Abstract
FtsH is a membrane-bound and energy-dependent metalloprotease in bacteria which is involved in the posttranslational control of the activity of a variety of important transcription factors and in the degradation of uncomplexed integral membrane proteins. For Bacillus subtilis, little is known about the target proteins of FtsH protease. Its gene is not essential, but knockout strains display a pleiotropic phenotype including sensitivity toward salt and heat stress, defects in sporulation and competence, and largely filamentous growth. Comparison of the intracellular proteomes of wild-type and ftsH knockout strains revealed that at least nine proteins accumulated in the absence of ftsH, four of which could be identified. Two of these proteins turned out to be members of the σW regulon. Accumulation of one of these σW-controlled proteins, the penicillin-binding protein PBP4*, was analyzed in more detail. We could show that PBP4* is not a proteolytic substrate of FtsH and that its overproduction is due to the enhanced transcription of its gene (pbpE) in ftsH null mutants. The filamentous growth phenotype of ΔftsH strains was abolished in a ΔftsH ΔpbpE double knockout. In ftsH wild-type strains with the pbpE gene under regulatable control, pbpE overexpression caused filamentation of the cells. DNA macroarray analysis revealed that most genes of the σW regulon are transcribed at elevated levels in an ftsH mutant. The influence of FtsH on σW-controlled genes is discussed.
FtsH belongs to the AAA (ATPases associated with diverse cellular activities) protein superfamily, and members of this family are involved in a wide variety of cellular processes, such as vesicle-mediated protein transport, cell cycle control, control of gene expression, and proteolysis (for recent reviews, see references 19 and 35). The AAA proteins constitute a subfamily of the Walker-type nucleoside triphosphatases and contain conserved ATPase domains, typically spanning 200 to 250 residues, referred to as AAA modules (6, 20). FtsH is a member of the AAA proteases, which occur in eubacteria and eukaryotic organelles such as mitochondria and chloroplasts, but not in archaea, and contain an AAA module and a zinc metalloprotease domain in a single polypeptide chain. Bacterial FtsH proteins are anchored via two transmembrane segments in the cytoplasmic membrane, with the short N- and the long C-terminal parts facing the cytoplasm. These metalloproteases are involved in the quality control of membrane-bound proteins and in the degradation of short-lived cytoplasmic regulatory proteins (for recent reviews, see references 16 and 26).
While several target proteins of the Escherichia coli FtsH metalloprotease have already been identified, they remained elusive in Bacillus subtilis. Therefore, we reasoned that comparing the proteomes of B. subtilis wild-type and ftsH knockout cells should reveal target proteins accumulating in the absence of the protease. It should be mentioned that, in contrast to that of E. coli, the B. subtilis ftsH gene is dispensable, though its absence results in a pleiotropic phenotype (9). Indeed, this proteomic approach led to the discovery of at least nine proteins present in elevated amounts in the ftsH null mutant, four of which could be identified by matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometry. Interestingly, two of these proteins belong to the σW regulon, and further experiments revealed that most mRNAs of this regulon are present at elevated levels in the ftsH knockout, suggesting that this membrane-anchored metalloprotease exerts a direct or indirect effect on the regulation of the σW regulon.
MATERIALS AND METHODS
Growth conditions, bacterial strains, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely grown aerobically at 37°C in Luria broth (LB). Ampicillin was included for all plasmid-bearing E. coli strains. Chloramphenicol, neomycin, tetracycline, and erythromycin were added at concentrations of 5, 10, 15, and 1 μg ml−1, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqlacZM15 Tn10] | Stratagene |
| DH10B | mcrA Δ(mrr hsdRMS mcrBC) φ80d lacZM15 ΔlacX74 deoR recA1 araD139 Δ(ara leu)7697 galU galK rpsL endA1 nupG | Bethesda Research Laboratories, Inc. |
| Bacillus subtilis | ||
| 1012 | leuA8 metB5 trpC2 hsrM1 | 23 |
| WW01 | ftsH::erm derivative of 1012 | 36 |
| ED04 | ftsH::tet in 1012 | 9 |
| MP01 | ftsH::cat in 1012 | 9 |
| sigW | sigW::neo allele in 1012 | 37 |
| SWV119 | abrB::tet | 30 |
| ΔpbpE | ΔpbpE::erm allele isolated from PS1805 and transformed into 1012 | 21 |
| BFS233 | pMUTIN inserted into yuaF; promoter of yuaF fused to lacZ | 27 |
| MSW01 | pX2-ftsH | This work |
| SZ01 | ΔpbpE::erm ftsH::cat | This work |
| SZ06 | ΔpbpE::erm amyE::PxylA-pbpE ftsH+ in 1012 | This work |
| SZ08 | ΔsigW::neo pX2-ftsH | This work |
| SZ09 | ΔpbpE::erm amyE::PxylA-pbpE ftsH::tet in 1012 | This work |
| TW51 | amyE::PyuaF-lacZ; derived from pPyuaF-bgaB, with bgaB replaced by lacZ | This work; PyuaF-bgaB from reference 37 |
| TW52 | amyE::PyuaF-lacZ ftsH::cat | This work |
| Plasmids | ||
| pQE30 | 3.5-kb vector allowing overproduction of proteins with six N-terminal histidine residues | 32 |
| pX | Carries xylose-regulatable expression cassette for insertion at the amyE locus | 14 |
| pX2 | Carries xylose-regulatable expression cassette to be fused in front of the gene of interest | 18 |
DNA manipulations and analysis.
