Abstract
Pyrococcus furiosus utilizes starch and its degradation products, such as maltose, as primary carbon sources, but the pathways by which these α-glucans are processed have yet to be defined. For example, its genome contains genes proposed to encode five amylolytic enzymes (including a cyclodextrin glucanotransferase [CGTase] and amylopullulanase), as well as two transporters for maltose and maltodextrins (Mal-I and Mal-II), and a range of intracellular enzymes have been purified that reportedly metabolize maltodextrins and maltose. However, precisely which of these enzymes are involved in starch processing is not clear. In this study, starch metabolism in P. furiosus was examined by biochemical analyses in conjunction with global transcriptional response data for cells grown on a variety of glucans. In addition, DNA sequencing led to the correction of two key errors in the genome sequence, and these change the predicted properties of amylopullulanase (now designated PF1935*) and CGTase (PF0478*). Based on all of these data, a pathway is proposed that is specific for starch utilization that involves one transporter (Mal-II [PF1933 to PF1939]) and only three enzymes, amylopullulanase (PF1935*), 4-α-glucanotransferase (PF0272), and maltodextrin phosphorylase (PF1535). Their expression is upregulated on starch, and together they generate glucose and glucose-1-phosphate, which then feed into the novel glycolytic pathway of this organism. In addition, the results indicate that several hypothetical proteins encoded by three gene clusters are also involved in the transport and processing of α-glucan substrates by P. furiosus.
Pyrococcus furiosus is an obligately anaerobic, heterotrophic, hyperthermophilic archaeon that was isolated from a shallow hydrothermal vent near Vulcano Island, Italy (12). The organism grows optimally near 100°C and is able to utilize a range of sugars as a primary carbon source (4, 11, 28, 29). These include cellobiose (28, 43), laminarin (43), chitin (16), maltose (29), barley glucan (3), and starch (29). However, the means by which these compounds are metabolized by P. furiosus are not well understood. In attempting to define the pathway by which starch, and one of its degradation products, maltose, serve as carbon sources, there are two complicating factors. First, the genome sequence of P. furiosus (36) contains an array of genes that encode putative glycosyl hydrolases and glycosyl transferases. With respect to starch, they include five enzymes that potentially hydrolyze α-glucans. Two of them (PF0477 and PF1935) have putative signal peptides and are presumably extracellular, while the other three (PF0272, PF0478, and PF1939) lack a signal peptide and are presumably intracellular. They are annotated as amylases (PF0272 and PF0477), amylopullulanases (PF1934 and PF1935), and cyclodextrin glucanotransferases (PF0478). The question is, are all of these enzymes involved in degrading starch? The pathway for utilizing maltose and maltodextrins, primary products of starch breakdown, is well defined in bacteria such as Escherichia coli (5). The P. furiosus genome contains homologs of virtually all of the genes involved in this process. However, the extent to which the pathways of maltose and/or maltodextrin metabolism differ between a mesophilic bacterium and a hyperthermophilic archaeon is not known.
The second complicating factor in understanding starch and maltose metabolism in P. furiosus is that, while the biochemical properties of several potentially relevant enzymes have been determined, their true physiological functions are typically far from clear. For example, the recombinant form of a putative extracellular α-amylase (PF0477), which produces maltooligosaccharides from starch, has been characterized (9, 25), and an α-amylase closely related to PF0477 was characterized from the related species, P. woesei (13, 18, 27). In addition, an extracellular starch-hydrolyzing amylopullulanase has been characterized from P. furiosus and from the related hyperthermophile Thermococcus litoralis (7). An intracellular α-amylase (PF0272) has also been purified from P. furiosus (30). This is puzzling since most α-amylases are extracellular enzymes and, although PF0272 reportedly hydrolyzes starch (30), this was not the case with close homologs purified from T. litoralis (46) and T. kodakaraensis (42). In fact, it has been proposed (46) that T. litoralis and P. furiosus use different enzymatic strategies to metabolize maltose. An additional issue is the enzyme that hydrolyzes maltose. An intracellular α-glucosidase is thought to be the enzyme that converts maltose to glucose in P. furiosus (8) but a homolog is not present in any bacterial pathway of maltose metabolism (5). On the other hand, P. furiosus contains two homologs of the mal transporter found in E. coli, and although both have been characterized (29), the extent to which they are differentially regulated in the presence of various sugars has not been studied.
Many questions therefore remain concerning the specific processes by which P. furiosus and related hyperthermophiles detect, process, and utilize starch and maltose. We have previously shown (38) by using whole-genome DNA microarrays that P. furiosus undergoes dramatic changes in its global transcriptional pattern when maltose rather than peptides is used as the primary carbon and energy source. We have expanded this approach here to ascertain how the organism responds to utilizing one of six different sugars as its carbon source. We have utilized the global transcriptional profiling data in combination with biochemical analyses and DNA sequencing to definitively describe the pathway by which P. furiosus metabolizes starch.
MATERIALS AND METHODS
Bioinformatics.
Three programs were used for signal peptide prediction; SignalP (http://www.cbs.dtu.dk/services/SignalP/), SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuisignal/sosuisignal_submit.html), and TargetP (http://www.cbs.dtu.dk/services/TargetP/). Proteins were considered to have a signal sequence if two of the three programs found a signal peptide. Classification of glycosyl hydrolases was done by utilizing the CAZY (Carbohydrate-Active enzymes [http://afmb.cnrs-mrs.fr/CAZY/]) database, and InterPro (http://www.ebi.ac.uk/interpro/) was used for functional motif or domain searches. For prediction of putative transmembrane domains (TMDs), three different programs were used: TSEG (http://www.genome.ad.jp/SIT/tsegdir/tseg_exe.html), SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosui_submit.html), and PRED_TMR2 (http://biophysics.biol.uoa.gr/PRED-TMR2/input.html). The number of TMDs was defined as the average number of the results from the three different programs.
Growth of Pyrococcus furiosus on glucans.
P. furiosus (DSM 3638) was cultured anaerobically at 90°C on sea salts medium (SSM) with 1 g of yeast extract/liter as described previously (38). No elemental sulfur was added to the medium for any growth condition. Maltose, laminarin, cellobiose, starch, and chitin were obtained from Sigma (St. Louis, MO), and barley glucan was obtained from Megazyme (Wicklow, Ireland). Growth on chitin was performed as described previously (16). Each glucan was added to the medium at a final concentration of 3.3 g/liter prior to inoculation. A 60-ml batch culture was used to inoculate 400 ml of SSM medium supplemented with each primary carbon source in a 1-liter Pyrex bottle. P. furiosus was grown at 90°C until early log phase, after which 1 ml of sample was removed periodically for cell density enumeration by epifluorescence microscopy with acridine orange staining (20).
Microarray design.
