Abstract
Group B streptococcus (GBS) remains a major cause of morbidity and mortality among newborn children. The bacterium is a commensal organism colonizing the rectum and the gastrointestinal and urogenital tracts of adults, but it can be transmitted to neonates by an ascending infection of the maternal genital tract or during parturition. We previously reported that a transposon insertion disrupting rpoE resulted in the decreased survival of the mutant in the neonatal rat sepsis model of GBS infection. rpoE encodes the δ protein, a subunit of RNA polymerase (RNAP) that has been characterized in Bacillus species. In this study, we confirm the association of the δ protein with purified GBS RNAP and show that it is expressed in strains representing all nine serotypes. Flow cytometric analysis of a reporter strain containing a transcriptional fusion of the rpoE promoter to gfp revealed that, in vitro, this gene is continuously expressed. Analysis of δ expression in the transposon mutant by quantitative Western blotting revealed a 10-fold reduction in relative abundance (which was linked to the attenuation in virulence that was observed for this mutant) compared to that for the wild-type strain. These data suggest that a minimum intracellular concentration of δ is necessary for this organism to cause disease.
Streptococcus agalactiae, or group B streptococcus (GBS), remains a leading cause of pneumonia, sepsis, and meningitis in newborns within the developed world (37). GBS infection can be separated into two distinct syndromes, one occurring among neonates of up to several days old (early onset disease) and the other among infants that are several weeks or months older (late-onset disease). Although the screening of at-risk obstetric patients, followed by the administration of intrapartum antibiotics, has drastically reduced the incidence of early onset disease (41), invasive GBS disease still accounts for significant morbidity and mortality in newborns (27, 43). Increasing numbers of reports of invasive disease among the elderly and in immunocompromised adults have also classified GBS as an important pathogen in these two populations (3). Recognition of the prevalence and severity of human neonatal and adult disease underscores the need for investigating the mechanisms that are important to the pathogenesis of GBS infections.
The GBS bacterium is a commensal organism in adults that colonizes the rectum and the urogenital and gastrointestinal tracts (13). To establish early onset disease, the bacterium must ascend from the acidic environment of the mother's vaginal mucosa to the lung epithelia of the neonate during birth and access the highly oxygenated environment of the blood (for a review, see reference 2). Thus, an important feature that is exhibited by this pathogen is an ability to survive and grow in diverse host environments that differ with respect to the physiological conditions and immune defenses. This ability to adapt likely involves an alteration in the pattern of gene expression. The regulation of gene expression in bacteria primarily occurs at the level of transcription and is controlled through the activity of the transcribing enzyme, RNA polymerase (RNAP). Typically, bacterial RNAP can coexist in two distinct forms: (i) the core enzyme comprised of β and β′ and two identical α subunits and (ii) as a holoenzyme which, in addition to the core, contains a dissociable σ subunit. Functionally, the core enzyme has a low binding affinity for any DNA sequence but is sufficient for transcriptional elongation and termination. The association of a σ factor with the core enzyme drastically increases the affinity for σ-specific promoter sequences, allowing for the initiation of transcription. The constitutively expressed sigma factors σ70 and σA are responsible for initiating the transcription of the majority of bacterial genes in gram-negative and gram-positive bacteria, respectively. However, the induction of competence, adaptation to temperature change, control of sporulation, and survival response to oxidative stress have all been attributed, in part, to the association of specific secondary σ factors with RNAP (14, 15, 28, 31).
A number of accessory proteins are also found in association with RNAP during various stages in transcription. In addition to the core subunits and σ, RNAP purified from Bacillus subtilis has also been shown to contain a novel protein designated δ, which is encoded by the rpoE gene (1, 24, 26). An interrogation of the published sequence databases suggests that rpoE is ubiquitous among gram-positive bacteria.
Activity for δ has thus far been characterized in only Bacillus spp. by using in vitro assays. δ has been shown to bind to the core of Bacillus subtilis RNAP (25). It is thought to play a role in maintaining transcriptional specificity. Association of δ with RNAP reduces binding to DNA templates containing nonbiologically relevant promoter sites (39) and inhibits transcription from weak promoters. In addition, depending on the template, δ has been implicated in enhanced promoter melting (20) and may be involved in RNAP recycling (24). These data suggest that δ acts as an allosteric modulator of RNAP conformation and lacks enzymatic activity.
The function of δ in vivo has not been established. We previously reported that a GBS mutant with a transposon insertion adjacent to rpoE exhibited decreased survival in a neonatal rat sepsis model (18), suggesting a role for δ in virulence. In this study, we confirm the association of the δ protein with purified GBS RNAP and show that it is widely expressed among the serotypes. Using a reporter assay, we also demonstrate that rpoE expression in vitro is growth-phase dependent. An analysis of δ expression in the transposon mutant by quantitative Western blotting revealed a 10-fold reduction in the relative abundance, which was linked to the attenuation in virulence that was observed for this mutant. These data suggest that the abundance of δ may be an important aspect of GBS virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. GBS strains were grown in Todd-Hewitt broth (Difco Laboratories) in 5% CO2 at 37°C. Escherichia coli strain MC1061 served as a host strain for cloning purposes. E. coli Origami was used as a host strain for the production of recombinant protein. E. coli was grown at 37°C under aeration in Luria broth (LB). When required, antibiotics were added to the medium as follows. For E. coli, ampicillin (Ap), 50 μg ml−1, spectinomycin (Sp), 100 μg ml−1, and erythromycin (Em), 500 μg ml−1, were used, and for GBS, 5 μg ml−1 Em or 100 μg ml−1 Sp was added.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| S. agalactiae | ||
| A909 | Serotype Ia, encapsulated, bloodstream isolate from septic newborn | 22 |
| DK14 | Serotype Ib, encapsulated | 5 |
| DK23 | Serotype II, encapsulated | 29 |
| COH1 | Serotype III, encapsulated, isolated from neonate with septicemia | 35 |
| CNCTC 1/82 | Serotype IV, encapsulated | 33 |
| CNCTC 10/84 | Serotype V, encapsulated | 44 |
| NT6 | Serotype VI, encapsulated | 44 |
| 87-603 | Serotype VII, encapsulated | Pat Ferrierib |
| JM9 | Serotype VIII, encapsulated | Pat Ferrieri |
| AJ200 | A909 ΔrpoE, allelic exchange mutant, Kmr | 19 |
| AJ8D3 | A909 Tn917stm::rpoE, transposon mutant, Emr | 18 |
| RN114 | AJ200 containing pRN021, complemented strain, Spr | This study |
| RN115 | AJ200 containing pLZ12, control strain, Spr | This study |
| RS020 | A909 bearing pRSgfpmut3, Emr | This study |
| RS021 | A909 bearing pRSPrpoE::gfp, Emr | This study |
| B. subtilis | ||
| MH5636 | 10× His tag fused to the 3′ end of rpoC (β′-subunit of RNAP) | 32 |
| E. coli | ||
| XL1-Blue | Host strain for general cloning | Stratagene |
| MC1061 | Host strain for propagation of Emr plasmids | Stratagene |
| Origami | IPTG inducible expression strain F-ompT hsdSB(rB mB)galλ(DE3)[pLysS Cmr] | Novagen |
| Plasmids | ||
| pLZ12 | Low-copy streptococcal expression vector, Spr | 8 |
| pRN021 | pLZ12 derivative containing rpoE promoter and coding sequence | This study |
| pDC125 | Streptococcal cloning vector, Emr, Cmr | 4 |
| pRSgfpmut3 | pDC125 derivative containing gfpmut3, Emr | This study |
| pBL26 | Source of gfpmut3 allele | Gift from B. Limbago |
| pRSPrpoE−gfp | pRSgfpmut3 derivative containing gfp under the control of the rpoE promoter | This study |
| pET32a | E. coli T7 expression vector, Apr | Novagen |
| pET32rpoE | Vector for over-expressing the δ protein, Apr | This study |
| pET32ck | pET32a derivative with trx removed, Apr | This study |
| pET32rpoD | Vector for over-expressing σA protein, Apr | This study |
| pET32rpoB | Vector for over-expressing β protein, Apr | This study |
Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Apr, ampicillin resistant; Spr, spectinomycin resistant.
