Abstract
In Neisseria gonorrhoeae and Neisseria meningitidis, we identified a gene that would encode a protein highly similar to NorM of Vibrio parahaemolyticus (Y. Morita et al., Antimicrob. Agents Chemother. 42:1778-1782, 1998). A nonpolar insertional mutation in either the gonococcal or meningococcal norM gene resulted in increased bacterial sensitivity to compounds harboring a quaternary ammonium on an aromatic ring (e.g., ethidium bromide, acriflavine hydrochloride, 2-N-methylellipticinium, and berberine). The presence of point mutations within the −35 region of a putative norM promoter or a likely ribosome binding site resulted in an increased resistance of gonococci and meningococci to the same compounds, as well as to norfloxacin and ciprofloxacin. Structure-activity relationship studies with putative NorM substrates have found that a cationic moiety is essential for NorM recognition.
Neisseria gonorrhoeae is a strict human pathogen that causes the sexually transmitted disease gonorrhea. Therefore, it has likely adapted strategies to survive host antimicrobial systems that exist on the mucosal surfaces that it infects. Recently, it was proposed that the utilization of efflux pumps was a mechanism by which gonococci could resist the antimicrobial action of several host-derived compounds such as antibacterial peptides (27) and long-chain fatty acids (15, 28). Bacterial efflux pumps also contribute to the development of resistance to clinically useful antibiotics (16).
Efflux systems are prevalent in most or all cells and can be classified into four families: the major facilitator (MF) family, the small multidrug resistance family, the resistance-nodulation-cell division (RND) family, and the ATP-binding cassette (ABC) family. The MF, small multidrug resistance, and RND families of transporters are typically energized by the proton motive force, and the ABC superfamily comprises ATP-dependent transporters (8, 23). Recently, three new multidrug efflux proteins have been identified: NorM from Vibrio parahaemolyticus; a NorM homologue in Escherichia coli, YdhE (20); and VmrA from V. parahaemolyticus (5). The NorM and YdhE proteins mediate resistance to a range of cationic dyes, aminoglycosides, and fluoroquinolones (20). The vmrA gene, cloned from V. parahaemolyticus, made E. coli resistant to 4′,6′-diamidino-2-phenylindole, tetraphenylphosphonium chloride (TPP), and ethidium bromide (Eb) when overexpressed (5). The NorM, YdhE, and VmrA proteins are Na+-drug antiporters (5, 21) and, by sequence analysis, are members of the MATE (multidrug and toxic compound extrusion) family, which contains more than 30 proteins present in all three kingdoms (4). Hydropathy analyses revealed that these proteins characteristically possess 12 putative transmembrane domains (TMs). Multiple sequence alignments indicated that their most highly conserved regions are located in the vicinity of TMs 5 and 6 and near the C terminus of TM 8 (4).
For N. gonorrhoeae, two efflux pumps have been identified and studied in detail. The mtr (multiple transferable resistance) system was originally described in 1973 (17). It belongs to the RND family of efflux pumps and exports hydrophobic agents including antibiotics, nonionic detergents, certain antibacterial peptides, bile salts, and gonadal steroidal hormones (7, 10, 11, 27). A second efflux pump has been recently described for gonococci: the FarA-FarB system belongs to the MF family and recognizes antibacterial long-chain fatty acids (15). Using the genome sequence information available online (www.genome.ou.edu) (22, 30), we identified an additional transporter possessed by N. gonorrhoeae and Neisseria meningitidis, which we called NorM because of its homology to NorM of V. parahaemolyticus. Genetic analysis indicated that NorM confers increased resistance to Eb, acriflavine hydrochloride (AFh), 2-N-methylellipticinium (NME), and berberine (BE) but not to other compounds recognized by the MtrC-MtrD-MtrE and FarA-FarB efflux pumps. Interestingly, a point mutation upstream of the norM gene, which results in its overexpression, provides decreased gonococcal susceptibility to the fluoroquinolones norfloxacin (NOR), ciprofloxacin (CIP), and benzalkonium chloride (BC).
Identification of norM in gonococci and meningococci.