DNA primers were designed for all 2,065 open reading frames originally identified in the genome sequence of P. furiosus DSM 3638 based on sequence information found online at The Institute for Genomic Research site (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database= ntpf01). DNA primers were designed with similar annealing temperatures and minimal hairpin formation using Genomax 2.0 (Informax, Bethesda, MD). All primers were obtained from IDTDNA (Coralville, IA). Probes were amplified in a PTC-100 Thermocycler (MJ Research, Inc., Waltham, MA) using Taq polymerase (Boehringer, Indianapolis, IN) and genomic DNA isolated from cell cultures according to previously published protocols (37). After amplification, PCR products were quantified, purified, and concentrated to a final concentration of 100 ng/μl by using QIAquick PCR purification kits (QIAGEN, Valencia, CA). Purified PCR products were resuspended in 50% dimethyl sulfoxide, randomized with respect to genome order, dispensed evenly into microarray printing plates (Genetix, London, United Kingdom), and printed onto ULTRAGAPS aminosilane-coated microscope slides (Corning, Corning, NY) with a QArray-Mini Arrayer (Genetix). The DNA was attached to the substrate by UV cross-linking in a GS GeneLinker UV Chamber (Bio-Rad, Hercules, CA) at 250 mJ and then incubated in an oven at 75°C for 2 h. The desiccated slides were stored at room temperature in the dark until use.
RNA isolation.
Cultures were inoculated with 0.5% (vol/vol) preculture. Cells were harvested in mid-exponential phase and centrifuged at 8,145 × g for 22 min at 4°C. Cells were then resuspended in ice-cold SSM, divided into aliquots in 2.0-ml Eppendorf tubes, and centrifuged at 13,800 × g for 30 s. The resulting pellets were resuspended in 85 μl of cold TE buffer. Cells were disrupted in 625 μl of lysis buffer (50 mM glucose, 10 mM EDTA, 25 mM Tris [pH 8.0]). The lysate was passed through a 20-gauge needle to shear the genomic DNA, after which 62.5 ml of 2 M sodium acetate (pH 5.2) was added. An equal volume of acidic phenol-chloroform (5:1) was then added, the aqueous phase was extracted, and the RNA was ethanol precipitated overnight at −20°C. After a wash with 70% ethanol, the RNA pellet was resuspended in 10 mM Tris (pH 8.5) and passed through fiber filter columns from the RNAqeuous RNA isolation kit (Ambion). Integrity of the RNA was confirmed by visual inspection on 1.0% native agarose gels, as well as by measuring the A260/A280 ratio with a DU 640 Spectrophotomer (Beckman Coulter, Inc., Fullerton, CA).
Generation of cDNA and hybridization.
First-strand cDNA synthesis from total RNA proceeded through Stratascript (Stratagene, La Jolla, CA) and random hexamer primers (Invitrogen Life Technologies, Carlsbad, CA). During first-strand cDNA synthesis, 5-[3-aminoallyl]-2′-deoxyuridine-5′-triphosphate (aa-dUTP; Sigma) was incorporated as previously described (40). The generated cDNA products were purified by using the QIAquick PCR purification kit (QIAGEN) and reacted with monoreactive cyanine-3 (Cy-3) and cyanine-5 (Cy-5) NHS-esters (Amersham Biosciences, Inc., Piscataway, NJ). Another round of purification was used to remove unincorporated dyes. Hybridizations and washes were performed as described previously (40). The slides were scanned by using the Scanarray 4000 scanner (Perkin-Elmer, Fremont, CA). Signal intensity data for all experiments were extracted by using Scanarray software (Perkin-Elmer).
Mixed model analyses.
Replication of treatments, arrays, dyes, and cDNA spots allowed the use of analysis of variance (ANOVA) models for data analysis (45). Loop designs were constructed, and reciprocal labeling was utilized for all samples so that dye effects could be estimated. Spot intensities obtained from Scanarray were imported directly into SAS (SAS Institute, Cary, NC), and low-intensity or low-quality spots flagged by Scanarray were removed before further analysis. After local background subtraction and log transformation of spot intensities, a linear normalization ANOVA model (45) was used to estimate global variation in the form of fixed (dye [D], treatment [T]) and random (array [A], block [B], and spot [S]) effects and random error using the model log2(yijklmn) = Ai + Dj + Tk + Ai(BlSm) + Ai(Bl) + ɛijklm. A gene-specific ANOVA model was then used to partition the remaining variation into gene-specific effects using the model rijklmn = μ + Ai + Dj +Tk + Ai(BlSm) + Ai(Bl) + γijklm. Information on the statistical significance of expression changes, fold changes, pairwise volcano plots, and hierarchical clustering for all of the genes included on the array is available online (www.che.ncsu.edu/extremophiles/page5.html).
Recombinant protein expression and purification.
Standard molecular biology techniques were performed as described previously (37). The gene corresponding to PF0272 was amplified by PCR from genomic DNA using the primer pairs 5′-CATGCCATGGGAGATAAAATTAACTTCATATTTG-3′ and 5′-ATAAGAATGCGGCCGCTCCCGAGGCTTCCTCAAATTTTAG-3′. An NcoI site was incorporated into the forward primer, and a NotI site was incorporated into the reverse primer (underlined) for cloning into pET-28b(+). A His-affinity tag of -AAALEHHHHHH was placed at the C terminus of PF0272, while the other proteins were expressed by using a modified form of the N-terminal His-tagged vector, pET24d. The vector was modified to have an N-terminal His tag (MAHHHHHHGS-) and BamHI and NotI sites were used for cloning (41). In brief, recombinant proteins were purified by using a column (5 ml) of Histrap nickel-affinity resin using an AKTA explorer (GE Healthcare, Piscataway, NJ). After application of the cell extract, the column was washed with five column volumes of 20 mM phosphate buffer (pH 7.0), containing 500 mM NaCl, 10 mM imidazole, 5% (vol/vol) glycerol, and 2 mM dithiothreitol. The absorbed protein was eluted with a gradient of 0 to 500 mM imidazole over 20 column volumes. The major protein peak was collected and concentrated to 10 ml by ultrafiltration (10-kDa cutoff, Amicon Ultrafilters; Millipore, Bedford, MA), diluted 15-fold in 20 mM Tris buffer (pH 8.0) containing 5% (vol/vol) glycerol and 2 mM dithiothreitol, and then applied to a column (5 ml) of Q Sepharose (GE Healthcare). The column was washed with 5 column volumes of the same buffer, and the bound protein was eluted with a 0 to 1 M NaCl gradient over 20 column volumes. The major protein was concentrated to 5 ml (10-kDa cutoff, Amicon) and applied to a 16/60 column of Superdex75 (GE Healthcare) equilibrated with the same Tris buffer. The major protein peak was collected and concentrated to a volume of 1 ml by ultrafiltration (10-kDa cutoff; Amicon). The protein concentration was estimated by the absorption at 280 nm using a calculated extinction coefficient. PF1256 was expressed by using the OverExpress C43(DE3) E. coli strain, which has been used for the production of membrane proteins (Avidis SA, St Beauzire, France). Cultures (5 liters) were grown in LB medium, and the expression of PF1256 was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an optical density at 600 nm of 1.0. Cells were harvested by centrifugation for 15 min at 10,000 × g and suspended in 100 ml of 10 mM imidazole buffer (pH 8.0), containing 20 mM sodium phosphate (pH 7.5), and 0.3 M NaCl. The cells were lysed by sonication for 5 min. Cell debris was removed by centrifugation for 30 min at 10,000 × g. The supernatant was then centrifuged at 125,000 × g for 2 h. The pellet was collected and then washed and centrifuged twice at 125,000 × g for 2 h using 20 mM phosphate buffer (pH 7.0) containing 300 mM NaCl and 2 mM dithiothreitol. The pellet was solubilized with β-octyl-d-glucopyranoside (OG, 0.8%) and stirred overnight at 4°C. The membrane fraction was recovered by centrifugation at 125,000 × g for 2 h. The recombinant protein was purified as described above for the other recombinant proteins except that all of the buffers contained OG (0.8%). DNA sequencing was performed by Integrated Biotech Laboratories (IBL) of the University of Georgia. The sequences of PF0478* and PF1935* have been deposited with respective accession numbers DQ192528 and DQ192527.