The strain was kindly provided by Pat Ferrieri, University of Minnesota, Minneapolis, MN.
Purification of RNAP.
RNAP was purified from GBS strains A909 and AJ200 and B. subtilis strain MH5636. For the isolation of RNAP from GBS, the strains were grown to mid-logarithmic phase (optical density at 600 nm [OD600] was 0.4 to 0.6) and harvested by centrifugation. Approximately 5 g of cells (wet weight) were resuspended in protoplast preparation buffer (0.3 M potassium phosphate buffer [pH 7], 40% sucrose) containing endo-N-acetylmuramidase (mutanolysin, 0.5 U/μl) (Sigma) and incubated at 37°C for 90 min. Protoplasts were collected by centrifugation, resuspended in lysis buffer (50 mM Tris-HCl [pH 8], 10 mM MgCl2, 0.1 mM dithiothreitol, 0.1 mM EDTA [pH 8], 10% glycerol, 1 mM phenylmethylsulfonyl fluoride), and lysed by sonication (Vibra Cell VC 750 sonicator; Sonics). Lysates were clarified by centrifugation at 23,000 × g for 20 min, and the undiluted supernatant was applied to a heparin-agarose column (10 ml bed volume) (Sigma). Heparin is a polyanion that mimics DNA in its overall binding properties; thus, it is commonly used as a ligand for the purification of various DNA binding proteins, including DNA-dependent RNA polymerase. The column was washed with 3 volumes of re-equilibration buffer (10 mM Tris-HCl [pH 8], 10 mM MgCl2, 0.1 mM dithiothreitol, 0.1 mM EDTA [pH 8], 10% glycerol), and proteins were eluted with a linear salt gradient of 0.1 to 1.0 M NaCl in the same buffer (20 ml total volume). Fractions that contained RNAP (based on the presence of the β/β′ subunit doublet) were pooled and concentrated, and the salt concentration was adjusted to 0.1 M NaCl by buffer exchange using an Amicon-Ultra filter device (Millipore). The concentrate was loaded onto a high-pressure liquid chromatography UNO Q1 column (Bio-Rad) that was equilibrated with pre-equilibration buffer, and proteins were eluted as described above by using an NaCl step gradient (50 ml total volume). Holoenzyme-containing fractions were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and finally loaded on a high-pressure liquid chromatography S-300 column (Amersham Biosciences) in re-equilibration buffer containing 200 mM NaCl. Peak fractions (based on absorbance at 280 nm) were pooled, concentrated, reconstituted in storage buffer (10 mM Tris-HCl [pH 8], 10 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 50% glycerol), and stored at −20°C. RNAP was isolated from B. subtilis strain MH5636 as described previously (32).
DNA manipulations.
Routine molecular biology techniques for cloning, sequencing, and PCR amplification were performed as previously described (36). Chromosomal DNA was isolated from GBS strains by using the method of Madoff et al. (30). Plasmid DNA was isolated from GBS by using a modified QIAGEN plasmid mini-prep procedure (19). DNA restriction and modification enzymes were used according to the manufacturer's recommendations (New England Biolabs). GBS was transformed by electroporation as described previously (10).
Generation of recombinant RNAP subunits.
Recombinant δ was expressed in E. coli by using the pET32a expression vector system (Novagen). A DNA fragment containing the entire coding sequence of rpoE was PCR amplified by using high-fidelity polymerase and the primers 5′-CATGCCATGGTATATGGATTAGAAAGAGAGGAATC and 5′-TTGCGGCCGCTTTTCTTGCTCGTTTTCC (underlining indicates either an EcoRI or an NotI restriction enzyme site) and A909 genomic DNA as a template. These primers were designed to include an NcoI or NotI restriction enzyme site in order to facilitate cloning into the pET32a plasmid vector. The resulting PCR product was digested with NcoI and NotI, ligated to similarly digested pET32a, and transformed directly into E. coli Origami (Novagen). The generation of the expected plasmid pET32rpoE was verified by restriction digest analysis and DNA sequencing of plasmids. To induce the expression of the thioredoxin (TRX) hexa-His-tagged-δ fusion protein (TRX-δ), E. coli Origami containing pET32rpoE was grown to an OD600 of 0.6, and isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM for 3 h. The TRX-δ fusion protein was purified under native conditions by using Ni-CAM HC agarose (Sigma) as the His tag affinity resin. TRX (containing the internal His tag) was cleaved from purified TRX-δ with enterokinase (Stratagene), which was used according to the manufacturer's instructions. Cleaved TRX and uncleaved TRX-δ were removed by further incubation with the Ni-CAM HC resin. The flowthrough containing pure δ was reconstituted in a phosphate-buffered saline solution (PBS; pH 7.0) containing 10% glycerol and stored in aliquots at −80°C until required for use.
Recombinant σA and β were generated as His-tagged fusion proteins. For the cloning of rpoD (σA) and rpoB (β), the trx coding sequence was first removed from pET32a by PCR, generating pET32ck. rpoD was amplified by using the primers 5′-CATGCCATGGCAGAGAAAAAAGGAAATAC and 5′-TTGCGGCCGCATCTTCCATGAAATCTTTAAGTTG (underlining indicates either an NcoI or an NotI restriction enzyme site), while rpoB was amplified by using the primers 5′-CGGAATTCTTGGCAGGACATGAAGTTCAG and 5′-TTGCGGCCGCATCTTCTTGAACGACTTCAGA (underlining indicates either an EcoRI or an NotI restriction enzyme site). Amplified products were digested with the appropriate restriction enzymes and ligated to similarly digested pET32ck. E. coli clones containing the correct vector were grown and induced as described above. Recombinant αA and β with carboxyl-terminal His tags were purified under native conditions through incubation with Ni-CAM HC agarose. Eluted protein fractions were reconstituted in PBS and stored as described above.
