Abstract
Class 1 integrons have strongly influenced the evolution of multiple antibiotic resistance. Diverse integrons have recently been detected directly in a range of natural environments. In order to characterize the properties of these environmental integrons, we sought to isolate organisms containing integrons from soils, which resulted in the isolation of Pseudomonas stutzeri strain Q. Further isolation efforts targeted at this species resulted in recovery of two other strains (P and BAM). 16S rRNA sequences and chromosome mapping showed that these three strains are very closely related clonal variants in a single genomovar of P. stutzeri. Only strains Q and BAM were found to contain an integron and an associated gene cassette array. The intI and attI components of these strains showed 99 and 90% identity, respectively. The structure of these integrons and their associated gene cassettes was similar to that reported previously for other integron classes. The two integrons contained nonoverlapping sets of cassette-associated genes. In contrast, many of the cassette-associated recombination sites in the two integrons were similar and were considered to constitute a distinct subfamily consisting of 59-base element (59-be) recombination sites (the Pseudomonas subfamily). The recombination activity of P. stutzeri integron components was tested in cointegrate assays. IntIPstQ was shown to catalyze site-specific recombination between its cognate attI site and 59-be sites from antibiotic resistance gene cassettes. While IntIPstQ did not efficiently mediate recombination between members of the Pseudomonas 59-be subfamily and other 59-be types, the former sites were functional when they were tested with IntI1. We concluded that integrons present in P. stutzeri possess recombination activity and represent a hot spot for genomic diversity in this species.
Integrons are genetic elements that act as scaffolds for the rearrangement of genes via site-specific recombination of gene cassettes. The first integrons were discovered as a result of investigations into the phenomenon of multiple antibiotic resistance (23, 43). These integrons came to be classified into three classes on the basis of sequence divergence in shared regions. Most subsequent biochemical characterization of the site-specific recombination system has been performed with class 1 integrons recovered from clinical isolates and exhibiting multiple antibiotic resistance (9, 10, 11, 12, 21, 22). All class 1 integrons possess the following key components: an integrase gene (intI1), a recombination site (attI1), and a promoter (Pc) for the transcription of cassette-associated genes (11, 28). These components encompass the basic functions needed for the acquisition and expression of a second type of mobile genetic element known as a gene cassette (22, 35). Gene cassettes typically consist of an open reading frame (ORF) associated with a recombination site known as a 59-base element (59-be) (5). IntI1 most commonly mediates recombination between 59-be sites and attI1, resulting in insertion of gene cassettes into attI1 (14), although recombination between two 59-be sites is also possible. As a result of multiple insertion and excision events, class 1 integrons can compile and shuffle arrays of gene cassettes.
Recent discoveries have shown that diverse integron-like structures are also found outside the clinical and antibiotic resistance context. Integron-like structures have been amplified from soil DNA samples and have been found in database searches of available genome sequences, including the sequences of Nitrosomonas europaea, Geobacter sulfurreducens, Vibrio cholerae, Shewanella oneidensis, Acidithiobacillus ferrooxidans, and Treponema denticola (31, 39). In almost all cases, classification of these structures as integrons is based solely on sequence analysis rather than on demonstration of function. For the purposes of this study we operationally defined an integron as a genetic element that, at a minimum, includes an intI homolog associated with a plausible attI recombination site and may include an associated, contiguous gene cassette array.
The principal integron components, namely, intI, attI, and Pc, show great sequence diversity. Although there are enough characteristic features, particularly in the arrangement of gene cassette arrays, to infer significant functional similarity with the well-characterized class 1 integrons, the extent of similarity has not been adequately tested. It has been shown that the net pool of gene cassettes present in bacterial communities contains unprecedented levels of genetic novelty (45). Therefore, the possibility that all integrons may share the flexible gene cassette acquisition and expression properties of class 1 and class 3 integrons has a wide range of implications. Do all integron-containing bacteria have general access to the extensive, mobile metagenome contained within gene cassettes?
In this study we began to focus on the various aspects of the diversity of the integron-gene cassette system that may influence movement of gene cassettes by diverse integrons. Collectively, the studies mentioned above showed that integrons exhibit significant diversity in terms of location, since they are found on mobile elements and in the chromosomes of ecologically diverse species, and in terms of the components involved in gene acquisition (IntI, attI, and 59-be sites). Despite the very early stage of exploration of integron-gene cassette diversity, there are notable patterns in the distribution of this diversity. Particular intI genes are always associated with specific attI sites, some integrons appear to be associated with certain species, and some 59-be sequence families are apparently characteristic of some integrons or species. These patterns raise a number of questions about integron biology. Do all members of some species contain fixed, chromosomal integrons? Does specificity of interactions among IntI, attI, and 59-be influence the flow of gene cassettes between either integrons or species? Do all integrons show the same flexibility in recombination reactions that is exhibited by the model class 1 integron system?
Here we describe isolation of integron-containing bacteria from soil samples. Examination of Pseudomonas stutzeri isolates revealed that integrons were present in some, but not all, strains. The integrase, attI, and 59-be components of these integrons all showed significant relationships to a chromosomal integron described for Pseudomonas alcaligenes. The associated cassette arrays from two strains were partially characterized. In the region examined, none of the cassettes were the same, suggesting that members of P. stutzeri exhibit a high degree of diversity with respect to mobile gene cassettes. Assays to determine the recombination activity of components of the P. stutzeri integron demonstrated that IntI, attI, and the 59-be sites associated with the adjacent gene cassettes are all active. These assays were performed by using various combinations of class 1 integron components. The activity of P. stutzeri integron components was found to be dependent on the types of recombination reaction partners available. Most notably, the P. stutzeri integrase did not efficiently integrate 59-be sites into a class 1 integron.
MATERIALS AND METHODS
Enrichment and isolation.
Soil samples were collected from a number of sites previously shown to contain gene cassettes (45). Enrichment cultures were established by inoculation of 1 g of soil into 50 ml of either Luria-Bertani (LB) medium or minimal medium. Samples used for isolation attempts were serially diluted in sterile media and spread plated onto 1.5% agar plates containing the same growth medium. The cultures were incubated for up to 1 week at 28°C. Isolated colonies that were representative of morphotypes observed on the plates were picked off and inoculated onto fresh plates to obtain pure cultures. Cultures were stored on slopes and by addition of 25% glycerol at −70°C. Isolates were presumptively identified by cloning and sequencing of the 16S rRNA gene.
Screening samples for integrons.
DNA samples from mixed or pure cultures were screened for the presence of integrons by PCR. Approximately 100 ng of a DNA sample was used as the template in PCR performed with various combinations of broad-specificity primers targeting intI or 59-be sequences, as shown in Fig. 1. Integron PCR experiments target intI and the proximal gene cassettes by using primers HS298 (or HS300) and HS286 (31). Cassette PCR experiments target gene cassettes within an array by using primers HS286 and HS287 (45). The amplification mixtures consisted of 10 μl of Genereleaser (Bioventures Inc., Murfreesboro, Tenn.), 50 pmol of each primer, each deoxynucleoside triphosphate at a concentration of 200 nM, 1.5 mM MgCl2, and 1.5 U of Taq DNA polymerase (Eppendorf) in the buffer supplied with the enzyme. After an initial denaturation step of 94°C for 3 min, 30 cycles of 94°C for 1 min, 60°C for 45 s, and 72°C for 90 s were performed. PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide.
