permease (18, 48), the sugar is not phosphorylated, and the galactose is degraded via the Leloir pathway (24).
The Leloir pathway is among the first metabolic pathways that were discovered and includes three enzymes, galactokinase (GalK; EC 2.7.1.6), hexose-1-phosphate uridylyltransferase (GalT; EC 2.7.7.12), and UDP-glucose 4-epimerase (GalE; EC 5.1.3.2), which are involved in the conversion of galactose to glucose 1-phosphate. Aldose 1-epimerase or mutarotase (GalM; EC 5.1.3.3) is an additional enzyme required for rapid lactose metabolism and was characterized more recently (7, 35, 40). GalM catalyzes the interconversion of the α- and β-anomers of galactose, and the presence of this enzyme was found to be essential for efficient lactose utilization in Escherichia coli, as cleavage of this β-galactoside by β-galactosidase yields glucose and β-d-galactose, while α-d-galactose is the sole substrate for GalK (7). Genes encoding Leloir pathway enzymes have been characterized in different LAB, and their molecular organization has been discussed (27).
Previous reports on the related strains Lactococcus lactis NCDO 712 and ML3 focused on characterization of their efficient lactose metabolic pathway (18, 47, 48). It was shown that lactose is transported in these strains via a phosphoenolpyruvate-dependent PTS and that enzymes of the tagatose 6-phosphate pathway, which are encoded by the lac operon that resides on a lactose plasmid, are involved in degradation of the galactose 6-phosphate (13, 34, 51). In addition, it was found that galactose was catabolized via two distinct routes, the tagatose 6-phosphate and the Leloir pathways (47, 48). Although it was shown that the lactose PTS was able to transport galactose with low affinity, an additional galactose PTS activity was detected in a derivative of strain ML3 lacking a lactose plasmid (18, 36). However, the well-studied plasmid-free NCDO 712 derivative Lactococcus lactis MG1363 (25) was found to utilize galactose via the Leloir pathway, which requires a galactose permease (18). A chromosomally encoded galactose permease system was partially characterized and was found to be highly specific for galactose and to have a low affinity for lactose, suggesting that the Leloir pathway is the major route for galactose utilization (48).
Here we describe molecular and functional characterization of the gal gene cluster of Lactococcus lactis MG1363, whose gene organization and CcpA-dependent expression were reported previously (27, 33). The gal genes identified encode galactose transport via a galactose permease (GalP; previously called GalA) (27, 33) that belongs to the galactoside-pentose-hexuronide (GPH) family of transporters, a mutarotase (GalM), and the Leloir pathway enzymes (GalK, GalT, and GalE). Gene inactivation experiments showed that the galP and galK genes are essential for growth on galactose, while galE gene disruption not only eliminated growth on galactose but also resulted in the appearance of a long-chain phenotype when the organism was grown on glucose.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are shown in Tables 1 and 2, respectively. E. coli MC1061 (12) was used as a host strain for cloning experiments and was grown in Luria broth-based medium with aeration at 37°C (44). MacConkey agar (Difco Laboratories, Detroit, Mich.) was used to detect sugar fermentation by E. coli strains. Lactococcus lactis strains were grown without aeration at 30°C in M17 broth (Merck, Darmstadt, Germany) supplemented with 1% sugar. For screening of the galactose-fermenting phenotype, Lactococcus lactis was plated on galactose indicator agar (GIA) plates containing Elliker medium (21) supplemented with 1% galactose and 0.004% bromocresol purple (Merck). Ampicillin (50 μg/ml), erythromycin (2.5 μg/ml), and chloramphenicol (10 μg/ml) were used when appropriate.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant featuresa | Source or reference(s) |
|---|---|---|
| Escherichia coli strains | ||
| MC1061 | 12 | |
| HB101 | GalK-deficient strain (carrying galK2 mutation) | 8 |
| DW2 | MelB-deficient strain (ΔmelB) | 58 |
| LE392 | GalK- and GalT-deficient strain (carrying galK2 and galT22 mutations) | 44 |
| CGSC4467 | GalT-deficient strain (carrying galT22 mutation) | CGSCb |
| CGSC4498 | GalE-deficient strain (carrying galE28 mutation) | CGSCc |
| Lactococcus lactis strains | ||
| MG1363 | Lac−, Gal+, plasmid-free and prophage-cured derivative of NCDO712 | 25 |
| MG1363ΔgalK | Lac−, Gal−, MG1363 derivative with 380-bp deletion in the coding sequence of galK, obtained after a replacement-recombination event using pNZ8402 | This studyd |
| MG1363::galP1 | Lac−, Gal−, Emr, MG1363 derivative obtained by insertion mutagenesis, galP gene disrupted by integration of pNZ8451 in the coding sequence | This studyd |
| MG1363::galP2 | Lac−, Gal− Emr, MG1363 derivative obtained by insertion mutagenesis, galP gene disrupted by integration of pNZ8452 in the coding sequence | This studyd |
| MG1363::galE | Lac−, Gal− Emr, MG1363 derivative obtained by insertion mutagenesis, galE gene disrupted by integration of pNZ8460 in the coding sequence | This studyd |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant featuresa | Source or reference(s) |
|---|---|---|
| pBR322 | Apr Tcr; cloning vector | 5 |
| pUC19 | Apr; cloning vector | 59 |
| pUC18Ery | Apr Emr; derivative of pUC18 carrying an ery gene | 50 |
| pUC19Ery | Apr Emr; derivative of pUC19 carrying an ery gene | 32 |
| pNZ8020 | Cmr; inducible expression vector carrying the nisA promoter | 14 |
| pNZ8400 | Apr; 2.4-kbp SstI fragment of Lactococcus lactis cloned in pUC19 | This studyb |
| pNZ8401 | Apr; pNZ8400 derivative with a ScaI/StuI 380-bp deletion in the galK reading frame | This studyb |
| pNZ8402 | Apr Emr; XbaI/PstI fragment of pUC19Ery containing the ery gene and cloned in pNZ8401 | This studyb |
| pNZ8410 | Apr; 7.3-kbp SalI/EcoRI fragment of Lactococcus lactis cloned in pBR322 | This studyb |
| pNZ8421 | Apr; 1.8-kbp EcoRI fragment of pNZ8410 containing galE cloned in pBR322, orientation 1 | This studyb |
| pNZ8422 | Apr; same as pNZ8421, orientation 2 | This studyb |
| pNZ8442 | Apr; 2.5-kbp SalI/NheI fragment of pNZ8410 containing galP cloned in pUC18 | This studyb |
| pNZ8451 | Apr Emr; 1.1-kbp internal HpaII fragment of galP cloned in pUC18Ery, orientation 1 | This studyb |
| pNZ8452 | Apr Emr; same as pNZ8452, orientation 2 | This studyb |
| pNZ8460 | Apr Emr; internal 0.8-kbp fragment of galE cloned in pUC18Ery | This studyb |
| pNZ8560 | Cmr; 1.8-kbp EcoRI fragment of pNZ8410 containing galE cloned in pNZ8020 | This studyb |
Emr, Apr, Cmr, and Tcr, resistance to erythromycin, ampicillin, chloramphenicol, and tetracycline, respectively.
