Abstract
Iron uptake systems were identified by global expression profiling of Magnetospirillum magneticum AMB-1. feo, tpd, and ftr, which encode ferrous iron transporters, were up-regulated under iron-rich conditions. The concomitant rapid iron uptake and magnetite formation suggest that these uptake systems serve as iron supply lines for magnetosome synthesis.
Iron is crucial in microbial metabolism. It exists in two redox states: the reduced Fe2+ soluble ferrous state and the oxidized Fe3+ ferric form, which is extremely insoluble (1). In magnetotactic bacteria, the formation of highly organized membrane-bound intracellular magnetites (Fe3O4) or greigites (Fe3S4) requires the acquisition of a large amount of iron several orders of magnitude greater than that required by Escherichia coli (5). Because of the large amount of iron to be transported, complex iron uptake systems that differ from known transport mechanisms in other microorganisms are expected to be operating in magnetotactic bacteria. Exceptional progress has been made in the isolation and characterization of functional genes and proteins involved in magnetosome synthesis (2, 11, 13, 21, 24), but at present, specific iron transport systems remain unidentified in magnetotactic bacteria.
Here we report the identification of iron uptake systems by global expression profiling of the magnetotactic bacterium Magnetospirillum magneticum AMB-1 grown under iron-rich and iron-deficient conditions. Our results indicate that despite the unusual high-iron requirement of M. magneticum AMB-1, it utilizes robust but simple iron uptake systems similar to those of other gram-negative bacteria. This robust ferrous iron uptake suggests a significant contribution to magnetite synthesis. This study is the first to identify specific iron uptake systems in the complex iron metabolism of magnetotactic bacteria. The data presented here may facilitate future studies on the mechanism of magnetosome formation.
To monitor iron uptake and magnetite formation, M. magneticum AMB-1 (ATCC 700264) was grown at 25°C under microaerobic conditions by sparging argon gas for 10 min into 500 ml of MSGM medium as previously described (5), with various iron concentrations of 0.1 to 300 μM. All iron measurements were performed by atomic absorbance spectrophotometry. Extracellular iron concentrations were measured at different time points in cell-free culture supernatants. For intracellular iron measurements, cells were disrupted by lysozyme treatment (20) and ultracentrifuged at 100,000 × g to separate the insoluble (magnetites) and soluble iron fractions.
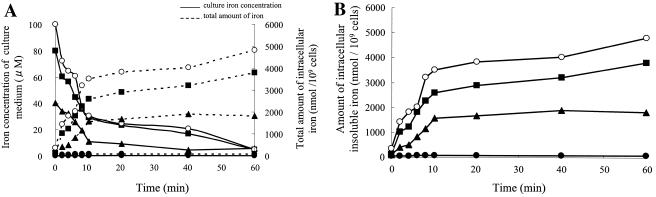
Iron was rapidly taken up in iron-rich cultures, and a corresponding increase of intracellular iron was observed within 10 min (Fig. 1A). Up to 70% of the initial iron concentration of the medium was taken up, and intracellular iron increased to 5,000 nmol/109 cells after 60 min. Insoluble iron in the cytoplasm, which mostly included magnetites, also increased within 10 min (Fig. 1B). These data indicate that the external iron was rapidly assimilated and formed into magnetite. Such rapid magnetite formation has also been observed in M. gryphiswaldense (26). Transmission electron microscopy confirmed the absence of magnetosomes in cells grown in 0.1 μM iron.
FIG.1.
Time course of the iron concentration in growth medium (MSGM) with initial iron concentrations of 0.1 (•), 40 (▴), 80 (▪), and 100 (○) μM. The amounts of total iron (A) and insoluble iron (magnetites) (B) from whole-cell extracts from three biological replicates were measured. Solid lines in panel A indicate the time course of iron concentrations in the culture medium. The total amount of iron is indicated by dotted lines.