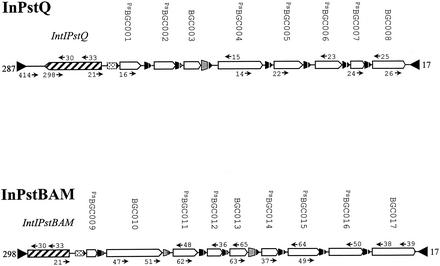
FIG. 1.
Genetic organization of the sequenced regions of InPstQ and InPstBAM (approximately to scale). The HS287 sequence at the left end of the InPstQ sequence represents a fortuitous mispriming event in a low-stringency PCR experiment. The approximate target sites for sequencing and mapping primers are indicated on the arrays. The left boundary of the integron-associated InPstQ and InPstBAM attI recombination sites is not precisely known. The regions containing these sites are indicated by dotted boxes. Cassette-associated recombination sites are indicated by broken triangles. The solid triangles represent members of the Pseudomonas subfamily of 59-be.
For enrichment cultures, samples were collected at weekly intervals, and DNA was extracted by a modification of the FastPrep (Bio 101, Vista, Calif.) bead-beating procedure as described previously (48). Samples that gave a positive integron PCR signal were selected for isolation attempts. One plate from each isolation attempt was used to test for the growth of integron-containing (In+) organisms on the plates. Cells were collected by washing the plate with 1 ml of sterile Tris-EDTA (TE) buffer (20 mM Tris [pH 8.0], 50 mM EDTA). Five hundred microliters of the bacterial suspension was used for DNA extraction, which was performed by the modified FastPrep procedure. For the plates that yielded a positive PCR signal individual colonies were then screened. A single colony was picked off with a sterile toothpick, placed into 50 μl of water, and lysed by boiling for 10 min. The suspension was then centrifuged for 1 min, and 5 μl of the supernatant was used directly as the template in a PCR.
Putative integron fragments were cloned with a pGEM-T kit (Promega) and were sequenced to confirm their identities. High-molecular-weight genomic DNA was purified from confirmed In+ strains by phenol-chloroform extraction (41). This DNA was used for further experiments to characterize the integron.
Cloning and sequencing of integrons.
The sequence of the P. stutzeri strain Q integron was obtained by PCR-mediated chromosome walking. In the first step PCR products were obtained by using the previously described primers HS298, HS286, and HS287 (31, 45), which target conserved sequences in the integron integrase gene and in the left and right domains of 59-be, respectively. After these integron or cassette PCR experiments were performed, the products were cloned into pGEM-T and sequenced. On the basis of the sequence information obtained, new primers were designed to target either cassette-associated genes or 59-be sequences.
The different components of the integron were linked after multiple rounds of PCR and sequencing. The integrity of the assembled integron sequence was confirmed by performing additional PCR with primers specific for the flanking genes and subsequent restriction mapping. All of the primers used for cloning, sequencing, and PCR are listed in Table 1, and their relative binding sites are shown in Fig. 1. Gene cassettes are traditionally named for their associated genes, but since most gene cassettes recovered outside the antibiotic resistance context contain hypothetical genes, we assigned a unique bacterial gene cassette number (BGC) to each cassette. Where information concerning the 59-be subfamily of a gene cassette is important below, gene cassettes are referred to by using the following notation: subfamilycassette, where cassette is the standard name or number of the gene cassette.
TABLE 1.
Primers used in this study
| Primer | Target | Sequence | Application |
|---|---|---|---|
| AJH14 | PsBGC004 | GAATCCCAACCACAAGCAGC | Mapping |
| AJH15 | PsBGC004 | GCGTGCTTGCGATCTGCTTGTAG | Mapping |
| AJH16 | PsBGC001 | GATAGTGGGTGTGAATTCGAAC | Mapping |
| AJH17 | Ps59-be | CCCAGYGARCGARGYGAGCG | Cassette PCR |
| AJH21 | intI | ATGAAGTAGTGTTTGAGCCG | Mapping |
| AJH22 | PsBGC005 | GTTCCTCGCATAAACTCACTG | Mapping |
| AJH23 | PsBGC006 | CTGCAGCTTTTATACGGATCGG | Mapping |
| AJH24 | PsBGC007 | CGACGAAGTGGGTAAGGTTG | Mapping |
| AJH25 | BGC008 | AAGCACCGCTTACTCCGACG | Mapping |
| AJH26 | BGC008 | GGGCATTAGGTCAGATGGC | Mapping |
| AJH27 | Ps59-be | GGCTGAAGCCVGCCCCTTARC | Cassette PCR |
| AJH30 | intI | CGATGTCTGGCTTCCTTTCG | Mapping |
| AJH36 | PsBGC012 | GTCCCAGTTAGCACCATACC | Mapping |
| AJH37 | PsBGC014 | CTAGATGCTTGGACAAGCG | Mapping |
| AJH38 | BGC017 | GATGTTCGTGTCGAACCTAC | Mapping |
| AJH39 | BGC017 | GCATGAACTAATGAGAGCTC | Mapping |
| HS298 | intI family | TGGATCCCACRTGNGTRTADATCATNGT | Integron PCR |
| HS286 | 59-be family | GGGATCCTCSGCTKGARCGAMTTGTTAGVC | Cassette PCR |
| HS287 | 59-be family | GGGATCCGCSGCTKANCTCVRRCGTTAGSC | Cassette PCR |
| HS401 | attI | CCCAAGCTTCCCAAACAGGTCGTCC | Subcloning attIA |
| HS405 | PsBGC001 | TTGGATCCTTGAGGTTGGCGTGCG | Subcloning attIA |
| HS413 | attI | AAGCTTGTTGTCATTTCCTTATTCC | Subcloning attIA |
| HS414 | attI | GGATCCATTACTATCGATCCAAAGG | Subcloning attIA |
| HS416 | PsBGC001 | AAGCTTCTCGTGACCATAATCG | Subcloning PsBGC001 59-be |
| HS417 | PsBGC002 | GGATCCATCGCGCTGTAGTTTCG | Subcloning PsBGC001 59-be |
| HS418 | PsBGC002 | AAGCTTAGCACATCAAAACCTG | Subcloning PsBGC002 59-be |
| HS419 | BGC003 | GGATCCATCGCACGCCAAGAGC | Subcloning PsBGC002 59-be |
| HS420 | BGC003 | GGATCCGAATATGAGTTGGGCAGG | Subcloning BGC003 59-be |
| HS421 | PsBGC004 | GTCGACAATGCAGTAGCTTATGGC | Subcloning BGC003 59-be |
Recombination activity of integron components.
