Abstract
Enterococci are opportunistic pathogens and among the leading causes of nosocomial infections. Enterococcus faecalis, the dominant species among infection-derived isolates, has recently been recognized as capable of forming biofilms on abiotic surfaces in vitro as well as on indwelling medical devices. A few bacterial factors known to contribute to biofilm formation in E. faecalis have been characterized. To identify additional factors which may be important to this process, we utilized a Tn917-based insertional mutagenesis strategy to generate a mutant bank in a high-biofilm-forming E. faecalis strain, E99. The resulting mutant bank was screened for mutants exhibiting a significantly reduced ability to form biofilms. One mutant, P101D12, which showed greater than 70% reduction in its ability to form biofilms compared to the wild-type parent, was further characterized. The single Tn917 insertion in P101D12 was mapped to a gene, bee-2, encoding a probable cell wall-anchored protein. Sequence information for the region flanking bee-2 revealed that this gene was a member of a locus (termed the bee locus for biofilm enhancer in enterococcus) comprised of five genes encoding three putative cell wall-anchored proteins and two probable sortases. Contour-clamped homogeneous electric field gel and Southern hybridization analyses suggested that the bee locus is likely harbored on a large conjugative plasmid. Filter mating assays using wild-type E99 or mutant P101D12 as a donor confirmed that the bee locus could transfer conjugally at high frequency to recipient E. faecalis strains. This represents the first instance of the identification of a mobile genetic element conferring biofilm-forming property in E. faecalis.
Biofilm formation is a dynamic process involving the attachment of bacteria to a biotic or abiotic surface followed by growth and maturation (2). Structurally, biofilms consist of single- or multispecies microbial communities encased in an extracellular polymeric matrix, which is mainly composed of carbohydrates. Existence as a biofilm confers a significant survival advantage to bacteria, increasing their resistance to stressful environmental conditions (7), rendering them severalfold more resistant to antimicrobial agents (4), and assisting them in evading the host immune system more effectively (45) than the planktonic cells. Thus, while an ability to form biofilms in itself may not be considered a virulence trait, since many nonpathogenic bacteria form biofilms, existence as biofilms has been found to facilitate survival and persistence of pathogens in the host (11). The Centers for Disease Control now estimates that 65% of human infections may be biofilm related (27). The pathogenesis of diseases like infective endocarditis, infectious kidney stones, and lung infections in cystic fibrosis has been attributed to biofilms (24).
Enterococci play dual roles in human ecology. They exist as commensals in the gastrointestinal tract or manifest themselves as opportunistic pathogens, and they are among the leading causes of nosocomial infections, causing bacteremia, urinary tract infections, and endocarditis (9, 30). Enterococci have been found to form biofilms on several medical devices implanted in patients, such as central venous catheters, urinary catheters, intrauterine devices, and prosthetic heart valves (3). A few factors contributing to the process of enterococcal biofilm formation have been identified, including the cell surface-localized enterococcal surface protein Esp (36, 41), the two-component quorum-sensing signal transduction system Fsr (12, 26), the secreted metalloprotease GelE (12, 16, 21), the sugar-binding transcriptional regulator BopD (13), and an autolysin and the enterococcal polysaccharide antigen Epa (21).
In an attempt to identify additional factors that may influence the process of biofilm formation in Enterococcus faecalis, a Tn917 mutant bank was generated in a high-biofilm-forming E. faecalis strain, E99. The region flanking the single Tn917 insertion in eight mutants which showed a significant reduction in the ability to form biofilms was characterized, and the transposon was found to be inserted within the open reading frames (ORFs) or the intergenic regions of a novel gene cluster, which we designate as the bee (biofilm enhancer in enterococcus) locus. In this study, we further characterized mutant P101D12, in which bee-2 was inactivated by a single Tn917 insertion. We show that the bee locus is likely carried on a large conjugative plasmid and is transferred at high frequency to recipient E. faecalis strains by a conjugal mating process, resulting in the enhancement of biofilm formation by the transconjugants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. faecalis strain E99 was isolated from the urine of a patient at the Veterans Administration hospital in Arkansas (provided by K. T. Madhusudhan). OG1RF (5) and JH2SS (14, 42), plasmid-free E. faecalis strains, were used as recipients for the filter mating assays. Escherichia coli XL1-Blue (Stratagene, La Jolla, CA) was used as a host for plasmid purifications. A list of all of the plasmids and E. faecalis strains used in this study is shown in Table 1. E. faecalis was cultured in Trypticase soy broth (TSB) supplemented with 0.5% glucose. Antibiotics (Sigma, St. Louis, MO) used for selection included kanamycin (25 μg/ml) for E99, kanamycin (1,000 μg/ml) and erythromycin (10 μg/ml) for E99(pTV1-OK), and erythromycin (10 μg/ml) for all of the Tn917 mutants. Strain OG1RF was selected using rifampin (25 μg/ml) and fusidic acid (10 μg/ml), whereas streptomycin (250 μg/ml) and spectinomycin (250 μg/ml) were used for JH2SS. Spectinomycin at 500 μg/ml for E. faecalis and 150 μg/ml for E. coli was used for the selection of strains containing pAT28-based constructs in the complementation assay. No significant differences were found in the rates of growth for the mutant and transconjugants compared to the growth rate of the wild-type strain in medium with or without appropriate antibiotics.
TABLE 1.
E. faecalis strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. faecalis strains | ||
| E99 | Clinical isolate | This study |
| P101D12 | E99 with a Tn917 insertion in bee-2, Ermr | This study |
| BLG1 | P101D12 complemented with pBLG1 expressing bee-2, Spcr | This study |
| BLG2 | P101D12 complemented with pBLG2 expressing bee-3, Spcr | This study |
| BLG12 | P101D12 complemented with pBLG12 expressing bee-2 and bee-3, Spcr | This study |
| OG1RF | Plasmid free, Rifr, Fusr | 5 |
| JH2SS | Plasmid free, Strr, Spcr | 14, 42 |
| POTC-1, POTC-2, and POTC-3 | Transconjugants derived from P101D12 and OG1RF mating, Rifr, Fusr, Ermr | This study |
| POJTC-1, POJTC-2, and POJTC-3 | Transconjugants derived from POTC-2 and JH2SS mating, Strr, Spcr, Ermr | This study |
| IG9, IIB3, and IIE7 | Transconjugants derived from E99 and OG1RF mating, Rifr, Fusr | This study |
| Tn917 mutant strains | ||
| P105H2 | Tn917 insertion in srt-1 | This study |
| P101A12 | Tn917 insertion in srt-1 | This study |
| P101D12 | Tn917 insertion in bee-2 | This study |
| P76D4 | Tn917 insertion in bee-1 | This study |
| P94C6 | Tn917 insertion in bee-1 | This study |
| P110B1 | Tn917 insertion in bee-3 | This study |
| P15D1 | Tn917 insertion upstream of srt-1 | This study |
| P77E12 | Tn917 insertion upstream of srt-1 | This study |
| Plasmids | ||
| pAT28 | Shuttle vector, Spcr | 44 |
| pTV1-OK | Temperature-sensitive plasmid containing Tn917, Kanr, Ermr | 10 |
| pBLG1 | pAT28 containing full-length bee-2 downstream of the aphA-3 promoter, Spcr | This study |
| pBLG2 | pAT28 containing full-length bee-3 downstream of the aphA-3 promoter, Spcr | This study |
| pBLG12 | pAT28 containing both bee-2 and bee-3 downstream of the aphA-3 promoter, Spcr | This study |
Biofilm assay.
