Abstract
YrzC has previously been identified as a repressor controlling ytmI expression via its regulation of YtlI activator synthesis in Bacillus subtilis. We identified YrzC as a master regulator of sulfur metabolism. Gene expression profiles of B. subtilis ΔyrzC mutant and wild-type strains grown in minimal medium with sulfate as the sole sulfur source were compared. In the mutant, increased expression was observed for 24 genes previously identified as repressed in the presence of sulfate. Since several genes involved in the pathways leading to cysteine formation were found, we propose to rename YrzC CymR, for “cysteine metabolism repressor.” A CymR-dependent binding to the promoter region of the ytlI, ssuB, tcyP, yrrT, yxeK, cysK, or ydbM gene was demonstrated using gel shift experiments. A potential CymR target site, TAAWNCN2ANTWNAN3ATMGGAATTW, was found in the promoter region of these genes. In a DNase footprint experiment, the protected region in the ytlI promoter region contained this consensus sequence. Partial deletion or introduction of point mutations in this sequence confirmed its involvement in ytlI, yrrT, and yxeK regulation. The addition of O-acetylserine in gel shift experiments prevented CymR-dependent binding to DNA for all of the targets characterized. Transcriptome analysis of a ΔcymR mutant and the wild-type strain also brought out significant changes in the expression level of a large set of genes related to stress response or to transition toward anaerobiosis.
Sulfur is a crucial atom in cysteine and methionine, as well as in several coenzymes and cofactors such as thiamine, biotin, or coenzyme A (CoA). Among these compounds, cysteine is important for the biogenesis of [Fe-S] clusters, for the catalytic sites of several enzymes, and for protein folding and assembly via the formation of disulfide bonds. Moreover, cysteine-derived proteins such as thioredoxin play a central role in protection against oxidative stress. Two major cysteine biosynthetic pathways have been described: the thiolation pathway requiring sulfide and the reverse transsulfuration pathway, which converts homocysteine to cysteine with the intermediary formation of cystathionine (49).
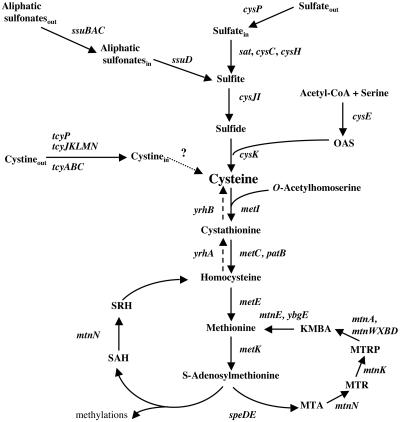
In Bacillus subtilis, the pathway of cysteine synthesis from sulfate has been characterized (Fig. 1). Sulfate is first transported into the cell via a sulfate permease, CysP, related to inorganic phosphate transporters (26). Sulfate is subsequently reduced to sulfide, probably in four steps involving the sequential action of ATP sulfurylase, adenosine 5′-phosphosulfate (APS) kinase, 3′-phosphoadenosine 5′-phosphosulfate (PAPS) reductase, and sulfite reductase (3, 27, 55). An O-acetylserine thiol-lyase, the cysK gene product, further condenses sulfide and O-acetylserine (OAS) to form cysteine (55). Several aliphatic sulfonates can be used as alternative sulfur sources for the synthesis of cysteine. They are taken up by a sulfonate ATP-binding cassette (ABC) transporter and then converted into sulfite by an FMNH2-dependent monooxygenase (56) (Fig. 1). Bacillus subtilis can also use methionine as the sole sulfur source, indicating efficient conversion of methionine into cysteine. The YrhA and YrhB proteins are involved in this conversion (S. Auger and M. F. Hullo, unpublished results). The transport of l-cystine has also been recently investigated. Three systems are present in B. subtilis: two ABC transporters, TcyABC and TcyJKLMN; and a symporter, TcyP (4). The TcyJKLMN and TcyP uptake systems are high-affinity transporters with apparent Km values for l-cystine of 2.5 μM and 0.6 μM, respectively. In addition, the TcyJKLMN system is involved in the uptake of cystathionine, S-methyl-cysteine, djenkolic acid, and other sulfur compounds (4, 47). The tcyJKLMN genes belong to a large operon (operon ytmI), which also encodes a riboflavin kinase, two putative flavin-dependent monooxygenases, a putative acetyltransferase, and a putative amidohydrolase (6, 47, 50, 51). The expression of the ytmI operon and the tcyP gene is regulated in response to sulfur availability, while the expression level of the tcyABC operon remains low under all conditions tested (4). Moreover, expression of the ytmI operon is induced by disulfide and oxidative stresses or in a strain depleted of thioredoxin A (23, 31, 48) and repressed by the Spx protein in sulfate-containing media (7). ytmI expression is controlled in response to sulfur availability by two different regulators, YtlI and YrzC (5, 6, 51). The YtlI regulator activates the transcription of the ytmI operon by direct binding to its promoter region. Expression of the ytlI gene itself is controlled by the negative regulator YrzC. A potential cis-acting target site for the YrzC protein has been identified just upstream from the −35 box of the ytlI promoter (5). However, direct binding of YrzC to the ytlI promoter region remains to be demonstrated. Interestingly, the cascade of regulation of ytmI-type operons involving YtlI and YrzC-like regulators seems to be conserved in Listeria species (5). YrzC shares similarities with regulators of the Rrf2 family, which includes IscR, the repressor of the iscRSUA operon of Escherichia coli involved in [Fe-S] cluster biogenesis. IscR when associated with a [2Fe-2S] cluster appears to repress iscRSUA expression (44). In Desulfovibrio vulgaris, inactivation of the rrf2 gene results in overexpression of the hmc operon, which encodes a redox protein involved in electron transport during hydrogen oxidation with sulfate as an electron acceptor (18). RirA of Rhizobium leguminosarum is crucial for the genetic response to iron availability (58). NsrR of Nitromonas europaea is a nitrite-sensitive repressor of the nirK gene encoding a nitrite reductase (2).
FIG. 1.
Biosynthesis and recycling pathways of sulfur-containing amino acids. The enzymes present in B. subtilis are indicated by the corresponding genes: cysP, sulfate permease; sat, ATP sulfurylase; cysC, APS kinase; cysH, APS-PAPS reductase; ssuBACD, aliphatic sulfonate uptake and degradation; cysJI, sulfite reductase; cysE, serine O-acetyltransferase; cysK, OAS-thiol-lyase; tcyP, tcyJKLMN, and tcyABC, cystine transporters; metI, cystathionine γ-synthase; metC and patB, cystathionine β-lyases; metE, methionine synthase; metK, S-adenosylmethionine synthetase; yrhA, putative cystathionine β-synthase; yrhB, putative cystathionine γ-lyase; speD, AdoMet decarboxylase; speE, spermidine synthase; mtnN, SAH/MTA nucleosidase; mtnK, MTR kinase; mtnA and mtnWXBD, gene products involved in the MTR-to-KMBA recycling pathway; mtnE and ybgE, aminotransferase. KMBA, 2-keto-4-methylthiobutyrate; MTA, methylthioadenosine; MTR, methylthioribose; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; SAH, S-adenosylhomocysteine; SRH, S-ribosylhomocysteine; AdoMet, S-adenosylmethionine.