The recombination activity of integron components (including IntIPstQ, attIPstQ, and 59-be from P. stutzeri strains) was assessed by performing conduction assays. These assays have been described in detail previously (14, 15). IntI proteins were supplied in trans by using plasmid pSU2056 (IntI1) or pMAQ711 (IntI and PstQ). The recombination sites to be assayed were supplied on separate plasmids. The test site was on a nontransferable plasmid, and potential recombination partner sites were located in In3, a class 1 integron located on the conjugative plasmid (pMAQ495). This plasmid was derived from R388 (1) by inactivation of IntI1 (13). As previously defined, recombination efficiency was measured by the frequency with which the test recombination site was transferred into a recipient cell via integration into a recombination site on the conjugative plasmid. All of the plasmids used and the relevant phenotypes are shown in Table 2.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Recombination site(s) | Phenotype | Reference |
|---|---|---|---|---|
| R388 | 33-kb IncW plasmid containing class 1 integron In3 | attI1, dfrB2 59-be, orfA 59-be | 1 | |
| pMAQ495 | R388 derivative with aphA inserted into intI1 | attI1, dfrB2 59-be, orfA 59-be | Tpr Sur Tra+ IntI1− | 13 |
| pACYC184 | Cloning vector | None | Cmr | 6 |
| pSU2056 | 1,176-bp RsaI-BamHI fragment of In2 in pUC9 | None | Apr IntI1+ | 30 |
| pMAQ28 | 400-bp Sau3A-HindIII fragment in pACYC184 (1) | aadB 59-be | Cmr | 22 |
| pMAQ702 | 166-bp HindIII-BamHI fragment of strain Q in pACYC184 (2) | attIPstQ | Cmr | This study |
| pMAQ707 | 238-bp HindIII-BamHI fragment of strain Q in pACYC184 (2) | BGC001 59-be | Cmr | This study |
| pMAQ708 | 333-bp HindIII-BamHI fragment of strain Q in pACYC184 (2) | BGC002 59-be | Cmr | This study |
| pMAQ709 | 354-bp BamHI-SalI fragment of strain Q in pACYC184 (2) | BGC003 59-be | Cmr | This study |
| pMAQ711 | 1,036-bp HindIII-BamHI fragment of strain Q in pUC9 | None | Apr IntI PstQ+ | This study |
The numbers in parentheses indicate the orientation of the cloned fragment with respect to pACYC184 as previously defined (14).
Analysis of cointegrates.
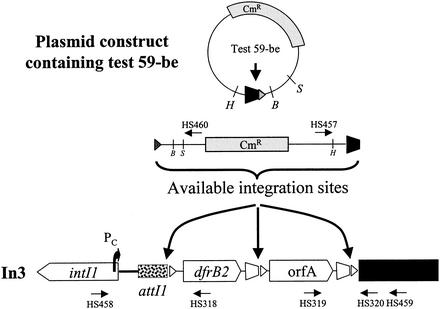
In3 contains three recombination sites. These are attI1 and the 59-be sites of dfrB2 and orfA (Fig. 2). Insertion of a test element at attI1 separates the dfrB2 gene from the Pc promoter, leading to a Tps phenotype. Consequently, the Tp phenotype was used as an indicator of insertion at attI1. However, to accurately and rapidly map cointegrates, a PCR-based strategy was used (Fig. 2). Two primers, one specific for a sequence within pACYC184 of the test element and one specific for a sequence within In3, were used to directly amplify cointegrate template DNA. The lengths of the derived PCR products were dependent on the insertion site. The primer pair commonly used was HS457 and HS459. As an example, the PCR product length involving pMAQ701 was 578 bp for an insertion at orfA, compared to 1,463 bp for an insertion at attI1. PCR products were also used as sequencing templates to identify the recombination crossover point. For HS457-HS459 products and insertion at orfA, right junctions were sequenced by priming with HS320. For insertion at attI1, the sequencing primer used was HS318. For some cointegrates the left junction was also amplified and sequenced, and this was achieved with HS458 and HS460 as the primers. The sequencing primers for these reactions were HS458 for insertion events at attI1 and HS319 for insertion events at orfA.
FIG. 2.
Schematic representation of primer sites used for mapping cointegrates resulting from the conduction assay (not to scale). Primer HS460 is located within the test construct. This primer is used with HS458 to map the left junction of any cointegrates formed. Similarly, primers HS457 and HS459 are used to map the right junction of cointegrates. Other primers shown are used to sequence the relevant junction. The promoter Pc is represented by a bent arrow.
Sequence analysis.
Bioinformatic analyses were conducted with BioManager provided by ANGIS (www.angis.org.au). Sequenced fragments were initially analyzed by using MAP (Genetics Computer Group, Madison, Wis.) to detect reading frames. Intergenic regions were manually examined for homology to 59-be sequences. BLASTP searches of both the nonredundant GenBank protein database and the National Center for Biotechnology Information microbial genome web site (www.ncbi.nlm.nih.gov/Microb.blast/unfinishedgenome) were performed for all inferred peptides. Equivalent BLASTN searches were performed with 59-be sequences recovered from the integron cassette arrays.
Phylogenetic analyses were performed by using programs from the Phylip package (18) within BioManager. The approaches used included distance matrix, parsimony, and maximum-likelihood methods for either nucleic acid or protein sequences (not all data are shown). Details of tree construction for the tree illustrated are given below (see Fig. 4). The sequence alignments included all positions between residues 40 and 308 (IntI1 numbering). For integron integrases the alignments were derived by using ClustalW. The outgroup Cre, XerC, and XerD sequences were manually added to the alignment by using the model alignment of the tyrosine recombinase family (32). Sequence regions which were present exclusively in the outgroup sequences (alignment gaps in all IntI sequences) were omitted from the analysis.
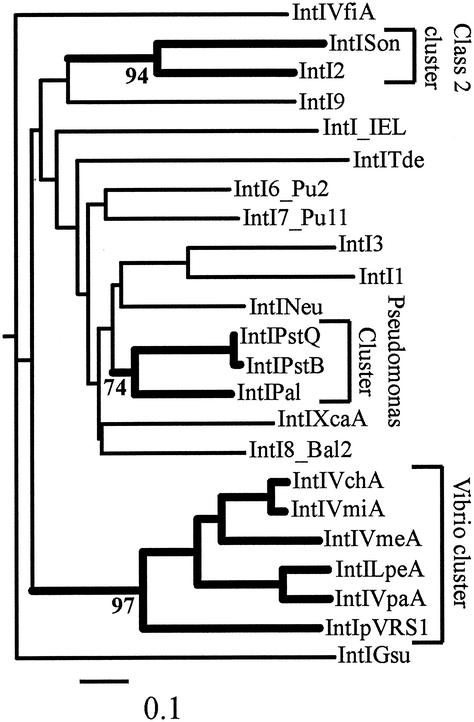
FIG. 4.
Evolutionary relationships of integrons based on IntI sequence analysis. There is no clear pattern of integron relationships based on IntI sequence for most known examples. The exceptions are the clades marked Vibrio cluster, Class 2 cluster, and Pseudomonas cluster. The numbers at the nodes leading to these groups indicate the levels of bootstrap support (100 replicates). Each of these groups is also supported by significant sequence conservation within the noncoding regions of the members. Evolutionary distances were calculated from an alignment of 278 positions (positions 40 to 308, IntI numbering) with PROTDIST by using the Dayhoff/PAM amino acid scoring matrix. A neighbor-joining tree was constructed by using NEIGHBOUR with XerD (Escherichia coli) as an outgroup. The sequences used and their source species (when they were not from mobile elements) are as follows: IntIVchA from Vibrio cholerae (accession no. af055586), IntIVmiA from Vibrio mimicus (af180939), IntIVmeA from Vibrio metschnikovii (ay014398), IntIVpaA from Vibrio parahaemolyticus (ay014399), IntILpeA from Listonella pelagia (ay014401), IntIVfiA from Vibrio fischeri (ay014400), IntI6_Pu2 (af314191), IntI1 (af071413), IntI2 (ap002527), IntI3 (d50438), IntI7_Pu11 (af314190), IntI8_Bal2 (af314189), IntIPal from P. alcaligenes (ay038186), IntI pRVS1 (aj277063), and IntIXcaA from Xanthomonas campestris (af324483). The sequences for IntIGsuA, IntISon, and IntITde were derived from preliminary genome sequence data for G. sulfurreducens, S. oneidensis, and T. denticola obtained from The Institute for Genomic Research web site (http://www.tigr.org). The sequence of IntINeuA was derived from the completed genome sequence of N. europaea, which was obtained from the DOE Joint Genome Initiative web site (www.jgi.doe.gov).