Preparation of antisera.
Antisera against recombinant δ, αA, and β were generated in female New Zealand White rabbits as a service from Lampire Biological Laboratories (Pipersville, PA). In brief, rabbits were immunized subcutaneously with 500 μg of recombinant protein that was emulsified in complete Freund's adjuvant for the first dose and in incomplete Freund's adjuvant for subsequent doses (days 21 and 42 after initial immunization). Serum was prepared from blood collected approximately 2 weeks after the last dose was given.
Preparation of GBS lysates and Western blot analysis.
GBS strains were grown to a designated OD600, pelleted, washed in PBS, and resuspended in lysis buffer (60 mM Tris-HCl [pH 6.8], 10% glycerol). Whole-cell lysates were prepared through the use of a FastPrep FP101 bead beater (Bio 101) by using 2× 45-s bursts at 4°C at full speed, followed by the addition of SDS (final concentration, 3.3% wt/vol). The lysates were boiled for 5 min, and insoluble material was pelleted by centrifugation. The protein content in the supernatant was quantified by using a bicinchoninic acid assay (Pierce). Samples were normalized for protein concentration, mixed with 2× SDS gel loading buffer (36), and heated at 95°C for 5 min prior to SDS-PAGE analysis (36). The proteins were transferred to a nitrocellulose membrane by using a semidry transfer chamber (Bio-Rad) for 25 min at 15 V. The membrane was blocked overnight at 4°C in a 5% (wt/vol) solution of nonfat dehydrated milk in PBS. The blots were incubated at room temperature with a 1:1,000 dilution of rabbit antisera (raised against the designated protein) for 2 h, followed by a 90-min incubation with a 1:3,000 dilution of Alexa Fluor-680 anti-rabbit immunoglobulin G (IgG) (Molecular Probes). Immunoreactive bands were visualized at 700 nm by using a LiCor infrared imager (LI COR Biosciences).
RNA isolation.
Total RNA was isolated from strains grown to an OD600 of 0.3 by using QIAzol (QIAGEN) according to the manufacturer's instructions, except that GBS cells were lysed through the use of a bead beater as described above. RNA samples were treated with DNase I (Promega) for 60 min at 37°C to remove any contaminating DNA and then purified by using an RNeasy mini kit (QIAGEN). RNA concentration was adjusted to 1 μg/ml, and samples were stored at −80°C until required for use.
Promoter mapping.
Rapid amplification of cDNA ends (RACE) was used to identify the 5′ end of the rpoE transcript in the wild-type strain A909. First-strand cDNA synthesis was carried out according to the protocol of the 5′-RACE system (version 2.0; Invitrogen) by using 2 μg of total RNA and the rpoE-specific primer 5′-CTAAACCTCTTCTTCCTC (GSP1), followed by dCTP tailing of the 5′ end of the cDNA. Tailed cDNA was PCR amplified by using the 5′ RACE-abridged anchor primer (AAP; Invitrogen) and rpoE-specific primer 2 5′-CTAAACCTCTTCTTCCTCTTCTTCC (GSP2) with high-fidelity polymerase (Bioline). The product that was generated served as a template for a second PCR by using the nested primers' abridged universal amplification primer (AUAP, Invitrogen) and rpoE-specific primer 3 5′-AAAGCATTGACACGTTTCTTCTTACG (GSP3). The final PCR product was sequenced and aligned to the published genome sequence by using Sequencher version 3.1 software (Gene Codes Corporation) to identify the transcription initiation start site, untranslated region, and promoter elements.
Construction of a complemented strain.
To confirm that we had identified the correct promoter, we constructed a complemented strain for testing in our animal infection model. rpoE and the promoter were amplified by high-fidelity PCR by using Bio-X-Act DNA polymerase (BioLine) and A909 chromosomal DNA as a template. Restriction enzyme sites for EcoR I were incorporated into the primers to facilitate cloning into pLZ12, a low-copy streptococcal shuttle vector that replicates at 6 to 9 copies per cell in GBS (7). The ligation mixture was transformed into E. coli XL1-Blue, and clones containing the correct construct were identified by PCR. Plasmid DNA was isolated from a positive clone and designated pRN021. Electrocompetent AJ200 was transformed with pRN021 (or pLZ12 as a control) and plated on medium containing Sp to select for the plasmid. The complemented strain, AJ200 containing pRN021, was designated RN114, and AJ200, containing the vector pLZ12, was designated RN115.
Animal infection studies.
Time-mated, barrier-sustained, female Sprague-Dawley rats were obtained from Charles River Laboratories. Fifty-percent-lethal-dose (LD50) assays were performed by using the neonatal rat sepsis model as previously described (18). All procedures were performed in accordance with the guidelines provided by the Children's Hospital and Regional Medical Center Institutional Animal Care and Use Committee.
Construction of a reporter plasmid.
To analyze rpoE expression, the native rpoE promoter was cloned upstream of the gfpmut3 allele to allow for monitoring of promoter activity by flow cytometric analysis of green fluorescent protein (GFP) expression. The streptococcus-E. coli shuttle vector, pDC125 (4), was used to construct a reporter plasmid. The original phoZ and cat reporter genes were removed from pDC125 by PCR, and ClaI and NotI sites were added to facilitate the cloning of inserts. A streptococcal ribosomal binding site (GGAGG) (9) was inserted into the vector 7 bp upstream of an ATG start codon that was contained within the cloned ClaI site to optimize the expression of GFP. The gfpmut3 allele, which has been optimized for bacterial codon usage (6), was PCR amplified by using pBL26 as a template. ClaI and NotI sites were incorporated into the 3′ and 5′ ends of gfpmut3, respectively. The resulting PCR product and plasmid were digested with ClaI and NotI and ligated together to generate pRSgfpmut3. A BamHI site was introduced into the gfp sequence of pRSgfpmut3 to facilitate the insertion of the rpoE promoter fragment. An EcoR I-BamHI fragment, containing the promoter and the first four codons of rpoE, was ligated to pRSgfpmut3 cut with the same enzymes, generating pRSPrpoE-gfp. This reporter construct contains an in-frame fusion of the rpoE promoter to gfp.
Flow cytometry.
Flow cytometric analysis was performed with a BD FACSCalibur (Becton Dickinson) equipped with a 488-nm argon laser and FlowJo software version 4.6.2. Overnight cultures of strains containing the reporter construct and the control vector were subcultured into fresh medium and grown to the indicated growth phases. Prior to analysis, cells were fixed at 4°C overnight in 2% (wt/vol) paraformaldehyde in PBS, washed, and finally resuspended in PBS. The PBS used in this study was filtered through a 0.22-μm-pore-size filter (Millipore) to minimize fluorescence. A total of 100,000 events were analyzed for each sample at constant parameter settings for each experiment.