Native protein purification and enzyme assays.
P. furiosus was grown in a 20-liter custom fermentor at 95°C in a medium containing casein and maltose as the primary carbon sources (32). The cellular cytosolic fraction was obtained as described previously (32) and was loaded onto a column (5 by 12 cm) of DEAE-Sepharose Fast Flow equilibrated with buffer A (50 mM Tris-HCl [pH 7.8], containing 2 mM dithiothreitol and 2 mM sodium dithionite). The column was eluted with a linear gradient (1.2 liter) of 0 to 0.6 M NaCl in buffer A, and 50-ml fractions were collected. The fraction showing 4-α-glucanotransferase and α-glucosidase started to elute as 0.4 M NaCl was applied to the column. The active fractions were combined (250 ml) and loaded onto a column (5 by 10 cm) of hydroxyapatite (Bio-Rad) equilibrated with buffer A. The column was eluted with a 1.0-liter linear gradient (0 to 0.5 M potassium phosphate) in buffer A with a flow rate of 4 ml/min. The fraction showing 4-α-glucanotransferase and α-glucosidase started to elute as 0.25 M and 0.35 M potassium phosphate were applied to the column, respectively. The standard reaction mixture (300 μl) for the 4-α-glucanotransferase assay contained the appropriate substrates in 100 mM morpholinepropanesulfonic acid (MOPS; pH 7.0) and recombinant PF0272 (20 μg). The substrates used (and their final concentrations) were maltose (20 mM), maltotriose (20 mM), maltotetraose (10 mM), maltopentaose (10 mM), maltohexaose (10 mM), and maltoheptaose (10 mM). The reaction was carried out at 90°C for the desired time (see Results), and the products of the reaction were applied to silica gel plates (Kiesel gel 60 F254; Merck, Rahway, NJ) with n-butanol-ethanol-water (5:3:2 by volume) as the mobile phase. The plates were dipped into ethanol containing H2SO4 (10% [vol/vol]) to visualize the carbohydrate spots. The hydrolysis of p-nitrophenyl-glucopyranoside (PNPG), the typical artificial substrate for α-glucosidase, was measured at 90°C by evaluating the formation of p-nitrophenol and monitoring the absorbance at 405 nm. The reaction mixture (500 μl) contained 10 mM PNPG in 100 mM MOPS buffer (pH 7.0). The reaction was started by the addition of recombinant PF1256 (80 μg).
RESULTS AND DISCUSSION
Starch utilization.
The hydrolysis of starch by P. furiosus takes place in the extracellular medium and the resulting maltodextrins and maltose are taken up in to the cell (6, 7). A total of five genes in the P. furiosus genome have been annotated as encoding amylases or enzymes that hydrolyze α-glucans (PF0272, PF0477, PF0478, PF1935, and PF1939). The first question to be addressed is which of these enzymes are actually utilized by P. furiosus to degrade starch. Previous studies have shown that P. furiosus, when grown on starch or maltooligosaccharides, produces an extracellular amylopullulanase (7). The gene for this enzyme was reported to encode a protein of 853 amino acids (10), but the corresponding gene in the genome sequence (PF1935) encodes 985 amino acids (Fig. 1). Moreover, the PF1935 protein is still smaller than the proteins encoded by the corresponding genes in Pyrococcus abyssi (NCBI accession number PAB0122, 1,362 amino acids) and T. litoralis (BAC10983, 1,089 amino acids, see Fig. 1). In order to clarify this, the region adjacent to PF1934 was amplified by PCR and the PCR products were sequenced (data not shown). These results indicated an error in the original genome sequence between PF1934 and PF1935. The correction combines PF1935 and PF1934 into one continuous gene, now termed PF1935*. The calculated molecular mass of PF1935* (124,395 Da, without the signal peptide) more closely corresponds to the estimate of 110 kDa for the amylopullulanase originally purified as PF1935 (6). As shown in Fig. 2, as determined by microarray analysis, the expression of what was originally annotated as PF1935 (and PF1934) is dramatically upregulated when starch or maltose is the carbon source, rather than chitin, cellobiose, laminarin, or barley glucan. Similarly, the expression of what was originally annotated as PF1935 (and PF1934) increased almost fivefold when P. furiosus cells were grown on maltose rather than peptides (Table 1). Thus, all of the expression as well as the biochemical data are consistent with what is now designated PF1935* being the major enzyme responsible for starch degradation.
FIG. 1.
Schematic representations of the different lengths of the amylopullulanase (PF1935*) and the α-amylase (PF0478*) of P. furiosus. The length of T. litoralis amylopullulanase (NCBI accession number BAC10983) is included for comparison.
FIG. 2.
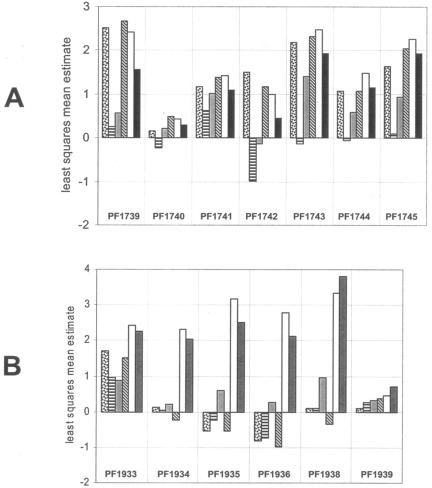
Relative expression patterns for the Mal-I (A) and Mal-II (B) operons on six different sugars. The height of each bar represents the least squares mean estimate for a given gene during growth on a given substrate, as calculated by mixed model analysis (see Materials and Methods). The average least-squares mean estimate across all included genes for growth on a single substrate is 0, where genes with positive least-squares mean estimates are above the average for that substrate and genes with negative least-squares mean estimates are below the average for that substrate. Differences between least-squares mean estimates for pairs of treatments represent log2-transformed fold changes. The sugars are represented as follows: laminarin (LA), dotted bar; barley glucan, horizontal bars; cellobiose, gray; chitin, diagonal bars; maltose, white; soluble starch, solid black. The expression of PF1937 in the Mal-II cluster is not included. PF1934 and PF1935 refer to the genes in the original genome annotation and not to PF1935*.
TABLE 1.