To relate the robust iron uptake to the corresponding gene expressions of M. magneticum AMB-1 cultured under different iron conditions, transcription profiles were obtained by standard DNA microarray (8, 18, 19). The sequence of each of the 4,492 genes obtained from M. magneticum AMB-1 (15), representing ∼99% of the total protein-coding capacity of the whole genome sequence, was determined and synthesized from the 60-mer region of minimal homology to other open reading frames analyzed with the BLAST program. The amino-activated oligonucleotides (40 pmol) were imprinted onto glass slides (TaKaRa-Hubble Slide Glass; TaKaRa Bio Inc., Shiga, Japan). Total RNA was extracted by the hot phenol acid method (10) and purified with an RNeasy Mini kit (QIAGEN, Hilden, Germany). The threshold value was determined by using RNA extracted from the strain AMB-1 cultures with various iron concentrations. Five micrograms of purified RNA was reverse transcribed with an RNA fluorescence labeling kit (TaKaRa Bio Inc.) by using random 6-mer primers and the fluorochromes Cy3 dUTP (for mRNA of cells grown with 20 to 300 μM iron) and Cy5 dUTP (for mRNA of cells grown with 0.1 μM iron). Cy3- and Cy5-labeled cDNAs were hybridized with oligonucleotides onto glass slides. The signal value of each spot was determined by the following formula: (signal of each spot − background)/sum of all the signal values from the 4,492 AMB-1 genes. The Cy3/Cy5 fluorescence ratio was plotted, and the distribution was determined. Most genes exhibited signals with ratios ranging from 0.7 to 1.5; hence, up-regulated genes were defined by a signal ratio of greater than 1.5 and down-regulated genes were defined by a signal ratio of less than 0.7. Relative gene expression is presented as the n-fold change in the fluorescence intensity of cDNA synthesized from total RNA derived from cells grown under several iron conditions (magnetosome-forming condition: cultures with a 20 to 300 μM initial iron concentration) compared to that of the reference condition (non-magnetosome-forming condition: culture with a 0.1 μM initial iron concentration).
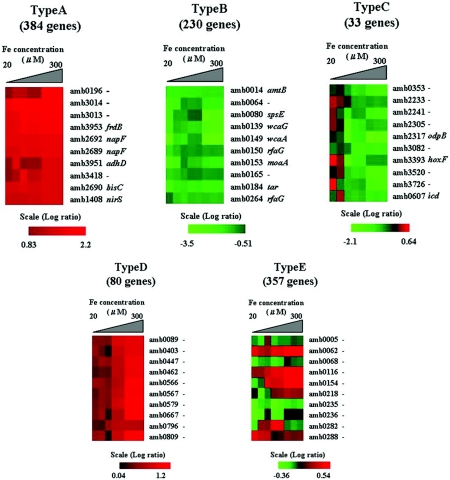
Global gene expression profiling of M. magneticum AMB-1 yielded five gene expression profile patterns (genes which have no uniform ratio were not categorized). Figure 2 shows the patterns categorized as types A, B, C, D, and E. Type A (384 genes) shows a pattern of genes consistently up-regulated in cells grown at initial iron concentrations of 20 to 300 μM with signal ratios 1.7-fold higher than genes from the iron-limiting condition (0.1 μM iron). Type B (230 genes) genes were consistently down-regulated in cells grown at iron concentrations of 20 to 300 μM with signal ratios less than 0.7-fold lower than genes in the cells grown at the low iron concentration. Type C genes (33 genes) were down-regulated above a 100 μM initial iron concentration, while type D genes (80 genes) were up-regulated above a 100 to 150 μM initial iron concentration. Type E genes (357 genes) were neither up-regulated nor down-regulated by any initial iron concentration, indicating that these genes are not responsive to the iron concentration. Figure 2 shows a partial list of genes.
FIG.2.