The biofilm assay on the Tn917 mutants was performed using flat-bottom polystyrene microtiter plates (Corning Inc., Corning, NY) as described previously (36, 41). The assay was done independently in triplicate, with 12 replicates per strain per assay. Statistical significance was tested using Student's t test. Wherever appropriate, multigroup comparisons were made by analysis of variance (ANOVA) using Tukey's test.
DNA manipulations.
Electrocompetent E. coli or E. faecalis (33) cells were prepared as described previously and transformed with specific plasmids using a Gene Pulser unit (Bio-Rad Laboratories, Hercules, CA). Nucleotide sequence information was obtained using an ABI3730 capillary sequencer at the Oklahoma Medical Research Foundation (Oklahoma City, OK). Takara LA Taq polymerase (TaKaRa Biomedicals, Shiga, Japan) was used for all PCR amplifications meant for cloning. Oligonucleotide primer sequences are listed in the supplemental material.
Generation of a Tn917 mutant library in E99.
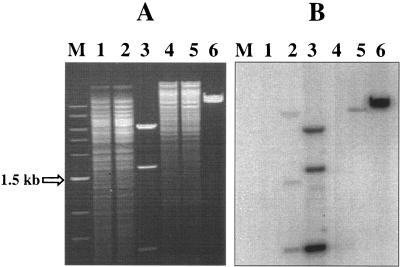
Plasmid pTV1-OK containing transposon Tn917, a temperature-sensitive origin of replication, and a kanamycin resistance determinant was used for performing mutagenesis (10). E. faecalis strain E99 was transformed with pTV1-OK by electroporation, and the transformants were selected on brain heart infusion (BHI) agar plates containing kanamycin (1,000 μg/ml) and erythromycin (10 μg/ml). E99(pTV1-OK) was grown at 30°C for 10 h, and appropriate dilutions were plated on prewarmed BHI agar plates supplemented with erythromycin at 10 μg/ml. The plates were further incubated at the nonpermissive temperature of 42°C for 48 h. The loss of plasmid pTV1-OK was confirmed by streaking the mutant colonies on BHI agar plates containing kanamycin (1,000 μg/ml). To generate a mutant bank in E99, 10,000 individual mutant colonies were archived. In order to confirm the presence of a single Tn917 insertion in the selected mutants, Southern analysis was performed on genomic DNA (2 μg) restricted with HindIII and EcoRI, employing a Tn917-specific probe (Fig. 1) generated using primers Tn917-1 and Tn917-2.
FIG. 1.
The presence of a single Tn917 insertion within the bee locus in mutant strain P101D12 was confirmed by Southern analysis using a Tn917-specific probe (a 1.9-kb internal region of Tn917 containing two HindIII restriction sites) as described previously (10). (A) Genomic DNA obtained from wild-type strain E99 and mutant P101D12 was restricted with HindIII (lanes 1 and 2) and EcoRI (lanes 4 and 5). (B) Southern hybridization was performed subsequently with the Tn917 probe obtained by using primers Tn917-1 and Tn917-2. The HindIII (lane 3)- and EcoRI (lane 6)-restricted plasmid pTV1-OK was used as the positive control. For size reference, a 1-kb DNA ladder (New England Biolabs, Beverly, MA) was used (lane M).
Characterization of the regions flanking the Tn917 insertion in P101D12.
Genomic DNA (2 μg) from P101D12 was restricted with EcoRI (New England Biolabs Inc., Beverly, MA), and the resulting fragments were self-ligated at 16°C using T4 DNA ligase (Promega, Madison, WI). To map the Tn917 insertion, an inverse PCR amplification was performed using a Takara LA PCR kit (Panvera Corp., Madison, WI) and outward-facing Tn917-specific primers SLT-L and SLT-R. The amplified fragment was purified from a 0.8% low-melting-point agarose gel using a Wizard Preps DNA purification system (Promega, Madison, WI), and the regions flanking the Tn917 insertion were sequenced with primers SLT-L and SLT-R. Homology searches were performed by BLAST analyses using National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and The Institute for Genomic Research (http://www.tigr.org) databases.
Dot blot analysis.
Dot blot analysis was performed on DNA isolated from 40 E. faecalis isolates. E99 DNA was used as the positive control. DNA from each strain was denatured in 0.4 N sodium hydroxide to a final concentration of 1 μg/ml and spotted onto a Zeta-probe GT genomic-tested blotting membrane (Bio-Rad, Hercules, CA) by placing 100 μl of sample into each well across the row of a 96-well dot blot apparatus (Bio-Rad, Hercules, CA). The membrane was rinsed thrice with Tris-EDTA buffer, pH 8.0. The DNA was then cross-linked to the membrane using a UV cross-linker (UVP Inc., Upland, CA). The membrane was probed at high stringency with the 1.4-kb bee locus-specific PCR product obtained from E99 DNA using primers P101D12-1 and P101D12-2 and radiolabeled with a Radprime DNA labeling system (Invitrogen, Carlsbad, CA).
Complementation of the transposon mutant.