In B. subtilis, several mechanisms of regulation are involved in the control of methionine and cysteine metabolism. The S-box transcription antitermination system controls the expression of genes participating in methionine uptake, biosynthesis, and recycling, in response to methionine availability (1, 16, 29, 46). In addition, two LysR-type regulators, CysL and YtlI, play a role in the regulation of sulfur metabolism. CysL positively controls expression of the cysJI operon encoding the sulfite reductase by binding to its promoter region (11). YtlI is a positive regulator of the ytmI operon, as discussed above (6). However, the key regulator controlling cysteine metabolism in this bacterium remains to be characterized. YrzC, which indirectly regulates the synthesis of the TcyJKLMN l-cystine ABC transporter, could play this role. To determine whether YrzC is a global regulator, the expression profiles of a wild-type B. subtilis strain and a ΔyrzC mutant grown with sulfate as the sole sulfur source were compared. Using this approach, we found that expression of several genes participating in cysteine metabolism was derepressed in a ΔyrzC mutant. Moreover, YrzC-dependent binding to the promoter region of seven genes or operons was observed. A cis-acting DNA motif required for this binding was characterized.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli DH5α [F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk−) phoA supE44 thi-1 gyrA96 relA1 λ−] was used for plasmid construction and for YrzC overproduction. The B. subtilis strains are listed in Table 1. Escherichia coli was grown in LB medium, and B. subtilis was grown in SP medium or minimal medium (6 mM K2HPO4, 4.4 mM KH2PO4, 0.3 mM trisodium citrate, 5 mM MgCl2, 0.5% glucose, 50 mg l-tryptophan liter−1, 22 mg ferric ammonium citrate liter−1, 0.1% l-glutamine) containing 1 mM K2SO4, 1 mM l-methionine, or 0.01, 0.1, or 1 mM l-cystine as the sole sulfur source. For the experiments involving expression of genes under the control of the xylA promoter, 0.5% fructose instead of glucose was used as a carbon source. l-Threonine (50 mg liter−1) and 1% xylose were added when required. Antibiotics were added to the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 5 μg ml−1; and spectinomycin, 100 μg ml−1. Solid media were prepared by addition of 20 g noble agar liter−1 (Difco). Standard procedures were used for transformation of E. coli (43) and B. subtilis (22).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotypea | Source or referenceb |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| BSIP1144 | trpC2 amyE::pΔAyrrT′-lacZ cat | pDIA5512→168 |
| BSIP1155 | trpC2 amyE::pΔDyrrT′-lacZ cat | pDIA5526→168 |
| BSIP1156 | trpC2 amyE::pΔByrrT′-lacZ cat | pDIA5527→168 |
| BSIP1161 | trpC2 amyE::pΔCyrrT′-lacZ cat | pDIA5536→168 |
| BSIP1215 | trpC2 amyE::pΔFytlI′-lacZ cat | 5 |
| BSIP1264 | trpC2 amyE::pΔIytlI′-lacZ cat | 5 |
| BSIP1309 | trpC2 amyE::pΔB(+20 G A)yrrT′-lacZ cat | Materials and Methods |
| BSIP1310 | trpC2 amyE::pΔB(+28 A T)yrrT′-lacZ cat | Materials and Methods |
| BSIP1311 | trpC2 amyE::pΔB(+29 T C)yrrT′-lacZ cat | Materials and Methods |
| BSIP1329 | trpC2 amyE::pΔAyxeK′-lacZ cat | pDIA5664→168 |
| BSIP1547 | trpC2 amyE::pΔB(+35 T G)yrrT′-lacZ cat | Materials and Methods |
| BSIP1549 | trpC2 amyE::pΔB(+13 A G)yrrT′-lacZ cat | Materials and Methods |
| BSIP1566 | trpC2 amyE::pΔByxeK′-lacZ cat | pDIA5740→168 |
| BSIP1585 | trpC2 amyE::pΔDyxeK′-lacZ cat | PDIA5650→168 |
| BSIP1638 | trpC2 amyE::pΔCyxeK′-lacZ cat | pDIA5651→168 |
| BSIP1793 | trpC2 ΔyrzC amyE::pΔFytlI′-lacZ cat | 5 |
| BSIP1794 | trpC2 ΔyrzC amyE::pΔAyrrT′-lacZ cat | pDIA5512→BSIP1798 |
| BSIP1798 | trpC2 ΔyrzC amyE::aphA3 lacZ | 5 |
| BSIP1807 | trpC2 ΔyrzC amyE::pΔFytlI′-lacZ cat thrC::pxylA-yrzC spc | pDIA5735→BSIP1793 |
| BSIP1816 | trpC2 ΔyrzC amyE::pΔAyxeK′-lacZ cat | pDIA5664→BSIP1798 |
| BSIP1817 | trpC2 amyE::p-yrzC′-lacZ cat | pDIA5738→168 |
cat is the pC194 chloramphenicol acetyltransferase gene; spc is a spectinomycin resistance gene.
Arrows indicate construction by transformation.
The loss of amylase activity was detected as previously described (53). β-Galactosidase specific activity was measured as described by Miller (30) with cell extracts obtained by lysozyme treatment. Protein concentrations were determined by the method of Bradford. One unit of β-galactosidase is defined as the amount of enzyme that produces 1 nmol of O-nitrophenol min−1 at 28°C. The mean values of at least two independent experiments are presented. Standard deviations are less than 20% of the mean.
Plasmids and strain constructions.
Plasmids from E. coli and chromosomal DNA from B. subtilis were prepared according to standard procedures. Restriction enzymes and phage T4 DNA ligase were used as specified by the manufacturers.
Nucleotides are numbered relative to the transcriptional start site of genes unless otherwise specified. Plasmid pAC6 (53) allowed the construction of transcriptional fusions between a series of 3′ deletions of the yxeK promoter region and the promoterless lacZ gene. The pΔA (nucleotides −104 to +130) [pΔA(−104; +130)], pΔB(−104; +63), pΔC(−104; +41), and pΔD(−104; +21) regions were amplified by PCR with the creation of EcoRI and BamHI sites. PCR products were inserted into pAC6 to give pDIA5664 (pΔA), pDIA5740 (pΔB), pDIA5651 (pΔC), and pDIA5650 (pΔD), respectively. These plasmids were linearized with ScaI, which allowed the insertion of the transcriptional lacZ fusions as a single copy at the amyE locus (Table 1).
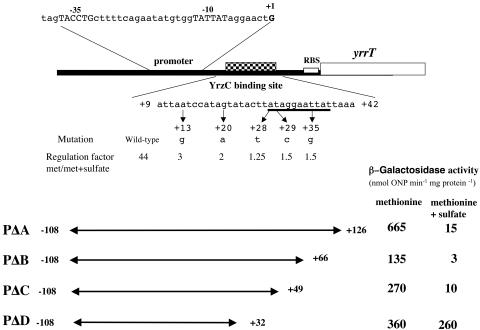
Transcriptional fusions between a series of 3′ deletions of the yrrT promoter region and the promoterless lacZ gene were also constructed. The pΔA(−108; +126), pΔB(−108; +66), pΔC(−108; +49), and pΔD(−108; +32) regions were amplified by PCR with the creation of EcoRI and BamHI sites. The PCR products were inserted into pAC6 to give pDIA5512 (pΔA), pDIA5527 (pΔB), pDIA5536 (pΔC), and pDIA5526 (pΔD), respectively. These plasmids were linearized with ScaI, which allowed the insertion of the transcriptional lacZ fusions as a single copy at the amyE locus (Table 1).