Plasmid DNA was purified on columns (Qiagen, Hilden, Germany), and PCR products were generated with DeepVent DNA polymerase as specified by the manufacturer (New England Biolabs). Cloning was carried out by standard methods (24).
Proteomics.
Protein extracts were prepared, and proteins were separated by two-dimensional (2-D) polyacrylamide gel electrophoresis (PAGE), as previously described (4). Individual protein spots were analyzed by MALDI-TOF mass spectrometry for identification.
Preparation of polyclonal antibodies against PBP4*.
The coding region of pbpE was amplified by PCR using appropriate primers, and the product was integrated into the vector plasmid pQE30, thereby fusing six histidine residues to the N terminus of PBP4*. The recombinant plasmid pQE30-pbpE was transformed into E. coli strain XL-1 Blue, and overexpression of the His-tagged protein was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After His-PBP4* was purified, it was sent to the “Elevage Scientifique des Dombes” company to be used for raising antibodies in rabbits. Polyclonal antibodies were used in Western blot experiments at the following dilutions: αHtpG, 1:10,000; αPBP4*, 1:10,000; αYdjF (a gift of A. Atalla), 1:10,000; and αAbrB, 1:2,000.
Western and Northern blotting and DNA macroarray analysis.
Samples were prepared for sodium dodecyl sulfate (SDS)-PAGE and immunoblot analysis as described previously (11). Preparation of high quality RNA, Northern blotting, and DNA macroarray analysis have been described previously (37). In summary, the analysis was carried out by using Panorama B. subtilis Gene Arrays and cDNA labeling primers (optimized for B. subtilis), all from Sigma Genosys. Two micrograms of total RNA was hybridized to 4 μl of cDNA labeling mix. Hybridization signal intensities were quantified with ArrayVision software (version 5.1; Imaging Research, St. Catherine's, Ontario, Canada). For normalization and background subtraction, the overall spot normalization function of ArrayVision was used. Each analysis was carried out using two independently isolated RNA preparations and different array batches. Means and deviations of expression level ratios from these different experiments were calculated using Microsoft Excel. Ratios were calculated only from spots with a signal-to-noise ratio larger than 3 in the ΔftsH experiments.
β-Galactosidase assays.
β-Galactosidase activities were assayed at 28°C as described previously (37) by using o-nitrophenyl-β-d-galactopyranoside as a substrate. All assays were repeated at least three times, and the replicates yielded comparable results.
RESULTS
At least nine proteins accumulate in an ftsH knockout.
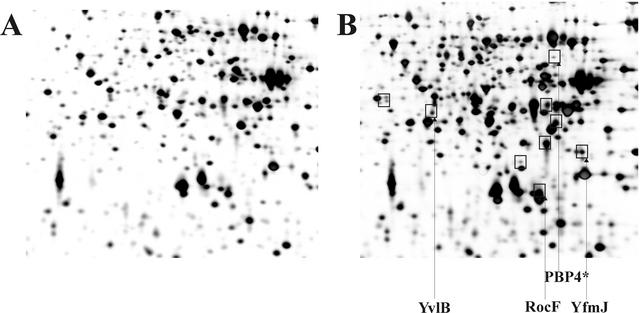
In contrast to E. coli, where several proteins have been identified as substrates for the FtsH protease, no target proteins for the B. subtilis integral membrane protease have been demonstrated so far. We reasoned that in the absence of FtsH, potential substrate proteins could accumulate. To detect these proteins, cellular extracts prepared from an ftsH wild-type strain and its isogenic ftsH::cat knockout strain were resolved by the 2-D gel electrophoresis technique. Careful comparison of the two patterns revealed at least nine proteins which accumulated in the absence of the proteolytic activity (Fig. 1). By use of MALDI-TOF mass spectrometry, four of these proteins present in elevated amounts in the ΔftsH strain were identified as PBP4*, RocF, YvlB, and YfmJ. While PBP4* is encoded by the pbpE gene and belongs to the group of penicillin-binding proteins (21), RocF is an arginase (10). The yvlB gene codes for a protein of unknown function, and yfmJ codes for a protein similar to quinone oxidoreductase. Most interestingly, the pbpE and yvlB genes belong to the σW regulon (12). These data suggest that the FtsH protease either affects the stability of one or more of these four proteins directly or affects their synthesis indirectly. To distinguish between these two possibilities, we have chosen PBP4* for more-detailed analysis of protein accumulation in ftsH knockout strains with respect to synthesis and stability.
FIG. 1.
Comparative proteomics of the B. subtilis wild-type strain 1012 (A) and its isogenic ftsH::cat knockout (B). Intracellular proteins of B. subtilis 1012 and MP01 (ftsH::cat) exponentially grown in minimal medium at 37°C were separated in a pH gradient of 4 to 7 in the first dimension. Gels were stained with silver nitrate. Proteins present in elevated amounts in the ftsH::cat strain are boxed.
Accumulation of PBP4* in B. subtilis is due to enhanced transcription of the pbpE gene.