To complement the biofilm defect in P101D12, 0.99-kb, 1.8-kb, and 2.6-kb regions (corresponding to full-length bee-2, full-length bee-3, and both bee-2 and bee-3, respectively) were amplified from E99 DNA using primer pairs E99-1/E99-3R, E99-7L/E99-4R, and E99-1/E99-4R. Primers E99-1 and E99-7L contained the recognition sequence for restriction endonuclease SacI, whereas primers E99-3R and E99-4R contained the recognition sequence for XbaI. The amplified products were restricted with SacI and purified from a 0.8% low-melting-point agarose gel. Primers Aph-1 (with an EcoRI recognition sequence) and Aph-2 (with a SacI recognition sequence) were used to amplify a 358-bp fragment containing the aphA-3 promoter (28) from the plasmid pTCV-aphA3. The amplification product was subsequently restricted with SacI, purified from a 0.8% low-melting-point agarose gel, and ligated to the SacI-restricted gene fragments using T4 DNA ligase (Promega, Madison, WI) at 16°C. One microliter of each of the ligation mixes was used in PCR amplification reactions with primer pairs Aph-1/E99-3R and Aph-1/E99-4R to enrich for the ligated aphA-3 and bee-2, aphA-3 and bee-3, and aphA-3 and bee-2-bee-3 products. All of the amplified products were then restricted with EcoRI and XbaI, purified from a low-melting-point agarose gel, and subsequently ligated to EcoRI/XbaI-restricted shuttle vector pAT28 (44) to generate plasmids pBLG1, pBLG2, and pBLG12. To verify the plasmid constructs, nucleotide sequence information was obtained from plasmids pBLG1, pBLG2, and pBLG12 using primer Aph-1. Mutant strain P101D12 was transformed with pBLG1, pBLG2, or pBLG12 to generate strains BLG1, BLG2, and BLG12. The abilities of the complemented strains to form biofilms were assessed using the 96-well-plate biofilm assay described above. Multigroup comparisons were made by ANOVA using Tukey's test to assess statistical significance.
RNA isolation and RT-PCR analysis.
Total RNA was isolated by a method described previously (32). Residual genomic DNA contamination was removed using a DNA-free DNase treatment and removal kit (Ambion, Austin, TX). Reverse transcription was performed using a Superscript first-strand synthesis system for reverse transcription (RT)-PCR (Invitrogen, Carlsbad, CA) as per the manufacturer's recommendations.
Filter mating assays.
Filter mating was carried out as described previously (6) with a donor/recipient ratio of 1:10. Overnight cultures of donor (0.5 ml; P101D12 or POTC-2) and recipient (4.5 ml; OG1RF or JH2SS) strains were mixed, and the cells were collected on a 0.2-μm cellulose nitrate membrane filter (Whatman International Ltd., Maidstone, England). The filter was then placed on a BHI agar plate bacteria side down and incubated at 37°C for 16 h. The cells from the filter and plate were recovered and resuspended in 1 ml BHI broth, and appropriate dilutions were plated on BHI agar plates containing suitable antibiotics. For the P101D12 (donor) and OG1RF (recipient) mating, the transconjugants were selected using rifampin, fusidic acid, and erythromycin. One of the transconjugants from the P101D12-OG1RF mating, POTC-2, served as the donor for the secondary filter mating, with JH2SS as the recipient. The transconjugants arising from the POTC-2 and JH2SS mating were selected using spectinomycin, streptomycin, and erythromycin. The transfer frequency was expressed as the number of transconjugants per recipient.
The filter mating between wild-type E99 as the donor and OG1RF as the recipient strain was carried out essentially as described above. However, due to the absence of a suitable antibiotic resistance marker for the transconjugants, appropriate dilutions of the bacterial cell suspension were plated on BHI agar plates containing rifampin and fusidic acid. This antibiotic selection allowed the recipient OG1RF as well as the transconjugants to grow. Individual colonies were picked, and an increase in the ability to form biofilms was then used as a screen for transconjugants. One hundred and eighty individual colonies were inoculated into the wells of 96-well plates containing TSB supplemented with 0.5% glucose. The crystal violet binding assay was then used to identify transconjugants that showed an enhanced ability to form biofilms compared to control strain OG1RF (recipient). After the initial identification of the transconjugants in a single-well assay, the biofilm assay was performed in 12 replicates per strain thrice independently to confirm the initial observation. The presence of the bee locus in three transconjugants identified by this screen was confirmed by PCR with primers P101D12-1 and P101D12-2 (data not shown) as well as by contour-clamped homogeneous electric field (CHEF) gel and Southern analysis using a bee locus-specific probe (see Fig. 7).
FIG. 7.
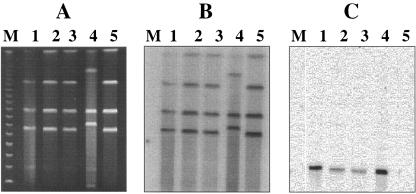
CHEF gel and Southern hybridization analysis of transconjugants obtained from filter mating experiments employing wild-type E99 as the donor and OG1RF as the recipient. (A) I-ceuI-restricted DNA from donor E99 (lane 1); three transconjugants, IG9, IIB3, and IIE7 (lanes 2, 3, and 4, respectively); and recipient strain OG1RF (lane 5). (B and C) Also shown are Southern hybridization analyses of I-ceuI-restricted DNA from donor strain E99 (lane 1), the transconjugants (lanes 2 to 4), and recipient strain OG1RF (lane 5) using a 23S rRNA gene probe (B) and a bee locus-specific probe (C). Bacteriophage lambda concatemers were used as molecular size markers (lane M; New England Biolabs, Beverly, MA).
CHEF gel analysis.
CHEF gel analysis was performed as described previously (1, 22, 40) with some modifications. A portion of the agarose plug containing total DNA from each strain was cut with a sterile razor and restricted with I-ceuI (New England Biolabs, Beverly, MA) at 37°C for 16 h. The plugs were washed in 1 ml dilute Tris-EDTA for 1 h at 37°C, then carefully loaded into the wells of a 1% pulsed-field certified agarose gel (Bio-Rad Laboratories, Hercules, CA) in 0.5× TBE (45 mM Tris HCl, 45 mM boric acid, 1 mM EDTA), and electrophoresed using a CHEF DRII device (Bio-Rad Laboratories, Hercules, CA) with the pulse time ramped from 5 to 120 s at 150 V for 40 h. The gels were then stained with ethidium bromide and photographed using a UVP gel documentation system (UVP Inc., Upland, CA) before the DNA was transferred to Zeta-probe GT genomic-tested blotting membrane by capillary or vacuum blotting. Southern hybridization analysis was then performed sequentially using either a 1.9-kb Tn917-specific probe generated using primers Tn917-1 and Tn917-2, a 1.4-kb bee locus-specific probe generated using primers P101D12-1 and P101D12-2, or a 1-kb probe specific for E. faecalis 23S rRNA genes generated using primers EF23sFor and EF23sRev.
Nucleotide sequence accession number.
The DNA sequence reported in this article has been deposited in the GenBank nucleotide sequence database under accession number DQ137124.
RESULTS
Effect of glucose on the ability of E. faecalis strain E99 to form biofilms.