A transcriptional fusion between the yrzC promoter region and the lacZ gene was constructed. A DNA fragment (nucleotides −209 to +24 from the translational start site of yrzC) flanked by EcoRI and BamHI sites was generated by PCR. The PCR product was inserted into pAC6 to give pDIA5738. This plasmid was linearized with ScaI, which allowed insertion of the transcriptional lacZ fusion as a single copy at the amyE locus (Table 1). Plasmid pXT (32) was used to express the yrzC gene under the control of a xylose-inducible promoter (pxylA). The complete coding sequence of yrzC (nucleotides −62 to +465 relative to the translational start site) was amplified by PCR with the creation of EcoRI and BamHI sites. The amplified fragments were inserted into the BamHI and EcoRI sites of pXT, producing pDIA5735. The absence of mutations in yrzC was confirmed by sequencing using the Thermo Sequenase kit (USB Corporation). The yrzC gene was then integrated by a double-crossing-over event at the thrC locus (Table 1).
Random PCR mutagenesis and characterization of mutants.
DNA fragments corresponding to the yrrT promoter region [pΔB(−104; +66)], were generated by PCR using pDIA5527 as a template and oligonucleotides IV80 (5′-GGGGAATTCGTTTTAGTACCTGCTTTTCAGAAT-3′) and IV37 (5′-GGGGGATCCTTGTACTGTTTGATCATAAG-3′). To introduce mutations, the purine/pyrimidine ratio in the reaction mixture was modified (1:10 dATP or dGTP). The DNA fragments were then cloned between the EcoRI and BamHI sites of pAC6. The ligation mixture was transformed into E. coli. Approximately 10,000 transformants were pooled, and total plasmid DNA was extracted, giving a library of mutagenized fragments containing the yrrT promoter region. Linearized plasmids were transformed into B. subtilis 168. Colonies having a Lac+ phenotype in the presence of methionine and sulfate as sulfur sources were retained for further studies. The yrrT promoter region amplified by PCR using oligonucleotides in the cat and lacZ genes and chromosomal DNA as a template was used for nucleotide sequencing reactions.
Handling of RNA.
Total RNA was isolated from B. subtilis wild-type (BSIP1215) and ΔyrzC (BSIP1793) strains grown in minimal medium with 1 mM sulfate as the sole sulfur source. For each strain, samples (10 ml) of four independent cultures were collected at an optical density at 600 nm (OD600) of 1.2 by centrifugation, frozen in liquid nitrogen, and stored at −80°C. Bacterial pellets were resuspended in 460 μl of buffer (22 mM Tris-HCl, pH 7.6, 65 mM EDTA) and transferred into tubes containing 500 μl of acid phenol, pH 4.5, and 0.4 g of 0.1-mm-diameter glass beads (Sigma). The cells were broken in a Fastprep apparatus (Bio101) twice for 30 s at maximum speed. The cells were then treated with Trizol according to the manufacturer's recommendations (Gibco-BRL). The resulting RNA preparations were incubated with RNase-free DNase I (DNA-free kit; Ambion). The amount of RNA was quantified by measuring the OD260. For primer extension experiments, total RNA was isolated from B. subtilis 168 grown in minimal medium in the presence of 1 mM methionine. Primers IV152 (5′-CCTGCTAAAAATACCCCTAATTT-3′) and SA54 (5′-AAGAGCCCTAAGATAAGTAAAAATAAA-3′) were used for primer extension on yxeK and tcyP, respectively. Primer extension experiments were performed as previously described (11).
cDNA probe synthesis and hybridization.
Hybridization probes were generated by mixing 2 μg of total RNA; 4 μl of B. subtilis gene-specific primers (Sigma-GenoSys Biotechnologies, Inc.); 3.3 μM dCTP; and 0.33 mM dATP, dGTP, and dTTP. After incubation at 80°C for 5 min, 40 μCi [α-33P]dCTP (3,000 Ci mmol−1) and 1.5 μl (50 U) avian myeloblastosis virus reverse transcriptase were added. Samples were incubated at 42°C for 2 h. RNA was degraded by alkaline hydrolysis, and unincorporated nucleotides were removed from the labeled cDNA by gel filtration through G-25 Sephadex columns (Roche).
Panorama B. subtilis gene arrays containing the 4107 B. subtilis coding sequences in duplicate were obtained from Sigma-GenoSys Biotechnologies. The arrays were prehybridized at 65°C for 5 h in 15 ml of hybridization solution: 5× SSPE (0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7), 2% sodium dodecyl sulfate, 1× Denhardt's reagent, and 100 mg of sheared salmon sperm DNA ml−1. The hybridization was carried out in 10 ml of hybridization solution containing the labeled cDNA for 16 h. Blots were washed in 0.5× SSPE-0.2% sodium dodecyl sulfate and were exposed to a PhosphorImager screen (Molecular Dynamics) for about 15 h.
Data analysis.
Exposed PhosphorImager screens were scanned with a pixel size of 50 μm on a 445SI PhosphorImager (Molecular Dynamics). The intensity of each dot was quantified with ArrayVision software (Imaging Research Inc.). To perform statistical data analyses, raw data were loaded into the GenoScript Database (http://genodb.pasteur.fr). Dot intensity was normalized according to mean values of the total intensities of all spots on each DNA array. The expression profiles of B. subtilis BSIP1215 and BSIP1793 strains grown with sulfate were compared by calculating the consistency of differential expression across replicate hybridizations by use of the Wilcoxon signed-rank and Welch tests. Differentially expressed genes were chosen with a P value of ≤0.05 and a ratio between the wild-type and mutant strain normalized data of ≥1.5. Only the genes found with both tests meeting the specified parameters were retained.
Overproduction of YrzC in E. coli.
E. coli DH5α was transformed with plasmid pDIA5735 carrying the yrzC gene under the control of the xylA promoter. Due to the absence of the XylR repressor, expression of yrzC in this strain was not controlled by xylose. E. coli DH5α carrying plasmid pXT (32) was used as a negative control. Both strains were grown at room temperature in 25 ml LB medium to late exponential phase, harvested by centrifugation at 5,000 × g for 10 min, and resuspended in 300 μl of 1× gel shift binding buffer (25 mM Na-phosphate, pH 7, 150 mM NaCl, 0.1 mM EDTA, 2 mM MgSO4, 1 mM dithiothreitol, 10% glycerol). After sonication, cell debris was removed by centrifugation at 13,000 × g at 4°C for 10 min. E. coli crude extracts were used directly for gel shift assays.
Gel mobility shift assays.