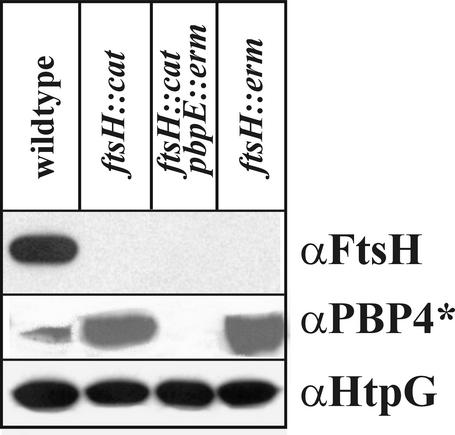
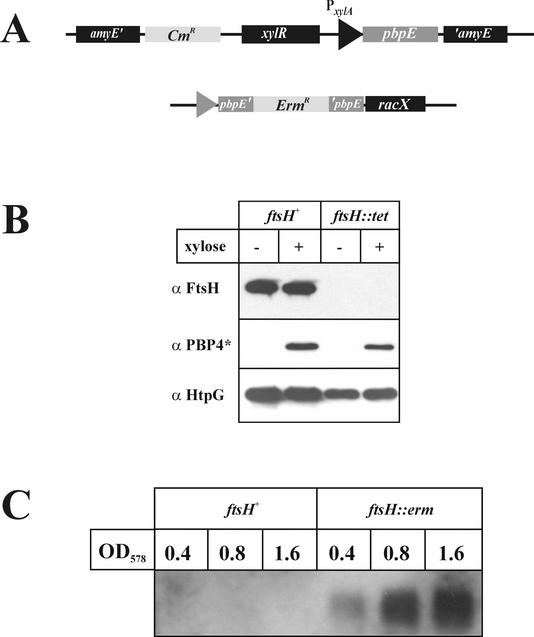
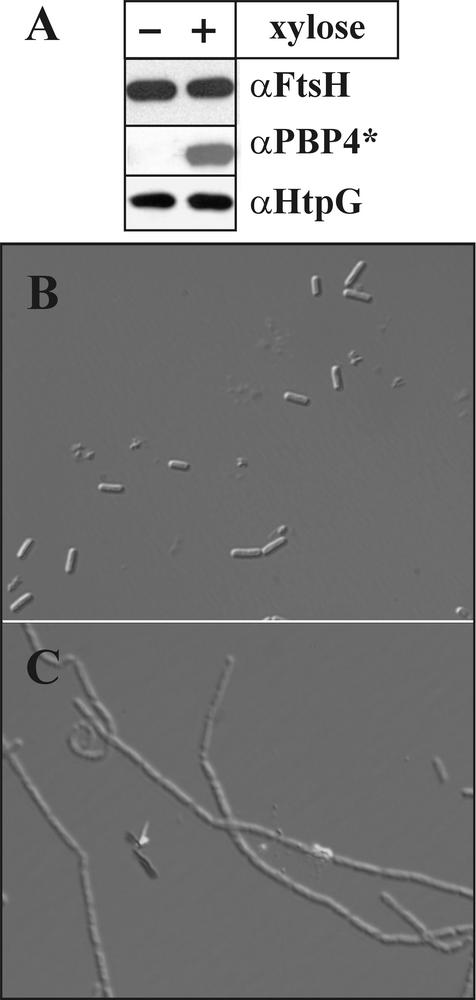
To elucidate whether the accumulation of PBP4* in the ΔftsH::cat strain is the result of enhanced stability or of increased synthesis, several experiments were carried out. First, the amounts of PBP4* in wild-type and ftsH knockout strains were determined by Western blot experiments. When cell extracts prepared from identical numbers of cells were probed with polyclonal antibodies raised against purified PBP4*, significantly greater amounts of the penicillin-binding protein were detected in the ftsH::cat and ftsH::erm strains (Fig. 2), corroborating the results obtained by 2-D gel electrophoresis. As expected, no PBP4* protein could be demonstrated in the pbpE knockout strain (Fig. 2). In in vitro experiments with purified FtsH protein incubated with purified His6-Pbp4*, no degradation of the penicillin-binding protein could be detected (data not shown). Therefore, to test whether Pbp4* accumulation was due to overexpression of pbpE at the transcriptional level, pbpE without its indigenous σW-controlled promoter was fused to the xylose-regulatable promoter PxylA (14) and ectopically integrated at the amyE locus (Fig. 3A). The pbpE gene at its chromosomal location was inactivated by insertion of an erythromycin resistance cassette. Two versions of that strain were constructed, one with ftsH (strain SZ06) and one without ftsH (strain SZ09). Both strains were grown to the mid-exponential-growth phase, and pbpE expression was induced by addition of 1% xylose. Extracts were prepared before and 90 min after addition of xylose and were probed with different antibodies. While FtsH is clearly absent from the ftsH::tet strain SZ09, comparable amounts of PBP4* are present in the presence and absence of the FtsH protease (Fig. 3B). As a control, we checked for HtpG, a heat-inducible protein (25); the amount of this protein was not significantly changed (Fig. 3B). These results suggested that PBP4* is not a substrate of FtsH.
FIG. 2.
Pbp4* accumulates in ftsH knockout strains. Immunoblot analysis detects increased amounts of PBP4* in ftsH null mutants. The different B. subtilis strains indicated were grown to the mid-exponential-growth phase in LB at 37°C. Proteins present in cell lysates were separated by SDS-PAGE, blotted, and developed with polyclonal antibodies against PBP4*, FtsH, or (as an internal control) HtpG.
FIG. 3.
Accumulation of Pbp4* in ftsH knockout strains is due to enhanced transcription of pbpE. (A) Schematic drawing showing fusion of the pbpE gene to the xylose-regulatable promoter PxylA. Comparable amounts of PBP4* are present in the presence and absence of ftsH. (B) Proteins from cells grown as described above were separated by SDS-PAGE and probed with three different antibodies as indicated. +, presence of xylose; −, absence of xylose. (C) Northern blot analysis reveals increased amounts of pbpE transcript in the ftsH knockout mutant. Total RNAs were prepared from B. subtilis 1012 and its ftsH::erm derivative at the OD578 indicated and were hybridized with digoxigenin-labeled pbpE antisense RNA.