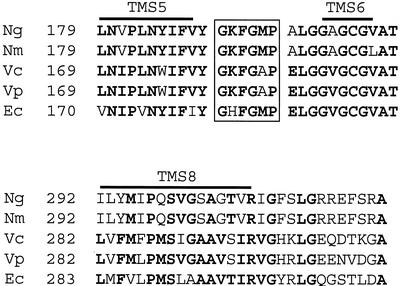
The FA1090 gonococcal genome sequence (www.genome.ou.edu) was screened for genes encoding putative efflux pumps. We identified an N. gonorrhoeae homologue of the norM gene of V. parahaemolyticus (20). This open reading frame was located between nucleotides 386944 and 388323 in the gonococcal genome sequence (www.genome.ou.edu). We amplified and sequenced this open reading frame from gonococcal strain FA19 with primers designed from the FA1090 genome sequence (strains used in this study are shown in Table 1). The predicted proteins from strains FA1090 and FA19 were 100% identical (data not presented). The putative gonococcal protein was 86 and 87% identical to the equivalent proteins encoded by meningococcal strains MC58 and Z2491, respectively (data not presented). The proteins belonging to the MATE family are divided into three distinctive clusters: cluster 1 includes bacterial efflux transporters such as NorM from Vibrio cholerae, cluster 2 includes proteins from fungi and plants, and cluster 3 includes proteins from Eubacteria-Archaea (4) and, recently published (5), the VmrA protein, a Na+-coupled multidrug efflux pump from V. parahaemolyticus. As observed by Miyamae et al. (19), all members of cluster 1 show remarkable conservation of the sequence GKFGXP (Fig. 1), which was not conserved in proteins from the other two clusters. The neisserial NorM homologues possessed the GKFGXP sequence, confirming that they belong to the NorM cluster of MATE proteins (Fig. 1).
TABLE 1.
Strains used
| Strain | Description | Reference or source |
|---|---|---|
| N. gonorrhoeae | ||
| FA19 | Wild type | P. F. Sparling |
| FA1090 | Wild type | J. G. Cannon |
| RD1 | FA19 mtrE::Km | 7 |
| GC525 | FA1090 mtrE::Km | This study |
| GC805 | GC525 −35/RBS | This study |
| GC806 | GC525 −35 | This study |
| GC663 | GC525::gyrA (S91F, D95G); parC (S88P, E91K) | This study |
| GC807 | GC663 −35/RBS | This study |
| GC808 | GC663 −35 | This study |
| BR54 | FA19 penA1 penB2 mtr-140 mtrD54 | 25 |
| 22-G | FA1090 −35 | This study |
| 1/8 B | FA1090 −35/RBS | This study |
| CR22 | FA19 −35 | This study |
| CR23 | FA19 −35/RBS | This study |
| CR24 | BR54 −35 | This study |
| CR25 | BR54 −35/RBS | This study |
| CR28 | FA19 norM::Km | This study |
| CR29 | BR54 norM::Km | This study |
| N. meningitidis | ||
| NMB | Wild type | 29 |
| M7 | NMB synX::Tn916 | 29 |
| CR26 | M7 −35 | This study |
| CR27 | M7 −35/RBS | This study |
| CR30 | NMB norM::Km | This study |
| CR31 | M7 norM::Km | This study |
FIG. 1.
Multiple sequence alignment of the most conserved regions of NorM with representative homologues. Ng, N. gonorrhoeae FA19; Nm, N. meningitidis Z2491; Vc, V. cholerae N16961; Vp, V. parahaemolyticus; Ec, E. coli. Amino acid numbering is shown at the beginning of the sequence presented. Identical residues (present in more than 50% of the five sequences) are presented in boldface. TMs are overlined. The GKFGXP sequences, characteristic of the MATE proteins from the NorM cluster, are boxed.
The neisserial NorM efflux pumps recognize antimicrobial cationic dyes.
In order to determine the function of the gonococcal and meningococcal NorM-like proteins, we used primers N6 and N7 (Table 2) to amplify the norM locus from gonococcal wild-type strain FA19. The corresponding PCR product was then cloned into pBAD-TOPO as described by the manufacturer (Invitrogen, Carlsbad, Calif.). A nonpolar, promoterless kanamycin resistance (Km) cassette (18) was then inserted into the NaeI restriction site, 443 bp after the start codon of norM. The resulting construct was transformed into gonococcal strains FA19 and BR54 as well as meningococcal strain NMB and its isogenic capsule-deficient mutant M7 (29). The gonococcal transformants were selected on gonococcal base (GCB) agar plates supplemented with 50 μg of kanamycin/ml while the meningococcal transformants were selected on brain heart infusion agar supplemented with fetal calf serum at 2.4% (vol/vol) and 80 μg of kanamycin/ml. The insertion of the Km cassette within norM was verified by PCR (data not presented). Reverse transcription-PCR (RT-PCR) studies confirmed that this nonpolar insertional mutation did not alter the transcription of the murB gene located downstream of norM (data not presented).