Northern hybridization.
Total RNA was separated on a 0.8% agarose gel and transferred to a positively charged nylon membrane (Hybond N+, Roche) by alkaline transfer as described previously (36). A probe that encompassed the complete rpoE open reading frame was generated by incorporating digoxigenin-11-dUTP into a PCR product by using the protocol provided by the manufacturer (Roche Diagnostics) and the following primer pair: 5′-ATGACAAAAAAACATCTTAAAACG and 5′-TTGCGGCCGCTTTTCTTGCTCGTTTTCC. The hybridization and wash conditions used were as specified by Roche Diagnostics. Gene-specific bands were detected by chemiluminescence using CPSD (Roche Diagnostics) as the alkaline phosphatase substrate.
RESULTS
δ is a subunit of RNA polymerase in GBS.
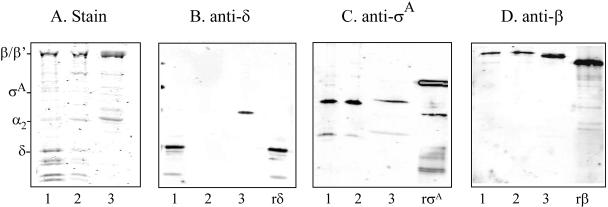
To identify the subunits of RNAP in GBS, native RNAP was purified from the wild-type strain A909 by using a modified affinity and size exclusion chromatography procedure that takes advantage of specific substrate binding and the size of the enzyme. Chromatographic steps included heparin affinity, anion exchange (UNO Q1), and size exclusion (S-300) chromatography. RNAP was isolated from B. subtilis MH5636 for comparison. As seen in Fig. 1A, SDS-PAGE analysis of the RNAP preparation from A909 revealed that it consists of subunits that are consistent with the expected sizes for β′ (135 kDa), β (135 kDa), α (34 kDa), σA (42 kDa), and δ (21 kDa). This is similar to what we observed for B. subtilis MH5636. These results are in agreement with the expected sizes for the gene products of rpoC, rpoB, rpoA, rpoD, and rpoE, respectively, based on the available GBS genome sequences (12, 40). As a control, we also isolated RNAP from AJ200, a mutant in which rpoE has been replaced with a kanamycin cassette by allelic exchange (19). As expected, while we detected subunits corresponding to β′, β, α, and σ, the δ protein was absent.
FIG. 1.
RNAP purification and subunit analysis. A total of 1 to 2 μg of purified RNAP from GBS strains A909 (1) and AJ200 (2) and B. subtilis strain MH5636 (3) were separated on a 12% SDS-polyacrylamide gel and stained with Coomassie blue (A). Subunit identity is shown on the left. Western blot analysis was performed on the RNAP preparations by using anti-δ (B), anti-σA (C), or anti-β (D) antisera. Recombinant δ (rδ), σA (rσA), and β (rβ) served as positive controls. Immunoreactive bands were detected by using an infrared imager after incubation with a rabbit anti-IgG infrared-labeled secondary antibody.
Western blot analysis was used to further confirm the association of δ with RNAP in GBS. Antiserum raised against purified recombinant GBS δ was used to probe the purified RNAP preparations from A909, AJ200, and B. subtilis MH5636. The antiserum reacted with a 21-kDa band corresponding to δ in the RNAP that was isolated from A909 (Fig. 1B). Although there was an approximately 21-kDa protein visible in the Coomassie-stained gel of the RNAP preparation from AJ200 (Fig. 1A), as expected, δ was not detected by Western blot analysis in this preparation (Fig. 1B). The antiserum did not recognize δ in the bacillus RNAP. This observation can be explained by the fact that there is a significant difference in the amino acid composition of δ between GBS and B. subtilis (33% identity and 61% similarity). Alternatively, there may be species-specific differences in the tertiary structure of this protein. Although the antisera cross-reacted with a 40-kDa protein in this RNAP preparation, antisera raised against bacillus δ also identified a similarly sized bacillus protein in addition to δ (data not shown). To confirm that we had correctly identified the other subunits of RNAP in GBS, Western blot analysis using antisera directed against σA (Fig. 1C) and the β subunit (Fig. 1D) was performed. Both subunits were detected in the RNAP preparations from the GBS strains that we examined. These data suggest that under these conditions, the sigma factor that is associated with RNAP is σA. The antisera also reacted with the σA and β subunits in RNAP from B. subtilis MH5636, which is consistent with the high degree of amino acid sequence homology that the B. subtilis σA (65% identity and 78% similarity) and β (68% identity and 82% similarity) subunits share with the GBS homologs.
δ is expressed among all serotypes of GBS.
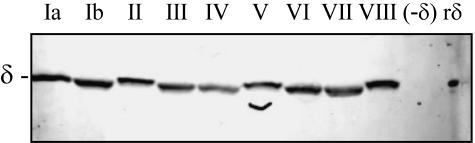
Western blot analysis was used to examine whether the expression of δ is limited to strains in serotype 1a. Whole-cell lysates of strains representing all nine serotypes were prepared. The anti-δ antiserum detected an approximately 21-kDa band corresponding to δ in all of the strains that were tested (Fig. 2). These data indicate that the expression of δ is not limited to A909.
FIG. 2.
Analysis of δ expression across the serotypes. Western blot analysis was performed on lysates of strains representing all nine clinically relevant serotypes by using antiserum raised against recombinant δ. The serotype of the strain is indicated above each lane. Ia, A909; Ib, DK14; II, DK23; III, COH1; IV, CNCTC 1/82; V, CNCTC 10/84; VI, NT6; VII, 87-603; VIII, M9; (-δ), AJ200; rδ, recombinant delta protein.
rpoE is expressed from a σA-dependent promoter.
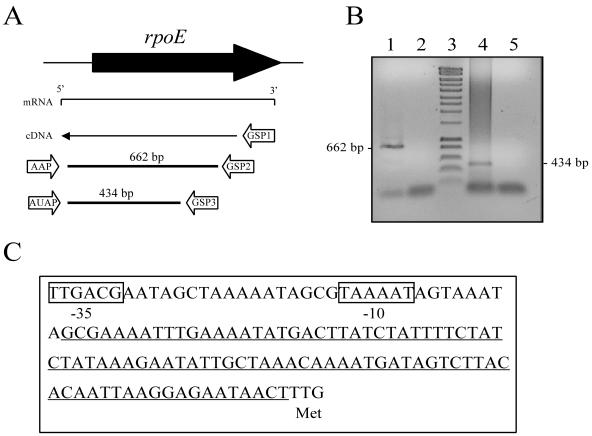
To identify the transcriptional start site and the promoter consensus elements, we mapped the 5′ end of the rpoE transcript by 5′-RACE PCR. The positions of the primers and predicted sizes of the products are shown in Fig. 3A. Sequencing of the PCR product that was generated by using the gene-specific GSP3 and AUAP anchor primers identified the transcriptional initiation site, which was located ∼109 bp upstream of the TTG start codon (Fig. 3C). Comparison of this sequence to the A909 genome sequence identified a putative −10 and −35 promoter region that was located 9 bp upstream of the 5′ untranscribed region. The predicted −35 (TTGACG) and −10 (TAAAAT) elements are consistent with what has been reported for σA-dependent promoters in B. subtilis (16), suggesting that rpoE is transcribed by the RNAP that is associated with σA in GBS.