If PBP4* does not represent a target protein for FtsH, the protease must influence its synthesis, most probably at the transcriptional level. This conjecture was tested by Northern blot analysis. Total RNAs were prepared at three different time points during the growth curves of the ftsH wild-type and ftsH::erm knockout strains and were probed with pbpE antisense RNA. While pbpE mRNA was not detectable in the presence of FtsH, significant amounts, increasing over time, were present in the ftsH null mutant (Fig. 3C). This result unequivocally demonstrates that FtsH influences the transcription of pbpE rather than the stability of PBP4*.
Overexpression of pbpE in an ΔftsH strain causes filamentous growth.
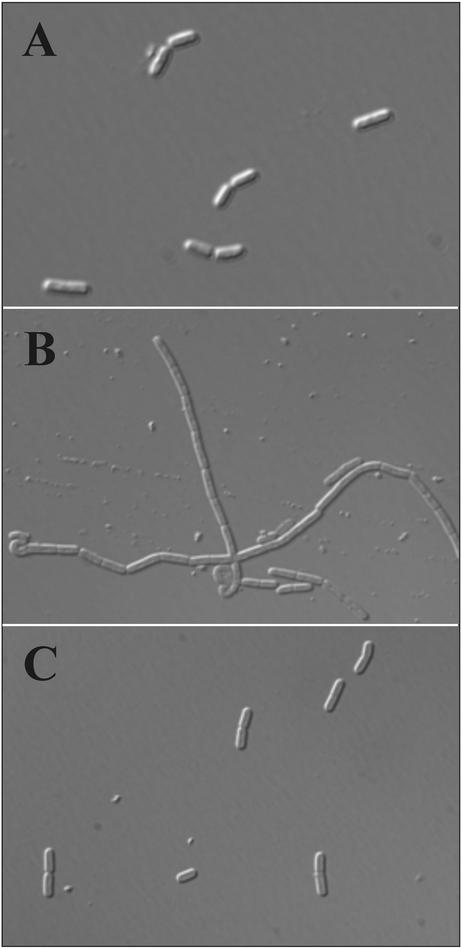
An imbalance in the synthesis of penicillin-binding proteins is reported to cause filamentous growth of bacteria (2). Therefore, we asked whether in B. subtilis the overproduction of PBP4* observed in the ftsH knockout might cause the filamentous-growth phenotype. First, we analyzed the phenotype of B. subtilis cells in the presence and absence of the wild-type ftsH and pbpE alleles. While wild-type cells were rod shaped (Fig. 4A), inactivation of ftsH resulted in filamentous growth (Fig. 4B). The absence of both ftsH and pbpE restored the wild-type cell morphology (Fig. 4C), indicating that enhanced expression of the pbpE gene is indeed responsible for the filamentous phenotype. If this conclusion is true, overexpression of pbpE in the presence of an intact ftsH allele should also result in filamentous growth. To test this idea, strain SZ06, allowing induction of pbpE by xylose (see Fig. 3A), was grown in the presence and absence of the inducer. In the absence of xylose, no Pbp4* was detectable in a Western blot and the cells exhibited a normal phenotype (Fig. 5A and B). Upon addition of xylose to the growth medium and Pbp4* overproduction (Fig. 5A), the cells grew largely in filaments (Fig. 5C). We conclude from these results that increased expression of pbpE due to either the absence of ftsH or artificial overproduction of PBP4* causes filamentous growth.
FIG. 4.
Accumulation of Pbp4* causes filamentous growth of B. subtilis cells. B. subtilis strains 1012 (wild type) (A), MP01 (ftsH::cat) (B), and SZ01 (ftsH::cat pbpE::erm) (C) were grown in LB medium to the mid-logarithmic-growth phase. Cells were analyzed by phase-contrast microscopy.
FIG. 5.
Overexpression of pbpE in the presence of the ftsH wild-type allele causes filamentous growth. (A) B. subtilis strain SZ06 (ftsH+ PxylA-pbpE) was grown either with or without addition of xylose to the medium to mid-logarithmic phase and was checked for the presence of FtsH, Pbp4*, and HtpG (internal control) by Western blotting. (B and C) Cells grown without (B) and with (C) xylose were then analyzed by phase-contrast microscopy.
The concentration of AbrB is not altered in a B. subtilis ftsH knockout.
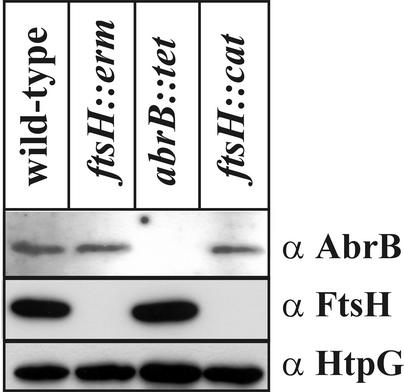
It has been reported that the pbpE gene is under the negative control of the transcriptional repressor AbrB (31). To analyze whether the influence of FtsH on pbpE transcription is indirect, due to an influence of FtsH on AbrB, cellular extracts of wild-type and isogenic ftsH and abrB knockout strains were tested for their amounts of AbrB protein by Western blotting. As can be seen in Fig. 6, the amounts of the repressor AbrB are identical in the absence and presence of FtsH. As an internal control, we also determined the amounts of the heat shock protein HtpG, which are identical in the extracts prepared from all four strains. We conclude from these results that FtsH does not influence the cellular level of AbrB and that pbpE induction in ftsH knockout strains is not due to altered amounts of AbrB.