See Materials and Methods and Fig. 1.
DNA isolation, library construction, and other manipulations.
Isolation of E. coli plasmid DNA and standard recombinant DNA techniques were performed by using established protocols (44). Total DNA was isolated from protoplasts of Lactococcus lactis as described previously (55). Large-scale isolation of E. coli plasmid DNA for nucleotide sequence analysis was carried out by performing isopycnic centrifugation in CsCl-ethidium bromide gradients. Restriction endonucleases and other enzymes used for DNA manipulations were used according to the instructions of the suppliers (GIBCO-BRL, Gaithersburg, Md.; New England Biolabs Inc., Beverly, Mass.; and Boerhinger GmbH, Mannheim, Germany). DNA fragments were recovered from agarose gels by using the GlassMAX system (GIBCO-BRL). Both E. coli and Lactococcus lactis were transformed by electroporation by using a Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.) as specified by the manufacturer in the case of E. coli and as previously described in the case of Lactococcus lactis (28, 57). A library of Lactococcus lactis MG1363 DNA was constructed in E. coli HB101 by inserting SstI fragments that were approximately 2 to 3 kb long into SstI-linearized pUC19, since it had been observed that a Lactococcus lactis galK-like gene was located on a 2.4-kb SstI fragment (R. D. Pridmore, unpublished data).
Nucleotide sequence analysis.
Nucleotide sequence analysis of double-stranded plasmid DNA was performed by the dideoxy chain termination method (45) by using an ALF automatic sequencer in combination with T7 DNA polymerase autoread kits (Pharmacia Biotech). Sequencing reactions were initiated by using fluorescein-labeled universal and reverse primers (in the case of pUC-based constructs) and were continued with synthetic primers in combination with fluorescein-dATP as recommended by the manufacturer (Pharmacia Biotech). The synthetic primers used for sequencing from the EcoRI and SalI sites of plasmid pBR322 were pBREco (5′-ATAAAGAAGACAGTCATA-3′) and pBRSal (5′-ATCATGACATTAACCTATA-3′), respectively. Sequence data were assembled and analyzed by using the PC/GENE program (version 6.70; IntelliGenetics) and the Clone Manager program (version 4.0; Scientific and Educational Software).
Construction of strains and plasmids.
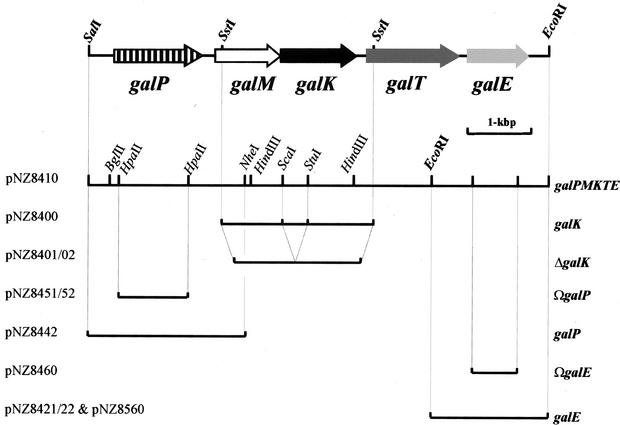
The relevant properties of the constructed strains and plasmids are shown in Tables 1 and 2. Plasmids pNZ8400 and pNZ8410 carrying part of the gal gene cluster and the complete gal gene cluster, respectively, were used for several subcloning experiments and for construction of the integration plasmids in E. coli (Fig. 1). Plasmid pNZ8401 is a derivative of pNZ8400 having a 380-bp ScaI/StuI deletion in the galK reading frame. Integration plasmid pNZ8402 was obtained after cloning of a pUC19Ery XbaI/PstI fragment containing the erythromycin resistance gene (ery) in the corresponding restriction sites of pNZ8401. In order to do this, plasmid pNZ8402 containing a deleted galK gene and the ery gene was introduced into Lactococcus lactis MG1363. Following transformation, erythromycin-resistant (Emr) colonies which contained a copy of the plasmid integrated in the chromosome were obtained, as confirmed by Southern hybridization (data not shown). These integrants were grown for at least 200 generations in the absence of erythromycin. Colonies sensitive to erythromycin and having a galactose-minus phenotype on GIA plates were analyzed by PCR, and Southern analysis confirmed that the intact galK gene was replaced by the deleted galK gene copy (ΔgalK) (Fig. 1). Integration plasmids pNZ8451 and pNZ8452, used for disruption of the galP gene, contained an internal HpaII fragment of galP cloned in the AccI site of plasmid pUC18Ery, in the same orientation as the orientation of the ery gene for pNZ8451 and in the opposite orientation for pNZ8452. For disruption of the galE gene, an integration vector designated pNZ8460 was constructed, and it contained an internal 0.8-kbp galE gene fragment that was PCR amplified, digested with restriction enzymes EcoRI and BamHI, and cloned in plasmid pUC18Ery (50) previously linearized with the same enzymes. The PCR primers used were primers DE1 (5′-GCACGAATTCAGTAACAGGTCATCGCGAGG-3′) and DE2 (5′-GCACGGATCCGCTACAAGTTCATCTGGATC-3′) carrying EcoRI and BamHI restriction sites (underlined). Plasmids pNZ8421 and pNZ8422 were pBR322 derivatives carrying a 1.8-kbp EcoRI fragment containing the complete galE gene and its upstream region cloned in the two possible orientations. The same fragment was used for construction of the lactococcal complementation plasmid pNZ8560 after cloning in the expression vector pNZ8020 (15). Plasmid pNZ8442 is a pUC18 derivative carrying the SalI/NheI fragment containing the complete galP gene and cloned between the pUC18 SalI and XbaI restriction sites.