Glucose in the growth medium has been found to influence the ability of E. faecalis strains to form biofilms (13, 26, 36) such that the presence of 0.5% (wt/vol) or greater amounts of glucose is required for biofilm enhancement. To assess if glucose is similarly necessary for E99 to form biofilms, we used the crystal violet binding assay described in Materials and Methods. In agreement with previous reports, E99 formed significantly more biofilms in the presence of 0.5%, 0.75%, and 1% glucose than in the presence of 0.25% glucose (P < 0.05) (data not shown).
Tn917 mutagenesis and characterization of the bee locus.
Tn917 mutagenesis identified eight mutants (Table 1) with significantly (>70%) reduced biofilm-forming ability compared to parent strain E99. In all of these mutants, the single Tn917 insertion mapped to the bee locus, and mutant P101D12 was chosen for further characterization. The presence of a single Tn917 insertion in P101D12, that within bee-2, was confirmed by inverse PCR and Southern analysis using a Tn917-specific probe (Fig. 1). The wild-type strain E99 (Fig. 1, lanes 1 and 4) was used as a negative control, and the purified plasmid pTV1-OK (Fig. 1, lanes 3 and 6) was used as a positive control. The bee locus (Fig. 2A) is comprised of three structural genes, designated bee-1, bee-2, and bee-3, which encode putative cell wall-anchored proteins, and two genes, srt-1 and srt-2, which encode probable sortases. The deduced amino acid sequences of Bee-1, Bee-2, and Bee-3 revealed the presence of the conserved LPxTG consensus sequence (20) commonly found in gram-positive cell wall-anchored surface proteins at their C termini. The 1,083-residue Bee-1 protein revealed some interesting structural features upon BLASTP analysis. While no repeat motifs were discernible, the region extending from residues 729 to 908 was identified by the conserved domain database search (18) to bear a low degree of similarity to the B domain of collagen-binding protein from Staphylococcus aureus (E = 1e−04) and Bacillus subtilis (E = 3e−05) (15, 35). The primary sequence of these collagen adhesins consists of a nonrepeat, collagen-binding A region followed by one to four 23-kDa B region repeats. The B domain has been suggested to serve as a “stalk” that projects the A regions from the bacterial cell surface to facilitate bacterial adherence to collagen. Residues 343 through 487 of Bee-1 also revealed a low degree of similarity (E = 6e−08) to the von Willebrand factor type A domain (47) that has been shown to bind to a variety of ligands, including collagen, laminin, and glycosaminoglycans.
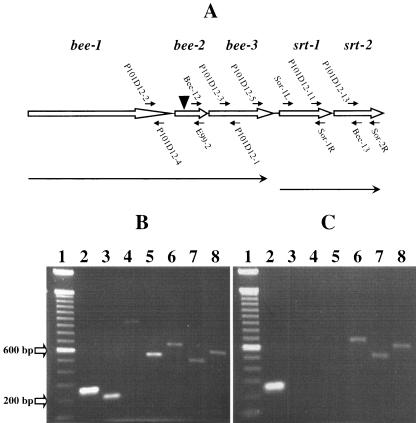
FIG. 2.
RT-PCR analyses to assess the expression and transcriptional linkage of genes within the bee locus in the wild-type strain E99 and mutant P101D12. (A) Schematic of the bee locus. The bee locus is comprised of three genes, bee-1, bee-2, and bee-3, encoding putative cell wall-anchored proteins, downstream of which are two ORFs, srt-1 and srt-2, encoding putative sortase enzymes. The locations of various primers used for the RT-PCR analysis are indicated. The position of the Tn917 insertion (▾) in mutant P101D12 is also indicated. Line arrows indicate putative transcriptional units. (B) Total RNA extracted from E99 was reverse transcribed using random hexamers and reverse transcriptase. An aliquot of the cDNA was then amplified by PCR with gene-specific primers. Primers P101D12-2/P101D12-4 (lane 2), P101D12-3/P101D12-1 (lane 3), P101D12-2/E99-2 (lane 4), Bee-12/P101D12-1 (lane 5), Sor-1L/Sor-1R (lane 6), P101D12-13/Sor-2R (lane 7), and P101D12-11/Bee-13 (lane 8) yielded 259-, 203-, 954-, 532-, 650-, 485-, and 581-bp amplification products, respectively. This implies that the genes bee-1, bee-2, and bee-3 may be cotranscribed as one transcriptional unit, with srt-1 and srt-2 cotranscribed independently. (C) Total RNA extracted from mutant strain P101D12 was used for RT-PCR. Primers P101D12-2/P101D12-4 (lane 2) yielded an expected 259-bp amplification product, indicating that bee-1 was being transcribed. However, no amplification products were obtained when primer pairs P101D12-3/P101D12-1 (lane 3), P101D12-2/E99-2 (lane 4), or Bee-12/P101D12-1 (lane 5) were used for amplification, suggesting that Tn917 insertion had abrogated not only the expression of bee-2 but also that of the downstream gene bee-3. Sor-1L/Sor-1R (lane 6), P101D12-13/Sor-2R (lane 7), and P101D12-11/Bee-13 (lane 8) yielded 650-, 485-, and 581-bp amplification products, respectively, implying that the Tn917 insertion in bee-2 did not abrogate expression of srt-1 and srt-2. For size reference, a 100-bp DNA ladder (Invitrogen, Carlsbad, CA) was used (lane 1).
While the 243-residue Bee-2 protein did not reveal any unique features or conserved domains except for putative signal sequence and the conserved LPxTG motif, the deduced 495-residue Bee-3 protein revealed a low degree of similarity to a conserved domain within outer membrane proteins from Leuconostoc mesenteroides.
The deduced amino acid sequences of both Srt-1 and Srt-2 revealed a high level of similarity to the conserved domains within sortase-type enzymes that cleave surface proteins at the LPxTG motif between threonine and glycine and catalyze the formation of an amide bond between the carboxyl group of threonine and the amino group of the cell wall cross-bridges (19). Table 2 lists the closest homologs of the predicted proteins encoded by the genes in the bee locus that were revealed using the BLASTP algorithm.
TABLE 2.