FIG. 6.
Cellular AbrB levels are not altered in B. subtilis ftsH knockout strains. B. subtilis 1012 (wild type), WW01 (ftsH::erm), SWV119 (abrB::tet), and MP01 (ftsH::cat) were grown to mid-logarithmic phase and submitted to Western blot analysis as described above.
Most genes controlled by the extracytoplasmic function sigma factor σW are overexpressed in an ftsH null mutant.
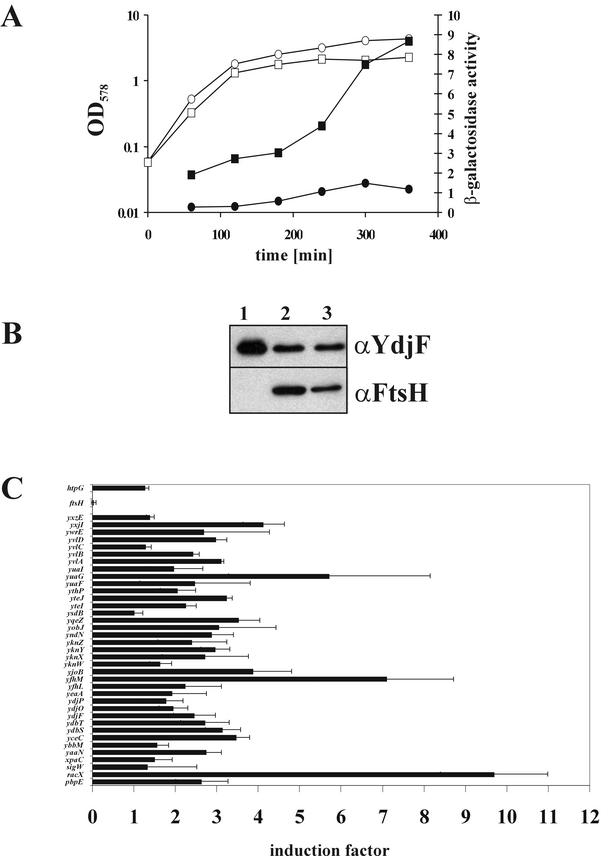
Since two of the four genes identified by the proteomic approach (pbpE and yvlB) are members of the σW regulon, we investigated whether additional genes of that regulon exhibit increased expression in the absence of ftsH. First, B. subtilis ftsH null and ftsH+ strains expressing a transcriptional fusion of the yuaF promoter to lacZ were tested for β-galactosidase activities at different growth stages. The yuaF promoter is one of the strongest σW-controlled promoters (37). It turned out that both the basal level of β-galactosidase activity and the level during stationary phase were enhanced in the ftsH null mutant over those in the isogenic wild-type strain (Fig. 7A). Second, using specific antibodies against the YdjF protein, we analyzed the expression of the σW-dependent gene ydjF in a strain where the promoter of ftsH has been replaced by the xylose-controllable promoter PxylA (Fig. 7B). In the absence of xylose in the growth medium, no FtsH protein could be visualized in cellular lysates and large amounts of YdjF protein were detected (Fig. 7B, lane 1). Upon xylose-induced ftsH expression or at wild-type levels of FtsH, the amount of YdjF protein was significantly reduced (Fig. 7B, lanes 2 and 3).
FIG. 7.
Genes controlled by σW are overexpressed in B. subtilis ftsH knockout strains. (A) Strains with a transcriptional fusion of the σW-controlled yuaF promoter to lacZ were grown at 37°C in LB medium. At different time points, samples were withdrawn and β-galactosidase activities were determined. Circles, B. subtilis TW51 (amyE::PyuaF-lacZ); squares, TW52 (amyE::PyuaF-lacZ ftsH::cat). Open symbols, OD578; solid symbols, β-galactosidase activity. (B) B. subtilis strain MSW01 (sigW+ pX2-ftsH) was grown in the absence (lane 1) or presence (lane 2) of xylose to mid-logarithmic phase and analyzed by Western blotting using antibodies against FtsH and YdjF. B. subtilis 1012 (wild type) grown without xylose (lane 3) was used as a control. (C) Schematic representation of DNA macroarray analysis of B. subtilis 1012 ftsH::cat compared to that of the wild type. DNA arrays of the entire B. subtilis genes were hybridized with cDNA probes generated from RNA extracted from cells of the two strains at an OD578 of 0.8. Mean results and standard deviations from two independent experiments are presented. Spot intensities were quantified as described previously (37), and the quotients of ftsH::cat/wild-type intensities for σW-controlled genes are given.
Both experiments suggest that in the absence of the FtsH metalloprotease, there is an upregulation of the whole σW regulon. Therefore, the global transcription pattern of a B. subtilis wild-type strain compared to that of the ftsH::erm knockout (WW01) at an optical density at 578 nm (OD578) of 0.8 was analyzed by the DNA macroarray technique. Indeed, it turned out that most genes of the σW regulon exhibited 1.5- to 3-fold-enhanced expression in the ftsH null mutant, while some were expressed up to 9-fold in the absence of the FtsH protein (Fig. 7C). It should be noted that σW-controlled genes such as xpaC, yvlC, and yxzE, which are obviously not induced in ΔftsH mutants, also respond poorly to the strong inducing conditions of an alkaline shock when assayed in DNA microarrays (37). The σW-controlled genes were reproducibly induced, as was also seen when the global transcriptional pattern was analyzed at ODs of 0.4 and 1.2 (data not shown). Probably because of the various phenotypic defects of ΔftsH strains, the expression pattern of various other genes altered to different extents, and without further experiments it is not possible to present a conclusive overall transcriptional pattern of ΔftsH strains.