FIG. 1.
Arrangement of the gal gene cluster of Lactococcus lactis MG1363 (accession no. AJ011653) and schematic diagram of the inserts of relevant plasmids.
Hybridization analysis.
DNA was transferred from a 0.8% agarose gel to GeneScreen Plus membranes (Du Pont, NEN Research Products, Boston, Mass.) as described previously (44, 46). Radioactively labeled oligonucleotide primers or DNA fragments were used as probes; these probes were obtained either by end labeling with [γ-32P]ATP or by nick translation with [α-32P]ATP (44).
Galactokinase and galactose uptake assays.
Galactokinase activity was measured by using a procedure adapted from the procedure described by Ajdić et al. (1). Cells of Lactococcus lactis were grown in M17 medium supplemented with the appropriate sugar (at a concentration of 1%) to an optical density at 600 nm of 1, washed twice with ice-cold 50 mM Tris-HCl (pH 7.3) buffer, and finally resuspended at a concentration of 30% in the same buffer. Cell lysates were prepared by disrupting the cells by using a Bead Beater (Biospec Products, Bartlesville, Okla.) and an equal volume of zirconia-silica beads (diameter, 0.1 mm). The protein concentration in the cell extract was determined by using Bradford's reagent (Bio-Rad Laboratories) (9). For galactokinase activity measurement, 20 μl of appropriately diluted cell extract was analyzed in a 100-μl reaction mixture containing 100 mM Tris-HCl (pH 7.3), 4 mM MgCl2, 3.2 μM NaF, 1.6 mM ATP, and 1 mM [14C]galactose (0.5 μCi ml−1; Amersham). The assay was started by adding the radiolabeled galactose, and galactose phosphorylation was determined at periodic intervals during 15 min of incubation at 30°C. Aliquots were transferred onto Whatman DE81 filter discs and air dried for 5 min. The filters, which bound sugar phosphates but not free galactose, were washed twice with water and oven dried. The radioactivity bound to the discs was determined by using a liquid scintillation counter (LS7500; Beckman Instruments Inc., Palo Alto, Calif.). Galactose uptake was measured essentially as described by Russel et al. (42).
SEM sample preparation.
Cells grown overnight were fixed in 0.2 M phosphate-buffered saline (pH 7.4) containing 2% glutaraldehyde for 30 min. Samples of cells were dehydrated by using a graded series of ethanol concentrations and were subsequently washed twice with absolute ethanol. The samples were critical point dried and coated with gold-palladium prior to observation with a Zeiss DSM950 scanning electron microscope (SEM).
Nucleotide sequence accession number.
The nucleotide sequence of the 7,309-bp SalI/EcoRI fragment containing the galactose gene cluster of Lactococcus lactis MG1363 has been deposited in the EMBL, GenBank, and DDBJ databases under accession number AJ011653.
RESULTS
Cloning, nucleotide sequence, and organization of the gal gene cluster of Lactococcus lactis.
A library of Lactococcus lactis subsp. cremoris MG1363 chromosomal DNA in pUC19 was used to complement the galK-deficient E. coli strain HB101. Transformants able to ferment galactose were selected on MacConkey agar plates containing galactose. A red colony, indicating galactose utilization, was isolated, and it contained a pUC19 derivative (pNZ8400) carrying a 2.4-kb SstI fragment derived from the Lactococcus lactis MG1363 chromosomal DNA, as shown by hybridization. Sequence analysis of the fragment revealed the presence of one complete gene, designated galK, preceded by an incomplete gene, designated galM, oriented in the same direction (Fig. 1). Comparison of the deduced amino acid sequences with the sequences available in the databases showed that there was significant homology between the complete GalK sequence and several galactokinase sequences, while the incomplete GalM sequence displayed homology with the sequences of mutarotases. Since the cloned fragment appeared to contain part of a gal gene cluster with a new gene order (galM-galK), we used a second step to clone a larger fragment using the same complementation strategy, based on preliminary restriction enzyme mapping of the chromosomal region (data not shown). An approximately 7.5-kbp SalI-EcoRI fragment was cloned and stably maintained in the medium-copy-number vector pBR322, resulting in pNZ8410. Sequencing of the insert in pNZ8410 revealed the presence of three additional genes, one located upstream from galM and designated galP and two situated downstream from galK and designated galT and galE, all oriented in the same direction (Fig. 1). GalT and GalE exhibited high sequence homologies with previously described GalT and GalE sequences, respectively, and the sequence of GalP showed homology with sequences of membrane proteins belonging to the GPH transporter family (38). This subfamily of transporters is called TC 2.A.2 in the recent transporter classification (TC) system (43). Thus, the galPMKTE gene cluster of Lactococcus lactis MG1363 (Fig. 1) encodes all the proteins required for transport and utilization of galactose via the Leloir pathway.
Amino acid homologies and identity of GalP with transport proteins.