FIG. 3.
Identification of the rpoE promoter. (A) Schematic diagram of the 5′-RACE PCR strategy, primers, and product sizes. (B) Amplicons were obtained in primary PCR by using primers AAP and GPSP2 (lane 1) and were obtained in nested PCR by using primers AUAP and GSP3 (lane 4). Lanes 2 and 5 contain the respective negative controls for these PCRs, and lane 3 contains a DNA marker. (C) Sequence of rpoE promoter and 5′-untranslated region. The predicted −35 and −10 consensus elements are boxed. The 5′-untranslated region is underlined. The methionine start codon (TTG) is also indicated.
Complementation of virulence in neonatal rat sepsis model.
The rpoE deletion mutant AJ200 was transformed with a low-copy vector containing the rpoE coding sequence and promoter elements that we identified by using the 5′-RACE technique. We compared the virulence of the complemented strain (RN114) to the wild-type strain in the neonatal rat sepsis model. AJ200 containing empty vector (RN115) was used as a control. In two separate experiments, providing rpoE expressed from the promoter that we identified on a low-copy vector restored virulence to wild-type levels (Table 2). The log of the LD50 for RN114 was similar to that for the wild-type strain A909, while the log of the LD50 for RN115 was 1.5 to 2 log units higher than that for the wild type. These data confirmed that we had correctly identified the native rpoE promoter. In addition, the virulence of the original transposon mutant AJ8D3 was compared to those of A909 and the complemented strain. The LD50 for AJ8D3 was 2 log units higher than those for A909 and RN114 (Table 2), confirming that rpoE expression is required for survival in the host.
TABLE 2.
Lethal dose values for complemented strains in neonatal rat sepsis model of infection
| Strain | Log LD50 for expt. no.
|
|
|---|---|---|
| 1 | 2 | |
| A909 (wt) | 4.91 | 5.6 |
| RN115 (ΔrpoE) | 6.99 | 7.11 |
| RN114 (ΔrpoE + rpoE) | 4.85 | 5.24 |
| AJ8D3 (A909 Tn917::rpoE) | 7.00 | |
LD50 values were calculated using the method of Reed and Muench (34).
Analysis of rpoE expression.
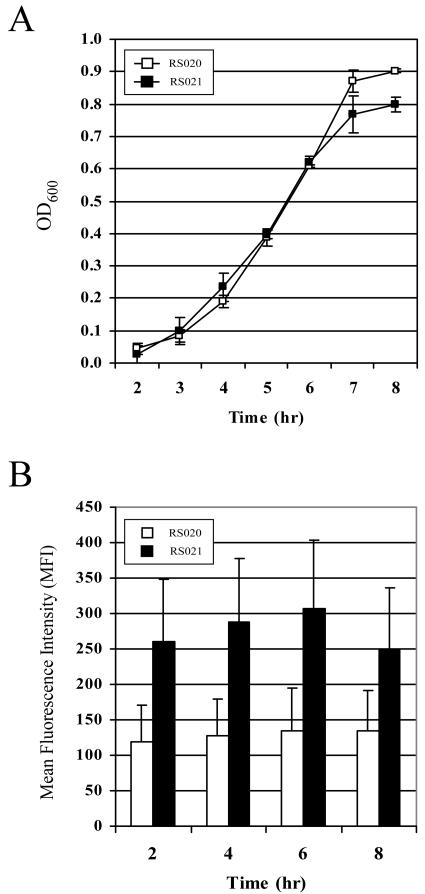
Once we confirmed that we had identified the native promoter for rpoE, we generated a reporter construct to characterize expression during growth in vitro. The expression of rpoE was analyzed by measuring the fluorescence of strains containing transcriptional fusions to gfpmut3. The promoter region for rpoE was cloned upstream of a promoterless gfpmut3 allele, generating pRSPrpoE-gfp. A909 was transformed with the reporter plasmid, generating strain RS021, or empty vector as a control, generating strain RS020. We first confirmed that the expression of GFP from the rpoE promoter did not affect growth of the strain. As shown in Fig. 4A, the expression of GFP did not significantly affect the growth of RS021 in laboratory medium relative to the control strain RS020. Flow cytometric analysis of GFP expression in RS021 indicated that rpoE was expressed during all stages of growth. GFP fluorescence increased from lag to early logarithmic phase, reached a maximum at the late logarithmic stage, and dropped off during stationary phase (Fig. 4B). These data are consistent with what we have observed by using microarray analysis of rpoE expression in A909 during growth in vitro (unpublished data), confirming that the pattern of expression that we observed with the plasmid construct is representative of gene expression occurring from the chromosomal locus.
FIG. 4.
Analysis of rpoE promoter activity using a reporter construct. (A) Growth of RS021 containing the rpoE reporter plasmid (▪) and the RS020 control strain (□). (B) Flow cytometric analysis of RS021 and RS020. GFP fluorescence is shown as the mean fluorescence intensity (MFI) of the bacterial population, which was calculated by using the FlowJo software version 4.6.2. The data presented are means ± standard error of the means and are representative of three experiments.
rpoE is transcribed in AJ8D3.