PFGE.
For pulsed-field gel electrophoresis (PFGE), cultures were grown for 36 h at 30°C in LB broth. Numbers of cells were determined by microscopy. A total of 1 × 109 cells were harvested by centrifugation and resuspended in 1 ml of modified cell suspension buffer (10 mM Tris [pH 7.2], 20 mM NaCl, 10 mM EDTA). One milliliter of 2% low-melting-point agarose was added to the suspended cells before 100-μl aliquots were cast in a block. Plugs were then incubated for 1 h at 37°C in 5 ml of lysis buffer (10 mM Tris [pH 7.2], 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 1 mg of lysozyme per ml). The buffer was then removed, and the plugs were washed in 25 ml of TE buffer. Proteinase digestion was performed overnight at 50°C in 5 ml of proteinase K reaction buffer (100 mM EDTA [pH 8.0], 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg of proteinase K per ml). The plugs were rinsed four times in wash buffer for 30 min. The second and third wash solutions contained 1 mM phenylmethylsulfonyl fluoride.
Restriction digestion was performed overnight in the agarose plugs by using 30 U of I-Ceu1, SpeI, or PacI in the buffer supplied with the enzymes. In all cases the plugs were equilibrated by incubation in 10 volumes of 1× restriction buffer for 1 h at room temperature prior to digestion in 3 volumes of fresh buffer.
Electrophoresis was performed in a CHEF DRIII apparatus (Bio-Rad). For SpeI digests the conditions were as follows: 6 V/cm; 18 h; switch times, 50 to 90 s; and reorientation angle, 120°. For I-Ceu1 and PacI digests the following conditions were used: 6 V/cm; 36 h; switch times, 50 to 200 s; and reorientation angle, 120°.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AY129391, AY129392, and AY129393.
RESULTS
Integron detection.
Our objective was to isolate integron-containing bacteria from soil for the purpose of recovery and characterization of integron classes present in natural environments. Detection of integrons in either the environment or bacterial isolates is complicated by the considerable diversity in the core integron sequence between classes, the associated gene cassette arrays, and the genomic context of the integron-gene cassette system. Generally, integrases belonging to different classes show only 50 to 60% sequence identity. However, in PCR experiments aimed at generic detection of integrons, new integron classes and new gene cassettes have been recovered from environmental DNA (31).
A total of 20 enrichment cultures were examined. Cassette PCR performed with HS286 and HS287 yielded PCR products for all of these cultures, although the yields and diversities of products varied. Cloning and sequencing of PCR products from six of these cultures confirmed that these products all included 59-be sequences (data not shown). This implied that integron-containing bacteria were present in all enrichment cultures. However, only two enrichments (Balmain soil LB broth and Lidsdale soil minimal medium enrichments) gave PCR products of the expected size (>1,200 bp) in integron PCR experiments. These putative integron fragments (intI and the first cassette) were cloned and sequenced. In practice, integrons have been assigned to different classes on the basis of sequence divergence of more than 5% in IntI. On this basis, both of the integrases described here represent new integron classes. The integrase obtained from the Lidsdale microcosm was designated IntI_IEL. The integrase obtained from the Balmain microcosm was subsequently found to be identical to IntIPstQ found in an isolate recovered from this microcosm (see below). The relationship of these integrases to integrases from other integron classes is described below.
Isolation of integron-containing bacteria.
Bacterial isolates were obtained from the two enrichments giving positive integron PCR results by direct plating onto solid media. Integron PCR was used to screen representative colony morphotypes from both enrichments. An integron-positive isolate was recovered from the Balmain enrichment; however, none of the isolates obtained from the Lidsdale minimal medium enrichment gave positive integron PCR results. The most likely explanation for the latter results is that the host bacterium either failed to grow or was present at a sufficiently low level that it did not appear on the isolation plates. Lidsdale cultures were not examined further.
A single colony giving positive integron PCR results was recovered in the initial isolation attempts from the Balmain LB broth microcosm (strain Q). Strain Q had a distinctive wrinkled colony morphology. Two additional isolates (P1 and P2) that did not give an integron PCR product but were morphologically indistinguishable from strain Q were recovered from the same plate. To further explore this finding, we repeated the enrichment and isolation process with the same soil sample and obtained two additional isolates (BAM17 and BAM21). These isolates also gave positive integron PCR results and had the distinctive wrinkled colony morphology. All five isolates from Balmain LB broth enrichments were used for further experiments.
Strain identification and genotypic characterization.
The 16S ribosomal DNAs of isolates Q, P1, P2, BAM17, and BAM21 were cloned and sequenced by PCR. All five sequences were found to be identical and showed 100% identity to the P. stutzeri JM300 sequence (accession no. X98607). The levels of identity to sequences of other strains of P. stutzeri ranged from 98 to 99.8%. Phylogenetic analysis of 16S ribosomal DNA sequences from a representative range of P. stutzeri genomovars and other γ-proteobacteria confirmed that the strains classified as P. stutzeri formed a monophyletic group that included our new isolates (data not shown).
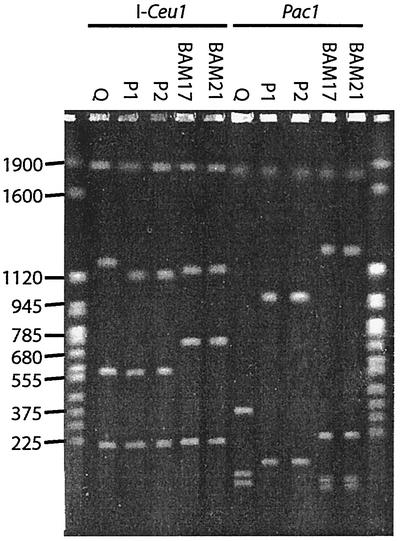
Despite the 16S rRNA identity, chromosomal digests of the five isolates showed that they represent three distinct genotypes (Fig. 3). With all three enzymes tested isolates P1 and P2 had the same profile, which was unique. Isolates BAM17 and BAM21 also had profiles that were identical and unique. Isolate Q had a distinct third profile. This same pattern of interstrain relationships was seen when the results of integron PCR, cassette PCR, and two-dimensional polyacrylamide gel electrophoresis of membrane proteins were examined (unpublished data). Since the matching isolates were recovered from the same plate in each case, it is probable that they represent sibling colonies and should not be considered independent strains. We therefore believe that the five isolates represent three distinct strains, which are referred to as Q, P (comprising P1 and P2), and BAM (BAM17 and BAM21) below. The I-Ceu1 digests contained four bands for each of the three strains. This enzyme is known to cut within the 23S rRNA of P. stutzeri and typically produces four chromosomal fragments for this species (19). I-Ceu1 fragments at approximately 210 and 1,900 kbp were obtained for all five strains (Fig. 2). PacI digestion resulted in three to five fragments for each strain. A fragment at 1,850 kbp was obtained for all of the strains. SpeI digestion resulted in 9 to 13 fragments, the majority of which were <450 kbp long. None of the fragments was obtained for all three strains, although several were obtained for two strains (data not shown). The integron-containing Q and BAM strains both produced a large 700-kbp fragment. The estimated genome sizes for these strains were 3.7 Mbp (P strains) to 3.9 Mbp (BAM and Q strains).