DNA fragments containing various ytlI promoter regions [pΔF(−130; +111), pΔF containing a point mutation (T-38A), or pΔI(−25; +111)] were amplified by PCR using pDIA5575, pDIA5692, and chromosomal DNA of B. subtilis BSIP1264 (Table 1) (5), respectively, as a template. PCRs were performed using IV141 (5′-GGGGAATTCAACAGCTCCGATGCATCTTC-3′) for pΔF or IV194 (5′-GGGAATTCTTTTATTGTTTATACTATAAAG-3′) for pΔI as the forward primer and IV47 (5′-AAGTTGGGTAACGCCAGGGTTT-3′) as the reverse primer. For yrrT, a PCR fragment containing the pΔA(−108; +126) promoter region was amplified using primers IV48 (5′-GGGAATTCATATGAAGTATAAGCTTTTTTGC-3′) and IV37 (5′-GGGGGATCCT-TGTACTGTTTGATCATAAG-3′) and pDIA5512 carrying the pΔAyrrT'-lacZ fusion as template. For yxeK, a PCR fragment containing the pΔA(−104; +130) promoter region was amplified using primers SA74 (5′-GGGGAATTCAATGCCGTTCCCTCATGGTCA-3′) and IV152 (5′-GGGGGATCCTGCTAAAAATACCCCTAATTT-3′) and pDIA5664 carrying the pΔAyxeK′-lacZ fusion as template. For cysK, a PCR fragment containing the promoter region from positions −168 to +82 was amplified using primers IV132 (5′-GGGAATTCTCATTTGATGAAGTAAAGGACC-3′) and IG43 (5′-GGGGGATCCATTCCCAATTAATTCAG-3′) and genomic DNA of B. subtilis 168 as template. For ssuB and tcyP, two PCR fragments containing the promoter region (−106 to +120 and −241 to +84, respectively) were amplified with primers SE8 (5′-ATAAGAATTCCTTCTCTATTTGCGAAACAAGCAG-3′) and SE9 (5′-AGCCGGATCCATCCCTCTCTTTATTGCCAACCC-3′) for ssuB and SA50 (5′-GGGGAATTCTTTTTTATGTACTAGCCTTTC-3′) and SA51 (5′-GGGGGATCCAGCCCTAAGATAAGTAAAA-3′) for tcyP. B. subtilis chromosomal DNA was used as a template. For ydbM, a PCR fragment containing the putative promoter region (positions −171 to +53 relative to the translational start site) was amplified using primers SE6 (5′-TATAGAATTCTTTATCTCGGCATGAAACAAGACC-3′) and SE7 (5′-CGCAGGATCCCCGATTTTCTCCATCCATTGCC-3′) with B. subtilis genomic DNA as template.
These PCR products were labeled using [γ-32P]ATP 5′-end-labeled specific primers. Unincorporated oligonucleotides were removed using the QIAquick PCR purification kit (QIAGEN). Protein-DNA complexes were formed in 10-μl volumes, by incubating the 32P-end-labeled DNA fragments (10,000 cpm) with different amounts of crude extracts of E. coli DH5α carrying either pDIA5735 (pXT-pxylA-yrzC) or pXT (negative control) in binding buffer (25 mM Na-phosphate buffer, pH 7, 150 mM NaCl, 0.1 mM EDTA, 2 mM MgSO4, 1 mM dithiothreitol, 10% glycerol) in the presence of 0.1 mg of poly(dI-dC) ml−1. The DNA binding assays were performed as previously described (5). When specified, OAS (Sigma) was added to the reaction mixture at different concentrations.
DNase I footprinting.
A labeled DNA fragment corresponding to the ytlI promoter region obtained for gel shift experiments was used to analyze the YrzC protected region in DNase I footprinting reactions. Constant amounts (50,000 cpm) of the 5′-end-labeled 241-bp top-strand DNA fragment of the ytlI promoter region (nucleotides −130 to +111) were incubated in 50 μl without protein or with various concentrations of E. coli crude extracts with or without the YrzC protein. The binding conditions were similar to those used for gel mobility shift assays. DNase I (at a final concentration of 0.002 U μl−1; Roche) was added to the reaction mixtures. After 20 or 30 s at 25°C, the DNase I was inactivated by adding 140 μl of stop solution [0.4 M Na-acetate, 2.5 mM EDTA, 50 μg ml−1 poly(dI-dC), 10 μg ml−1 glycogen]. For each reaction, DNA was then subjected to ethanol precipitation followed by electrophoresis in a 7 M urea-polyacrylamide gel containing 7% acrylamide in Tris-borate-EDTA buffer. The sequence ladder (G+A) was produced as previously described (28).
RESULTS
Phenotype of strains inactivated for yrzC or overproducing YrzC.
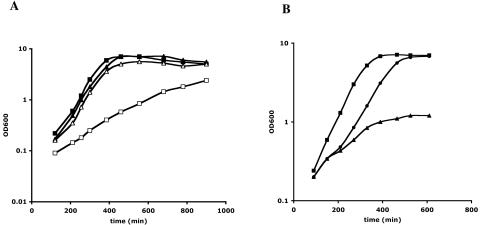
We have previously identified YrzC as a repressor controlling the expression of the ytmI operon via its regulation of YtlI activator synthesis (5). To determine whether this regulator could have a more global role in the regulation of sulfur metabolism, strains inactivated for yrzC or expressing yrzC under the control of a xylose-inducible promoter were grown in the presence of various sulfur sources. A yrzC+ strain (BSIP1215) and a ΔyrzC mutant (BSIP1793) grew similarly in the presence of 1 mM methionine (Fig. 2A). The growth rate of the ΔyrzC mutant strain (65 min) was slightly reduced compared to that of the wild-type strain (55 min) with 1 mM sulfate (data not shown). In the presence of 1 mM cystine, the growth rate of the ΔyrzC mutant decreased markedly, as evidenced by its doubling time of 160 min instead of 55 min for the yrzC+ strain (Fig. 2A). The same phenotype was found in the presence of lower cystine concentrations (100 μM and 10 μM) (data not shown).
FIG. 2.
Growth phenotype of a ΔyrzC mutant in the presence or absence of a yrzC gene expressed under the control of the xylA promoter. (A) Comparison of the growth of yrzC+ (triangles) and ΔyrzC (squares) strains in the presence of 1 mM methionine (closed) or 1 mM cystine (open). (B) Growth of strain BSIP1807 (ΔyrzC pxylA-yrzC) in the presence of 1 mM methionine (squares); 1 mM methionine and 1% xylose (triangles); or 1 mM methionine, 1% xylose, and 10 μM cystine (circles). In the latter case, cystine was added after 150 min of growth. For BSIP1807 containing pxylA-yrzC at the thrC locus, minimal medium contained threonine (50 mg liter−1) and fructose instead of glucose.
The yrzC gene was also expressed under the control of the xylose-inducible xylA promoter. The pxylA-yrzC gene was inserted at the thrC locus of the ΔyrzC mutant BSIP1793, giving strain BSIP1807 (Table 1). The expression of the pΔFytlI′-lacZ fusion present in BSIP1807 was then tested in the presence or absence of 1% xylose. With methionine as the sole sulfur source, the expression of this fusion was 88 U mg of protein−1 without xylose and 15 U mg of protein−1 with xylose. This result indicated that overexpression of yrzC led to the ytlI gene repression by the YrzC protein. The addition of xylose to the medium also strongly reduced the growth rate and growth yield of this strain with methionine as the sole sulfur source (Fig. 2B), while no growth defect was observed in the presence of sulfate and xylose (data not shown). However, the growth defect of BSIP1807 with methionine and xylose was abolished by the addition of 10 μM or 100 μM cystine (Fig. 2B) (data not shown). This phenotype was then due to cysteine depletion (see Discussion). All of these data suggested that YrzC could play a central role in the regulation of sulfur metabolism.
YrzC plays a major role in the regulation of expression of sulfur metabolism genes.
The role of YrzC in the control of sulfur metabolism was further investigated by the use of DNA arrays. The global gene expression profiles of B. subtilis ΔyrzC and wild-type strains grown on minimal medium with 1 mM sulfate as the sole sulfur source were compared. cDNAs were generated using RNAs extracted from exponentially growing cells and hybridized to DNA arrays as described in Materials and Methods. For each condition, eight data sets generated from four independent cultures and RNA extractions (http://genodb.pasteur.fr) were used to perform statistical data analyses. A total number of 202 genes showed a ≥1.5-fold change in transcription level combined with a P value of ≤0.05 using both the Wilcoxon and Welch tests (Tables 2 to 4) (see the supplemental material).