Comparison of the predicted amino acid sequences of the gal gene products with previously described equivalent protein sequences showed a high degree of identity. The enzymes of the Leloir pathway showed the greatest predicted similarities to the enzymes identified in related gram-positive bacteria; GalK exhibited 59% identity to Streptococcus thermophilus GalK (54), GalT exhibited 53% identity to Lactobacillus casei GalT (2), and GalE exhibited 61% identity to S. thermophilus GalE (40). Lactococcus lactis subsp. cremoris MG1363 GalM exhibited a high level of identity (86%) only with the predicted enzyme from the related strain Lactococcus lactis subsp. lactis IL-1403 (6). Lower levels of identity were observed when GalM was compared to E. coli GalM (31%) (7) and also to any other GalM from a gram-positive bacterium; S. thermophilus GalM (40) exhibited 26% identity. Low levels of conservation similar to those of Lactococcus lactis GalM were not observed among bacteria when the other Leloir enzymes (GalK, GalT, and GalE) were considered.
Analysis of the deduced sequence of Lactococcus lactis GalP revealed a level of identity of 51% with the galactose permease (GalP) recently characterized by Djordjevic and coworkers for the heterofermentative LAB Lactobacillus brevis (19). The galactose permease-encoding gene of Lactococcus lactis was therefore designated galP instead of galA, which was the designation used previously (26, 27, 33). Another putative lactose permease, which is designated LacS and exhibits 96% identity with GalP, is present in Lactococcus lactis strain IL-1403 (6). GalP is also 45 and 44% identical to the lactose permeases (LacS) of Leuconostoc lactis (52) and S. thermophilus (39), respectively. Both Leuconostoc lactis LacS (639 amino acids) and S. thermophilus LacS (634 amino acids) are chimeric, as they possess a carrier domain homologous to the melibiose carrier (MelB) of E. coli (60) and an extra enzyme IIA-like domain attached to the C-terminal end of the carrier domain (39, 52). In contrast, Lactococcus lactis MG1363 GalP (462 amino acids), Lactococcus lactis IL-1403 LacS (462 amino acids), and Lactobacillus brevis GalP (474 amino acids) lack this C-terminal enzyme IIA-like domain and are composed of a carrier domain that is approximately the same size as E. coli MelB (462 residues for GalP, compared with 469 residues for MelB). The level of identity between the GalP and MelB proteins was 26%. Prediction of membrane-spanning helices revealed the presence of 12 putative helices in the GalP protein, supporting the hypothesis that its putative function is as a transmembrane protein.
Complementation of mutations in E. coli by Lactococcus lactis gal genes.
In order to show the functionality of the gal genes, we complemented several E. coli gal-deficient strains using pNZ8400 or pNZ8410 or some of their derivatives (Fig. 1). The complementation test was performed on MacConkey agar plates supplemented with 1% galactose or melibiose. Plasmid integrity in the complementing colonies was confirmed by restriction enzyme analysis of the plasmid contents (data not shown). All Lactococcus lactis gal genes tested (galK, galT, and galE) could complement the galactose utilization defect in the corresponding E. coli gal mutant (Table 3). GalE complementation was observed with both pNZ8421 and pNZ8422, two plasmids that carry the galE gene in opposite orientations, suggesting that a functional E. coli promoter was present in front of the galE gene. As expected, pNZ8401, which had a deletion in the galK gene, in contrast to pNZ8400 (Fig. 1), did not show complementation of galactose utilization in E. coli HB101, which also confirmed the functionality of galK. Because we did not have E. coli mutants that were deficient in galactose transport, we were not able to investigate the ability of GalP to transport galactose in E. coli. However, we demonstrated that GalP was able to complement the chromosomal mutation in the MelB melibiose transport system of E. coli DW2 (Table 3). MelB also is a member of the GPH (TC 2.A.2) transporter family. Thus, the Lactococcus lactis galP gene was able to complement the E. coli melB gene, indicating that the encoded transmembrane protein GalP is also involved in transport of the α-galactoside melibiose.
TABLE 3.
Complementation of lesions in E. coli genes with Lactococcus lactis genes
Functional analysis of the galP and galK genes in Lactococcus lactis.
We investigated the ability of lactococcal cells to accumulate galactose and to exhibit galactokinase activity when they were grown on glucose, galactose, maltose, or a glucose-galactose mixture. In the wild-type strain Lactococcus lactis MG1363, three different levels of activity were detected, and they were subject to strong regulation (Table 4). Whereas the levels of galactose uptake and the galactokinase activities were very low on glucose, large increases were observed on galactose. A third, intermediate level of expression was detected in cells grown on maltose or on a galactose-glucose mixture (Table 4). These results suggest that expression of both galK and galP is subject to glucose repression (catabolite repression) or galactose activation or both.
TABLE 4.
Galactose uptake and galactokinase activities in wild-type and mutant strains grown on different sugars
| Sugar(s)a | MG1363
|
MG1363::galP2
|
MG1363ΔgalK galactokinase activityc | ||
|---|---|---|---|---|---|
| Gal uptakeb | Galactokinase activityc | Gal uptakeb | Galactokinase activityc | ||
| Glu | 0.56 ± 0.05 | 0.44 | 0.29 ± 0.07 | <0.1 | <0.1 |
| Gal | 102.7 ± 12.2 | 281 | NGd | NG | NG |
| Glu + Gal | 2.12 ± 0.06 | 74 | 0.30 ± 0.03 | <0.1 | NDe |
| Mal | 2.52 ± 0.18 | 55 | 0.63 ± 0.05 | <0.1 | <0.1 |
Glu, glucose; Gal, galactose; Mal, maltose.
Expressed as nanomoles of labeled galactose accumulated per minute per milligram of protein. The values are the means ± standard deviations for at least triplicate determinations.
Specific activity in cell extracts, expressed as nanomoles of phosphorylated galactose per minute per milligram of protein. The values are means for duplicate determinations.
NG, no growth.
ND, not determined.