Homologies of the predicted proteins encoded by the bee locus genes determined by BLASTP (NCBI, NLM)
| Protein | Length (amino acids) | BLASTP hits | Accession no. | Organism | % Identity | % Similarity | Expect (E) value |
|---|---|---|---|---|---|---|---|
| Bee-1 | 1,083 | Predicted OMPa | ZP_00062640 | Leuconostoc mesenteroides | 25 | 38 | 2e−48 |
| Bee-2 | 243 | Predicted OMP | ZP_00062638 | Leuconostoc mesenteroides | 33 | 48 | 4e−17 |
| Bee-3 | 495 | Predicted OMP | ZP_00062638 | Leuconostoc mesenteroides | 39 | 54 | 3e−80 |
| Srt-1 | 398 | Sortase | EAN10600 | Enterococcus faecium | 50 | 71 | 2e−75 |
| Srt-2 | 373 | Sortase | EAN10837 | Enterococcus faecium | 45 | 66 | 2e−75 |
OMP, outer membrane protein.
Gene expression and transcriptional linkage analysis in E99 and P101D12.
Primer pairs P101D12-2/P101D12-4, P101D12-3/P101D12-1, P101D12-2/E99-2, and Bee-12/P101D12-1 were used to detect gene transcripts corresponding to bee-1, bee-3, bee-1 and bee-2, and bee-2 and bee-3, respectively. As shown in Fig. 2B, all of the PCR amplifications yielded products of expected sizes: 259 bp (lane 2), 203 bp (lane 3), 954 bp (lane 4), and 532 bp (lane 5), respectively. This suggested that bee-1, bee-2, and bee-3 may be transcriptionally linked. No amplification product was detected with primers P101D12-2 and Sor-1R or with P101D12-5 and Sor-1R, implying that srt-1 is not transcriptionally linked to bee-3. However, as shown in Fig. 2B, PCR amplifications using the primer pairs Sor-1L/Sor-1R, P101D12-13/Sor-2R, and P101D12-11/Bee-13 yielded expected 650-bp (lane 6), 485-bp (lane 7), and 581-bp (lane 8) products, respectively, implying that srt-1 and srt-2 are expressed and likely cotranscribed. Reverse transcription reactions performed in the absence of reverse transcriptase to control for residual genomic contamination did not yield any amplification products.
RT-PCR analysis was also performed on RNA from the mutant P101D12 to investigate the effect of the Tn917 insertion on the expression of bee-2 and the genes downstream. As shown in Fig. 2C, primer pairs P101D12-3/P101D12-1 (lane 3), P101D12-2/E99-2 (lane 4), and Bee-12/P101D12-1 (lane 5) failed to yield amplification products, suggesting that the Tn917 insertion in bee-2 had abrogated the expression of bee-2 as well as the downstream gene bee-3. Expected 259-bp (Fig. 2C, lane 1), 650-bp (Fig. 2C, lane 6), 485-bp (Fig. 2C, lane 7), and 581-bp (Fig. 2C, lane 8) products were obtained using primers P101D12-2/P101D12-4, Sor-1L/Sor-1R, P101D12-13/Sor-2R, and P101D12-11/Bee-13, indicating that the Tn917 insertion did not abrogate the expression of the upstream gene bee-1 or the two downstream putative sortase genes srt-1 and srt-2.
Complementation of P101D12.
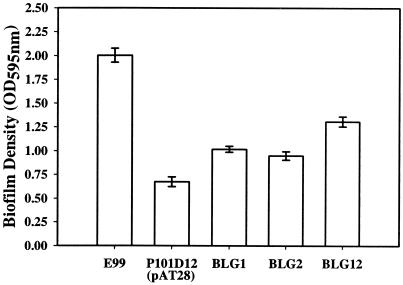
In order to confirm that the decrease in the biofilm-forming ability of mutant strain P101D12 was due to the insertion of Tn917 in the bee locus and not due to any other polar effect of the insertion, complementation studies were performed. RT-PCR analysis of strain P101D12 indicated that the insertion of Tn917 in bee-2 had abrogated the expression of the downstream gene bee-3 but not that of genes srt-1 and srt-2. P101D12 was therefore transformed with shuttle vector pAT28 (44) alone (vector control) or pAT28 harboring the full-length bee-2 gene, the full-length bee-3 gene, or both the bee-2 and bee-3 genes downstream of a constitutive aphA-3 promoter (28) to generate strains BLG1, BLG2, and BLG12, respectively. As shown in Fig. 3, strains BLG1 and BLG2 formed significantly more biofilms than the mutant strain P101D12 containing vector alone (ANOVA; Tukey's test, P < 0.05). A significantly higher biofilm-forming ability of BLG12 (which was complemented with both bee-2 and bee-3) compared to BLG1 (which was complemented with only bee-2) and BLG2 (which was complemented with only bee-3) was also observed (ANOVA; Tukey's test, P < 0.01).
FIG. 3.
Biofilm formation by wild-type strain E99, mutant P101D12(pAT28), and complemented strains BLG1 (P101D12 transformed with pAT28 harboring bee-2 downstream of the aphA-3 promoter), BLG2 (P101D12 transformed with pAT28 harboring bee-3 downstream of the aphA-3 promoter), and BLG12 (P101D12 transformed with pAT28 harboring bee-2 and bee-3 downstream of the aphA-3 promoter). The y axis represents the optical density (OD) of dissolved crystal violet measured at 595 nm. The error bars represent the mean ± standard error.
Transfer of the bee locus to other E. faecalis strains.
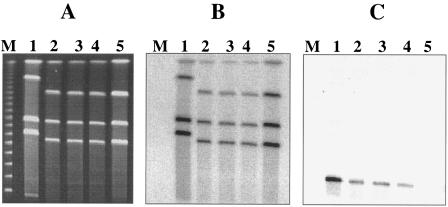
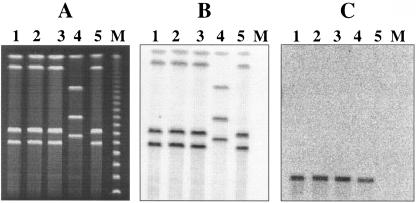
The nucleotide sequence downstream of the probable sortase gene srt-2 revealed the presence of genes encoding putative transposase and resolvase (data not shown), suggesting that the bee locus could be present on a mobile genetic element and thereby could horizontally transfer to other E. faecalis strains. Strain P101D12, which had a Tn917 insertion in bee-2, was filter mated with E. faecalis strain OG1RF. The absence of bee locus genes in recipient OG1RF was confirmed using Southern hybridization analysis. Erythromycin resistance encoded by Tn917 was used as a marker for selection of the transconjugants. Transconjugants arose at a frequency of 4 × 10−2/recipient (3.2 × 10−1/donor). As shown in Fig. 4A, I -ceuI-restricted DNA from three randomly chosen transconjugants, POTC-1 (lane 1), POTC-2 (lane 2), and POTC-3 (lane 3); the donor, P101D12 (lane 4); and the recipient, OG1RF (lane 5), was separated using CHEF gel electrophoresis and analyzed by Southern hybridization using 23S rRNA genes (Fig. 4B) and Tn917-specific probes (Fig. 4C). Four bands from the transconjugants as well as P101D12 and OG1RF hybridized to the 23S rRNA gene probe, indicating that these were chromosomal fragments. However, the Tn917-specific probe hybridized to a distinctly smaller band with an apparent size of ∼80 kb in the transconjugants and the donor strain P101D12 (Fig. 4C, lanes 1, 2, 3, and 4). This band was absent in recipient strain OG1RF (Fig. 4C, lane 5), implying that the bee locus is not located on the chromosome but likely harbored on a large conjugative plasmid that has transferred from the donor to the recipients.