TABLE 2.
Sulfur metabolism genes differentially expressed in B. subtilis ΔyrzC (BSIP1793) compared to the wild-type strain (BSIP1215) in the presence of sulfate as the sole sulfur sourcea
| Gene name and synonym | Function/similarity | Transcriptome analysis
|
Met/SO4 regulationb | |
|---|---|---|---|---|
| ΔyrzC/wild-type expression ratio | P value | |||
| cysK | OAS thiol-lyase | 3.21 | 8.10−3 | + |
| ydbMc | Similar to butyryl-CoA dehydrogenase | 1.93 | 4.10−3 | + |
| mtnN (yrrU, mtnA) | SAH/MTA nucleosidase | 2.37 | <1.10−4 | + |
| yrhA | Putative cystathionine β-synthase | 4.28 | 1.10−4 | + |
| yrhB | Putative cystathionine γ-lyase | 3.20 | <1.10−4 | + |
| Transporters and associated genes | ||||
| ytmI | Predicted acetyltransferase | 3.98 | 1.10−4 | + |
| tcyJ (ytmJ) | l-Cystine ABC transporter (binding protein) | 4.27 | 2.10−4 | + |
| tcyK (ytmK) | l-Cystine ABC transporter (binding protein) | 4.82 | 1.10−4 | + |
| tcyL (ytmL) | l-Cystine ABC transporter (permease) | 4.00 | 1.10−4 | + |
| tcyM (ytmM) | l-Cystine ABC transporter (permease) | 3.35 | 1.10−4 | + |
| tcyN (ytmN) | Similar to amino-acid ABC transporter (ATP-binding protein) | 4.49 | <1.10−4 | + |
| ytmO | Similar to monooxygenase | 5.05 | 1.10−4 | + |
| ytnI | Putative glutaredoxin-like protein (GrxC) | 3.34 | 1.10−4 | + |
| ytnJ | Monooxygenase | 4.88 | 1.10−4 | + |
| ribR | Riboflavin kinase | 4.50 | <1.10−4 | + |
| hipO | Hippurate hydrolase | 8.25 | <1.10−4 | + |
| ytnM | Permease-like protein | 5.59 | <1.10−4 | + |
| tcyP (yhcL) | Similar to sodium-glutamate symporter, l-cystine transporter | 2.84 | 1.10−4 | + |
| yxeK | Similar to monooxygenase | 1.67 | 1.10−4 | + |
| yxeL | Predicted acetyltransferase | 1.75 | 1.10−4 | + |
| ssuA (ygbA) | Aliphatic sulfonate ABC transporter (binding lipoprotein) | 3.69 | 1.10−4 | + |
| ssuC (ygaM) | Aliphatic sulfonate ABC transporter (permease) | 2.28 | 1.10−4 | + |
| ssuD (ygcA) | Aliphatic sulfonate monooxygenase | 5.89 | 1.10−4 | + |
| ygaN | Unknown | 3.42 | 1.10−4 | + |
| Sbox family | ||||
| metC (yjcJ) | Cystathionine β-lyase | 0.65 | 3.10−3 | |
| met E | Cobalamin-independent methionine Synthase | 0.62 | 8.10−3 | − |
| yoaB | Similar to α-ketoglutarate permease | 0.48 | 3.10−4 | − |
| mtnB (ykrY) | MTRu-1-P dehydratase | 0.64 | 4.10−3 | − |
| mtnD (ykrZ) | Aci-reductone dioxygenase | 0.64 | 1.10−3 | − |
The results obtained are representative of eight hybridizations from four independent cultures. The data sets generated were loaded into the GenoScript Database (http://genodb.pasteur.fr).
+ and − indicate a higher and a lower expression level on methionine than on sulfate, respectively (1).
The ydbM gene encoding a protein similar to butyryl-CoA dehydrogenase was included in this table. Indeed, the level of ydbM expression was high with methionine and repressed with sulfate (1). ydbM was also found to be a target of YrzC in this study. Its possible role related to sulfur metabolism remains to be determined.
TABLE 4.
Genes related to the transition to anaerobiosis differentially expressed in B. subtilis ΔyrzC (BSIP1793) compared to the wild-type strain (BSIP1215) in the presence of sulfatea
| Gene name and synonym | Function/similarity | Transcriptome analysis
|
Transition toward anaerobiosisb | |
|---|---|---|---|---|
| ΔyrzC/wild-type expression ratio | P value | |||
| csn | Chitosanase | 2.22 | 4.10−3 | + |
| hemH | Ferrochelatase | 1.77 | 1.10−4 | + |
| hemY | Protoporphyrinogen IX and Coproporphyrinogen III oxidase | 2.14 | 1.10−4 | + |
| yjlC | Unknown | 0.55 | 5.10−3 | − |
| yjlD | Similar to NADH dehydrogenase | 0.43 | 1.10−4 | − |
| yvaX | Unknown | 1.58 | 4.10−2 | + |
| yvfV | (Fe-S) protein | 0.47 | 2.10−4 | −/0c |
| ywfI | Unknown | 1.67 | 7.10−4 | + |
| yxaK | Similar to Ser/Thr protein kinase | 3.50 | 1.10−4 | + |
| Citric acid cycle and related pathways | ||||
| citB | Aconitase | 0.50 | 1.10−4 | − |
| citC, icd | Isocitrate dehydrogenase | 0.54 | 1.10−4 | − |
| citZ | Citrate synthase II | 0.52 | 2.10−4 | − |
| odhA | 2-Oxoglutarate dehydrogenase (EI subunit) | 0.36 | 2.10−3 | |
| odhB | 2-Oxoglutarate dehydrogenase (EII subunit) | 0.39 | 1.10−4 | |
| sucC | Succinyl-CoA synthetase (beta subunit) | 0.49 | 1.10−4 | |
| sucD | Succinyl-CoA synthetase (alpha subunit) | 0.59 | 1.10−4 | |
| pycA | Pyruvate carboxylase | 0.40 | 2.10−4 | |
| Fermentation pathways | ||||
| lctE, ldh | l-Lactate dehydrogenase | 0.36 | 1.10−4 | + |
| lctP | l-Lactate permease | 0.55 | 3.10−3 | + |
| Antibiotic production | ||||
| sbo | Subtilosin A | 6.96 | <1.10−4 | + |
| albA (ywiA) | Antilisterial bacteriocin (subtilosin) production | 5.80 | 1.10−4 | + |
| sunA | Sublancin 168 lantibiotic precursor | 1.53 | 2.10−2 | + |
The results obtained are representative of eight hybridizations from four independent cultures. The data sets generated for each condition were loaded into GenoScript.
Data from Ye et al. (57) and Nakano et al. (36); + and − respectively indicate a higher and lower level of expression under anaerobic conditions compared to aerobic conditions.
Repressed under nitrite or nitrate respiration and no change in expression level during fermentation.