Regulation of key genes in P. furiosus potentially related to starch and maltose metabolism
| Gene | Annotation | Cellular locationa | Sugars that upregulate expressionb | Upregulation (fold) on maltose or peptidesc |
|---|---|---|---|---|
| PF0132 | α-Glucosidase | Intracellular | None | None |
| PF0272 | 4-α-Glucanotransferase | Intracellular | ST, MA | MA (26.1) |
| PF0312 | ADP-dependent glucokinase | Intracellular | None | MA (2.1) |
| PF0477 | α-Amylase | Extracellular | None | PEP (5.5) |
| PF0478* | Cyclomaltodextrin glucanotransferase | Extracellular | None | None |
| PF0588 | Phosphoglucose mutase | Intracellular | None | MA (1.6) |
| PF1256 | Homolog of Thermococcus hydrothermalis α-glucosidase | Membrane | None | None |
| PF1535 | Maltodextrin phosphorylase | Intracellular | ST, MA | MA (2.5) |
| PF1739 | Maltose/trehalose-binding protein (Mal-I) | Membrane anchored | None | MA (6.0) |
| PF1740 | Maltose/trehalose transport (Mal-I) | Membrane | None | MA (11.8) |
| PF1741 | Maltose/trehalose transport (Mal-I) | Membrane | None | MA (10.5) |
| PF1742 | Trehalose synthetase | Intracellular | None | MA (10.7) |
| PF1743 | Maltose/trehalose transcriptional regulator | Intracellular | None | None |
| PF1744 | Maltose transport ATPase (Mal-I) | Intracellular | None | None |
| PF1933 | Maltodextin transport ATPase (Mal-II) | Intracellular | None | MA (2.4) |
| PF1935*d | Amylopullulanase | Extracellular | ST, MA | MA (4.9) |
| PF1936 | Maltodextrin transport (Mal-II) | Membrane | ST, MA | MA (5.4) |
| PF1937 | Maltodextrin transport (Mal-II) | Membrane | None | MA (5.9) |
| PF1938 | Maltodextrin-binding protein (Mal-II) | Membrane anchored | ST, MA | MA (4.4) |
| PF1939 | Maltodextrinase/cyclodextrinase/pullulanase | Intracellular | None | None |
As predicted by analysis for membrane domains. Those listed as extracellular may be anchored to the membrane.
Regulation of expression by starch (ST) or maltose (MA) is indicated. See Fig. 2 and 3 for details of the extent of regulation. The expression of none of the genes was upregulated by any of the other four sugars (laminarin, barley glucan, cellobiose, and chitin).
Genes that are significantly upregulated (>2-fold, P < 0.01) by maltose (MA) or peptides (PEP). Data obtained from Schut et al. (38).
The microarray results for PF1935* correspond to those of the originally annotated PF1935. See the text for details.
What are the catalytic activities and physiological functions of the other four genes annotated as encoding α-glucan-hydrolyzing enzymes in P. furiosus? In the case of PF0477, this does indeed appear to be an extracellular amylase. The recombinant form has been characterized by two different groups (9, 25) and was shown to hydrolyze starch to maltooligosaccharides. However, although there have been several reports on the extracellular amylase activity of P. furiosus cells grown on starch or maltose (6, 7, 25, 27), the native enzyme (PF0477) has not been characterized. Accordingly, our DNA microarray analyses show that PF0477 is not expressed to any significant extent when starch, maltose, or any other of the six sugars are used as a carbon source (consequently, PF0477 could not be included in the mixed model analysis). The function of PF0477 is therefore not clear at present, although it does not appear to be involved in starch metabolism. In fact, its expression is dramatically upregulated when cells are grown on peptides (Table 1), a result that presumably correlates with the amylase activity measured in peptide-grown cells (38). Although the significance of this remains to be elucidated, one possibility is that when polysaccharides become available during peptide-dependent growth, PF0477 generates oligosaccharides, which then induce the corresponding pathways to convert the oligosaccharides to glucose.
The protein encoded by the adjacent gene, PF0478, is also annotated as an amylase. However, it is more similar to proteins that hydrolyze linear polysaccharides and synthesize cyclic forms. Specifically, PF0478 shows 64 and 39% sequence identity with the extracellular cyclomaltodextrin glucanotransferases (CGTases) of Thermococcus kodakaraensis KOD1 (35) and Thermococcus sp. strain B1001 (19), respectively. Surprisingly, PF0478 is not predicted to contain a signal sequence by the SignalP, TargetP, and SOSUI programs. Resequencing of PF0478 and the surrounding region revealed an additional base pair that was not present in the genome sequence, which results in an N-terminal extension to the protein. The corrected PF0478, termed PF0478*, now consists of 722 amino acids, which is 32 amino acids longer than the original (see Fig. 1). Moreover, the protein is now predicted to contain a signal sequence and, hence, to be an extracellular protein. However, PF0478* appears not to be directly involved in the metabolism of starch or maltose or any of the other sugars under study. As shown in Fig. 3, its expression is not affected by starch or maltose, and it appears to be unregulated. Similarly, its expression was not significantly affected when cells were grown on maltose rather than peptides (Table 1). The function of PF0478* is therefore unknown at present.
FIG. 3.
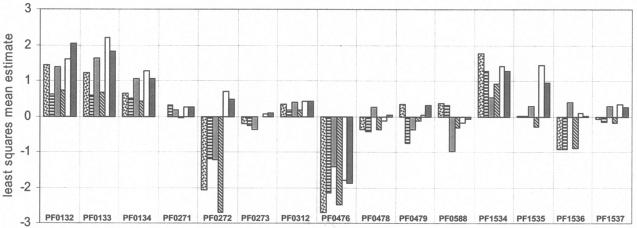
Relative expression patterns of additional genes potentially involved in starch and maltose metabolism. See the legend to Fig. 2 for details and bar definitions.
The recombinant version of the fourth amylase-like gene in P. furiosus, PF1939, has been characterized (47). The enzyme showed cyclodextrinase and pullulanase activity, as well as α-amylase activity. The function of PF1939 is also unclear, but it is not regulated by any of the six sugars, nor is it upregulated in maltose-grown relative to peptide-grown cells (Table 1), and it seems unlikely to be directly involved in starch or maltose degradation. We propose that PF1939 is the intracellular counterpart of PF0478* and hydrolyzes cyclodextrins generated by PF0478* to glucose and maltodextrins.
The product of the fifth amylase-like gene in P. furiosus, PF0272, has been purified from native biomass, and it was reported to be an intracellular amylase (30). As shown in Fig. 3, its expression is dramatically upregulated on starch and maltose relative to cellobiose, chitin, barley glucan, and laminarin. In fact, its expression is one of the most dramatically upregulated when cells are grown on maltose rather than peptides (Table 1). However, as discussed below, PF0272 does not appear to catalyze starch degradation, even though it does play a key role in starch and maltodextrin metabolism.
All of the evidence suggests, therefore, that a single enzyme, an amylopullulanase (PF1935*), is the major starch-hydrolyzing enzyme in P. furiosus. This is similar to the situation in bacteria that can use starch as a carbon source for growth, such as Klebsiella pneumoniae (39). On the other hand, such an enzyme is absent from E. coli, since this organism cannot utilize starch.
Uptake of maltodextrins and maltose.