The chromosomal copy of the galP gene was inactivated by single-crossover integration of the nonreplicating plasmid pNZ8451 or pNZ8452 (Fig. 1). These two plasmids carry an internal fragment of the galP gene cloned in two orientations, which allows integration of the erythromycin resistance-encoding gene (ery) of pUC18Ery in both directions. As the ery gene is not followed by a terminator and has its own promoter, readthrough transcription allows expression of the downstream genes provided that the ery gene is cloned in the same orientation (50). Therefore, pNZ8451 integration in galP allows expression of the downstream genes (galMKTE), while pNZ8452 integration probably leads to a loss of expression of these downstream genes. Strains were selected in which pNZ8451 (Lactococcus lactis MG1363::galP1) or pNZ8452 (Lactococcus lactis MG1363::galP2) was integrated, and these strains were analyzed by PCR and by Southern hybridization (data not shown). This analysis confirmed that the plasmids were integrated in the galP gene, which resulted in disruption of the gene into two truncated parts. The phenotypes of the different integrants were analyzed on GIA plates and in M17 liquid medium containing galactose. Both Lactococcus lactis MG1363::galP1 and MG1363::galP2 appeared to be unable to use galactose as a sole carbon source. These results show that the galP gene is necessary for galactose metabolism in Lactococcus lactis MG1363. Enzymatic assays in which Lactococcus lactis MG1363::galP2 was used showed that there was a strong decrease in the ability to import galactose concomitant with a complete loss of detectable galactokinase activity (Table 4). In contrast, Lactococcus lactis MG1363::galP1 showed significant galactokinase activity when it was grown on glucose (data not shown), confirming the expected readthrough from the ery gene, as reported previously for Lactococcus lactis (50). The absence of galactokinase activity in strain Lactococcus lactis MG1363::galP2 suggests that a functional promoter downstream of the galP gene is not present and therefore that an operon-like organization of the gal genes is likely.
A 380-bp deletion was introduced into the galK gene by a standard two-step homologous recombination procedure (Fig. 1). The resulting strain, Lactococcus lactis MG1363ΔgalK, was not able to use galactose as a carbon source, as determined both on GIA plates and in M17 liquid medium. Furthermore, no galactokinase activity could be detected in this strain after growth on maltose (Table 4).
Inactivation of chromosomal galE by gene replacement and effect on the phenotype of the recombinants.
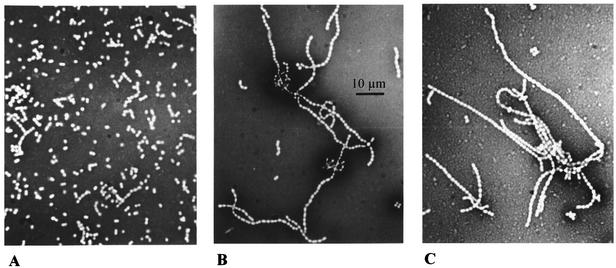
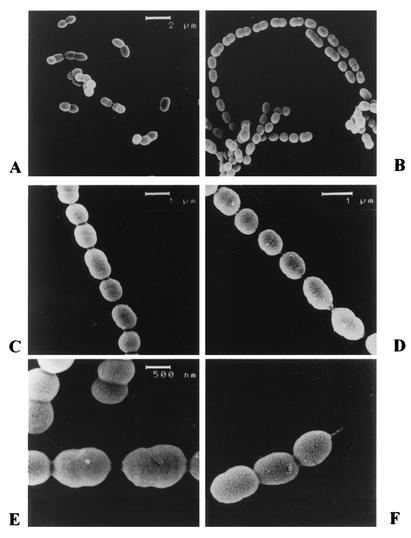
An internal fragment of the galE gene was PCR amplified and cloned in plasmid pUC18Ery. The resulting nonreplicating plasmid, pNZ8460 (Fig. 1), was introduced into Lactococcus lactis MG1363 by electroporation, and recombinant strains were selected after plating on M17 agar plates supplemented with glucose and containing erythromycin. Integration of pNZ8460 into the galE gene following a single recombination event was confirmed by Southern hybridization (data not shown), and a clone showing the expected genotype was designated Lactococcus lactis MG1363::galE and was analyzed further. As expected, Lactococcus lactis MG1363::galE was not able to use galactose as a sole carbon source. Moreover, the galE mutant strain showed a strongly reduced growth rate when it was grown in a liquid medium containing glucose or maltose as the carbon source (data not shown). This growth defect was characterized by cell flocculation, followed by sedimentation of the cells in the bottoms of the culture tubes. The mutant cells were examined by light microscopy, and the results suggested that the growth defect was linked to formation of long chains of cells containing up to 50 cocci (Fig. 2). Analysis of the cells by SEM revealed a normal shape but the presence of a possible peptidoglycan bridge between unseparated cells (Fig. 3). This could reflect an unfinished separation mechanism, while the elongation and septation steps were not affected, as observed by transmission electron microscopy (data not shown). In order to fully associate the phenotype of the galE mutant with the galE disruption and not with a possible polar effect of insertion of the knockout plasmid pNZ8460, genetic complementation of the galE gene was performed. We constructed a plasmid that contained the intact galE gene and was able to replicate in lactococcal cells. Introduction of this plasmid, pNZ8560, into the galE mutant strain resulted in restoration of the ability of the cells to grow on galactose and in the disappearance of the long-chain phenotype when cells were grown on glucose (data not shown). No nisin induction was required for such genetic complementation when the pNZ8020 expression vector was used, suggesting that a functional promoter was present upstream of the galE gene. Remarkably, disappearance of the long-chain phenotype was also observed when galE mutant cells were grown on a mixture of glucose and galactose. The galactose was probably used by the cells to restore a sufficient level of UDP-galactose even when the gal genes were expressed at a low level because of the presence of glucose (catabolite repression of the gal genes). Similarly, when cells of the galE mutant were grown on maltose, they showed the long-chain phenotype, which was not observed during growth on a mixture of maltose and galactose. A minor amount of galactose appeared to be necessary for such substrate complementation, as 0.005% galactose was enough to prevent the long-chain phenotype in the presence of 1% glucose. This finding is consistent with the previous observation that the gal genes were not fully repressed in the presence of glucose and with the observation that galactose transport and galactokinase activities were detectable in a culture grown on a mixture of glucose and galactose (Table 4).
FIG. 2.