TABLE 2.
Oligonucleotides used
| Oligonucleotide | Sequence (5′ to 3′) | Usea |
|---|---|---|
| N3 | CCCACATCAAAATCATGCCG | cDNA synthesis and RT-PCR (349-bp 3′ norM ATG) |
| N4 | ATGCTGCTCGACCTCGACC | RT-PCR (ATG overlap) |
| N6 | TCGGTATCGGATGGGTTGC | pBADΩnorM construction |
| N7 | TTTGCCGCAACGCATCACG | pBADΩnorM construction; RT-PCR (269-bp 5′ norM ATG) |
| N8 | CATTGTCGCCACGCCGCAAC | 22-G norM promoter mapping product |
| N9 | CTTGACCTGCGCTTCGACCGAC | 22-G norM promoter mapping product |
| N11 | CGGTCAGCAGGCGGATTTCTTTCAGG | norM promoter mapping |
| N20 | GCGATTATGTGGAAGGCACA | qRT-PCR for norM |
| N21 | ACCAACATAATCAGGCGCG | qRT-PCR for norM |
| N12 | AATTTCTGCTGTCGGCTTGG | qRT-PCR for rmp |
| N13 | ACATGCAATCAGAGCCTCACG | qRT-PCR for rmp |
| G1 | GTCCGCCATGGCAGGTTTCTCGACAAAC | 5′ gyrA outside primer |
| G2 | CATACGGACGATGGTGCCGTAAACTGCGAAATCGCCGTGGGGGTG | 3′ SOE primer resulting in gyrA (Ser91Phe and Asp95Gly) mutations (mutated codons underlined) |
| G3 | CACCCCCACGGCGATTTCGCAGTTTACGGCACCATCGTCCGTATG | 5′ SOE primer resulting in gyrA (Ser91Phe and Asp95Gly) mutations (mutated codons underlined) |
| G4 | CAACTTGAATTCGTTGACCTGATAGGG | 3′ gyrA outside primer |
| P1 | CGTGGCGGATAAATACCAATTC | 5′ parC outside primer |
| P2 | CATGCGCACCATCGCTTTATAGGCGGGACTGTCGCCGTGCGG | 3′ SOE primer resulting in parC (Ser88Pro and Glu91Lys) mutations (mutated codons underlined) |
| P3 | CCGCACGGCGACAGTCCCGCCTATAAAGCGATGGTGCGCATG | 5′ SOE primer resulting in parC (Ser88Pro and Glu91Lys) mutations (mutated codons underlined) |
| P4 | GGGCCTCCAGCGTCGGTTTCTTCAACAG | 3′ parC outside primer |
SOE, splicing by overlap extension.