P. furiosus contains two homologs of the so-called mal transporter that transports maltose in E. coli. One is the Mal-I transporter (PF1739 to PF1741, PF1744), which in P. furiosus has been shown to recognize and transport maltose and trehalose but not maltotriose or longer maltooligosaccharides (29). The second transporter (Mal-II; PF1933, PF1936 to PF1938) is specific for maltooligosaccharides, but it does not transport maltose and trehalose. Accordingly, we found that most of the genes of the Mal-II cluster are specifically upregulated on maltose and starch (PF1934 to PF1936 and PF1938) but not on any of the other four sugars (Fig. 2B). PF1933 is also induced by starch and maltose but less specifically, and PF1937 was not expressed at all on the six sugars tested. The genes of Mal-I are also upregulated on the sugars, laminarin, cellobiose, and chitin but not on barley glucan (Fig. 2A). Mal-II is therefore the major transporter of starch degradation products, whereas Mal-I is an additional transporter for maltose (and trehalose).
Although Mal-II is induced only by maltose and starch, consistent with it being a specific maltodextrin transporter, Mal-I is induced by every sugar investigated, although it only transports maltose and trehalose. This type of transporter is relatively rare, since the only other known examples of an ABC transporter able to transport maltose and trehalose are found in T. litoralis (21) and Sinorhizobium meliloti Agl (44). Mal-I cannot transport sucrose, while the S. meliloti Agl system transports sucrose, as well as maltose and trehalose.
Degradation of maltodextrins and maltose.
The two major intracellular enzymes for the degradation of maltose and maltodextrins in E. coli are MalQ (4-α-glucanotransferase) and MalP (maltodextrin phosphorylase). Both of these proteins have homologs in P. furiosus: PF0272 and PF1535, respectively. As mentioned above, the expression of PF0272 is one of the most dramatically upregulated within the genome when cells are grown on maltose rather than peptides (Table 1). PF0272 is also highly upregulated on maltose and starch relative to the other four sugars tested (Fig. 3). The same is true for PF1535 (Fig. 3), although it is less upregulated in maltose-grown versus peptide-grown cells (2.5-fold) than is PF0272 (26-fold).
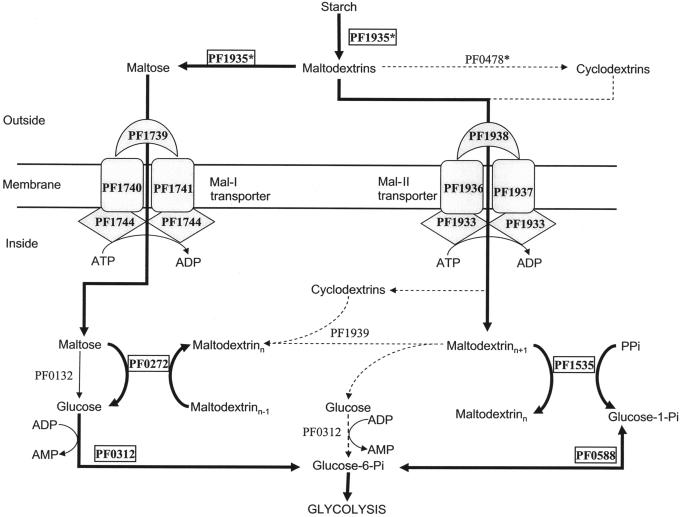
Thus, as shown in Fig. 4, we propose that PF0272 generates glucose from maltodextrins and maltose, while PF1535 converts maltodextrins to glucose-1-phosphate. Note that this is contrary to what was previously proposed for P. furiosus by us (8) and others (46). The previous pathway was based in part on the assumption that PF0272 functions as an intracellular α-amylase (30, 31). The protein does show high sequence similarity to the amylase from an anaerobic bacterium (46), and both the native and recombinant forms of the P. furiosus enzymes were reported to be able to convert starch to glucose, maltose, and maltodextrins. Since these data are not consistent with the proposed role of PF0272 (Fig. 4), we also cloned the gene and the obtained the recombinant protein. As shown in Fig. 5A (lane 1), recombinant PF0272 is not able to hydrolyze starch to maltooligosaccharides under our experimental conditions. However, it did exhibit 4-α-glucanotransferase activity (Fig. 5A) in the presence of glucose and maltose (lanes 2 and 3), although not with trehalose, cellobiose, or sucrose. Therefore, we propose that PF0272 is not an α-amylase but a 4-α-glucanotransferase. In further support of this assignment, the protein shows between 64 and 69% sequence similarity with the 4-α-glucanotransferases of T. kodakaraensis and T. litoralis, respectively (24, 42).
FIG. 4.
Proposed pathway for starch and maltose metabolism in P. furiosus. The bold arrows indicate the major pathway. Dotted arrows indicate possible minor or alternative pathways.
FIG. 5.
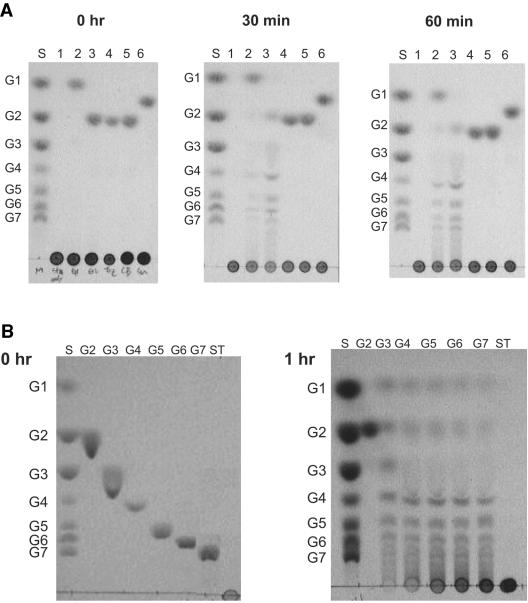
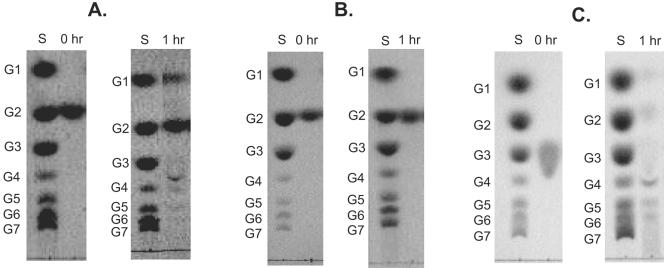
Substrate specificity of recombinant PF0272 (4-α-glucanotransferase). Starch hydrolysis (A) and 4-α-glucanotransferase (B) activities were determined by using thin-layer chromatograms after recombinant PF0272 (20 μg) was incubated at 90°C in 300 μl of 100 mM MOPS (pH 7.0) for the indicated times with the following substrates. (A) Each reaction contained 1% (wt/vol) soluble starch with the following additions: lane 1, none; lane 2, 20 mM glucose; lane 3, 20 mM maltose; lane 4, 20 mM trehalose; lane 5, 20 mM cellobiose; and lane 6, 20 mM sucrose. (B) Lanes: 1, 20 mM maltose (G2); 2, 20 mM maltotriose (G3); 3, 10 mM maltotetraose (G4); 4, 10 mM maltopentaose (G5); 5, 10 mM maltohexaose and 10 mM maltoheptaose (G7); and 6, 1.0% (wt/vol) starch (ST). A total of 2 μl was added to each lane. Lane S contains the standard sugars.