To summarize, these experiments show that the FtsH protease exerts an influence on the expression of the σW regulon which is either direct or indirect.
Cells carrying a sigW ftsH double knockout display a growth defect.
It is possible that the absence of FtsH activity generates a certain stress for B. subtilis cells, which in turn induces the σW regulon, helping the cells to overcome the stressful situation. Therefore, we tested whether induction of the σW regulon is of crucial importance for the survival of ftsH knockout strains. Cells carrying single knockouts in sigW or ftsH are viable, though the latter exhibit a pleiotropic phenotype as already mentioned. However, when the ftsH knockout was recombined into a sigW null mutant strain, cell colonies remained small and reached a final OD of only about 0.5 in liquid culture. This suggests that the combination of both null alleles severely affects viability.
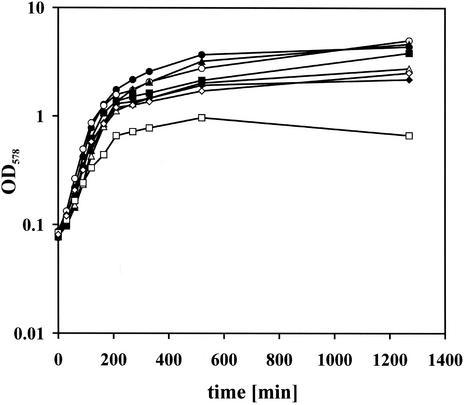
To analyze this in more detail, we crossed the sigW null allele into strain MSW01, where expression of ftsH is xylose dependent. The resulting strain SZ08 was grown overnight in the presence of 1.5% xylose, and cells were washed twice with LB to remove the xylose and were then used to inoculate LB medium without xylose. To fully deplete FtsH, these cells were grown to early-stationary phase and used to inoculate cultures with and without xylose to an OD578 of 0.08. As controls, strain MSW01 with the xylose-regulatable ftsH and wild-type sigW background, the sigW::neo strain, and the isogenic wild-type B. subtilis strain were treated in the same manner. The B. subtilis wild-type and sigW::neo strains grew independently of the addition of xylose (Fig. 8). Strain MSW01 reached about half of the final OD when xylose was omitted, showing that FtsH depletion is harmful to the cells. The growth of strain SZ08 was impaired more severely upon depletion of FtsH, as the strain reached a final OD of about 0.8 (Fig. 8), showing that induction of the σW regulon in the absence of FtsH is crucial for survival of the cells under standard laboratory conditions.
FIG. 8.
Cells of a B. subtilis sigW knockout display a growth defect upon depletion of FtsH protein. Growth of B. subtilis strains 1012 (wild type) (circles), MSW01 (sigW+ pX2-ftsH) (triangles), 1012 sigW::neo (sigW knockout; ftsH+) (diamonds), and SZ08 (sigW knockout; pX2-ftsH) (squares) was monitored in the presence (solid symbols) and absence (open symbols) of xylose added to the growth medium. Cultures were inoculated from early-stationary-phase cultures without xylose to an OD578 of 0.08.
DISCUSSION
The ATP- and Zn2+-dependent, membrane-anchored metalloprotease FtsH is not essential in B. subtilis, though knockouts display a pleiotropic phenotype including a complete inability to sporulate, severe growth defects under heat and salt stresses, and largely filamentous growth (9). Here, we report a new phenotype, which is upregulation of most genes of the σW regulon in ftsH null mutants.
The present study was originally aimed at identifying substrate proteins recognized by the FtsH protease. So far, no such proteins have been unambiguously identified in B. subtilis, though it has been reported that the B. subtilis SpoVM protein is degraded by the E. coli FtsH protease and strongly inhibits proteolysis of σ32 in vitro (7). This result is reminiscent of those obtained with the λ CIII protein, which also inhibits the proteolytic activity of FtsH and is later degraded (29). It may be assumed that the 26-amino-acid SpoVM peptide will also be degraded by B. subtilis FtsH, which has to be proven experimentally.
Our screen was based on the assumption that FtsH target proteins synthesized under physiological conditions may be present at elevated levels in the absence of the protease. To investigate this possibility, the proteomes of a wild-type and an ftsH knockout strain were compared. It turned out that at least nine proteins were present in increased amounts and that four of them could be identified by MALDI-TOF mass spectrometry. These proteins accumulate in the ftsH null mutant either because they are true substrate proteins or, alternatively, because ftsH is involved in the regulation of their synthesis. Two of these proteins (PBP4* and YvlB) are members of the σW regulon (12), and we have chosen PBP4* for further analyses to find out why this protein accumulates in the absence of the protease. PBP4* is similar to E. coli PBP4, which has been demonstrated to cleave peptidoglycan peptide cross-links (15). However, B. subtilis PBP4* does not have a classical signal peptide and is found mainly in the cytoplasmic fraction.
It has already been reported that ftsH influences the level of penicillin-binding proteins. While the amount of PBP2A was increased in an ftsH mutant, the amounts of PBP2B and PBP4 were decreased (17). In general, penicillin-binding proteins are involved in peptidoglycan synthesis and cell shape determination during cell division, sporulation, and germination (1). After identifying PBP4* as a protein overproduced in the absence of ftsH, we asked whether the increased amount of PBP4* might be responsible for filamentous growth. While cells of a pbpE ftsH double-knockout strain indeed failed to form filaments, overexpression of pbpE in an ftsH wild-type strain led to the same phenotype. Therefore, elevated amounts of PBP4* are the causative agent inducing filamentous growth. How this phenotype is induced by PBP4* is completely unknown, mainly because the function of this protein remains elusive. On the other hand, depletion of PBP2B results in filamentation, too (8).