FIG. 3.
Variation in chromosomal digests of P. stutzeri strains. DNA was digested with I-Ceu1 or PacI. The following PFGE conditions were used: 1% agarose; 0.5× TBE; switch times, 70 to 120 s; 21 h; 6 V/cm; reorientation angle, 120°. The molecular weight markers used were Saccharomyces cerevisiae chromosomes; the sizes of bands (in kilobases) are indicated on the left. The sibling P strains do not have a detectable integron and have a significantly smaller genome than the strains possessing an integron. The image was prepared from a scanned photograph by using Adobe Photoshop 7.0.
Previous studies of the P. stutzeri species complex have shown that it has exceptional genetic diversity (37). Various data, including 16S rRNA sequence (2, 20) and chromosome architecture (19) data, correlate with subgroups termed genomovars. Given the identical 16 rRNA sequences of strains Q, P, and BAM and the similar pattern generated by restriction digestion, these three strains are almost certainly members of the same genomovar. No evidence for the presence of plasmids in any of the three strains was obtained with undigested samples electrophoresed in PFGE gels (data not shown).
Compilation of integron sequences.
The integron sequences of strains Q and BAM17 were assembled from a series of overlapping fragments generated by PCR (Fig. 1). In the case of strain Q a fortuitous mispriming event with HS287 downstream of intI resulted in recovery of the complete core integron sequence. Thus, the compiled 5,957-bp sequence contained an intact intI gene, a putative attI site, and an array of eight gene cassettes. This integron was designated InPstQ. The compiled sequence (7,275 bp) from strain BAM17 included a partial intI gene, a putative attI site, and nine gene cassettes, and this integron was designated InPstBAM.
Since multiple copies of gene cassettes may be present in an integron array (8, 23, 24) or gene cassettes may be free circular molecules (9), the arrangement of cassettes within the integrons of both strains was confirmed by PCR performed with primers targeting intI (AJH21) and the terminal gene cassette in the array (AJH25 in InPstQ and AJH38 in InPstBAM) and subsequent restriction mapping of the products (Fig. 1; also data not shown). Both cassette arrays described here include only the cassettes that we could identify as contiguous. Both strains possess additional cassettes recoverable by cassette PCR. These are presumably contained in the same array, but their order is unknown, as is the total number of cassettes in the array.
Relationship to other integrons.
Because of the mobility of gene cassettes, the relationship of integrons is not reflected by their gene cassette arrays. However, the core components of integrons represent stable genetic units whose sequence relationships reflect the evolutionary history of the integrons. Here, comparative sequence analyses were performed for both the coding regions of integrons (intI) and the noncoding regions of integrons (sequences between the start codon of intI and the core site of attI). Collectively, these analyses indicated that the integrons recovered from our P. stutzeri strains are related to a chromosomal integron in P. alcaligenes ATCC 55044 (47).
In phylogenetic analyses of IntI sequences a group comprised of integrons found in Pseudomonas species was observed with all methods of analysis. The level of support for this group in bootstrap analyses was, however, only moderate (Fig. 4). The relationship of IntINeu to this Pseudomonas cluster is worthy of comment. This sequence consistently branches closest to the Pseudomonas cluster, but it is derived from a chromosomal integron in a very different bacterial species, N. europaea, a member of the β-proteobacteria. IntI_IEL showed no significant relationship to any other integron integrase sequence.
Comparative analysis of the noncoding regions of integrons confirmed the relationship predicted from the IntI analysis. All three clusters observed in IntI trees showed significant sequence relationships in the noncoding regions. It is not possible to perform phylogenetic analyses equivalent to those done with IntI using the noncoding region. The sequence is too variable; the length ranges from 114 bp (IntI3) to 296 bp (IntISon) in different integrons. However, significant conservation of this region (71 of 137 bases are identical) in the three Pseudomonas integrons does indicate that there was a recent common origin (Fig. 5). The corresponding noncoding region adjacent to IntINeu shows no similarity to the noncoding region in the Pseudomonas cluster integrons.
FIG. 5.
Sequence alignment for the noncoding region of Pseudomonas cluster integrons. The sequences shown span the region from the base adjacent to the start codon of intI to the insertion point of the first gene cassette. This region contains the integron-associated attI recombination site. Bases that are conserved (Cons) in all three sequences are shown in boldface type below the alignment.
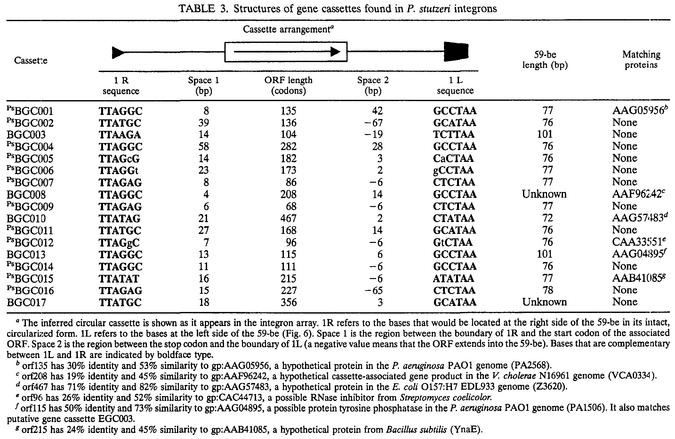
Cassette-associated genes.
All 17 cassettes characterized had the type A arrangement, containing a single ORF in the forward orientation (45). In common with the vast majority of previously described gene cassettes, no promoter sequences were evident in these cassettes, and in most cases there was no room for a promoter (Table 3). There were six cassette-associated genes that gave database matches, but only two of the matches were to families containing proteins whose functions are known. orf96 is a member of a family of proteins that includes the RNase inhibitor BARSTAR. Orf115 showed a significant relationship to a protein-tyrosine phosphatase family (Table 3).
TABLE 3.
Structures of gene cassettes found in P. stutzeri integrons
The inferred circular cassette is shown as it appears in the integron array. 1R refers to the bases that would be located at the right side of the 59-be in its intact, circularized form. 1L refers to the bases at the left side of the 59-be (Fig. 6). Space 1 is the region between the boundary of 1R and the start codon of the associated ORF. Space 2 is the region between the stop codon and the boundary of 1L (a negative value means that the ORF extends into the 59-be). Bases that are complementary between 1L and 1R are indicated by boldface type.
orf135 has 30% identity and 53% similarity to gp:AAG05956, a hypothetical protein in the P. aeruginosa PAO1 genome (PA2568).
orf208 has 19% identity and 45% similarity to gp:AAF96242, a hypothetical cassette-associated gene product in the V. cholerae N16961 genome (VCA0334).
orf467 has 71% identity and 82% similarity to gp:AAG57483, a hypothetical protein in the E. coli O157:H7 EDL933 genome (Z3620).
orf96 has 26% identity and 52% similarity to gp:CAC44713, a possible RNase inhibitor from Streptomyces coelicolor.
orf115 has 50% identity and 73% similarity to gp:AAG04895, a possible protein tyrosine phosphatase in the P. aeruginosa PAO1 genome (PA1506). It also matches putative gene cassette EGC003.
orf215 has 24% identity and 45% similarity to gp:AAB41085, a hypothetical protein from Bacillus subtilis (YnaE).