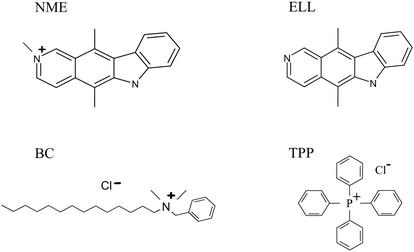
We then determined the susceptibilities of strain FA19 and its insertional mutant (CR28) to compounds that are substrates for the MtrC-MtrD-MtrE efflux pump (AFh, Eb, Triton X-100 [TX-100], and antibacterial peptide LL-37), for the FarA-FarB efflux system (palmitic acid [PA]), or for the NorM efflux pump of V. parahaemolyticus (CIP, NOR, BE, and streptomycin [STR]). Also tested were ellipticine (ELL) and a methylated derivative containing a quaternary ammonium (NME) (Pfizer) (Fig. 2). Strains FA19 and CR28 (FA19 norM::Km) did not differ in their susceptibilities to TX-100, LL-37, PA, CIP, NOR, ELL, and STR. In contrast, strain CR28 was four- to sixfold more sensitive than parental strain FA19 to AFh, BE, NME, and Eb (Table 3). To eliminate activity of the MtrC-MtrD-MtrE efflux system, we introduced the norM insertional mutation into strain BR54 (25). BR54 is a derivative of strain FA140 (like FA19 but penA1 penB2 mtrR140 mtrD54) that has a 10-bp deletion in its mtrD gene that results in a nonfunctional MtrC-MtrD-MtrE efflux pump (31). The insertional mutant of strain BR54 (strain CR29) displayed increased susceptibilities to AFh, BE, and Eb (Table 3). In addition, one quaternary ammonium compound (BC) and two anionic dyes (orange II and eosin [EO]), as well as TPP, Tween 80, and rhodamine B, were tested against these strains. None of these compounds appeared to be substrates of the NorM efflux pump in gonococci (data not presented). Similar to the results obtained with gonococci, the norM::Km mutants of meningococcal strains NMB (CR30) and M7 (CR31) were more susceptible to AFh, BE, NME, and Eb than were their respective parental strains (Table 3). Testing a wide variety of compounds allowed us to determine that a common characteristic of NorM substrates is the presence of a quaternary ammonium such as those in Eb, Afh, BE, and NME. This requirement is most notable when one compares the susceptibility profiles of gonococci against NME and ELL. The inability of TPP to be exported by NorM demonstrates that a positive charge per se is not sufficient to define efflux substrates (Fig. 2).
FIG. 2.
Structural attributes of NME compared to those of ELL, BC, and TPP.
TABLE 3.
Susceptibilities of gonococcal and meningococcal strains to diverse antimicrobial agents
| Strain | MIC (μg/ml)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFh | Eb | BE | TX-100 | LL-37 | PA | CIP | NOR | STR | NME | ELL | TPP | BC | |
| Gonococci | |||||||||||||
| FA19 | 0.5 | 2 | 2.5 | 150 | 6.25 | 50 | 0.0025 | 0.00002 | 40 | 1 | 2 | 50 | 1.75 |
| CR28 | 0.125 | 0.5 | 1.25 | 150 | 6.25 | 50 | 0.0025 | 0.00002 | 40 | ≤0.25 | 2 | 50 | 1.75 |
| BR54 | 0.25 | 1 | 1.25 | 25 | 0.78 | 50 | 0.0025 | 0.00002 | >160b | ≤0.25 | 0.5 | 12.5 | NDa |
| CR29 | 0.0625 | 0.125 | 0.625 | ND | 0.78 | 50 | 0.0025 | 0.00002 | ND | ND | ND | 12.5 | ND |
| Meningococci | |||||||||||||
| NMB | 0.5 | 12 | 80 | 50 | ND | ND | 0.01 | 0.00009 | ND | 24 | 2 | 200 | ND |
| CR30 | 0.25 | 6 | 40 | 50 | ND | ND | 0.01 | 0.00009 | ND | 8 | 2 | 200 | ND |
| M7 | 0.5 | 12 | 80 | 50 | ND | ND | 0.01 | 0.00009 | ND | 24 | 2 | 200 | ND |
| CR31 | 0.25 | 6 | 40 | 50 | ND | ND | 0.01 | 0.00009 | ND | 16 | 2 | 200 | ND |
ND, not determined.
BR54 is resistant to STR due to the str-7 allele (26).
Selection of mutants resistant to NME.
To identify chromosomal mutations that result in resistance to NorM substrates, an NME-resistant mutant was isolated at 2× the MIC from an ethyl methanesulfonate-mutagenized pool of strain GC525. GC525 was constructed by transforming FA1090 with genomic DNA from strain RD1, which is isogenic to strain FA19 but contains a Km cassette inserted into mtrE (7). To determine if the mutation responsible for resistance was located in norM, a 1.5-kb PCR product was generated with primers N8 and N9 (Table 2). The ability of this PCR product to confer resistance in the parent strain demonstrated that the mutation responsible for resistance was in or near norM. DNA sequence analysis of this product and the NME-resistant backcross mutant, 22-G, identified a single C-to-T mutation in the putative −35 promoter element, resulting in a CTGACG-to-TTGACG change (data not presented). To identify other mutations in this region that may result in resistance, the mutant template was subjected to PCR-mediated mutagenesis (14) with primers N8 and N9. This mutant pool of PCR products was then used as donor DNA for transformation experiments that used strain 22-G as the recipient. The results of these experiments identified a strain, 1/8 B, that was twofold less sensitive to NME than was strain 22-G, suggesting that an additional mutation had been introduced that resulted in a further increase in NorM-mediated resistance to this antimicrobial agent. DNA sequence analysis of this strain identified an A-to-G mutation 7 bp upstream of the ATG codon resulting in a TGAA-to-TGGA alteration of the putative ribosome binding site (RBS) in addition to the previously noted −35 mutation (data not presented).