FIG. 4.
CHEF gel and Southern hybridization analysis of transconjugants obtained from filter mating experiments employing mutant P101D12 as the donor and OG1RF as the recipient. (A) CHEF gel of I-ceuI-restricted DNA from three primary transconjugants, POTC-1, POTC-2, and POTC-3 (lanes 1, 2, and 3, respectively); donor strain P101D12 (lane 4); and recipient strain OG1RF (lane 5). (B and C) Also shown are Southern hybridization analyses of I-ceuI-restricted DNA from the transconjugants (lanes 1 to 3), donor strain P101D12 (lane 4), and recipient strain OG1RF (lane 5) using a 23S rRNA gene probe (B) and a Tn917-specific probe (C). Bacteriophage lambda concatemers were used as molecular size markers (lane M; New England Biolabs, Beverly, MA).
The possibility of secondary transfer of the bee locus to another E. faecalis recipient strain, JH2SS, was investigated using one of the primary transconjugants (POTC-2) from the P101D12 and OG1RF mating as the donor. JH2SS also lacked bee locus genes, as was confirmed by Southern hybridization analysis. The secondary transfer frequency for the bee locus was found to be similar to the primary transfer frequency at 3.8 × 10−2/recipient (2.92 × 10−1/donor). As shown in Fig. 5A, I-ceuI-restricted DNA from three randomly chosen transconjugants, POJTC-1 (lane 1), POJTC-2 (lane 2), and POJTC-3 (lane 3); the donor, POTC-2 (lane 4); and the recipient, JH2SS (lane 5), was separated by CHEF gel electrophoresis and analyzed by Southern hybridization using 23S rRNA genes (Fig. 5B) and Tn917-specific probes (Fig. 5C). Four bands from the transconjugants as well as POTC-2 and JH2SS hybridized to the 23S rRNA gene probe, indicating that they were chromosomal. Similar to that observed with the P101D12/OG1RF transconjugants, the Tn917-specific probe hybridized to a distinct smaller band with an apparent size of ∼80 kb in transconjugants POJTC-1, POJTC-2, and POJTC-3 and donor strain POTC-2 (Fig. 5C, lanes 1, 2, 3, and 4). This band was absent in recipient strain JH2SS (Fig. 5C, lane 5).
FIG. 5.
CHEF gel and Southern hybridization analysis of transconjugants obtained from filter mating experiments employing primary transconjugant POTC-2 as the donor and JH2SS as the recipient. (A) I-ceuI-restricted DNA from three secondary transconjugants, POJTC-1, POJTC-2, and POJTC-3 (lanes 1, 2, and 3, respectively); donor POTC-2 (lane 4); and recipient JH2SS (lane 5). (B and C) Also shown are Southern hybridization analyses of I-ceuI-restricted DNA from the transconjugants (lanes 1 to 3), donor strain POTC-2 (lane 4), and recipient strain JH2SS (lane 5) using a 23S rRNA gene probe (B) and a Tn917-specific probe (C). Bacteriophage lambda concatemers were used as molecular size markers (lane M; New England Biolabs, Beverly, MA).
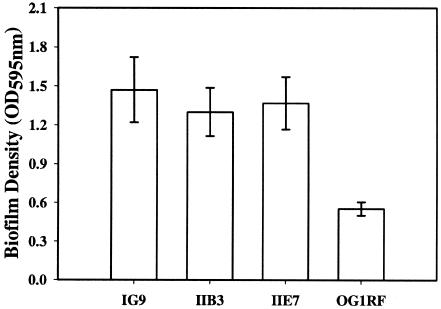
In order to investigate whether the acquisition of the bee locus was accompanied by an enhancement in the biofilm-forming ability of the transconjugants, the wild-type E99 strain was filter mated with OG1RF. Although no antibiotic resistance marker was available for the bee locus, based on our previously observed bee locus transfer frequencies of ∼4 transconjugants per 100 recipient cells, we screened a total of 180 colonies comprised of both recipient OG1RF as well as transconjugants by the crystal violet binding biofilm assay as described in Materials and Methods. As shown in Fig. 6, three transconjugants, IG9, IIB3, and IIE7, which showed an enhancement in biofilm-forming ability compared to parent strain OG1RF (ANOVA; Tukey's test, P < 0.01), were identified. PCR on genomic DNA prepared from IG9, IIB3, and IIE7 with primers P101D12-1/P101D12-2 revealed the presence of a 1.4-kb region corresponding to the 3′ end of bee-1, full-length bee-2, and the 5′ end of bee-3. In addition, CHEF gel followed by Southern hybridization analysis of I-ceuI-restricted DNA from E99 (Fig. 7, lane 1), IG9, IIB3, IIE7 (Fig. 7, lanes 2, 3, and 4), and OG1RF (Fig. 7, lane 5) using a bee locus probe revealed the presence of the expected extrachromosomal band with an apparent size of ∼80 kb in E99 and the transconjugants. These results are in agreement with those obtained from CHEF gel and Southern hybridization analyses of transconjugants obtained using P101D12/OG1RF and POTC-2/JH2SS mating pairs (Fig. 4 and 5).
FIG. 6.
Biofilm assay on three transconjugants, IG9, IIB3, and IIE7, obtained by filter mating wild-type E99 with E. faecalis strain OG1RF. Crystal violet binding was used to measure the 24-h biofilm densities of IG9, IIB3, IIE7, and parent strain OG1RF grown in TSB supplemented with 0.5% glucose. The error bars represent the mean ± standard error.
In separate experiments, we also compared the biofilm-forming abilities of transconjugants POTC-1, POTC-2, and POTC-3 to recipient strain OG1RF using the crystal violet binding assay. As expected, owing to the Tn917 insertion within bee-2, no significant difference in the biofilm-forming abilities of these strains was noted. These results provide strong evidence that the biofilm phenotype imparted to transconjugants IG9, IIB3, and IIE7 (obtained using E99 as the donor and OG1RF as the recipient) is attributable to the bee locus and not a result of cotransfer of some other unknown element.