Given that the mechanism of gene cassette formation is unknown, matches with other gene cassettes or with Pseudomonas genes are also of interest. Two of the cassette ORFs showed a relationship to previously described gene cassette ORFs. Orf208 exhibited similarity to the inferred product of a cassette-associated gene from the V. cholerae N16961 integron. This gene cassette has a 59-be of the VCR type (7), which is not related to the Pseudomonas subfamily 59-be (see below). Orf115 exhibited similarity to a hypothetical protein encoded by a gene cassette (EGC003) cloned from soil (45). Although the host integron(s) and 59-be sequence of EGC003 are unknown, we observed that EGC003 originated from the same soil sample as the P. stutzeri strains isolated in this study. Two cassettes showed strong matches with hypothetical proteins from Pseudomonas aeruginosa PAO1 (46). These were Orf135 (which matched PA2568) and Orf115 (which matched PA1506). Neither PA2568 nor PA1506 is cassette associated, and P. aeruginosa PAO1 does not contain an integron.
59-be recombination sites.
From the two cassette arrays a total of 15 complete 59-be recombination sites were identified; 7 of these sites were in InPstQ, and 8 were in InPstBAM. The integration of circular gene cassettes into a cassette array results in the native, circular gene cassette being opened within the 59-be site. As a result of integration, the last six base pairs of the 59-be, located within the 1R site (Fig. 6), are at the front of the integrated gene cassette (Table 3). It follows that the 59-be sites observed in linear arrays are hybrids between two adjacent gene cassettes. This is readily recognizable since the imperfect inverted repeat structure of 59-be sites means that 1R is generally complementary to 1L with reference to the inferred circular form of a cassette but is not necessarily complementary to the observed integrated form. As shown in Table 3, the complementarity of the 1L and 1R sites for the inferred circular forms of the P. stutzeri integron gene cassettes is consistent with the hypothesis that these arrays were compiled by IntI-mediated site-specific recombination.
FIG. 6.
Alignment of the inferred circular forms of PsBGC 59-be sequences with representative 59-be sequences and the 59-be structural model. Five bases flanking the 59-be site are shown at each end of the alignment. The gray shading indicates the 1L, 2L, 1R, and 2R regions proposed to be IntI binding domains (44). The Position line indicates sites that are considered equivalent in all known sequences; a position at which there is a lowercase letter characteristically complements a position indicated by the corresponding uppercase letter (also indicated by arrowheads). Plus signs indicate positions that characteristically disrupt the inverted repeat. An 18-bp insertion at m-n, including the 17-bp PAR signature (47), is diagnostic for the Pseudomonas subfamily. Conserved bases within this sequence are shaded. An insertion at the same site in BGC003 does not conform to the signature and is considered heterologous. The BGC013 59-be is an illegitimate sequence that is missing 11 bp with respect to the structural model (indicated by asterisks). The adjacent 9 bp of this element (integrated form) is unlikely to belong to the authentic BGC013 59-be site and is indicated by lowercase letters. The sources of 59-be sequences not described in this paper are as follows: aacA7, accession no. U13880; orfC5ab, accession no. AY035340; PsorfX, accession number AJ251519; PsorfI, accession no. AJ223604; PsSonBGC, accession no. AE014299; PsPAR 1, accession no. AY038186; PsPAR 26, accession no. AY038186; and PsPAR 5, accession no. AY038186.
The variability of 59-be sites within the InPst cassette arrays was relatively low (compared to the variability in arrays in class 1 integrons), and 12 of the 15 sites comprised a closely related subfamily of 59-be recombination sites exhibiting a minimum of 68% sequence identity. The structure of these 59-be sites is distinctive in that the sites contain an insertion between positions m and n in the left portion of the site compared with the majority of the previously characterized 59-be sites (Fig. 6). Similar 59-be sequences were recently reported by Vaisvila et al. for the integron In55044 located in the P. alcaligenes chromosome (47). These authors designated all the cassette-associated recombination sites in In55044 PAR elements (P. alcaligenes repeat). Of the 32 PAR elements, 30 contained a conserved sequence motif designated the PAR signature. This sequence very closely corresponds to the m-n insertion (Fig. 6). On the basis of our expanded data set, we believe that 59-be contains the PAR signature if the sequence between positions m and n exhibits more than 90% identity to the consensus sequence CGYTCRCYYCGYTCRCT. The term PAR was also used to refer to elements without the PAR signature (47). Here, to avoid confusion, we refer to all the gene cassettes possessing 59-be sites with this characteristic signature as Pseudomonas subfamily bacterial gene cassettes (PsBGC). Three of the gene cassettes identified in our study, BGC003, BGC010, and BGC013, are not members of the Pseudomonas subfamily. In the case of BGC010 and BGC013 59-be sites, there is no insertion at m-n, and in the case of BGC003 there is an insertion, but it is clearly not homologous to the PAR signature (Fig. 6).
BLASTN searches of sequence databases with representative 59-be sequences from the P. stutzeri integrons were performed. These searches revealed three instances in which 59-be sites belonging to the PsBGC subfamily were detected in unrelated integrons (Fig. 6). These were orfN in the class 1 integron In31 of P. aeruginosa isolate 101/1477 (27) (accession number AJ223604 refers to this cassette as orfI in In101), orfO in an unnamed class 1 integron in Acinetobacter baumanni BM4426 (34) (accession number AJ251519 refers to this cassette as orfX), and one site in an integron-like cassette array in the S. oneidensis genome (25). The genes associated with these elements all encoded hypothetical proteins whose functions are not known. Although Pseudomonas subfamily elements are not unique to Pseudomonas spp., it is apparent that these elements are not strongly represented in other currently known integrons.
Of the three unaffiliated 59-be sites, two did show significant matches with sites of previously described gene cassettes. The BGC010 59-be exhibited a significant match (83% identity) with the recently described orfC5ab 59-be from a class 9 integron in V. cholerae SXTET (26). The BGC013 59-be was of particular interest since the leftmost 86 bases of the element exhibited 83% identity to the 59-be of a gene cassette encoding the aminoglycoside resistance gene aacA7 (4). Positions S to H with respect to the conserved structural model of 59-be have apparently been deleted, and the remaining 15 bp do not show significant homology to the aacA7 59-be (Fig. 6). It is likely that the BGC013 sequence, as observed, does not contain an intact 59-be site. It is possible that the observed sequence resulted from an illegitimate excisive recombination event that removed an unknown gene cassette(s) from between BGC013 and PsBGC014 in this array, failing to reconstitute a 59-be site.
Recombination activity.