The question arises as to the substrate range of PF0272 and its mechanism of action in vivo. As shown in Fig. 5B, the enzyme is not able to hydrolyze maltose directly, although it will use maltose, as well as glucose, as a glucan acceptor (Fig. 5A). In this respect PF0272 resembles MalQ of E. coli. Mutants of E. coli lacking this gene cannot grow on maltose since the smallest substrate utilized by MalQ is maltotriose (34). Boos and Shuman (5) suggested that the primers needed for E. coli MalQ are bound to the purified enzyme, and thus it is able to process maltose, even though it needs a glucan acceptor. To investigate whether this was also the case for P. furiosus, a cell extract of cells grown in the presence of maltose was found to exhibit 4-α-glucanotransferase activity even with maltose as the substrate (Fig. 6). However, the maltose-dependent activity was lost after the protein responsible for the activity was subjected to two steps of column chromatography, a finding consistent with the loss of primers during the purification (Fig. 6). The nature of the glucan that serves as the primer for PF0272 in vivo is not known. Possible sources of the primer are precursors or breakdown products of glycogen or other endopolysaccharides (17).
FIG. 6.
Substrate specificity of native 4-α-glucanotransferase. Thin-layer chromatograms of cytoplasmic fraction (A) and, fractions (B and C) exhibiting 4-α-glucanotransferase activity after two chromatography steps (DEAE-Sepharose and hydroxyapatite, see Materials and Methods). The fractions were incubated with 20 mM maltose (A and B) or 20 mM maltotriose (C) in 100 mM MOPS (pH 7.0) at 90°C for the indicated times. Lane S contains standard sugars.
In contrast to PF0272 and E. coli MalQ, the 4-α-glucanotransferase of T. litoralis was reported to convert maltose to glucose and maltodextrins (46). The structure of the T. litoralis enzyme was recently solved (22), and it consists of two domains. The catalytic domain I shows high sequence similarity (79%) to that of PF0272, but the other domain (of unknown function) is much less similar (45%). It is not clear whether the apparent difference in substrate specificity between the P. furiosus and T. litoralis enzymes is related to the differences in the nature of their second domains.
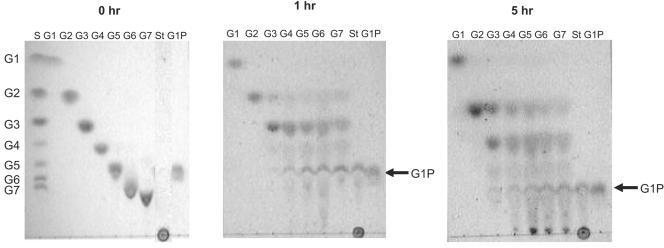
We propose that the maltodextrins generated by PF0272 are converted to glucose-1-phosphate by PF1535, which functions as a maltodextrin phosphorylase, using inorganic phosphate as the phosphate donor (see Fig. 4). The PF1535 protein shows 70% identity with the MalP of T. litoralis (46). As shown in Fig. 7, PF1535 can use G4 or longer maltodextrins as substrates for producing glucose-1-phosphate, while E. coli MalP uses G5 or longer as substrates. Glucose-1-phosphate is converted to glucose-6-phosphate by phosphoglucose mutase (PF0588), and the glucose produced by PF0272 is converted to glucose-6-phosphate by PF0312, a novel ADP-dependent glucokinase (26). The expression of both PF0312 and PF0588 is slightly upregulated (1.6- and 2.1-fold, respectively) when cells are grown on maltose rather than peptides, whereas these genes are not differentially regulated to any significant extent by any of the six sugars (Table 1 and Fig. 3).
FIG. 7.
Substrate specificity of recombinant PF 1535 (maltodextrin phosphorylase). Thin-layer chromatograms are shown after recombinant PF1535 (20 μg) was incubated in 100 mM potassium phosphate buffer (pH 7.0) with 20 mM glucose (G1), 20 mM maltose (G2), 20 mM maltotriose (G3), 10 mM maltotetraose (G4), 10 mM maltopentaose (G5), 10 mM maltohexaose and 10 mM maltoheptaose (G7), and 1.0% starch (St) for the indicated periods. Glucose-1-phosphate (G1P) was also applied as a standard and its point of migration is indicated by the arrow.
The pathway shown in Fig. 4 does not include a role for an α-glucosidase, the enzyme that converts maltose to two glucose molecules. Most α-glucosidases characterized thus far belong to glycosyl hydrolase family 4 (GH4) or GH31, but the P. furiosus genome lacks a hydrolase of this type. However, Costantino et al. (8) purified from P. furiosus biomass a protein that showed α-glucosidase activity, and this was identified by using N-terminal sequencing as the product of gene PF0132. This protein does not show sequence similarity to any known α-glucosidase or any characterized protein and represents a new family of glycosyl hydrolases. The physiological function of this protein is unclear, however, since the N-terminal sequence of the sucrose α-glucohydrolase (invertase) that was purified from P. furiosus (2) also corresponds to PF0132. A specific role for PF0132 in starch and maltose metabolism seems unlikely since it appears to be constitutively expressed (Fig. 3). It is not differentially regulated by any of the six sugars studied, including maltose, nor does it show a change in regulation when maltose rather than peptides are the primary carbon source (Table 1). Moreover, the pathway shown in Fig. 4 does not specifically require an enzyme that produces glucose from maltose. PF0132 seems to be expressed at a high level regardless of the carbon source, suggesting that it is an essential enzyme that has an as-yet-unknown role in general carbohydrate metabolism regardless of the growth substrate.
Alternative pathways for starch and maltose metabolism.
Although the pathway for starch to glucose conversion that is shown in Fig. 4 is consistent with the biochemical and transcriptional data, there are several other enzymes annotated in the genome that might a priori be expected to play a role in starch metabolism. For example, the P. furiosus genome contains a gene annotated as encoding a cyclomaltodextrin glucanotransferase (CGTase; PF0478*). This catalyzes the formation of cyclodextrins from starch and shares high sequence similarity with amylases (23) but appears to be expressed at low levels and is not regulated by any starch or maltose (Table 1). Similarly, as mentioned above, the intracellular cyclomaltodextrinase in P. furiosus (PF1939) is not regulated by maltose and starch (Table 1). Hashimoto et al. (19) have proposed that Thermococcus sp. strain B1001 possesses a unique starch utilization pathway, which includes extracellular synthesis, transmembrane uptake, and intracellular degradation of cyclodextrins. P. furiosus has homologs (Mal-II transporter, PF1939, and PF0478*) of the genes involved in cyclodextrin metabolism in Thermococcus sp. strain B1001. However, we conclude that cyclodextrin formation is not the major pathway of starch degradation in P. furiosus because these enzymes are not differentially regulated in response to starch or maltose with respect to other sugars or to peptides.
Finally, in addition to the α-glucosidase discussed above (PF0132), the P. furiosus genome contains another putative α-glucosidase encoded by PF1256. This shows high sequence identity (43%) to the α-glucosidase that was recently identified in Thermococcus hydrothermalis by functional complementation of a Saccharomyces cerevisiae mal11 mutant (15). Surprisingly, no similarity was found between the T. hydrothermalis enzyme and any known glycosyl hydrolase. However, our analyses indicate that PF1256 and T. hydrothermalis α-glucosidase are membrane bound since they are predicted to contain four and three transmembrane domains, respectively (predicted by SOSUI). This proved correct in the case of PF1256 because the recombinant form was associated with the membrane fraction of E. coli (data not shown). The partially purified recombinant protein did not hydrolyze maltose or trehalose, nor did it exhibit glycosyl hydrolase activity (using p-nitrophenyl-α-d-glucopyranoside as the substrate). Regardless of function, it seems unlikely that PF1256 is involved in starch or maltose metabolism since it appears to be expressed at a low level and it was not differentially regulated by any one or more of the six sugars nor during growth on maltose and peptides (Table 1).