The −35 and −35/RBS mutations were transformed into strains FA19, BR54, GC525, and M7, and transformants were selected on GCB agar supplemented with 4, 2, 1, and 24 μg of Eb/ml, respectively. The levels of susceptibility to diverse compounds of the resulting transformants were then tested: Eb, NME, and AFh, which originally gave evidence of being exported by the neisserial NorM; NOR, CIP, and STR, which were not exported by the neisserial NorM but were exported by NorM from V. parahaemolyticus; TX-100, which is exported by the MtrCDE efflux pump but not by the neisserial NorM protein; and BC, TPP, and EO. As expected, the −35 mutation conferred intermediate resistance to Eb and AFh but not to STR and TX-100 in the three neisserial backgrounds (gonococcal strains FA19 and BR54 and meningococcal strain M7). The −35/RBS mutations conferred a higher level of resistance to Eb and AFh but not to STR and TX-100 than did the −35 mutation alone (Table 4). Surprisingly, the −35 and −35/RBS mutations also conferred decreased susceptibilities to NOR, CIP, and BC compared to those conferred by FA19.
TABLE 4.
Effects of −35 and −35/RBS mutations on susceptibilities of gonococcal and meningococcal strains to diverse antimicrobial agents
| Strain | MIC (μg/ml)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eb | AFh | NOR | CIP | NME | ELL | STR | TX-100 | BC | TPP | EO | |
| Gonococci | |||||||||||
| FA19 | 2 | 0.5 | 0.00002 | 0.0025 | 1 | 2 | 40 | 150 | 1.75 | 50 | 20 |
| CR22 | 16 | 2 | 0.00016 | 0.005 | >32 | 2 | 40 | 150 | 7 | 50 | 20 |
| CR23 | >32 | 4 | 0.00032 | 0.01 | >32 | 2 | 40 | 150 | 14 | 50 | 20 |
| BR54 | 1 | 0.25 | 0.00002 | 0.0025 | ≤0.25 | 0.5 | >160 | 25 | NDa | ND | ND |
| CR24 | 8 | 1 | 0.00004 | 0.0025 | 16 | 1 | >160 | 25 | ND | ND | ND |
| CR25 | 12 | 2 | 0.00016 | 0.01 | 32 | 1 | >160 | 25 | ND | ND | ND |
| Meningococci | |||||||||||
| M7 | 12 | 0.5 | 0.00009 | 0.01 | 24 | 2 | 80 | 50-100 | ND | ND | ND |
| CR26 | >32 | 4 | 0.00016 | 0.01 | >32 | 2 | 80 | 50-100 | ND | ND | ND |
| CR27 | >32 | 4 | 0.00032 | 0.02 | >32 | 2 | 80 | 50-100 | ND | ND | ND |
ND, not determined.
To verify that the increased cationic dye and fluoroquinolone resistance observed for the −35 and −35/RBS mutants of strain FA19 was related to the product of the norM gene, we inserted a nonpolar Km cassette into the norM gene of mutants CR22 (FA19 −35) and CR23 (FA19 −35/RBS) and studied their levels of susceptibility to AFh, Eb, NOR, NME, and CIP. These norM mutants showed increased susceptibilities to these compounds identical to those of the FA19 norM::Km mutant of strain FA19 (data not presented). These results confirmed that the decreased antimicrobial susceptibility phenotype of mutant strains CR22 (FA19 −35) and CR23 (FA19 −35/RBS) was due to the NorM efflux pump and not a polar effect on a downstream gene.
Importance of cationicity in NorM substrate recognition.