The level of expression of genes involved in sulfur metabolism was considerably modified in a ΔyrzC mutant (Table 2). In this mutant, an increase in expression (from 1.67- to more than 8-fold) was observed for 24 genes previously identified as repressed in the presence of sulfate (1). Among those were the cysK gene encoding an OAS-thiol-lyase (55), the mtnN yrhAB genes involved in the conversion of methionine to cysteine (1, 45; unpublished data), and the ydbM gene encoding a putative butyryl coenzyme A (CoA) dehydrogenase. Likewise, the expression of several genes involved in the transport and assimilation of sulfur compounds was higher in the ΔyrzC mutant: tcyP encoding a cystine symporter and genes of the ytmI, ssu, and yxeK operons (Fig. 1). Seven out of the nine gene products of the yxeK operon exhibit sequence similarities to proteins of the ytmI operon. The yxeK operon probably participates in the transport and degradation of an unknown sulfur compound different from cystine. Surprisingly, no changes in ytlI expression appeared in the transcriptome experiments, although derepression of ytlI was observed in a ΔyrzC mutant using a ytlI′-lacZ fusion (5). This probably was caused by the low level of expression of this regulatory gene. The strong increase in expression of the ytmI operon could be due to ytlI derepression, as previously demonstrated (5).
In contrast, five members of the S-box regulon showed a reduced level of expression in the ΔyrzC mutant: metC and metE, encoding cystathionine β-lyase and methionine synthase, respectively; yoaB, encoding a protein similar to an α-ketoglutarate permease; and mtnBD, involved in the methionine recycling pathway via methylthioribose (1, 46) (Fig. 1).
YrzC binding to the ytlI promoter region.
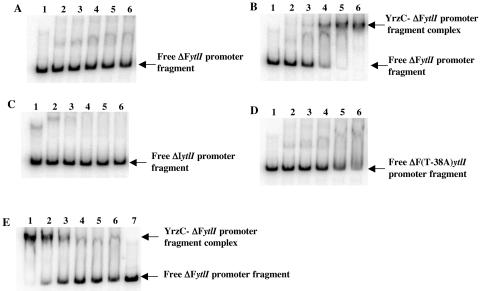
Transcriptome analysis indicated a major role for YrzC in the regulation of genes repressed during growth in the presence of sulfate. To determine whether YrzC controls these genes by binding to their promoter region, we performed gel shift DNA binding assays. A cis-acting target required for sulfate-dependent repression of ytlI has been previously identified just upstream from the −35 box of this gene (5). We tested the ability of YrzC to interact with this sequence. Crude extracts of E. coli DH5α carrying pDIA5735, which contains the yrzC gene, were prepared and used in mobility shift DNA-binding assays. Crude extracts of E. coli DH5α carrying the pXT vector were used as a control. A complex was formed by adding increasing amounts of E. coli crude extracts containing YrzC to a DNA fragment containing the wild-type pΔF ytlI promoter region (positions −130 to +111) (Fig. 3B). No complex was formed using either crude extracts of E. coli DH5α carrying pXT (Fig. 3A) or a DNA fragment containing the pΔI ytlI promoter region (positions −25 to +111) deleted from the cis-acting target (Fig. 3C). Moreover, YrzC-dependent binding to a pΔF ytlI promoter region containing a point mutation (T-38A) leading to constitutive ytlI expression (5) was less efficient (Fig. 3D). It seems extremely unlikely that YrzC could modulate the synthesis or the activity of an E. coli protein, which in turn might precisely interact with the ytlI promoter region necessary for sulfate-dependent repression in B. subtilis. Indeed, due to the absence of E. coli proteins sharing more than 31% identity with YrzC, the conservation of a cascade of regulation between B. subtilis and E. coli is highly improbable. This strongly supports the idea that YrzC binds to the ytlI promoter region. However, since these experiments were performed with E. coli crude extracts, we cannot exclude that a component present in these extracts could help YrzC binding. The formation of a DNA-protein complex between YrzC and the ytlI promoter region required the DNA sequence located between positions −130 and −25 and the T base at position −38 located within the cis-acting target (5).
FIG. 3.
Binding of the YrzC repressor to different fragments of the ytlI promoter region in mobility shift assays. Gel mobility shift experiments were performed by incubating crude extracts of E. coli DH5α carrying either pXT (A) or pDIA5735 (pXT-pxylA-yrzC) (B, C, D, and E) with 5′-radiolabeled DNA fragments containing different ytlI promoter regions: (A, B, E) ΔF(−130; +111), (C) ΔI(−25; +111), (D) ΔF containing a point mutation(T-38A). (A, B, C, D) Lanes 1, free probes; lanes 2 to 6, increasing amounts of E. coli DH5α crude extracts (1, 2.5, 5, 7.5, and 10 μg protein, respectively). (E) Lanes 1 to 6, 7.5 μg of protein from E. coli DH5α crude extracts producing YrzC and increasing amounts of OAS (0, 0.05, 0.1, 0.2, 0.5, and 1 mM, respectively); lane 7, free probe.
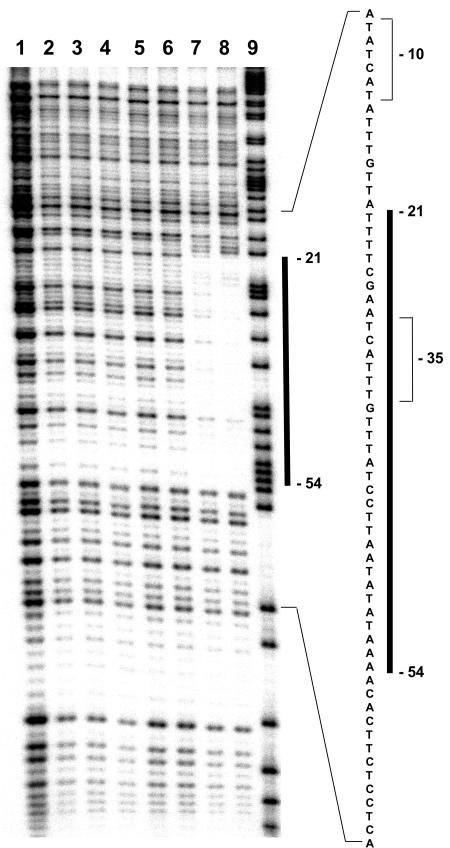
DNase I footprint experiments were then carried out to determine the precise location of the YrzC binding site within the ytlI promoter region. Comparison of the sequence patterns produced in the absence of YrzC (Fig. 4, lanes 2 to 4) and in the presence of saturating concentrations of YrzC (Fig. 4, lanes 7 to 8) located the protected region between positions −54 and −21 upstream from the transcriptional start site of the ytlI gene (Fig. 4). This region contained the target required for sulfate-dependent repression of ytlI identified by deletions and site-directed mutagenesis. However, the region protected by YrzC seems to be larger.
FIG. 4.
DNase I footprint of E. coli crude extracts containing YrzC on the ytlI promoter region. The 241-bp PCR fragment representing the coding strand of the ytlI promoter region (positions −130 to +111) was 5′-end labeled and incubated in separate reactions without protein (lane 1), in the presence of 20 μg, 30 μg, or 40 μg of crude extracts without YrzC (lanes 2, 3, and 4), or in the presence of 10 μg (lane 5), 20 μg (lane 6), 30 μg (lane 7), or 40 μg (lane 8) of the E. coli crude extracts containing YrzC and subjected to DNase I digestion. Lane 9, G+A sequencing ladder. The position of the protected region (positions −54 to −21) is marked by a vertical bar. Numbers indicate the distance from the transcription initiation site of the ytlI operon. The −10 and −35 boxes are indicated by brackets.