Light micrographs of the wild-type strain Lactococcus lactis MG1363 (A) and the galE mutant strain Lactococcus lactis MG1363::galE (B and C). Both strains were grown overnight in M17 medium supplemented with glucose. Bar = 10 μm.
FIG. 3.
SEM micrographs of the wild-type strain Lactococcus lactis MG1363 (A) and the galE mutant strain Lactococcus lactis MG1363::galE (B to F). Both strains were grown overnight in M17 medium supplemented with glucose.
DISCUSSION
To investigate the genetics of galactose utilization via the Leloir pathway in Lactococcus lactis, we cloned and characterized the chromosomal gal gene cluster, which has the order galP-galM-galK-galT-galE. This cluster organization is novel compared to the organization of previously described gal gene clusters and features the presence of a galactose permease gene (galP) located upstream of the genes for the Leloir pathway, reflecting the order of galactose utilization. The functionality of the gal gene products was established based on complementation studies, transport experiments, and analysis of strains having mutations in the gal gene cluster.
A unique feature of this gal gene cluster is the presence of the galP gene, encoding a galactose permease. The mutant strains Lactococcus lactis MG1363::galP1 and MG1363::galP2 were constructed and used to show the relationship between galactose accumulation and the presence of a functional galP gene. Galactose transport was strongly reduced in the mutant strains but was not completely eliminated. The observed residual uptake could be related to nonspecific transporters. The functionality of GalP was also confirmed with the complementation in E. coli DW2 of a transporter belonging to the same family, the melibiose transport protein MelB. These results corroborate previous observations which suggested the presence in Lactococcus lactis ATCC 7962 of a galactose permease with higher affinity for galactose than for lactose (31). The low capacity of Lactococcus lactis MG1363 to transport lactose has also been reported previously (17). Moreover, a DNA fragment containing a putative lactose permease-encoding gene was described, but it was characterized only by restriction enzyme digestion, which showed that it is clearly different from the galP gene described here (41). Hence, it seems likely that Lactococcus lactis MG1363 contains a lactose transport system with low efficiency that differs from the highly efficient galactose permease that is encoded by the galP gene. Based on sequence comparisons and functionality data, the GalP galactose permease of Lactococcus lactis MG1363 was classified as a new member of the GPH family of transporters (38), which is also called TC 2.A.2 (43), and clustered with the recently characterized GalP protein of Lactobacillus brevis (19). While sequence identity comparisons predict that the GalP proteins belong to the LacS subfamily of the GPH family (38), these proteins do not contain the C-terminal extra enzyme IIA-like domain which is a characteristic and functionally important feature of the LacS subfamily (38). Thus, Lactococcus lactis GalP and Lactobacillus brevis GalP are the only members of a new subfamily in the GPH family of transporters and probably have the same properties.
Comparison of the organization of the gal genes with the organization of previously described clusters revealed some similarities but also important differences (27). Among other gram-positive bacteria, high degrees of identity were found when the Leloir enzymes, GalK, GalT, and GalE, were considered (more than 50% identity). The gene order galK-galT-galE has been observed in five different LAB or closely related bacteria (S. thermophilus, Streptococcus mutans, Streptococcus salivarius, Streptococcus equi, and Lactococcus lactis MG1363) (27, 49). The complementation studies performed with specific E. coli mutants demonstrated the functionality of the Leloir enzyme genes, galK, galT, and galE. We show here that a galM gene is present in the gal gene cluster of Lactococcus lactis MG1363. So far, this gene has also been identified in other LAB (1, 2, 40). However, its function has been studied only in E. coli, where it is related to efficient lactose utilization (7). The presence of galM in a gal cluster like that of Lactococcus lactis MG1363 may reflect possible involvement of GalM in efficient galactose metabolism. Construction of Lactococcus lactis mutant strains deficient in GalP, GalK, or GalE showed the functionality of the corresponding genes and also confirmed that the Leloir pathway is the only functional pathway for galactose utilization in Lactococcus lactis MG1363.
A remarkable feature of the gal gene cluster of Lactococcus lactis subsp. cremoris MG1363 is that it does not consist of a combination of gal and lac genes. Genes related to lactose metabolism are often found together with gal genes in LAB (18, 27). This is the case for Lactobacillus helveticus, in which the galE gene is separated from the galKTM genes and is associated with the lacL and lacM genes encoding the subunits of the β-galactosidase, and for S. thermophilus, in which there is an association among the gal operon, the lactose permease gene (lacS), and the β-galactosidase gene (lacZ) (18). Remarkably, another lactococcal strain, Lactococcus lactis ATCC 7962, was found to have a different organization, in which galT and galE are separated by two inserted lac genes, lacA encoding a putative thiogalactosidase and lacZ encoding the β-galactosidase (53). This organization is also found in Lactococcus lactis subsp. lactis IL-1403, in which the predicted gene order is lacS-galM-galK-galT-lacA-lacZ-galE (6). Thus, the gal gene cluster of Lactococcus lactis MG1363 appears to be highly specific for galactose, containing all the structural genes needed for galactose utilization via the Leloir pathway in an uninterrupted form at the same locus. This is not the case in other dairy LAB, in which the gal gene clusters are parts of lactose utilization systems or do not contain galactose-specific transporters (27). The gal operon of Lactobacillus casei also contains only gal genes (galKETRM), but it lacks a galactose permease (2).