Transcription profiles of genes by microarray of the whole-genome sequence of M. magneticum AMB-1. Red indicates positive, and green indicates negative, as shown in the scale bars (log2 ratio). The genes were categorized into five groups based on their expression at different iron concentrations (20 to 300 μM). Type A shows genes that were up-regulated above a 20 μM initial iron concentration. Type B genes were down-regulated above a 20 μM initial iron concentration. Type C genes were down-regulated above a 100 μM initial iron concentration. Type D genes were up-regulated above a 100 to 150 μM initial iron concentrations. Type E genes were not affected at any initial iron concentration. Microarray experiments were demonstrated with three biological replicates and used duplicate microarray slides (different lot number). The remaining genes which did not fall into any category showed a very inconsistent expression profile. The genes did not show distinct down- or up-regulation at any specific iron concentration. Slides were scanned with a ScanArray Lite apparatus (model 900-3011538000; PerkinElmer Japan Co., Ltd.). Visualization of expression pattern data was done with Vector Xpression Software (InforMax Inc., Frederick, Md.).
Interestingly, two distinct patterns were observed in relation to iron uptake and magnetosome synthesis. High-affinity ferrous iron transport genes were up-regulated in magnetosome-forming cells grown under iron-rich conditions, and ferric iron transport genes were down-regulated under these conditions (Table 1). The ftr, tpd, and feo genes are known to be expressed under low-oxygen conditions when ferrous iron remains stable and predominates over ferric iron (1, 7, 9, 12, 14). In this study, these gene expressions are consistent with the microaerobic culture conditions in which the cells were grown. This codependence of high iron assimilation and microaerobic conditions for magnetosome synthesis to occur is in agreement with earlier findings on M. magnetotacticum (5) and M. gryphiswaldense (26, 27). On the other hand, ferric iron transport genes, which include fepA, tonB, exbB, and exbD (1), were down-regulated under iron-rich conditions. This trend is consistent with the mode of ferric iron uptake in other bacteria. Two copies, cirA, encoding a ferric-siderophore outer membrane receptor, and fepC, encoding an inner membrane ferric-siderophore transporter (1), however, were up-regulated under high-iron conditions. In M. magnetotacticum MS-1, more siderophores were produced under iron-rich conditions than under iron-limited conditions (25). In M. magneticum AMB-1, we have shown that the initial high concentration of iron is rapidly assimilated from the medium within only 4 h after inoculation, reaching levels comparable to those of iron-deficient cultures, thereby triggering siderophore excretion (6). Microorganisms require a minimum effective iron concentration of ∼0.01 μM for growth but ∼1.0 μM for optimal growth (22). This range is much higher in magnetotactic bacteria, at least 6 μM (6), because of their high iron requirement for magnetosome synthesis. Ferric siderophores may not be directly involved in magnetosome synthesis but may contribute in ensuring that the high iron supply demanded by magnetotactic bacteria is acquired.
TABLE 1.
Expression of genes involved in ferrous and ferric ion transport and nitrogen respiration in M. magneticum AMB-1 at various initial iron concentrations
Red, up-regulated; green, down-regulated.
COG score indicates the result of homology search by BLAST performed against COG database.
Additionally, higher transcript levels of nitrate reductase (amb2686, amb2687, amb2690) and ferric reductase (amb3335) genes were obtained under iron-rich, magnetosome-forming conditions. In M. magnetotacticum, ferric reductase was apparently required for magnetite production (23). We have previously reported that when M. magneticum AMB-1 utilizes nitrate as its sole nitrogen source, magnetites are produced, with ferric iron serving as the terminal electron acceptor (16, 17). Iron reduction and magnetosome formation with respiratory nitrate reduction were inhibited by dicumarol, an inhibitor of quinone (17), indicating that iron reduction is coupled with respiratory nitrate reduction during magnetosome synthesis.
In order for magnetosome production to proceed in magnetotactic bacteria, two main precise physiological conditions are required: (i) a narrow range of low oxygen concentrations and (i) an extraordinary amount of iron to be assimilated. Although these have been clearly demonstrated by several groups (3-5, 26), these findings must be augmented with specific iron transport systems to further elucidate the unique process of magnetosome synthesis.
Nucleotide sequence accession number.
The microarray data and nucleotide sequence information obtained in this study were submitted to the National Center for Biotechnology Information GEO database and assigned accession no. GSE3914.