Identification of targets of YrzC.
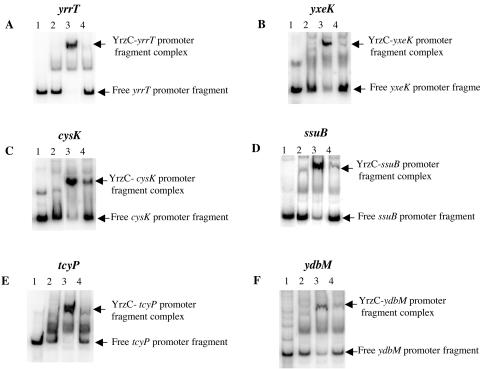
Additional genes involved in sulfur metabolism and derepressed in a ΔyrzC mutant (Table 2) were tested in gel shift mobility assays. In the case of operons, the region upstream of the first gene of the operon was used: ssuB for the ssu operon (54), yrrT for the yrrT mtnN yrhAB operon (S. Auger, unpublished results), and yxeK for the corresponding operon (1). Crude extracts of E. coli DH5α carrying pDIA5735 (pxylA-yrzC) were added to different radiolabeled DNA fragments containing the promoter regions of either yrrT, yxeK, cysK, ssuB, tcyP, or ydbM. Crude extracts of E. coli DH5α carrying pXT were used as a negative control. One main DNA-protein complex was obtained with all of the probes tested when crude extracts containing YrzC were added (Fig. 5, lanes 3), while this complex was not formed with the control (Fig. 5, lanes 2). YrzC-dependent binding to the yrrT, yxeK, cysK, ssuB, tcyP, and ydbM promoter regions was observed.
FIG. 5.
Binding of the YrzC repressor to DNA fragments containing the yrrT, yxeK, cysK, ssuB, tcyP, or ydbM promoter region in gel mobility shift assays. Gel mobility shift experiments were performed by incubating crude extracts of E. coli DH5α carrying either pXT or pDIA5735 (pXT-pxylA-yrzC) with 5′-radiolabeled DNA fragments containing different promoter regions. (A) yrrT promoter region (−108 to +126). (B) yxeK promoter region (−104 to +130). (C) cysK promoter region (−168 to +82). (D) ssuB promoter region (−106 to +120). (E) tcyP promoter region (−241 to +84). (F) ydbM promoter region (−226 to +59 from the translational start site). Lanes 1, free probes; lanes 2, E. coli DH5α carrying pXT (7.5 μg protein); lanes 3, E. coli DH5α carrying pDIA5735 (7.5 μg protein); lanes 4, E. coli DH5α carrying pDIA5735 (7.5 μg protein) and OAS at 0.05 mM (panels A, B, and C) or 0.2 mM (panels D, E, and F).
Characterization of the yxeK and yrrT promoter regions required for sulfate-dependent repression.
An 11-bp motif, AT(A/T)ATTCCTAT, found in the promoter region of the ytlI gene of B. subtilis, Listeria monocytogenes (locus tag lmo2352), and L. innocua (locus tag lin2446) has been proposed to be necessary for the sulfate-dependent repression of ytlI (5). In the yrrT and yxeK promoter regions, a sequence rather similar to this motif was present on the complementary strand (Fig. 6 and 7, underlined sequences). To test whether this motif could be involved in the regulation of the yrrT or the yxeK operon, different promoter regions of these two operons were fused to the lacZ gene (Fig. 6 and 7). These fusions were introduced as single copies at the amyE locus of B. subtilis 168. The expression of the pΔA(−104; +130), pΔB(−104; +63), and pΔC(−104; +41) yxeK′-lacZ fusions was 7- to 20-fold higher in the presence of methionine than in the presence of sulfate (Fig. 6). In contrast, the expression of the pΔD(−104; +21) yxeK′-lacZ fusion was even higher with sulfate than with methionine (Fig. 6). The DNA fragment located between nucleotides +21 and +41 including the 11-bp motif is therefore necessary for the sulfate-dependent repression of yxeK transcription.
FIG. 6.
Effect of the sulfur source on the expression of different yxeK′-lacZ transcriptional fusions. The transcription start site of the yxeK operon (+1), which was mapped by primer extension (see Materials and Methods), is indicated. The −10 (TACATT) and −35 (TTGACA) regions similar to the consensus sequence of σA-dependent promoters are in uppercase. The white box and checkered box correspond to the ribosome-binding sites (RBS) and the YrzC binding site, respectively. The 11-bp motif complementary to that identified in the ytlI promoter region (5) is underlined. Deletion end points of the different fusions with the lacZ gene are numbered with respect to the transcriptional start site of yxeK. Cells were grown in minimal medium in the presence of sulfate or methionine at a final concentration of 1 mM. The β-galactosidase activities were obtained from cultures in mid-exponential growth phase. The mean values of at least two independent experiments are presented. Standard deviations are less than 20% of the mean.
FIG. 7.
Effect of sulfur source on expression of different yrrT′-lacZ transcriptional fusions. The transcription start point of the yrrT operon (+1) is indicated (S. Auger, unpublished results). The −35 and −10 regions for this operon are in uppercase. The white box and checkered box correspond to the ribosome-binding sites (RBS) and the YrzC binding site, respectively. The 11-bp motif complementary to that identified in the ytlI promoter region (5) is underlined. Deletion end points of the different fusions with the lacZ gene are numbered with respect to the transcriptional start sites of yrrT. Cells were grown in minimal medium in the presence of 1 mM methionine or 1 mM methionine plus 1 mM sulfate. The β-galactosidase activities were obtained from cultures in the mid-exponential growth phase. The mean values of at least two independent experiments are presented. Standard deviations are less than 20% of the mean. Mutations in the YrzC binding site are also indicated. For each point mutation, the regulation factor was obtained by calculating the β-galactosidase activity of the wild-type or mutated pΔB yrrT′-lacZ fusion in cells grown on methionine versus methionine plus sulfate.
For the yrrT operon, expression of the pΔA(−108; +126) yrrT′-lacZ, pΔB(−108; +66) yrrT′-lacZ, and pΔC(−108; +49)yrrT′-lacZ fusions was strongly reduced in the presence of methionine plus sulfate as compared to the level observed with methionine (Fig. 7). In contrast, the pΔD(−108; +32) yrrT′-lacZ fusion was constitutively expressed. The DNA fragment located between nucleotides +32 and +49 covering a part of the 11-bp conserved motif was necessary to observe sulfate-dependent repression. The expression of pΔA(−108; +126) yrrT′-lacZ and pΔA(−104; +130) yxeK′-lacZ fusions was then also tested in a ΔyrzC mutant after growth in the presence of methionine or sulfate. These fusions were constitutively expressed in a ΔyrzC mutant (data not shown). This result confirmed that YrzC is a repressor controlling yxeK and yrrT transcription.