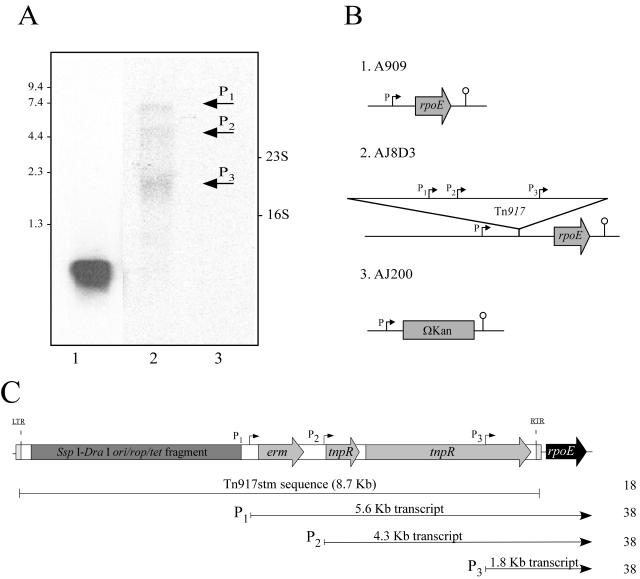
rpoE was first identified as being required for virulence of GBS during a signature-tagged transposon mutagenesis (STM) screen utilizing a modified Tn917 (Tn917stm) (18). The transposon mutant AJ8D3 was severely attenuated for virulence and had a single Tn917stm insertion adjacent to rpoE. Sequencing of genomic DNA isolated from AJ8D3 subsequently revealed that the transposon had inserted between the −10 promoter element and the transcription initiation site (data not shown). To examine whether rpoE was expressed in AJ8D3, Northern blot analysis was conducted on total RNA isolated from this mutant and from A909 for comparison. Using a probe that was specific for rpoE, we detected a single transcript in RNA from A909 that we estimated to be 550 bp in size, indicating that rpoE is transcribed as a monocistronic message (Fig. 5A). In contrast, multiple transcripts were detected in the AJ8D3 RNA. Three primary transcripts were detected (Fig. 5A). However, none were large enough to represent a transcript that was being initiated from the native promoter now located 8.6 kb upstream of the coding sequence for rpoE. These data indicate that transcription of rpoE in AJ8D3 is being initiated from promoters within the transposon. In addition, the total band intensity of the rpoE-specific transcripts detected in AJ8D3 was considerably reduced compared to that of the single transcript detected in A909, suggesting that the strength of the nonnative promoters may be weaker than the native one. Control experiments using a probe for rpsL and the same RNA samples demonstrated that equal amounts of total RNA from the isogenic strains were used in the analysis (data not shown).
FIG. 5.
Transcriptional analysis of rpoE in isogenic strains. (A) Northern blot of RNA prepared from A909 (1), AJ8D3(2), and AJ200 (3) using a digoxigenin-labeled DNA probe that was specific for rpoE. The positions of size markers (in kilobases) and the 23S and 16S rRNAs are indicated. (B) Schematic diagram of the rpoE locus in the isogenic strains. (C) Promoter location within Tn917stm. The location of promoter regions within Tn917stm are indicated. Predicted transcript sizes that were initiated from these three promoters sites are given in kilobases. Shaded box, pBR322 ori/rop/tet region; gray arrows, erythromycin gene (erm), resolvase (tnpR), and transposase (tnpR); black arrow, rpoE gene. LTR, left terminal repeat; RTR, right terminal repeat. Promoter regions within are represented as P1, P2, and P3. Numbers on the right are reference numbers.
The 5′-RACE approach was used to identify the transcription initiation sites for rpoE in AJ8D3. Consistent with our Northern analysis, we obtained several amplification products from the 5′-RACE PCR (data not shown). Previous sequencing of Tn917, together with transcription studies in Enterococcus faecalis transposon mutants, identified three functional promoters (P1, P2, and P3) (Fig. 5C) that were responsible for initiating the transcription of Tn917 genes (38). The promoter P1 (−35 [TTGATA] and −10 [TATAAT]) is located upstream of the erm gene, promoter P2 (−35 [TTAATG] and −10 [TATAAT]) is immediately upstream of the resolvase gene tnpR and promoter P3 (−35 [ATGCCA] and −10 [TATAAA]) is located within the transposase (tnpA). Based on the size of the transcripts and the sequence of the 5′-RACE PCR products that we obtained from AJ8D3, it is likely that the transcription of rpoE is being initiated from these previously identified promoters in Tn917.
Abundance of δ in AJ8D3 is reduced compared to that of the wild-type strain.
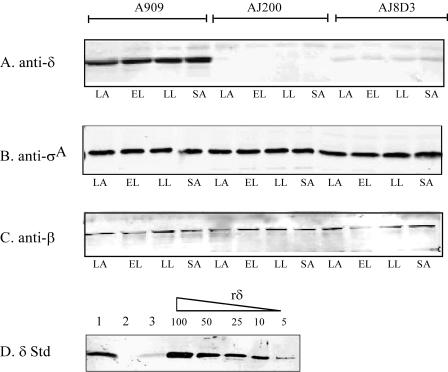
Since we detected transcription of rpoE from nonnative promoters in AJ8D3, we sought to determine whether these messages were translated into protein. We compared the expression of δ in whole-cell lysates from A909 and AJ8D3 during growth in vitro by Western blot analysis using the antiserum directed against δ. In order to estimate the relative quantity of δ, a standard curve of serially diluted recombinant protein was prepared and analyzed in parallel with the lysates. As shown in Fig. 6A, the amount of δ in A909 appeared constant throughout all phases of growth, indicating continual expression. Interestingly, we also detected δ in AJ8D3 at all growth phases tested. However, the amount of δ protein in AJ8D3 was significantly reduced. Comparison of the band intensity to the standard curve (Fig. 6D) indicated that at all stages of growth, there is at least 10-fold less δ present in AJ8D3 compared to that in A909. As expected, no δ was detected in AJ200. As a control to demonstrate equal protein loading between samples, we repeated the analysis by using antisera directed against σA (Fig. 6B) and the β subunit (Fig. 6C). Western blotting using anti-σA and anti-β antisera indicated that the levels of these two subunits remained constant in the three strains throughout growth. These data demonstrated that the transcription of rpoE from the nonnative promoters in Tn917 results in a reduction in δ protein levels in AJ8D3. This finding provides a likely explanation for the attenuated virulence that was observed for this mutant and suggests that a specific level of δ is required for virulence of GBS.
FIG. 6.
Immunoblot analysis of δ, σA, and β subunits during growth. A909, AJ200, and AJ8D3 were grown to lag (LA) (OD600 = 0.1), early log (EL) (OD600 = 0.3), late-log (LL) (OD600 = 0.3) and stationary (SA) phases (OD600 = 1.0). Lysates were normalized for total protein (10 μg) and subjected to SDS-PAGE. Western blots were performed by using antisera directed against δ (A), σA (B), and β (C). Protein levels were probed by using polyclonal sera raised against the recombinant protein. (D) Estimation of δ levels. A total of 10 μg of A909 (1), AJ200 (2), and AJ8D3 (3) lysates were loaded together with 100 ng, 50 ng, 25 ng, 10 ng, and 5 ng of purified δ. Blots were probed with antisera directed against δ. Immunoreactive bands were detected by using an infrared imager after incubation with a rabbit anti-IgG infrared-labeled secondary antibody, and band intensity was compared between strains. rδ, recombinant delta protein.
DISCUSSION
Bacterial RNAP is considered to play a pivotal role in global changes in gene expression. Although the core enzyme is a highly conserved heteromeric structure (ββ′α2), the composition of the holoenzyme can differ considerably among bacterial species and with the environmental conditions. A significant amount of information regarding RNAP composition and the mechanics of transcription has been generated. However, most of this information is based on the analysis of only a few model organisms, including E. coli and B. subtilis. Our knowledge of RNAP composition in streptococci, particularly in GBS, remains in its infancy. Yet, the elucidation of the composition of the transcriptional machinery is of fundamental importance for understanding the mechanisms that this microbe utilizes to control the transcription process.