The data provided here confirm that galactose metabolism in Lactococcus lactis MG1363 is strongly regulated and subject to catabolite repression. The involvement of Lactococcus lactis catabolite control protein CcpA in control of gal gene expression has been shown recently (33). This study also showed that the galP promoter contains a consensus cre site. Galactokinase and galactose uptake assays performed under different growth conditions demonstrated that the two activities are similarly regulated. This observation is consistent with the transcriptional organization of the gal genes that showed the presence of a galPMKTE polycistronic transcript which was present only in galactose-grown cells (26; Grossiord, unpublished data). The lack of GalK activity in the Lactococcus lactis MG1363::galP2 mutant also confirms the possible operon organization of the Lactococcus lactis MG1363 gal gene cluster. However, notably, the complementation studies performed with Lactococcus lactis suggested that an additional functional promoter is present in the gal gene cluster and is located in front of the galE gene; this suggestion is supported by the role of galE in glucose-grown cells and is analogous with previous observations that indicated the role of galE in more general metabolic properties (1, 4, 40). A promoter was mapped upstream from the galE gene in Lactococcus lactis NCDO2054 (53) and was also characterized in Lactococcus lactis MG1363 (26). In Lactococcus lactis MG1363, the role of the galE gene product in general metabolism was observed when the galE mutant was grown on glucose, which resulted in the long-chain phenotype. The inability of the galE mutant cells to separate after division is reminiscent of the phenotype of Lactococcus lactis deficient in AcmA, the major autolysin (10). A similar observation was described by Duwat et al. (20) for a strain deficient in dltD, a gene involved in d-alanylation of the lipoteichoic acids (LTAs). Evaluation of the biological functions and the fate of UDP-galactose in the cells (14) showed that it is used in Lactococcus lactis for substitution of LTAs, which are molecules of polyglycerol-phosphate that are linked to the membrane and extend through the cell wall (22). LTAs have been shown to play a key role in the regulation of autolysis and are often branched with d-alanine or, in some cases, with monosaccharides, such as glucose or galactose (23). The composition of the LTAs has been studied previously, and these molecules have been shown to contain galactose in Lactococcus lactis NCDO712 (23), the parental strain of Lactococcus lactis MG1363 (25). The level of alanine substitution in the LTAs of Bacillus subtilis was found to be important for the function of these molecules in the regulation of autolysis (56). Similarly, galactose substitution in the LTAs in Lactococcus lactis could link this process, which requires UDP-galactose, with the regulation of autolysis.
In conclusion, we present evidence that the galPMKTE gene cluster is functional and allows efficient galactose utilization via the Leloir pathway in Lactococcus lactis MG1363. The organization of this cluster is unique among the known gal-lac gene clusters, and there is a galP gene encoding a galactose permease which is a new member of the GPH family of transporters. We also demonstrate the essential role of the galE gene product for normal growth, which is related to the function of UDP-galactose in cell wall biosynthesis.
Acknowledgments
The excellent technical assistance of Jean-Pierre Bossy and Marc Ravallec with SEM and transmission electron microscope analyses of the galE mutant is gratefully acknowledged. We are grateful to David Pridmore for sharing his results for the Lactococcus lactis gal system. We thank Richard van Kranenburg and Monique Zagorec for fruitful suggestions and Michiel Kleerebezem for critically reading the manuscript.
Part of this work was supported by contracts ERB-CHGT-CT-93-0459 and BIOT-CT-96-0498 from the Commission of European Communities. B.P.G. also received a grant from the Institut Yoplait International.
REFERENCES
- 1.Ajdić, D., I. C. Sutcliffe, R. R. B. Russel, and J. J. Ferretti. 1996. Organization and nucleotide sequence of the Streptococcus mutans galactose operon. Gene 180:137-144. [DOI] [PubMed] [Google Scholar]
- 2.Bettenbrock, K., and C.-A. Alpert. 1998. The gal genes for the Leloir pathway of Lactobacillus casei 64H. Appl. Environ. Microbiol. 64:2013-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisset, D. L., and R. L. Anderson. 1974. Lactose and d-galactose metabolism in group N streptococci: presence of enzymes for both the d-galactose-1-phosphate and d-tagatose-6-phosphate pathways. J. Bacteriol. 117:318-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar, F., R. L. Rodrigez, P. J. Greene, M. C. Betlach, H. L. Heyneker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouffard, G. G., K. E. Rudd, and S. L. Adhya. 1994. Dependence of lactose metabolism upon mutarotase encoded in the gal operon in Escherichia coli. J. Mol. Biol. 244:269-278. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, H. B., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-469. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haetrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buttin, G. 1963. Mécanismes régulateurs dans la biosynthèse des enzymes du métabolisme du galactose chez Escherichia coli K12. La synthèse induite de la galactokinase et l'induction simultanée de la séquence enzymatique. J. Mol. Biol. 7:164-182. [DOI] [PubMed] [Google Scholar]
- 12.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 13.Crow, V. L., G. P. Davey, L. E. Pearce, and T. D. Thomas. 1983. Plasmid linkage of the d-tagatose 6-phosphate pathway in Streptococcus lactis: effect on lactose and galactose metabolism. J. Bacteriol. 153:76-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delcour, J., T. Ferrain, M. Deghorian, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Leeuwenhoek 76:159-184. [PubMed] [Google Scholar]
- 15.de Ruyter, P. G. G. A., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vos, W. M. 1996. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek 70:223-242. [DOI] [PubMed] [Google Scholar]
- 17.de Vos, W. M., and G. Simons. 1988. Molecular cloning of lactose genes in dairy lactic streptococci: the phospho-β-galactosidase and β-galactosidase genes and their expression products. Biochimie 70:461-473. [DOI] [PubMed] [Google Scholar]
- 18.de Vos, W. M., and E. E. Vaughan. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217-237. [DOI] [PubMed] [Google Scholar]
- 19.Djordjevic, G., J. H. Tchieu, and M. H. Saier, Jr. 2001. Genes involved in control of galactose uptake in Lactobacillus brevis and reconstitution of the regulatory system in Bacillus subtilis. J. Bacteriol. 183:3224-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliker, P. R., A. Anderson, and G. H. Hannessen. 1956. An agar culture medium for lactic streptococci and lactobacilli. J. Dairy Sci. 39:1611-1612. [Google Scholar]
- 22.Fischer, W. 1988. Physiology of lipoteichoic acids in bacteria, p. 233-302. In A. H. Rose and D. W. Tempest (ed.), Advances in microbial physiology. Academic Press, London, United Kingdom. [DOI] [PubMed]
- 23.Fischer, W., P. Rösel, and H. U. Koch. 1981. Effect of alanine ester substitution and other structural features of lipoteichoic acids on their inhibitory activity against autolysins of Staphylococcus aureus. J. Bacteriol. 146:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey, P. A. 1996. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10:461-470. [PubMed] [Google Scholar]
- 25.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossiord, B. P. 1998. Métabolisme du galactose par la voie de Leloir: l'opéron gal de Lactococcus lactis. Ph.D. thesis. École Nationale Supérieure Agronomique, Montpellier, France.