Additional genes whose expression is differentially regulated on starch and maltose.
Although the major pathway shown in Fig. 4 is sufficient to convert extracellular starch to intracellular glucose-6-phosphate, the DNA microarray analyses indicate that there are several other gene products that are involved in the metabolism of starch and maltose. Those differentially upregulated on one or both sugars relative to laminarin, barley glucan, cellobiose, and chitin are listed in Table 2. They include three gene clusters, each containing two genes. There is evidence that all three clusters may be involved in binding and transporting starch and/or maltose. In fact, one of the clusters, PF1109 and PF1110, represents the most highly upregulated genes on both starch and maltose. Both genes are annotated as encoding hypothetical proteins, but both are predicted by InterPro to contain carbohydrate binding family 9 (CBD9) domains. The CBD9 domain can bind cellulose, xylan, and a range of soluble di- and monosaccharides and is found in cellulose- and xylan-degrading enzymes (33). However, in P. furiosus these two genes are highly downregulated on cellobiose and barley glucan. One of these proteins (PF1109) is predicted to be membrane associated; however, a potential role in α-glucan metabolism remains unclear. The second gene cluster contains PF1748 and PF1749, which are both predicted to encode transmembrane permeases (ABC transporters). These are also upregulated specifically on maltose and starch and, apparently coincidentally, are adjacent, but not linked, to the Mal-I operon. The third gene cluster contains PF0261 and PF0262, and this is upregulated by maltose but not starch. Both genes are predicted to be membrane associated, and PF0262 is a member of the AcrB family of multidrug efflux transporters.
TABLE 2.
Additional genes significantly upregulated by maltose and/or starcha
| Gene | Annotation | Predicted cellular location | Upregulationb |
|---|---|---|---|
| PF0126 | DNA repair protein rad25 | Intracellular | MA |
| PF0133 | Conserved hypothetical protein | Intracellular | MA ST |
| PF0261 | Conserved hypothetical protein | Extracellular/membrane anchored | MA |
| PF0262 | Multidrug efflux transporter | Transmembrane | MA |
| PF1109 | Hypothetical protein | Extracellular/membrane anchored | MA ST |
| PF1110 | Hypothetical protein | Intracellular | MA ST |
| PF1468 | Hypothetical protein | Intracellular | ST |
| PF1748 | Permease protein, ABC transporter | Transmembrane | MA ST |
| PF1749 | Permease protein, ABC transporter | Transmembrane | MA ST |
| PF2056 | SSU ribosomal protein S15P | Intracellular | MA ST |
| PF2061 | Putative ABC transporter | Intracellular | ST |
These data therefore suggest that the recognition and transport of starch and maltose is not simply the responsibility of the Mal-I and Mal-II systems and that other proteins are involved. On the other hand, from the transcriptional analyses, there appears to be no need to invoke anything but a very simple pathway for the utilization of starch, maltodextrins and maltose. This involves only three enzymes, amylopullulanase (PF1935*), 4-α-glucanotransferase (PF0272), and maltodextrin phosphorylase (PF1535). These generate glucose and glucose-1-phosphate, which then feed into the novel glycolytic pathway of this organism (Fig. 4). Close homologs of these enzymes are also found in Thermococcus kodakaraensis, the only other starch-metabolizing, hyperthermophilic archaeon whose genome sequence is available (1, 14). Current efforts with P. furiosus are focused on the roles of open reading frames that respond to other α-glucan substrates to determine their possible roles in carbohydrate utilization, in addition to deciphering the mechanisms of β-glucan processing by this archaeon.
Acknowledgments
This study was supported in part by grants from the National Science Foundation (BES-0317886 to R.M.K., BES-0317911 and MCB-0129841 to M.W.W.A.) and the Department of Energy (DE-FG02-96ER20219 to R.M.K. and DE-FG05-95ER20175 to M.W.W.A.).
REFERENCES
- 1.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badr, H. R., K. A. Sims, and M. W. W. Adams. 1994. Purification and characterization of sucrose α-glucohydrolase (invertase) from the hyperthermophilic archaeon Pyrococcus furiosus. Syst. Appl. Microbiol. 17:1-6. [Google Scholar]
- 3.Bauer, M. W., L. E. Driskill, W. Callen, M. A. Snead, E. J. Mathur, and R. M. Kelly. 1999. An endoglucanase, EglA, from the hyperthermophilic archaeon Pyrococcus furiosus hydrolyzes β-1,4 bonds in mixed-linkage (1→3),(1→4)-β-d-glucans and cellulose. J. Bacteriol. 181:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, M. W., S. B. Halio, and R. M. Kelly. 1996. Proteases and glycosyl hydrolases from hyperthermophilic microorganisms. Adv. Protein Chem. 48:271-310. [DOI] [PubMed] [Google Scholar]
- 5.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, S. H., H. R. Costantino, and R. M. Kelly. 1990. Characterization of amylolytic enzyme-activities associated with the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl. Environ. Microbiol. 56:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, S. H., and R. M. Kelly. 1993. Characterization of amylolytic enzymes, having both alpha-1,4 and alpha-1,6 hydrolytic activity, from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis. Appl. Environ. Microbiol. 59:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costantino, H. R., S. H. Brown, and R. M. Kelly. 1990. Purification and characterization of an alpha-glucosidase from a hyperthermophilic archaebacterium, Pyrococcus furiosus, exhibiting a temperature optimum of 105 to 115°C. J. Bacteriol. 172:3654-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, G., C. Vieille, A. Savchenko, and J. G. Zeikus. 1997. Cloning, sequencing, and expression of the gene encoding extracellular alpha-amylase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl. Environ. Microbiol. 63:3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, G., C. Vieille, and J. G. Zeikus. 1997. Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl. Environ. Microbiol. 63:3577-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driskill, L. E., K. Kusy, M. W. Bauer, and R. M. Kelly. 1999. Relationship between glycosyl hydrolase inventory and growth physiology of the hyperthermophile Pyrococcus furiosus on carbohydrate-based media. Appl. Environ. Microbiol. 65:893-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 13.Frillingos, S., A. Linden, F. Niehaus, C. Vargas, J. J. Nieto, A. Ventosa, G. Antranikian, and C. Drainas. 2000. Cloning and expression of alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in the moderately halophilic bacterium Halomonas elongata. J. Appl. Microbiol. 88:495-503. [DOI] [PubMed] [Google Scholar]
- 14.Fukui, T., H. Atomi, T. Kanai, R. Matsumi, S. Fujiwara, and T. Imanaka. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galichet, A., and A. Belarbi. 1999. Cloning of an alpha-glucosidase gene from Thermococcus hydrothermalis by functional complementation of a Saccharomyces cerevisiae mal11 mutant strain. FEBS Lett. 458:188-192. [DOI] [PubMed] [Google Scholar]