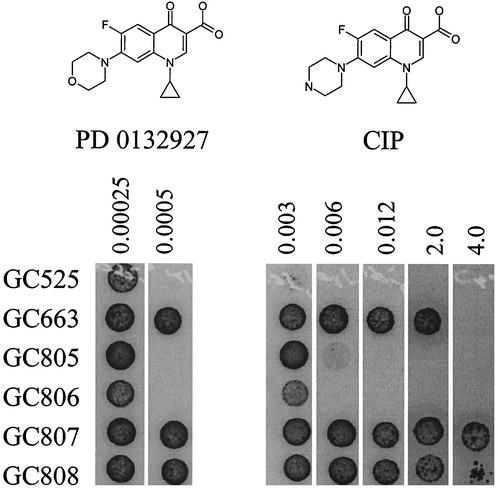
At physiological pH, it is expected that the piperazine ring of both CIP and NOR is positively charged. Combining this with the observation that all of the NorM substrates identified in this study are cationic, we hypothesized that removing the positive charge on this fluoroquinolone side chain would reduce or eliminate its ability to be exported by NorM. Agar dilution MICs indeed demonstrated that PD 0132927-0000, an oxazine analog of CIP, is not a substrate for NorM in either the −35 or −35/RBS mutant strain backgrounds (Fig. 3). These results provide strong evidence that a charged moiety on fluoroquinolones is extremely important for NorM recognition in N. gonorrhoeae and are in accordance with the observed increase in the efflux of NME relative to ELL.
FIG. 3.
Agar dilution of CIP and PD 0132927 against isogenic N. gonorrhoeae strains harboring −35 and −35/RBS norM promoter mutants in wild-type (GC805 and GC806) and quinolone-resistant (GC807 and GC808) backgrounds. Five-microliter spots containing 1 × 104 to 4 × 104 CFU were incubated for 24 h. No growth was observed on plates containing 8 μg of CIP per ml.
NorM can further increase CIP resistance in gonococci.
Although a single chromosomal mutation can result in an eightfold decrease in susceptibility to NOR, it is important to note that the gonococcus is exquisitely susceptible to quinolones and that this change represents an increase in the MIC of only 0.00014 μg/ml. This suggests that quinolones are poor substrates for the gonococcal NorM efflux system. Chen et al. (5) found that null mutations of ydhE (norM homologue) in an E. coli AcrAB-negative background also resulted in a slight increase in susceptibility to NOR relative to that of the parent strain. The largest increase in NOR resistance observed by overexpression of any norM homologue in an E. coli AcrAB-negative background was 0.25 μg/ml (19, 20). It is noteworthy that a bexA-negative mutant of Bacteroides thetaiotaomicron showed an increase in sensitivity of 96 and 8 μg/ml to NOR and CIP, respectively, compared to that of the parent, suggesting that this NorM homologue does indeed contribute to the intrinsic resistance to quinolones in B. thetaiotaomicron.
To assess if the modest NorM-mediated decrease in sensitivity to CIP observed for quinolone-sensitive strains was also observed for quinolone-resistant strains, we constructed a high-level quinolone-resistant strain and introduced the −35/RBS and norM mutations into its chromosome. This is an important question because it is unlikely that the two- to fourfold decrease in susceptibility to CIP observed for a wild-type background would provide any selective advantage in the clinic since susceptibility to CIP is 0.01 μg/ml, well below the MIC breakpoint (>1 μg/ml) for N. gonorrhoeae. Strains containing known quinolone resistance gyrA and parC alleles (2) were generated via natural transformation with PCR products containing mutations resulting in GyrA (S91F, D95G) and ParC (S88P, E91K) mutant enzymes. Strain GC663 was generated by cotransformation of the gyrA and parC mutant alleles into GC525 followed by selection at 1 μg of CIP/ml. Genotypes were confirmed by restriction analyses of PCR products. Strains GC807 and GC808 were generated by transforming strain GC663 with the −35 and −35/RBS mutations, respectively, to generate an isogenic set of quinolone-resistant strains harboring the norM promoter mutations. Agar dilution MICs of these strains were determined and demonstrated that a slight but measurable decrease in susceptibility to CIP was associated with the −35/RBS mutation (Fig. 3). Because this phenotype resulted in a twofold change, it is possible that strains with susceptibilities just below the breakpoint could acquire a selective advantage with the −35 and −35/RBS norM promoter mutations.
Transcriptional analysis of norM.