In this study, we used a well-documented multistage purification procedure to isolate RNAP from the cytosol of GBS. To our knowledge, this represents the first detailed description of RNAP isolation and subunit composition in GBS. The RNAP that we isolated consisted of subunits corresponding to α, β, β′, and δ, which are consistent with RNAP composition analyses reported for other gram-positive bacterial species (7, 17, 28, 39). The preparation also contained a subunit corresponding to σA, the primary sigma factor in gram-positive bacteria. Interestingly, following size exclusion chromatography, the final GBS RNAP preparation also contained a small number of protein bands in addition to the α, β, β′, σA, and δ subunits, indicating that other potential RNAP-associated proteins may have been isolated. Analyses of the available genome sequences have identified genes encoding two alternative sigma factors in addition to σA (12, 40). Thus, these additional protein bands may represent alternative sigma factors. Alternatively, proteins sharing a similar binding affinities to the various columns used in the chromatography steps may have also copurified, a feature that is indicative of isolating a multisubunit enzyme (17, 39). However, without further analysis, it is impossible to determine whether these proteins represent additional sigma factors, breakdown products of the core, or copurifying contaminating proteins.
The δ protein is a subunit of RNAP found in only gram-positive bacteria. This ∼21 kDa protein was originally identified in B. subtilis (26) but has also been demonstrated to copurify with RNAP in Staphylococcus aureus and Streptococcus pneumoniae (7, 28). Here we provide evidence that the δ homolog in GBS also associates with RNAP following purification.
GBS strains are grouped into serotypes on the basis of capsular polysaccharide on the bacterial surface. A number of virulence traits, particularly surface proteins, have been reported to be serotype specific (for a review, see reference 23). Western blot analysis of a panel of strains representing all of the nine serotypes confirmed that δ was expressed in all of the strains tested and not limited to one serotype. Taken together with the observation that an rpoE homolog is present in every gram-positive bacterial genome currently sequenced (19), it appears that δ is highly conserved among gram-positive bacteria.
The putative promoter for rpoE was identified by using a 5′-RACE PCR approach. The expression of rpoE from this promoter on a low-copy plasmid restored the virulence of the deletion mutant to wild-type levels, confirming that we had correctly identified the promoter. Reporter constructs were then used to monitor rpoE expression from this promoter during growth in vitro. The expression of rpoE was continual throughout growth, but it appeared to reach a maximal level during exponential growth. A similar pattern of expression has been reported for rpoE in Bacillus subtilus (25), suggesting that this pattern may be conserved among gram-positive bacteria.
δ was initially linked to the virulence of GBS when a mutant with a transposon insertion adjacent to rpoE was identified in an STM screen (18). In the current study, we demonstrated that rpoE is transcribed in the transposon mutant AJ8D3 but that transcription is initiated from multiple promoters within the transposon and not from the native promoter. Additionally, the overall amount of rpoE transcript was substantially reduced compared to that of the wild-type strain. The modified transposon used in the STM screen (Tn917stm) contains at least five known promoters. Three promoters were originally reported during the sequencing and analysis of Tn917 (38), and modification of the transposon for STM introduced two additional promoters (18). The results presented here indicate that the transcription of rpoE can occur from these promoters, although this appears to be less efficient than from the native promoter. Western blot analysis demonstrated that this message is translated into δ protein. However, δ is present in reduced abundance of ∼10-fold relative to the wild-type strain. We have previously reported that AJ8D3 is as attenuated in the neonatal rat sepsis model as it is in the rpoE deletion mutant that does not express any δ (19). Based on these collective observations, we conclude that a critical amount of δ is required within the cell for virulence but that the levels of the protein in AJ8D3 do not reach this threshold. The decreased amount of δ in AJ8D3 is likely a result of the low abundance of the rpoE transcript. It is also possible that transcripts that were initiated from the promoters in the transposon are less stable or are translated less efficiently.
It is not yet known what role δ plays in the virulence of GBS. Functional activity for δ has been demonstrated in Bacillus subtilis by using in vitro assays (20, 24, 39). The generation of rpoE mutants of Bacillus subtilis has been reported (21, 25), though they lack a well-defined phenotype. More recently, it has been suggested that rpoE may have a direct or indirect role in Bacillus subtilis sporulation by affecting the transcription of genes that are required at specific stages (11). A mutation in rpoE has also been reported to affect the ability of S. aureus to recover from nutrient starvation (42).
If the activities demonstrated for bacillus δ can be extrapolated to GBS, then maintaining the transcriptional specificity and efficient recycling of RNAP would appear to have a profound effect on virulence. These functions may allow this organism to adapt to environmental change more efficiently, making it a more successful pathogen. It seems unlikely that δ has global effects on transcription since rpoE is not required for viability and mutants have only a limited number of phenotypic changes (11, 19, 25, 42). In GBS, δ may impact the expression of a subset of genes that are required at a critical stage for survival in our animal model. Our data suggest that the relative abundance of δ is of critical importance for virulence of this organism. Further study is needed to determine how δ influences RNAP activity and whether it affects gene expression in vivo.
Acknowledgments
We thank J. D. Helmann for generously providing B. subtilis strain MH5636 and antiserum directed against bacillus δ as well as Donald Chaffin for technical assistance with the HPLC.
This work was funded by the Streptococcal Initiative of the National Institutes of Health, grant no. BWH 811501/N01-AI-75326 and R01AI52299-01.
REFERENCES
- 1.Achberger, E. C., M. D. Hilton, and H. R. Whiteley. 1982. The effect of the delta subunit on the interaction of Bacillus subtilis RNA polymerase with bases in a SP82 early gene promoter. Nucleic Acids Res. 10:2893-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. J., and M. W. Edwards. 2005. Group B streptococcal infections, p. 1091-1156. In J. S. Remington, J. O. Klein, C. B. Wilson, and C. J. Baker (ed.), Infectious diseases of the fetus and newborn infant, 6th ed. Elsevier Saunders, Philadelphia, Pa.
- 3.Blancas, D., M. Santin, M. Olmo, F. Alcaide, J. Carratala, and F. Gudiol. 2004. Group B streptococcal disease in nonpregnant adults: incidence, clinical characteristics, and outcome. Eur. J. Clin. Microbiol. Infect. Dis. 23:168-173. [DOI] [PubMed] [Google Scholar]
- 4.Chaffin, D. O., and C. E. Rubens. 1998. New vectors for blue/white screening and in vitro expression (IVET) in gram-positive bacteria. American Society for Microbiology Conference on Streptococcal Genetics, Vichy, France.