Detection of the bee locus in other E. faecalis strains.
Dot blot hybridization was used to assess the presence of the bee locus in a collection of archived E. faecalis isolates. A 1.4-kb region corresponding to the 3′ end of bee-1, full-length bee-2, and the 5′ end of bee-3 was used as a probe for the high-stringency hybridization. Out of a total of 40 randomly selected E. faecalis isolates that were screened, this gene cluster was detected in only 2 geographically unrelated clinical isolates (data not shown). DNA from wild-type strain E99 was spotted as a positive control for the hybridization assay.
DISCUSSION
Surface proteins play an important role in the multistep and multifactorial processes of biofilm formation. The ability of bacterial cells to persist as a biofilm poses a therapeutic challenge by limiting the antimicrobial treatment options available. In E. faecalis, very few surface-associated proteins, including aggregation substance (8), Esp (31), and Ace (29), have been extensively characterized. Notably, several studies have shown that expression of Esp enhances biofilm formation in E. faecalis (21, 37, 41). Aggregation substance was found to influence biofilm formation by promoting bacterial cell-cell interaction due to positive cooperativity (46). However, the genome of E. faecalis strain V583 (25) reveals the presence of 41 putative cell wall-anchored proteins and three sortase-like enzymes. Seventeen of these 41 putative cell wall-anchored proteins were found to contain tandem immunoglobulin-like folds commonly found in staphylococcal MSCRAMMs (34). While the functions of most of these proteins are presently unknown, it is possible that some of them may play a role in biofilm formation.
In this study, we identified a novel gene cluster, the bee locus, comprising genes encoding three putative cell surface-anchored proteins, Bee-1, Bee-2, and Bee-3, and two sortase-like enzymes, Srt-1 and Srt-2. This locus is not present in the E. faecalis genome strain V583 (25). The deduced protein sequences of Bee-1, Bee-2, and Bee-3 revealed the presence of a signal sequence at the amino termini and an LPxTG motif followed by a hydrophobic domain at the C termini. Thus, there is strong evidence to support the hypothesis that Bee-1, Bee-2, and Bee-3 are anchored to the bacterial cell wall by a sortase-dependent mechanism. Furthermore, BLASTP analyses revealed that the proteins Bee-1, Bee-2, and Bee-3 bear extensive similarities to predicted cell wall-anchored proteins of unknown functions from Leuconostoc mesenteroides (Table 2).
The bee locus includes two ORFs in tandem, downstream of bee-3, that encode putative sortases. This kind of sortase-substrate clustering might represent an independent functional unit encoding cell surface-associated proteins along with sortase enzymes exclusively dedicated to the cell wall anchoring of those proteins. This hypothesis is supported by the observation that the Tn917 mutant P101A12, which has a Tn917 insertion in srt-1 (Table 1), is positive for the expression of enterococcal surface protein, Esp, as detected by enzyme-linked immunosorbent assay using antibodies specific to the N-terminal domain of Esp (data not shown). Studies have identified genes encoding sortase enzymes juxtaposed with genes encoding their substrates in the genomes of other gram-positive organisms, including E. faecalis, Streptococcus agalactiae, and Corynebacterium diphtheriae (17, 23, 34, 43). Interestingly, immunogold electron microscopy revealed that some of the surface-exposed proteins encoded at these loci formed pilus-like structures extending from the bacterial cell surface in S. agalactiae and C. diphtheriae (17, 43). Similarly, a gene cluster comprised of three genes encoding surface-anchored proteins of unknown function is present immediately upstream of three sortase-like proteins in the genome of virulent Streptococcus pneumoniae (39). It remains to be seen whether the bee locus similarly represents a gene cluster involved in the expression of pilus-like structures extending from the cell surface in E. faecalis.
The nucleotide sequence flanking the Tn917 insertion in at least eight mutants that were attenuated in the ability to form biofilms revealed that Tn917 had inserted within different regions of the ORFs or the intergenic regions of the bee locus. The identification of several independent mutants with decreased ability to form biofilms, all of which had an insertion in the bee locus, suggested that the observed phenotype was specifically due to the inactivation of one or more genes within the bee locus rather than any other nonspecific polar effect of the insertion. However, RT-PCR analysis of RNA purified from the mutant P101D12, which had an insertion in gene bee-2, revealed that the Tn917 insertion not only interfered with the expression of bee-2 but also had a polar effect on the expression of the downstream gene bee-3. Complementation in trans with either bee-2 or bee-3 alone significantly increased the ability of P101D12 to form biofilms. Complementation of the mutant strain with a plasmid constitutively expressing both bee-2 and bee-3, however, further enhanced the biofilm-forming ability of P101D12, suggesting that both Bee-2 and Bee-3 may be important for biofilm formation. The exact mechanism by which these two putative cell wall-associated proteins may enhance biofilm formation remains to be resolved. Interestingly, complementation of the defect in trans did not result in restoration of biofilm-forming ability to wild-type levels. Although the reasons for this remain unclear at this time, it is possible that overexpression of the Bee proteins driven by a heterologous promoter leads to an incorrect surface display disturbing the optimal stoichiometric interaction between these proteins at the cell surface.
Sequence information downstream of srt-2 revealed the presence of genes putatively encoding transposition functions. A site-specific recombinase belonging to the resolvase family (exhibiting 80% identity and 92% similarity to a resolvase identified in Staphylococcus epidermidis strain RP62A; GenBank ID 57854758) and a probable transposase (with 59% identity and 79% similarity to the transposase found on Staphylococcus aureus transposon Tn552, which encodes β-lactam resistance; GenBank ID 33390965) suggested the location of the bee locus on a mobile genetic element such as a transposon. Filter mating experiments, however, revealed that this locus is able to transfer to recipient E. faecalis strains at high frequencies, typical for conjugative plasmids. To determine the nature of the genetic element harboring the bee locus, we employed a CHEF gel approach previously used to examine genome diversity in enterococci (22) and to assign a chromosomal location to the vanG operon in E. faecalis (1). The enzyme I-ceuI is an intron-encoded endonuclease that specifically recognizes unique sequences present within rrn operons (22). E. faecalis strains were postulated to contain four rrn operons based on mapping experiments (22), and this was subsequently confirmed by the complete genome sequence of strain V583 (25). CHEF gel analysis of I-ceuI-digested DNA from E. faecalis followed by Southern hybridization analysis with 23S rRNA gene probes would therefore be expected to identify the four chromosomal fragments. Our Southern hybridization results with a 23S rRNA gene probe, shown in Fig. 4, 5, and 7, clearly identify these chromosomal fragments from donors, recipients, and transconjugants. Additionally, the banding pattern of the I-ceuI fragments also distinguishes between donor and recipient strains, such that in all cases described, it is clear that the transconjugants (obtained from independent mating experiments) were derived from the recipient. Probes specific to either the bee locus or Tn917 hybridized to the same extrachromosomal band with an apparent size of ∼80 kb (Fig. 4, 5, and 7). These results therefore support our argument that the bee locus is harbored on a large extrachromosomal element with an apparent size of ∼80 kb. Our results showing high-frequency transfer of this element in filter mating experiments strongly suggest that the bee locus is harbored on a conjugative plasmid. No transconjugants were detected when mating experiments were performed in broth, suggesting that the conjugative plasmid may not be pheromone responsive.