To determine if InPstQ was capable of assembling gene cassettes, the IntIPstQ recombinase and the attI recombination site were tested for recombination activity in cointegration assays. The potential target recombination sites for attIPstQ were attI1, dfrB2 59-be, and orfA 59-be (Fig. 1) contained in the class 1 integron In3. The frequency of conduction in these experiments was approximately 4 orders of magnitude above the background frequency (Table 4). These data indicate that both IntIPstQ and attIPstQ are functional. The participating recombination sites in In3 were determined by PCR mapping of 24 individual cointegrates (from six independent crosses). A marked preference for insertion at the orfA 59-be was found (23 cointegrates). The remaining cointegrate had a PCR product length consistent with insertion at the dfrB2 59-be. These insertion sites were confirmed by sequencing cointegrates for five (independent) predicted orfA recombination events, as well as the predicted dfrB2 insertion (data not shown). In each of these cases the sequence was consistent with a recombination crossover point within the attIPstQ and 59-be core sites, as is the case for site-specific recombination reactions catalyzed by both IntI1 and IntI3 (15, 36).
TABLE 4.
Recombination activity of integron components from P. stutzeri strain Q
| Integrase | Test plasmid | Test element | Fragment length (bp)a | Range | Cointegration frequencyb |
|---|---|---|---|---|---|
| IntI1 | pMAQ28 | aadB 59-be | 202/198 | 4.5 × 10−3-1.6 × 10−2 | 1.1 × 10−2 (5)c |
| IntI1 | pMAQ707 | PsBGC001 | 124/114 | 2.1 × 10−4-2.5 × 10−3 | 1.3 × 10−3 (6) |
| IntI1 | pMAQ708 | PsBGC002 | 150/183 | 6.8 × 10−5-3.6 × 10−4 | 2.1 × 10−4 (4) |
| IntI1 | pMAQ709 | BGC003 | 216/138 | 1.7 × 10−3-7.7 × 10−3 | 4.7 × 10−3 (2) |
| IntI1 | pACYC184 | None | NA | 8.8 × 10−7-4.5 × 10−6 | 2.9 × 10−6 (4) |
| IntIPstQ | pMAQ702 | attI | 121/19 | 9.2 × 10−3-5.0 × 10−2 | 3.1 × 10−2 (7) |
| IntIPstQ | pMAQ28 | aadB 59-be | 202/198 | 2.5 × 10−6-8.0 × 10−6 | 4.7 × 10−6 (4) |
| IntIPstQ | pMAQ707 | PsBGC001 | 124/114 | 3.1 × 10−6-9.8 × 10−6 | 5.6 × 10−6 (4) |
| IntIPstQ | pMAQ708 | PsBGC002 | 150/183 | 1.6 × 10−7-2.6 × 10−6 | 1.2 × 10−6 (4) |
| IntIPstQ | pMAQ709 | BGC003 | 216/138 | 5.4 × 10−7-2.9 × 10−6 | 2.0 × 10−6 (4) |
| IntIPstQ | pACYC184 | None | NA | 1.6 × 10−7-3.4 × 10−6 | 1.0 × 10−6 (7) |
The numbers are the numbers of base pairs on either side of the predicted recombination crossover point.
Averages. The numbers in parentheses are the numbers of independent assays.
Control value reported by Collis et al. (14).
Conduction assays with the target recombination sites described above were also performed to test if the 59-be sites of PsBGC001, PsBGC002, and BGC003 from P. stutzeri strain Q were functional recombination sites. Surprisingly, when tested with IntIPstQ, these three elements were inactive since the cointegration efficiency was not significantly greater than that of a no-element control (Table 4). As IntI1 is known to recognize a number of disparate 59-be sites, we tested the ability of this integrase to recognize PsBGC001, PsBGC002, and BGC003. With IntI1 as the integrase all three 59-be sites were found to be active, and the conduction frequencies were 2 to 3 orders of magnitude above the background frequency (Table 4).
DISCUSSION
It is not yet possible to directly assess the abundance and diversity of integron-containing organisms in nature. Crude estimates of abundance may be based on the incidence of integrons in genome sequences. At the time of this study, 7 of the 106 partial or complete genome sequences available for searching through the National Center for Biotechnology Information contained integrons. Given that viable bacterial counts are typically >106 cells g of soil−1, even allowing for biases in the bacterial genome sample set, an estimate of 103 In+ bacteria g of soil−1 is very conservative. This estimate is consistent with the abundance and diversity of components of the integron-gene cassette system that can be directly recovered from soil DNA samples (31, 45). Thus, although the functional significance of integron-gene cassette diversity is largely unknown, it is evident that integrons and gene cassettes are sufficiently common to strongly influence bacterial evolution.
Diversity of integrons.
The simplest means of recognizing integron diversity is on the basis of sequence divergence in the intI gene. The levels of pairwise IntI sequence identity of known integrons range from ca. 50 to 99%. The partial integron recovered from the Lidsdale enrichment clearly represents a new class of integrons that forms a distinctive line of descent within the IntI radiation (Fig. 4).
The integrons recovered from the P. stutzeri Q and BAM strains exhibit high levels of identity and are considered representatives of a single, new class. Comparative sequence analyses of both coding and noncoding regions of integrons indicate that the two InPst integrons, together with In55044 found in P. alcaligenes ATCC 55044, comprise a distinct subfamily of integrons. The support for a monophyletic Pseudomonas group in IntI trees was only moderate (74% bootstrap support). This reflects poor resolution of the IntINeu sequence in these trees, probably because of the incomplete nature of current integron integrase sequence databases. An analysis of the noncoding region between intI and the attI core site provided unambiguous support for a monophyletic group that includes the Pseudomonas cluster integrons but not the integron from N. europaea.
There are notable parallels between the Pseudomonas group of integrons and some of the integrons found in Vibrio species (39). In both cases, organisms that are related as determined by 16S rRNA phylogeny possess closely related integrons, as determined by both IntI and noncoding region analyses, and are strongly associated with distinctive 59-be subfamilies. This suggests the possibility that there is a degree of fidelity among certain integrons, gene cassettes, and bacterial species. Interestingly, of the three P. stutzeri strains examined here, one did not contain an integron and the two integron-containing strains had nonoverlapping gene cassette contents. Clearly, further investigation of this issue with respect to population structure in both Vibrio and Pseudomonas requires strain collections that are much larger than those that are currently available and extensive testing of organisms that do not belong to these genera.
Recombination activity.
The integrative and excisive recombination activities of the integron-gene cassette system involve an integron integrase and two types of recombination sites. All components of InPstQ tested here were shown to participate in site-specific recombination when appropriate combinations of reaction partners were present. These partners include both the integrase protein and the attI site since IntIPstQ mediates recombination between attIPstQ and at least the two different 59-be sites located in In3. In addition, 59-be sites found in P. stutzeri-associated cassettes are active since all three elements tested were recognized by IntI1.
The fact that IntIPstQ was apparently unable to recognize any of the P. stutzeri- associated 59-be sites was possibly an artifact of the conduction assay. Data from experiments with the extensively studied integron IntI1 have clearly demonstrated that there are strong partner site preferences (14). While other explanations are also possible, the simplest explanation is perhaps that IntIPstQ has a rigid requirement for recombining elements with its own attI site, an option not available in the present assay system since the only available attI site in In3 is attI1. Further experiments are required to clarify this question. Notwithstanding the fact that IntIPstQ may have strong partner site preferences, it is still clear that this protein is capable of recognizing members of the 59-be family, a phenomenon seen with all other integron integrases that have been tested for this activity. These include IntI3 (15), IntISon (17), and IntIVchA (39). Consequently, it is likely that gene cassettes can be captured by a disparate range of integron-containing organisms.