- 5.Chmouryguina, I., A. Suvorov, P. Ferrieri, and P. P. Cleary. 1996. Conservation of the C5a peptidase genes in group A and B streptococci. Infect. Immun. 64:2387-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 7.Deora, R., and R. Misra. 1995. Purification and characterization of DNA dependent RNA polymerase from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 208:610-616. [DOI] [PubMed] [Google Scholar]
- 8.de Vos, W. M. 1986. Genetic improvement of starter streptococci by the cloning and expression of the gene coding for a nonbitter proteinase, p. 465-472. In E. Magnien (ed.), Biomolecular engineering in the European community: achievements of the research programme (1982-1986) final report. Martinus Nijhoff, Lancaster, England.
- 9.Ferretti, J. J., and R. Curtiss (ed.). 1987. Streptococcal genetics. American Society for Microbiology, Washington, D.C.
- 10.Framson, P., A. Nittayajarn, J. Merry, P. Youngman, and C. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, H., and A. I. Aronson. 2004. The delta subunit of RNA polymerase functions in sporulation. Curr. Microbiol. 48:401-404. [DOI] [PubMed] [Google Scholar]
- 12.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 13.Gotoff, S. P. 2002. Group B streptococcal infections. Pediatr. Rev. 23:381-386. [PubMed] [Google Scholar]
- 14.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 15.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helmann, J. D. 2003. Purification of Bacillus subtilis RNA polymerase and associated factors. Methods Enzymol. 370:10-24. [DOI] [PubMed] [Google Scholar]
- 18.Jones, A. L., K. M. Knoll, and C. E. Rubens. 2000. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol. Microbiol. 37:1444-1455. [DOI] [PubMed] [Google Scholar]
- 19.Jones, A. L., R. H. V. Needham, and C. E. Rubens. 2003. The delta subunit of RNA polymerase is required for virulence of Streptococcus agalactiae. Infect. Immun. 71:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juang, Y. L., and J. D. Helmann. 1995. Pathway of promoter melting by Bacillus subtilis RNA polymerase at a stable RNA promoter: effects of temperature, delta protein, and sigma factor mutations. Biochemistry 34:8465-8473. [DOI] [PubMed] [Google Scholar]
- 21.Lampe, M., C. Binnie, R. Schmidt, and R. Losick. 1988. Cloned gene encoding the delta subunit of Bacillus subtilis RNA polymerase. Gene 67:13-19. [DOI] [PubMed] [Google Scholar]
- 22.Lancefield, R. C., M. McCarty, and W. N. Everly. 1975. Multiple mouse-protective antibodies directed against group B streptococci. Special reference to antibodies effective against protein antigens. J. Exp. Med. 142:165-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindahl, G., M. Stalhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez de Saro, F. J., A. Y. Woody, and J. D. Helmann. 1995. Structural analysis of the Bacillus subtilis delta factor: a protein polyanion which displaces RNA from RNA polymerase. J. Mol. Biol. 252:189-202. [DOI] [PubMed] [Google Scholar]
- 25.Lopez de Saro, F. J., N. Yoshikawa, and J. D. Helmann. 1999. Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis. J. Biol. Chem. 274:15953-15958. [DOI] [PubMed] [Google Scholar]
- 26.Losick, R., and J. Pero. 1976. Bacillus subtilis RNA polymerase and its modification in sporulating and phage-infected bacteria. Adv. Enzymol. Relat. Areas Mol. Biol. 44:165-185. [DOI] [PubMed] [Google Scholar]
- 27.Luck, S., M. Torny, K. d'Agapeyeff, A. Pitt, P. Heath, A. Breathnach, and A. B. Russell. 2003. Estimated early-onset group B streptococcal neonatal disease. Lancet 361:1953-1954. [DOI] [PubMed] [Google Scholar]
- 28.Luo, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madoff, L. C., S. Hori, J. L. Michel, C. J. Baker, and D. L. Kasper. 1991. Phenotypic diversity in the alpha C protein of group B streptococci. Infect. Immun. 59:2638-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madoff, L. C., J. L. Michel, E. W. Gong, D. E. Kling, and D. L. Kasper. 1996. Group B streptococci escape host immunity by deletion of tandem repeat elements of the alpha C protein. Proc. Natl. Acad. Sci. USA 93:4131-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manganelli, R., L. Fattorini, D. Tan, E. Iona, G. Orefici, G. Altavilla, P. Cusatelli, and I. Smith. 2004. The extra cytoplasmic function sigma factor σE is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 72:3038-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi, Y., and F. M. Hulett. 1998. PhoP-P and RNA polymerase σA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1197. [DOI] [PubMed] [Google Scholar]
- 33.Rabinowitz, S., M. Ferne, and S. T. Halfon. 1980. Group B streptococcal serotypes associated with clinical infection. Lab. Pract. 29:733-734. [Google Scholar]
- 34.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 35.Rubens, C. E., H. V. Raff, J. C. Jackson, E. Y. Chi, J. T. Bielitzki, and S. L. Hillier. 1991. Pathophysiology and histopathology of group B streptococcal sepsis in Macaca nemestrina primates induced after intraamniotic inoculation: evidence for bacterial cellular invasion. J. Infect. Dis. 164:320-330. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schrag, S. J., and A. Schuchat. 2004. Easing the burden: characterizing the disease burden of neonatal group B streptococcal disease to motivate prevention. Clin. Infect. Dis. 38:1209-1211. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiegelman, G. B., W. R. Hiatt, and H. R. Whiteley. 1978. Role of the 21,000 molecular weight polypeptide of Bacillus subtilis RNA polymerase in RNA synthesis. J. Biol. Chem. 253:1756-1765. [PubMed] [Google Scholar]
- 40.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velaphi, S., J. D. Siegel, G. D. Wendel, Jr., N. Cushion, W. M. Eid, and P. J. Sanchez. 2003. Early-onset group B streptococcal infection after a combined maternal and neonatal group B streptococcal chemoprophylaxis strategy. Pediatrics 111:541-547. [DOI] [PubMed] [Google Scholar]
- 42.Watson, S. P., M. Antonio, and S. J. Foster. 1998. Isolation and characterization of Staphylococcus aureus starvation-induced, stationary-phase mutants defective in survival or recovery. Microbiology 144:3159-3169. [DOI] [PubMed] [Google Scholar]
- 43.Weisner, A. M., A. P. Johnson, T. L. Lamagni, E. Arnold, M. Warner, P. T. Heath, A. Efstratiou, G. Balfour, H. Tighe, L. A. O'Connell, M. Cafferkey, N. Q. Verlander, A. Nicoll, A. C. McCartney, C. Simon, H. Schroder, D. Weisner, M. Bruck, and U. Krieg. 2004. Characterization of group B streptococci recovered from infants with invasive disease in England and Wales. Clin. Infect. Dis. 38:1203-1208. [DOI] [PubMed] [Google Scholar]
- 44.Wilkinson, H. W. 1977. Nontypable group B streptococci isolated from human sources. J. Clin. Microbiol. 6:183-184. [DOI] [PMC free article] [PubMed] [Google Scholar]