Acknowledgments
This work was funded in part by Grant-in-Aid for Specially Promoted Research 13002005 from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Arakaki, A., J. Webb, and T. Matsunaga. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 278:8745-8750. [DOI] [PubMed] [Google Scholar]
- 3.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 4.Bazylinski, D. A., R. B. Frankel, and H. W. Jannasch. 1988. Anaerobic magnetite production by a marine, magnetotactic bacterium. Nature 334:518-519. [Google Scholar]
- 5.Blakemore, R. P., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calugay, R. J., H. Miyashita, Y. Okamura, and T. Matsunaga. 2003. Siderophore production by the magnetic bacterium Magnetospirillum magneticum AMB-1. FEMS Microbiol. Lett. 218:371-375. [DOI] [PubMed] [Google Scholar]
- 7.Dubbels, B. L., A. A. DiSpirito, J. D. Morton, J. D. Semrau, J. N. Neto, and D. A. Bazylinski. 2004. Evidence for a copper-dependent iron transport system in the marine, magnetotactic bacterium strain MV-1. Microbiology 150:2931-2945. [DOI] [PubMed] [Google Scholar]
- 8.Ducey, T. F., M. B. Carson, J. Orvis, A. P. Stintzi, and D. W. Dyer. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felice, M. R., I. De Domenico, L. Li, D. M. Ward, B. Bartok, G. Musci, and J. Kaplan. 2005. Post-transcriptional regulation of the yeast high affinity iron transport system. J. Biol. Chem. 280:22181-22190. [DOI] [PubMed] [Google Scholar]
- 10.Gilman, M. 1998. Preparation of RNA from eukaryotic and prokaryotic cells. Curr. Protocols Mol. Biol. 1:4.3.1-4.3.2. [Google Scholar]
- 11.Grunberg, K., E. C. Muller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schuler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komeili, A., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marlovits, T. C., W. Haase, C. Herrmann, S. G. Aller, and V. M. Unger. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. USA 99:16243-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsunaga, T., Y. Okamura, Y. Fukuda, A. T. Wahyudi, Y. Murase, and H. Takeyama. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 12:157-166. [DOI] [PubMed] [Google Scholar]
- 16.Matsunaga, T., T. Sakaguchi, and F. Tadokoro. 1991. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl. Microbiol. Biotechnol. 35:651-655. [Google Scholar]
- 17.Matsunaga, T., and N. Tsujimura. 1994. Respiratory inhibitors of a magnetic bacterium Magnetospirillum sp. AMB-1 capable of growing aerobically. Appl. Microbiol. Biotechnol. 39:368-371. [Google Scholar]
- 18.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 19.Methe, B. A., J. Webster, K. Nevin, J. Butler, and D. R. Lovley. 2005. DNA microarray analysis of nitrogen fixation and Fe(III) reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miura, T., and S. Mizushima. 1969. Separation and properties of outer and cytoplasmic membranes in Escherichia coli. Biochim. Biophys. Acta 193:268-276. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, C., J. G. Burgess, K. Sode, and T. Matsunaga. 1995. An iron-regulated gene, magA, encoding an iron transport protein of Magnetospirillum sp. strain AMB-1. J. Biol. Chem. 270:28392-28396. [DOI] [PubMed] [Google Scholar]
- 22.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 23.Noguchi, Y., T. Fujiwara, K. Yoshimatsu, and Y. Fukumori. 1999. Iron reductase for magnetite synthesis in the magnetotactic bacterium Magnetospirillum magnetotacticum. J. Bacteriol. 181:2142-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamura, Y., H. Takeyama, and T. Matsunaga. 2001. A magnetosome-specific GTPase from the magnetic bacterium Magnetospirillum magneticum AMB-1. J. Biol. Chem. 276:48183-48188. [DOI] [PubMed] [Google Scholar]
- 25.Paoletti, L. C., and R. P. Blakemore. 1986. Hydroxamate production by Aquaspirillum magnetotacticum. J. Bacteriol. 167:73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuler, D., and E. Baeuerlein. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180:159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuler, D., and E. Baeuerlein. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166:301-307. [DOI] [PubMed] [Google Scholar]