In order to map the promoter of norM, we performed primer extension analysis with primer N3 as described previously (24), by using RNAs extracted from strains FA19 and CR22 (FA19 −35) grown in GCB broth (3). We did not detect a transcription start point signal with RNA extracted from strain FA19. In contrast, we detected a transcription start point signal with RNA extracted from its transformant strain, CR22 (FA19 −35), that corresponded to a C nucleotide located 68 bp upstream of the translational start codon of norM. By sequence analysis, a putative norM promoter was identified in strain CR22 (FA19 −35): TTGACG for the −35 sequence and TATATA for the −10 sequence, with a spacing of 17 bp. In strain FA19, the putative −35 sequence would be CTGACG, instead of TTGACG.
To understand the effect of the putative promoter and RBS mutations on expression of norM, real-time quantitative RT-PCR (qRT-PCR) was performed with primers specific to norM and an internal control, the rmp gene (9), with cDNA from FA19, CR22 (FA19 −35), and CR23 (FA19 −35/RBS). Real-time RT-PCR was performed on a ABI Prism 7700 sequence detection system (Applied Biosystems) with primers N20 and N21 to amplify the norM transcript and primers N12 and N13 to amplify rmp as an internal control. The relative expression of the two genes was determined by both the comparative CT method and the standard curve method as described by the manufacturer (user bulletin 2, ABI Prism 7700 sequence detection system; Applied Biosystems). cDNA for qRT-PCR was prepared with the TaqMan RT kit (Applied Biosystems) with 1 μg of RNA as template and a random hexamer according to the manufacturer's directions. Twenty-five-microliter-total-volume qRT-PCRs were performed with the SYBR Green PCR master mix (Applied Biosystems) by a four-step cycling protocol as described by the manufacturer (user bulletin 2). For standard curve generation twofold serial dilutions of 250 to 31 ng of FA19 genomic DNA were used. qRT-PCRs were performed with RNA as template to confirm the absence of contaminating genomic DNA. All data points are averages from at least three independent reactions. We determined that the expression of norM relative to rmp showed an approximately 40-fold (40.9 ± 2.7) increase in strain CR22 over that in parental strain FA19 (data not presented). The addition of the putative RBS mutation showed no significant increase in transcription relative to the −35 mutation alone (44.8 ± 2.9), suggesting that the increase in efflux activity associated with the RBS mutation is likely due to an increase in translation.
This study showed that chromosomal mutations resulting in the overexpression of norM provided decreased susceptibility to the fluoroquinolones CIP and NOR. This may be of interest given the emerging problem of quinolone-resistant isolates of gonococci (1, 6, 12, 13). Our studies have shown that mutations in both the −35 sequence and putative RBS can result in a modest but reproducible twofold decrease in gonococcal susceptibility to CIP in a high-level quinolone-resistant strain. Thus, while these mutations are not likely to be significant in an otherwise wild-type sensitive strain, they could be significant in strains expressing a level of CIP susceptibility that is near the MIC breakpoint. Finally we also emphasize that the presence of the NorM efflux pump in gonococci and meningococci should be noted during the design and testing of new antimicrobials bearing structural similarities to cationic dyes, quaternary ammonium compounds, and quinolones.
Acknowledgments
We thank D. Stephens (Emory University School of Medicine) for providing meningococcal strains NMB and M7, P. F. Sparling and J. G. Cannon (University of North Carolina—Chapel Hill) for providing strains FA19 and FA1090, respectively, T. Hrobowski for technical assistance, and L. Pucko for editorial help. We are grateful to the Gonococcal Genome Sequencing Project (supported by NIH grant AI-38399) of the University of Oklahoma (B. A. Roe, S. P. Lin, L. Song, X. Yuan, S. Clifton, T. Dulcey, L. Lewis, and D. W. Dyer) for providing the sequence of the FA1090 genome online.
This work was supported by PHS grant AI-21150 (W.M.S.) and AI-37945 (W.M.S.) from the NIH. W. Shafer is the recipient of a Senior Research Career Scientist Award from the VA Medical Research Service.