Evolutionary significance of integrons.
The species concept for prokaryotes has long been debated (38). It is now widely accepted that a large part of bacterial diversity arose via lateral gene transfer (33). With this background, our observations take on new significance in the species-genome concept (3). In this model, species contain a core set of genes that are very rarely transferred and whose sequence reflects the evolutionary history of the cell. They also include a much larger auxiliary set of genes that is distributed across the population. This gene set may more easily transfer between members of the population and occasionally may transfer to different species. In an evolutionary context this presents an interesting dilemma for cells containing integrons. The ability of an integron to assemble useful sets of genes is restricted by the extent of contact between integrons and gene cassettes. However, we might also expect that the usefulness of an integron to a host population is determined by the ability of the population to restrict export of gene cassettes to competing populations. The apparent predisposition of some integron and gene cassette subfamilies for particular genera is highly significant in this regard.
By virtue of the activity (23) and prevalence (31, 40) of integrons and the diversity of the associated gene cassette pool (45), integrons clearly have the potential to exert tremendous influence over the evolution of bacterial genomes. The core components of integrons comprise a very simple structure that has been observed to reside within a range of higher genetic elements, including insertion sequences, transposons, constins (conjugative transposons), plasmids, and chromosomes (7, 16, 26, 29, 30, 42). This diversity of genomic context, in conjunction with the ability of at least some integrons to act as a scaffold for multigene assembly, makes it increasingly likely that we will observe the impact of integrons at all scales of bacterial genome evolution.
Acknowledgments
This work was supported by the Macquarie University Research Innovation Fund and by the Australian Research Council.
We thank Iain Morris and Amy Ramsden for experimental assistance during portions of this work; Ruth Hall, Bridget Mabbutt, and Helena Nevalainen for helpful discussions; and Elisabeth Raleigh for sharing some data prior to publication.
REFERENCES
- 1.Avila, P., and F. de la Cruz. 1988. Physical and genetic map of the IncW plasmid R388. Plasmid 20:155-157. [DOI] [PubMed] [Google Scholar]
- 2.Bennasar, A., R. Roselló-Mora, J. Lalucat, and E. R. B. Moore. 1996. 16S rRNA gene sequence analysis relative to genomovars of Pseudomonas stutzeri and proposal of Pseudomonas balearica sp. nov. Int. J. Syst. Bacteriol. 46:200-205. [DOI] [PubMed] [Google Scholar]
- 3.Boucher, Y., C. L. Nesbo, and W. F. Doolittle. 2001. Microbial genomes: dealing with diversity. Curr. Opin. Microbiol. 4:285-289. [DOI] [PubMed] [Google Scholar]
- 4.Bunny, K. L., R. M. Hall, and H. W. Stokes. 1995. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloroamphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob. Agents Chemother. 39:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron, F. H., D. J. Groot Obbink, V. P. Ackerman, and R. M. Hall. 1986. Nucleotide sequence of the AAD(2") aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 14:8625-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, C. A., L. Purins, P. Kaewrakon, and P. Manning. 1997. VCR repetitive sequence elements in the Vibrio cholerae chromosome constitute a mega-integron. Mol. Microbiol. 26:1137-1138. [DOI] [PubMed] [Google Scholar]
- 8.Clark, C. A., L. Purins, P. Kaewrakon, T. Focareta, and P. A. Manning. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146:2605-2612. [DOI] [PubMed] [Google Scholar]
- 9.Collis, C. M., and R. M. Hall. 1992. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6:2875-2885. [DOI] [PubMed] [Google Scholar]
- 10.Collis, C. M., and R. M. Hall. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents and Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collis, C. M., G. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 13.Collis, C. M., M.-J. Kim, H. W. Stokes, and R. M. Hall. 1998. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 14.Collis, C. M., G. D. Recchia, M.-J. Kim, H. W. Stokes, and R. M. Hall. 2001. Efficiency of recombination reactions catalyzed by the class 1 integron integrase IntI1. J. Bacteriol. 183:2535-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collis, C. M., M.-J. Kim, S. R. Partidge, H. W. Stokes, and R. M. Hall. 2002. Characterization of the class 3 integron and the site-specific recombination system it determines. J. Bacteriol. 184:3017-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. C. Alves, A. M. do Amaral, M. C. Bertolini, L. E. A. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Clapina, R. M. B. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. S. Ferreira, R. C. C. Ferreira, M. I. T. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. M. Lemos, M. V. F. Lemos, E. C. Locali, M. A. Machado, A. M. B. N. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. M. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. M. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. D. Sena, C. Silva, R. F. de Souza, L. A. F. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. D. Tezza, M. T. dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 17.Drouin, F., J. Mélançon, and P. H. Roy. 2002. The IntI-like tyrosine recombinase of Shewanella oneidensis is active as an integron integrase. J. Bacteriol. 184:1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1993. PHYLIP: phylogeny inference package. University of Washington, Seattle.
- 19.Ginard, M., J. Lalucat, B. Tümmler, and U. Römling. 1997. Genome organization of Pseudomonas stutzeri and resulting taxonomic and evolutionary considerations. Int. J. Syst. Bacteriol. 47:132-143. [DOI] [PubMed] [Google Scholar]
- 20.Guasp, C., E. R. B. Moore, J. Lalucat, and A. Bennasar. 2000. Utility of internally transcribed 16S-23S rDNA spacer regions for the definition of Pseudomonas stutzeri genomovars and other Pseudomonas species. Int. J. Syst. E vol. Microbiol. 50:1629-1639. [DOI] [PubMed] [Google Scholar]
- 21.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 22.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 23.Hall, R. M., C. M. Collis, M.-J. Kim, S. R. Partridge, G. D. Recchia, and H. W. Stokes. 1999. Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870:68-80. [DOI] [PubMed] [Google Scholar]
- 24.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Doson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nature Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 26.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J.-M. Frere, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levesque, C., S. Brassard, J. Lapointe, and P. H. Roy. 1994. Diversity and relative strength of tandem promoters for the antibiotic resistance genes of several integrons. Gene 142:49-54. [DOI] [PubMed] [Google Scholar]
- 29.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez, E., and F. de la Cruz. 1990. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 9:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. H. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 32.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 34.Ploy, M.-C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 36.Recchia, G. D., H. W. Stokes, and R. M. Hall. 1994. Characterisation of specific and secondary recombination sites recognized by the integron DNA integrase. Nucleic Acids Res. 22:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rius, N., C. Fusté, C. Guas, J. Lalucat, and J. G. Lorén. 2001. Clonal population structure of Pseudomonas stutzeri, a species with exceptional genetic diversity. J. Bacteriol. 183:736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 39.Rowe-Magnus, D. A., A. M. Guerot, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. USA 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 43.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 44.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 45.Stokes, H. W., A. J. Holmes, B. S. Nield, M. P. Holley, K. M. H. Nevalainen, B. C. Mabbutt, and M. R. Gillings. 2001. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 67:5240-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 47.Vaisvila, R., R. D. Morgan, J. Posfai, and E. A. Raleigh. 2001. Discovery and distribution of super-integrons among pseudomonads. Mol. Microbiol. 42:587-601. [DOI] [PubMed] [Google Scholar]
- 48.Yeates, C., and M. R. Gillings. 1998. Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27:49-53. [Google Scholar]