- 16.Gao, J., M. W. Bauer, K. R. Shockley, M. A. Pysz, and R. M. Kelly. 2003. Growth of hyperthermophilic archaeon Pyrococcus furiosus on chitin involves two family 18 chitinases. Appl. Environ. Microbiol. 69:3119-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruyer, S., E. Legin, C. Bliard, S. Ball, and F. Duchiron. 2002. The endopolysaccharide metabolism of the hyperthermophilic archaeon Thermococcus hydrothermalis: polymer structure and biosynthesis. Curr. Microbiol. 44:206-211. [DOI] [PubMed] [Google Scholar]
- 18.Grzybowska, B., P. Szweda, and J. Synowiecki. 2004. Cloning of the thermostable alpha-amylase gene from Pyrococcus woesei in Escherichia coli: isolation and some properties of the enzyme. Mol. Biotechnol. 26:101-110. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto, Y., T. Yamamoto, S. Fujiwara, M. Takagi, and T. Imanaka. 2001. Extracellular synthesis, specific recognition, and intracellular degradation of cyclomaltodextrins by the hyperthermophilic archaeon Thermococcus sp. strain B1001. J. Bacteriol. 183:5050-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horlacher, R., K. B. Xavier, H. Santos, J. DiRuggiero, M. Kossmann, and W. Boos. 1998. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 180:680-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura, H., S. Fushinobu, M. Yamamoto, T. Kumasaka, B. S. Jeon, T. Wakagi, and H. Matsuzawa. 2003. Crystal structures of 4-alpha-glucanotransferase from Thermococcus litoralis and its complex with an inhibitor. J. Biol. Chem. 278:19378-19386. [DOI] [PubMed] [Google Scholar]
- 23.Janecek, S., E. A. MacGregor, and B. Svensson. 1995. Characteristic differences in the primary structure allow discrimination of cyclodextrin glucanotransferases from alpha-amylases. Biochem. J. 305(Pt. 2):685-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon, B. S., H. Taguchi, H. Sakai, T. Ohshima, T. Wakagi, and H. Matsuzawa. 1997. 4-α-Glucanotransferase from the hyperthermophilic archaeon Thermococcus litoralis: enzyme purification and characterization, and gene cloning, sequencing, and expression in Escherichia coli. Eur. J. Biochem. 248:171-178. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen, S., C. E. Vorgias, and G. Antranikian. 1997. Cloning, sequencing, characterization, and expression of an extracellular alpha-amylase from the hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli and Bacillus subtilis. J. Biol. Chem. 272:16335-16342. [DOI] [PubMed] [Google Scholar]
- 26.Kengen, S. W., J. E. Tuininga, F. A. de Bok, A. J. Stams, and W. M. de Vos. 1995. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Biol. Chem. 270:30453-30457. [DOI] [PubMed] [Google Scholar]
- 27.Koch, R., A. Spreinat, K. Lemke, and G. Antranikian. 1991. Purification and properties of a hyperthermoactive alpha-amylase from the archaeobacterium Pyrococcus woesei. Arch. Microbiol. 155:572-578. [Google Scholar]
- 28.Koning, S. M., M. G. Elferink, W. N. Konings, and A. J. Driessen. 2001. Cellobiose uptake in the hyperthermophilic archaeon Pyrococcus furiosus is mediated by an inducible, high-affinity ABC transporter. J. Bacteriol. 183:4979-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koning, S. M., W. N. Konings, and A. J. Driessen. 2002. Biochemical evidence for the presence of two alpha-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1:19-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laderman, K. A., K. Asada, T. Uemori, H. Mukai, Y. Taguchi, I. Kato, and C. B. Anfinsen. 1993. Alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus: cloning and sequencing of the gene and expression in Escherichia coli. J. Biol. Chem. 268:24402-24407. [PubMed] [Google Scholar]
- 31.Laderman, K. A., B. R. Davis, H. C. Krutzsch, M. S. Lewis, Y. V. Griko, P. L. Privalov, and C. B. Anfinsen. 1993. The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J. Biol. Chem. 268:24394-24401. [PubMed] [Google Scholar]
- 32.Ma, K., R. Weiss, and M. W. Adams. 2000. Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182:1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Notenboom, V., A. B. Boraston, D. G. Kilburn, and D. R. Rose. 2001. Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry 40:6248-6256. [DOI] [PubMed] [Google Scholar]
- 34.Palmer, T. N., B. E. Ryman, and W. J. Whelan. 1976. The action pattern of amylomaltase from Escherichia coli. Eur. J. Biochem. 69:105-115. [DOI] [PubMed] [Google Scholar]
- 35.Rashid, N., J. Cornista, S. Ezaki, T. Fukui, H. Atomi, and T. Imanaka. 2002. Characterization of an archaeal cyclodextrin glucanotransferase with a novel C-terminal domain. J. Bacteriol. 184:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, and D. M. Dunn. 2001. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Schut, G. J., S. D. Brehm, S. Datta, and M. W. Adams. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185:3935-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz, M. 1996. The maltose regulon, p. 1482-1502. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 40.Shockley, K. R., D. E. Ward, S. R. Chhabra, S. B. Conners, C. I. Montero, and R. M. Kelly. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 69:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Story, S. V., C. Shah, F. E. Jenney, Jr., and M. W. Adams. 2005. Characterization of a novel zinc-containing, lysine-specific aminopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 187:2077-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tachibana, Y., S. Fujiwara, M. Takagi, and T. Imanaka. 1997. Cloning and expression of the 4-alpha-glucanotransferase gene from the hyperthermophilic archaeon Pyrococcus sp. KOD1, and characterization of the enzyme. J. Ferment. Bioeng. 83:540-548. [Google Scholar]
- 43.Voorhorst, W. G., Y. Gueguen, A. C. Geerling, G. Schut, I. Dahlke, M. Thomm, J. van der Oost, and W. M. de Vos. 1999. Transcriptional regulation in the hyperthermophilic archaeon Pyrococcus furiosus: coordinated expression of divergently oriented genes in response to beta-linked glucose polymers. J. Bacteriol. 181:3777-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willis, L. B., and G. C. Walker. 1999. A novel Sinorhizobium meliloti operon encodes an alpha-glucosidase and a periplasmic-binding-protein-dependent transport system for alpha-glucosides. J. Bacteriol. 181:4176-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolfinger, R. D., G. Gibson, E. D. Wolfinger, L. Bennett, H. Hamadeh, P. Bushel, C. Afshari, and R. S. Paules. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 46.Xavier, K. B., R. Peist, M. Kossmann, W. Boos, and H. Santos. 1999. Maltose metabolism in the hyperthermophilic archaeon Thermococcus litoralis: purification and characterization of key enzymes. J. Bacteriol. 181:3358-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, S. J., H. S. Lee, C. S. Park, Y. R. Kim, T. W. Moon, and K. H. Park. 2004. Enzymatic analysis of an amylolytic enzyme from the hyperthermophilic archaeon Pyrococcus furiosus reveals its novel catalytic properties as both an alpha-amylase and a cyclodextrin-hydrolyzing enzyme. Appl. Environ. Microbiol. 70:5988-5995. [DOI] [PMC free article] [PubMed] [Google Scholar]