It is not surprising that the bee locus genes were detected in only two other strains among our archived isolates. While it is possible that a higher frequency of occurrence of this locus may become evident upon screening more isolates, the probability remains that this locus is a recent acquisition in Enterococcus faecalis. The latter possibility is strengthened by the observation that the bee locus is likely carried on a large conjugative plasmid. A number of recent comparative genomic hybridization studies (38, 39) have revealed extensive genetic heterogeneity, even among the same serotype, and the absence of a number of gene clusters among closely related strains, suggesting that they are absent or significantly divergent in these strains. Interestingly, a majority of the loci that differed in these strains encoded surface proteins or were related to pathogenesis (39), suggesting that such differences may contribute to differences in virulence. The existence of a bee locus on a conjugative element with an ability to transfer at high frequency and confer on recipients a high-biofilm phenotype may define the emergence of E. faecalis strains ideally suited to persist and disseminate virulence traits in the nosocomial environment.
Supplementary Material
Acknowledgments
This work was supported, in part, by Public Health Services Grant AI 059673 (N.S.) from the National Institutes of Health.
We thank K. T. Madhusudhan for providing us with E. faecalis strain E99. We are grateful to Michael S. Gilmore, Phillip S. Coburn, and G. T. Dolan for helpful discussions.
Footnotes
These authors contributed equally to this work.
REFERENCES
- 1.Depardieu, F., M. G. Bonora, P. E. Reynolds, and P. Courvalin. 2003. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 50:931-948. [DOI] [PubMed] [Google Scholar]
- 2.Donlan, R. M. 2001. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 33:1387-1392. [DOI] [PubMed] [Google Scholar]
- 3.Donlan, R. M. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 8.Galli, D., F. Lottspeich, and R. Wirth. 1990. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4:895-904. [DOI] [PubMed] [Google Scholar]
- 9.Gilmore, M. S., D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.). 2002. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 10.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall-Stoodley, L., and P. Stoodley. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7-10. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hufnagel, M., S. Koch, R. Creti, L. Baldassarri, and J. Huebner. 2004. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J. Infect. Dis. 189:420-430. [DOI] [PubMed] [Google Scholar]
- 14.Ike, Y., D. B. Clewell, R. A. Segarra, and M. S. Gilmore. 1990. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J. Bacteriol. 172:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 16.Kristich, C. J., Y. H. Li, D. G. Cvitkovitch, and G. M. Dunny. 2004. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186:154-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in Group B Streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 18.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 20.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 21.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72:3658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oana, K., Y. Okimura, Y. Kawakami, N. Hayashida, M. Shimosaka, M. Okazaki, T. Hayashi, and M. Ohnishi. 2002. Physical and genetic map of Enterococcus faecium ATCC 19434 and demonstration of intra- and inter-specific genomic diversity in enterococci. FEMS Microbiol. Lett. 207:133-139. [DOI] [PubMed] [Google Scholar]
- 23.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9:97-102. [DOI] [PubMed] [Google Scholar]
- 24.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 25.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 26.Pillai, S. K., G. Sakoulas, G. M. Eliopoulos, R. C. Moellering, Jr., B. E. Murray, and R. T. Inouye. 2004. Effects of glucose on fsr-mediated biofilm formation in Enterococcus faecalis. J. Infect. Dis. 190:967-970. [DOI] [PubMed] [Google Scholar]
- 27.Potera, C. 1999. Forging a link between biofilms and disease. Science 283:1837-1839. [DOI] [PubMed] [Google Scholar]
- 28.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 29.Rich, R. L., B. Kreikemeyer, R. T. Owens, S. LaBrenz, S. V. Narayana, G. M. Weinstock, B. E. Murray, and M. Hook. 1999. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J. Biol. Chem. 274:26939-26945. [DOI] [PubMed] [Google Scholar]
- 30.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 31.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shepard, B. D., and M. S. Gilmore. 1999. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shepard, B. D., and M. S. Gilmore. 1995. Electroporation and efficient transformation of Enterococcus faecalis grown in high concentration of glycine. Methods Mol. Biol. 47:217-226. [DOI] [PubMed] [Google Scholar]
- 34.Sillanpaa, J., Y. Xu, S. R. Nallapareddy, B. E. Murray, and M. Hook. 2004. A family of putative MSCRAMMs from Enterococcus faecalis. Microbiology. 150:2069-2078. [DOI] [PubMed] [Google Scholar]
- 35.Snodgrass, J. L., N. Mohamed, J. M. Ross, S. Sau, C. Y. Lee, and M. S. Smeltzer. 1999. Functional analysis of the Staphylococcus aureus collagen adhesin B domain. Infect. Immun. 67:3952-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tendolkar, P. M., A. S. Baghdayan, M. S. Gilmore, and N. Shankar. 2004. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 72:6032-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tendolkar, P. M., A. S. Baghdayan, and N. Shankar. 2003. Pathogenic enterococci: new developments in the 21st century. Cell. Mol. Life Sci. 60:2622-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 40.Thal, L. A., J. Silverman, S. Donabedian, and M. J. Zervos. 1997. The effect of Tn916 insertions on contour-clamped homogeneous electrophoresis patterns of Enterococcus faecalis. J. Clin. Microbiol. 35:969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomich, P. K., F. Y. An, and D. B. Clewell. 1980. Properties of erythromycin-inducible transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 141:1366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 44.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 46.Waar, K., H. C. van der Mei, H. J. Harmsen, J. E. Degener, and H. J. Busscher. 2002. Enterococcus faecalis surface proteins determine its adhesion mechanism to bile drain materials. Microbiology 148:1863-1870. [DOI] [PubMed] [Google Scholar]
- 47.Whittaker, C. A., and R. O. Hynes. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13:3369-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.