Western blot experiments using two different ftsH knockout strains confirmed the results obtained by the 2-D PAGE analysis, namely, enhanced levels of PBP4*. To distinguish between degradation of PBP4* and increased overproduction due to the involvement of ftsH in the regulation of expression of pbpE on the transcriptional level, the pbpE gene was fused to a controllable promoter. Upon induction of the ectopically induced pbpE, comparable amounts of the penicillin-binding protein were detectable in the presence and absence of ftsH. This result strongly argues against PBP4* being a target protein of FtsH. This assumption is confirmed by the finding that purified FtsH was unable to degrade PBP4* in an in vitro assay (data not shown).
Since FtsH is not involved in the degradation of PBP4*, it must influence its expression. This hypothesis could be confirmed by Northern blot analysis. While significant amounts of the pbpE transcript could be seen with the ftsH null mutant, this transcript was not detectable in the wild-type strain. Therefore, the absence of FtsH induces transcription of pbpE. Since, in addition to PBP4*, YvlB, another product of the σW regulon, is also present in increased amounts, we asked whether additional σW-dependent genes exhibit enhanced expression in an ftsH null mutant; this was shown for ydjF and yuaF. Global analysis of all protein-coding genes of the B. subtilis genome by the DNA macroarray technique revealed that most genes of the regulon are indeed transcribed at elevated levels. Taken together, these findings show that ftsH somehow influences expression of the σW regulon.
Negative regulation of the pbpE gene by the global repressor protein AbrB has been described previously (31). In addition, an interaction between the σW regulon and AbrB has been revealed recently. It was shown that AbrB binds not only to the promoter region of pbpE but also to other genes of the σW regulon, including the sigW operon itself (22). Since recent work also suggested an influence of FtsH on abrB transcription (9), we tested for the level of AbrB protein in ftsH knockout strains. In the case of an influence of FtsH on AbrB and therefore on σW-controlled genes, reduced levels of AbrB in ftsH knockout strains would be expected. By use of specific antibodies against AbrB, it could be shown that its levels are comparable in the presence and absence of the FtsH protease, excluding the possibility that altered levels of AbrB cause overexpression of σW-controlled genes in ftsH knockout strains.
What could be the role of ftsH in expression of the σW regulon? σW is one of seven extracytoplasmic function sigma factors of B. subtilis, and work carried out over recent years has identified a minimum of 30 promoters (controlling about 60 genes) recognized by σW (5, 12). The mechanism that induces the σW regulon is unknown, but it has been shown that the σW regulon is activated early in the stationary phase (13), is strongly induced by a sudden imposition of alkali stress (37), and, most interestingly, is induced by antibiotics such as vancomycin which inhibit peptidoglycan biosynthesis (5). Most probably, the signal that activates σW is sensed and transduced by a membrane-bound anti-sigma factor (RsiW) encoded by the ybbM gene, which lies downstream of sigW, and this signal is linked to cell wall damage or stress induced by antimicrobial compounds.
The role of FtsH in the expression of the σW regulon might be direct or indirect. First, E. coli FtsH is known to degrade the heat shock sigma factor σ32 and is therefore involved in modulating the heat stress response (3, 33). Similarly, it is possible that FtsH degrades σW when it is present in an uncomplexed form in the cytoplasm and/or when it is sequestered to the membrane via the RsiW anti-sigma factor. Second, the influence of FtsH might be indirect and comparable to that of the E. coli Cpx extracytoplasmic stress response system, which has been described most recently to be induced in the absence of FtsH (28); in this case, the absence of FtsH would result in the accumulation of compounds that in turn would induce the σW regulon. The mechanism of resolution of this stress is elusive, because the functions of most of the σW-controlled genes are unknown. The importance of an induction of σW-controlled genes in ftsH knockout is underlined by the fact that a sigW ftsH double-knockout strain displays severe growth defects. The compounds accumulated might be either uncomplexed membrane proteins or penicillin-binding proteins that alter cell wall integrity. A screen to isolate mutants with increased σW activity had resulted in three different types of mutants, related to (i) genes of transport, (ii) sugar metabolism, and (iii) antibiotic biosynthesis (34). It was speculated that upregulation of sigma factor activity was due to the inability of the cell to export toxic compounds. An influence of B. subtilis FtsH on export has never been shown in detail, but it may well be that certain transport processes are impaired because of the accumulation of nonnative proteins in the membrane. The 2-D gel electrophoresis technique used in this work is not able to resolve membrane proteins. Therefore, most of the proteins belonging to the σW regulon did not appear, as most of them are membrane bound (12). For the same reason, other true FtsH substrate proteins that might accumulate in the membrane could not be identified.
Acknowledgments
We thank Peter Setlow and J. D. Helmann for strains, Mark Strauch and Akram Atalla for polyclonal antibodies raised against AbrB and YdjF, respectively, and Karin Angermann for excellent technical assistance.