REFERENCES
- 1.Anonymous. 2000. Surveillance of antibiotic resistance in Neisseria gonorrhoeae in the WHO Western Pacific region. The WHO Western Pacific Region Gonococcal Antimicrobial Surveillance Programme. Commun. Dis. Intell. 24:269-271. [DOI] [PubMed] [Google Scholar]
- 2.Belland, R. J., S. G. Morrison, C. Ison, and W. M. Huang. 1994. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 4:371-380. [DOI] [PubMed] [Google Scholar]
- 3.Biran, D., N. Brot, H. Weissbach, and E. Z. Ron. 1995. Heat shock-dependent transcriptional activation of the metA gene of Escherichia coli. J. Bacteriol. 177:1374-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M. H., I. T. Paulsen, and R. A. Skurray. 1999. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31:394-395. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., Y. Morita, M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2002. VmrA, a member of a novel class of Na+-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J. Bacteriol. 184:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deguchi, T., M. Yasuda, M. Nakano, E. Kanematsu, S. Ozeki, Y. Nishino, T. Ezaki, S. Maeda, I. Saito, and Y. Kawada. 1997. Rapid screening of point mutations of the Neisseria gonorrhoeae parC gene associated with resistance to quinolones. J. Clin. Microbiol. 35:948-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delahay, R. M., B. D. Robertson, J. T. Balthazar, W. M. Shafer, and C. A. Ison. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127-2133. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman, M. M., and I. Pastan. 1993. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu. Rev. Biochem. 62:385-427. [DOI] [PubMed] [Google Scholar]
- 9.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 171:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 11.Hagman, K. E., C. E. Lucas, J. T. Balthazar, L. A. Snyder, M. Nilles, R. C. Judd, and W. M. Shafer. 1997. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143:2117-2125. [DOI] [PubMed] [Google Scholar]
- 12.Knapp, J. S., R. Ohye, S. W. Neal, M. C. Parekh, H. Higa, and R. J. Rice. 1994. Emerging in vitro resistance to quinolones in penicillinase-producing Neisseria gonorrhoeae strains in Hawaii. Antimicrob. Agents Chemother. 38:2200-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knapp, J. S., J. A. Washington, L. J. Doyle, S. W. Neal, M. C. Parekh, and R. J. Rice. 1994. Persistence of Neisseria gonorrhoeae strains with decreased susceptibilities to ciprofloxacin and ofloxacin in Cleveland, Ohio, from 1992 through 1993. Antimicrob. Agents Chemother. 38:2194-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kok, R. G., D. A. D'Argenio, and L. N. Ornston. 1997. Combining localized PCR mutagenesis and natural transformation in direct genetic analysis of a transcriptional regulator gene, pobR. J. Bacteriol. 179:4270-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, E.-H., and W. M. Shafer. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33:839-845. [DOI] [PubMed] [Google Scholar]
- 16.Levy, S. B. 1992. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 36:695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maness, M. J., and P. F. Sparling. 1973. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128:321-330. [DOI] [PubMed] [Google Scholar]
- 18.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita, Y., K. Kodama, S. Shiota, T. Mine, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1998. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita, Y., A. Kataoka, S. Shiota, T. Mizushima, and T. Tsuchiya. 2000. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J. Bacteriol. 182:6694-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, et al. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 23.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouquette, C., J. B. Harmon, and W. M. Shafer. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol. Microbiol. 33:651-658. [DOI] [PubMed] [Google Scholar]
- 25.Sarubbi, F. A., F. P. Sparling, E. Blackman, and E. Lewis. 1975. Loss of low-level antibiotic resistance in Neisseria gonorrhoeae due to env mutations. J. Bacteriol. 124:750-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shafer, W. M., L. F. Guymon, I. Lind, and P. F. Sparling. 1984. Identification of an envelope mutation (env-10) resulting in increased antibiotic susceptibility and pyocin resistance in a clinical isolate of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 25:767-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafer, W. M., X.-D. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shafer, W. M., W. L. Veal, E.-H. Lee, L. Zarentonelli, J. T. Balthazar, and C. Rouquette. 2001. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis. J. Mol. Microbiol. Biotechnol. 3:219-225. [PubMed] [Google Scholar]
- 29.Swartley, J. S., and D. S. Stephens. 1994. Identification of a genetic locus involved in the biosynthesis of N-acetyl-d-mannosamine, a precursor of the (α2→8)-linked polysialic acid capsule of serogroup B Neisseria meningitidis. J. Bacteriol. 176:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, et al. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 31.Veal, W. L., A. Yellen, J. T. Balthazar, W. Pan, B. G. Spratt, and W. M. Shafer. 1998. Loss-of-function mutations in the mtr efflux system of Neisseria gonorrhoeae. Microbiology 144:621-627. [DOI] [PubMed] [Google Scholar]