- 27.Grossiord, B. P., E. E. Vaughan, E. J. Luesink, and W. M. de Vos. 1998. Genetics of galactose utilisation in lactic acid bacteria. Lait 78:77-84. [Google Scholar]
- 28.Holo, H., and I. F. Nes. 1989. High frequency transformation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hols, P., M. Kleerebezem, A. N. Schanck, T. Ferain, J. Hugenholtz, J. Delcour, and W. M. de Vos. 1999. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 17:588-592. [DOI] [PubMed] [Google Scholar]
- 30.Hutkins, R. W., and H. A. Morris. 1987. Carbohydrate metabolism by Streptococcus thermophilus: a review. J. Food Prot. 50:876-884. [DOI] [PubMed] [Google Scholar]
- 31.Kashket, E. R., and T. H. Wilson. 1973. Proton-coupled accumulation of galactoside in Streptococcus lacis 7962. Proc. Natl. Acad. Sci. USA 70:2866-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leenhouts, K. J., J. Kok, and G. Venema. 1990. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl. Environ. Microbiol. 56:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luesink, E. J., R. E. M. A. van Herpen, B. P. Grossiord, O. P. Kuipers, and W. M. de Vos. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789-798. [DOI] [PubMed] [Google Scholar]
- 34.McKay, L. L., L. A. Walter, W. E. Sandine, and P. R. Elliker. 1969. Involvement of phosphoenolpyruvate in lactose utilization in group N streptococci. J. Bacteriol. 99:603-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mollet, B., and N. Pilloud. 1991. Galactose utilization in Lactobacillus helveticus: isolation and characterization of the galactokinase (galK) and the galactose-1-phosphate uridyl transferase (galT) genes. J. Bacteriol. 173:4464-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, Y. H., and L. L. McKay. 1982. Distinct galactose phosphoenolpyruvate-dependent phosphotransferase system in Streptococcus lactis. J. Bacteriol. 149:420-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poolman, B. 1993. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12:125-148. [DOI] [PubMed] [Google Scholar]
- 38.Poolman, B., J. Knol, C. van der Does, P. J. F. Henderson, W. J. Liang, G. Leblanc, T. Pourcher, and I. Mus-Veteau. 1996. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 19:911-922. [DOI] [PubMed] [Google Scholar]
- 39.Poolman, B., T. J. Royer, S. E. Mainzer, and B. F. Schmidt. 1989. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J. Bacteriol. 171:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poolman, B., T. J. Royer, S. E. Mainzer, and B. F. Schmidt. 1990. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDP glucose 4-epimerase. J. Bacteriol. 172:4037-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross, R., F. O'Gara, and S. Condon. 1989. Cloning of chromosomal genes of Lactococcus by heterologous complementation: partial characterisation of a putative lactose transport gene. FEMS Microbiol. Lett. 61:183-188. [DOI] [PubMed] [Google Scholar]
- 42.Russel, R. R. B., J. Aduse Opoku, I. C. Sutcliffe, L. Tao, and J. J. Ferretti. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J. Biol. Chem. 267:4631-4637. [PubMed] [Google Scholar]
- 43.Saier, M. H., Jr. 2000. A functional-phylogenic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:254-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sanger, F. S., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. 1979. Lactose metabolism in Streptococcus lactis: phosphorylation of galactose and glucose moieties in vivo. J. Bacteriol. 140:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. 1980. Galactose transport systems in Streptococcus lactis. J. Bacteriol. 144:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaillancourt, K., S. Moineau, M. Frenette, C. Lessard, and C. Vadeboncoeur. 2002. Galactose and lactose genes from the galactose-positive bacterium Streptococcus salivarius and the phylogenetically related galactose-negative bacterium Streptococcus thermophilus: organization, sequence, transcription, and activity of the gal gene products. J. Bacteriol. 184:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Kranenburg, R., J. D. Marugg, I. I. van Swam, J. W. Norwin, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 51.van Rooijen, R. J., S. van Schalkwijk, and W. M. de Vos. 1991. Molecular cloning, characterization, and nucleotide sequence of the tagatose 6-phosphate pathway gene cluster of the lactose operon of Lactococcus lactis. J. Biol. Chem. 266:7176-7181. [PubMed] [Google Scholar]
- 52.Vaughan, E. E., S. David, and W. M. de Vos. 1996. The lactose transporter in Leuconostoc lactis is a new member of the LacS subfamily of galactoside-pentose-hexuronide translocators. Appl. Environ. Microbiol. 62:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaughan, E. E., R. D. Pridmore, and B. Mollet. 1998. Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J. Bacteriol. 180:4893-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaughan, E. E., P. van den Bogaard, P. Catzedu, O. P. Kuipers, and W. M. de Vos. 2001. Activation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183:1184-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vos, P., M. van Asseldonk, F. van Jeveren, R. Siezen, G. Simons, and W. M. de Vos. 1989. A maturation protein is essential for the production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J. Bacteriol. 171:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wecke, J., M. Perego, and W. Fischer. 1996. d-Alanine deprivation of Bacillus subtilis teichoic acids is without effect on cell growth and morphology but affects the autolytic activity. Microb. Drug Resist. 2:123-129. [DOI] [PubMed] [Google Scholar]
- 57.Wells, J. M., P. W. Wilson, and R. W. F. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, D. M., and T. H. Wilson. 1987. Cation specificity for sugar substrates of the melibiose carrier in Escherichia coli. Biochem. Biophys. Acta 904:191-200. [DOI] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 60.Yazyu, H., S. Shiota-Niiya, T. Shimamoto, H. Kanazawa, M. Futai, and T. Tsuchiya. 1984. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J. Biol. Chem. 259:4320-4326. [PubMed] [Google Scholar]