This work was financially supported by the European Union (project QLRT-1999-01455) and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 2.Begg, K. J., T. Tomoyasu, W. D. Donachie, M. Khattar, H. Niki, K. Yamanaka, S. Hiraga, and T. Ogura. 1992. Escherichia coli mutant Y16 is a double mutant carrying thermosensitive ftsH and ftsI mutations. J. Bacteriol. 174:2416-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaszczak, A., C. Georgopoulos, and K. Liberek. 1999. On the mechanism of FtsH-dependent degradation of the σ32 transcriptional regulator of Escherichia coli and the role of the DnaK chaperone machine. Mol. Microbiol. 1:157-166. [DOI] [PubMed] [Google Scholar]
- 4.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 5.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. W. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 6.Confalonieri, F., and M. Duguet. 1995. A 200-amino acid ATPase module in search of a basic function. Bioessays 17:639-650. [DOI] [PubMed] [Google Scholar]
- 7.Cutting, S., M. Anderson, E. Lysenko, A. Page, T. Tomoyasu, K. Tatematsu, T. Tatsuta, L. Kroos, and T. Ogura. 1997. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J. Bacteriol. 179:5534-5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, R. A., E. J. Harry, and J. Errington. 2000. Role of penicillin-binding protein PBP2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 35:299-311. [DOI] [PubMed] [Google Scholar]
- 9.Deuerling, E., A. Mogk, C. Richter, M. Purucker, and W. Schumann. 1997. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol. Microbiol. 23:921-933. [DOI] [PubMed] [Google Scholar]
- 10.Gardan, R., G. Rapoport, and M. Debarbouille. 1995. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J. Mol. Biol. 249:843-856. [DOI] [PubMed] [Google Scholar]
- 11.Homuth, G., M. Heinemann, U. Zuber, and W. Schumann. 1996. The genes lepA and hemN form a bicistronic operon in Bacillus subtilis. Microbiology 142:1641-1649. [DOI] [PubMed] [Google Scholar]
- 12.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function σ factor, σW. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 13.Huang, X. J., K. L. Fredrick, and J. D. Helmann. 1998. Promoter recognition by Bacillus subtilis σW: autoregulation and partial overlap with the σX regulon. J. Bacteriol. 180:3765-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, L., A. Mogk, and W. Schumann. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71-76. [DOI] [PubMed] [Google Scholar]
- 15.Korat, B., H. Mottl, and W. Keck. 1991. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol. Microbiol. 5:675-684. [DOI] [PubMed] [Google Scholar]
- 16.Langer, T. 2000. AAA proteases: cellular machines for degrading membrane proteins. Trends Biochem. Sci. 25:247-251. [DOI] [PubMed] [Google Scholar]
- 17.Lysenko, E., T. Ogura, and S. M. Cutting. 1997. Characterization of the ftsH gene of Bacillus subtilis. Microbiology 143:971-978. [DOI] [PubMed] [Google Scholar]
- 18.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6:575-597. [DOI] [PubMed] [Google Scholar]
- 20.Patel, S., and M. Latterich. 1998. The AAA-team: related ATPases making it and breaking it in the cell. Trends Cell Biol. 8:65-71. [PubMed] [Google Scholar]
- 21.Popham, D. L., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpE operon, which codes for penicillin-binding protein 4* and an apparent amino acid racemase. J. Bacteriol. 175:2917-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian, Q., C. L. Lee, J. D. Helmann, and M. A. Strauch. 2002. AbrB is a regulator of the σW regulon in Bacillus subtilis. FEMS Microbiol. Lett. 211:219-223. [DOI] [PubMed] [Google Scholar]
- 23.Saito, H., T. Shibata, and T. Ando. 1979. Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol. Gen. Genet. 170:117-122. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schulz, A., S. Schwab, S. Versteeg, and W. Schumann. 1997. The htpG gene of Bacillus subtilis belongs to class III heat shock genes and is under negative control. J. Bacteriol. 179:3103-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumann, W. 1999. FtsH—a single-chain charonin? FEMS Microbiol. Rev. 23:1-11. [DOI] [PubMed] [Google Scholar]
- 27.Schumann, W., S. D. Ehrlich, and N. Ogasawara. 2001. Functional analysis of bacterial genes. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 28.Shimohata, N., S. Chiba, N. Saikawa, K. Ito, and Y. Akiyama. 2002. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells 7:653-662. [DOI] [PubMed] [Google Scholar]
- 29.Shotland, Y., S. Koby, D. Teff, N. Mansur, D. A. Oren, K. Tatematsu, T. Tomoyasu, M. Kessel, B. Bukau, T. Ogura, and A. B. Oppenheim. 1997. Proteolysis of the phage lambda CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol. Microbiol. 24:1303-1310. [DOI] [PubMed] [Google Scholar]
- 30.Strauch, M. A. 1995. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J. Bacteriol. 177:6727-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strauch, M. A. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J. Bacteriol. 177:6999-7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stüber, D., H. Matile, and G. Garotta. 1990. System for high level production in E. coli and rapid purification of recombinant proteins: application to epitope mapping, preparation of antibodies and structure-function analysis, p. 121-152. In I. Lefkovits and B. Pernis (ed.), Immunological methods. Academic Press, Orlando, Fla.
- 33.Tomoyasu, T., J. Gamer, B. Bukau, M. Kanemori, H. Mori, A. J. Rutman, A. B. Oppenheim, T. Yura, K. Yamanaka, H. Niki, S. Hiraga, and T. Ogura. 1995. Escherichia coli FtsH is a membrane-bound, ATP-dependent protease which degrades the heat-shock transcription factor σ32. EMBO J. 14:2551-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner, M. S., and J. D. Helmann. 2000. Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the σX and σW factors in Bacillus subtilis. J. Bacteriol. 182:5202-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vale, R. D. 2000. AAA proteins: lords of the ring. J. Cell Biol. 150:F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wehrl, W., M. Niederweis, and W. Schumann. 2000. The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation. J. Bacteriol. 182:3870-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]