We also performed PCR random mutagenesis on a 174-bp fragment corresponding to the pΔB(−108 to +66) yrrT promoter region (see Materials and Method). Five mutants that showed derepressed expression of the lacZ reporter gene in the presence of methionine and sulfate on plates were isolated. The DNA sequence of the corresponding yrrT promoter regions was determined, showing that each mutant had a single modification (Fig. 7). The level of β-galactosidase in these mutants was determined in methionine-grown cells in the presence or absence of sulfate. The contribution of the different nucleotides to sulfate-dependent repression was estimated by calculating the ratio of β-galactosidase activity measured after growth with methionine and methionine plus sulfate (Fig. 7). Mutations in the yrrT promoter region at positions +28 (A→G), +29 (G→A), and +35 (T→G) led to a nearly constitutive expression of the corresponding fusions. This was expected for modifications in the binding site of a repressor. These modifications are located within the 11-bp motif also present in the promoter region of yxeK and ytlI (in the opposite direction in the latter case). In addition, a pΔB(−108; +66) yrrT′-lacZ fusion containing a replacement of an A by a T at position +13 or a G by an A at position +20 was only two- to threefold repressed by the addition of sulfate compared to the repression observed with the wild-type promoter (Fig. 7). Surprisingly, these two additional mutations are located upstream of the 11-bp conserved DNA sequence, indicating that the motif necessary for YrzC repression is larger.
Identification of a common motif in the promoter region of YrzC targets.
Using deletion and mutagenesis experiments, cis-acting regions important for sulfate-dependent repression of the ytlI, yrrT, and yxeK genes were determined (Fig. 6 and 7) (5). We also showed that YrzC interacted in gel shift mobility assays with the promoter region of ytlI, yrrT, yxeK, cysK, ssuB, tcyP, and ydbM (Fig. 3 to 5). Alignment of the promoter region of these seven targets of YrzC led to the identification of a conserved common motif, TAAWNCN2ANTWNAN3ATMGGAATTW (Fig. 8). Six nucleotides of this 27-bp motif were present in all of these genes, and nine additional bases were found in six promoters. This region corresponds to the sequence protected by YrzC in the DNase footprint experiment (Fig. 4). The transcriptional start sites of all these genes with the exception of ydbM have been characterized (Fig. 6 to 7) (5, 54, 55; S. Auger, unpublished results). The motif is located downstream of the transcriptional start sites of yrrT, yxeK, and cysK. For ytlI, ssuB, and tcyP, this sequence is centered around the −35 box (Fig. 8). The 27-bp motif is located from positions −44 to −18 and −46 to −20 relative to the transcriptional start sites of ssuB and tcyP, respectively, while this motif is present on the complementary strand in the opposite direction for ytlI (positions −21 to −47 relative to the transcriptional start site of ytlI). Deletions or point mutations in this motif led to a decrease of sulfate-dependent repression of the ytlI, yxeK, and yrrT genes (5; this study). This consensus sequence is likely to include the YrzC binding site.
FIG. 8.
Identification of a common motif in the nucleotide sequence of YrzC target genes. Alignment of the promoter regions of the cysK, ssuB, tcyP, yrrT, yxeK, ydbM, and ytlI genes. The term −ytlI means that the sequence corresponds to the complementary sequence of the ytlI promoter sequence. The consensus sequence is established according to the IUPAC code: W represents T or A, M represents C or A, and N represents any base (A, C, G, or T). A, T, G, and C correspond to bases present in six to eight promoters. The nucleotides underlined in the yrrT and ytlI promoter region correspond to mutations leading to significant derepression with sulfate (less than fivefold residual repression). The −35 regions of tcyP, ssuB, and ytlI (TTTACT on the complementary strand) are boxed.
OAS, a negative effector of the binding of YrzC to the ytlI promoter region.
The signaling pathway modulating the YrzC-dependent regulation in response to sulfur availability is still unknown. The expression of a transcriptional fusion between the yrzC promoter region and the lacZ gene was tested after growth in the presence of different sulfur sources. No significant changes in the level of expression of the pyrzC-lacZ fusion were observed in the presence of cystine, sulfate, or methionine (data not shown). This indicated that YrzC was not regulated in response to sulfur availability at the synthesis level but rather by direct control of its activity. The addition of OAS to a minimal medium containing sulfate or cysteine resulted in derepression of the ssuB and ytlI genes (5, 54). This suggests that OAS or a derivative could modulate YrzC binding to DNA. OAS was therefore tested in gel mobility shift experiments with crude extracts containing YrzC (7.5 μg of proteins) and a DNA fragment carrying the wild-type pΔF ytlI promoter region (positions −130 to +111) (Fig. 3E). The addition of increasing concentrations of OAS ranging from 0.05 mM to 1 mM to the binding assay resulted in the release of the probe from the protein-DNA complex. In the presence of 0.2 mM OAS, the formation of this complex was strongly reduced (Fig. 3E, lane 4). The negative effect of OAS on YrzC repressor activity was further confirmed for other YrzC targets: yrrT, yxeK, cysK, ssuB, tcyP, and ydbM (Fig. 5). A significant dissociation of the protein-DNA complex was observed in the presence of 0.05 or 0.2 mM OAS (Fig. 5, lanes 4).
Pleiotropic effect of the yrzC inactivation.
Transcriptome analysis of a ΔyrzC mutant strain in comparison with a wild-type strain suggested a more pleiotropic role of YrzC than merely the control of sulfur metabolism genes. Significant changes of expression were observed for several cellular functions and metabolic pathways, including genes involved in sporulation, chemotaxis and mobility, transcriptional and translational machinery, lipid metabolism, and transport (see the supplemental material). The most striking results are probably the observed variations in expression level of a large set of genes related to stress response (Table 3) and to transition toward anaerobiosis (Table 4).
TABLE 3.
Genes related to stress response differentially expressed in B. subtilis ΔyrzC mutant compared to the wild-type strain in the presence of sulfate as the sole sulfur sourcea
| Gene name and synonym | Function/similarity | Transcriptome analysis
|
σB promoterb | |
|---|---|---|---|---|
| ΔyrzC/wild-type expression ratio | P value | |||
| recA | Multifunctional SOS repair regulator | 0.64 | 1.10−4 | |
| yoeB | Unknown; member of the PerR regulon | 0.25 | 1.10−4 | |
| yumC | Similar to thioredoxin reductase | 0.65 | 8.10−3 | |
| Heat shock-related genes | ||||
| dnaK | Class I heat-shock protein (molecular chaperone) | 0.46 | 1.10−4 | |
| grpE | Heat-shock protein (activation of DnaK) | 0.52 | 3.10−4 | |
| lonA | Class IV heat-shock ATP-dependent protease | 0.56 | 3.10−2 | |
| ftsH | Cell-division protein/general stress protein | 0.65 | 9.10−3 | |
| σB regulonc | ||||
| rsbV | Anti-anti-σB factor | 0.64 | 2.10−2 | + |
| rsbW | Anti-σB factor | 0.22 | 1.10−4 | |
| sigB | RNA polymerase general stress σ factor | 0.30 | 1.10−4 | |
| rsbX | Indirect negative regulator of σB activity | 0.38 | 1.10−4 | |
| csbB | Stress response protein, similar to glycosyl transferases | 0.54 | 1.10−3 | |
| ctc | General stress protein | 0.27 | 1.10−4 | + |
| dps | Stress- and starvation-induced gene | 0.46 | 1.10−4 | + |
| gspA | General stress protein | 0.38 | 1.10−4 | + |
| ybyB | Unknown | 0.26 | 1.10−4 | Probable |
| ydaG | Similar to general stress protein | 0.63 | 1.10−4 | + |
| ydaP | Similar to pyruvate oxidase | 0.48 | 2.10−4 | + |
| yfhK | Similar to cell division inhibitor | 0.54 | 1.10−4 | + |
| yfkM | Predicted member of the DJ-1/PfpI family | 0.49 | 2.10−2 | + |
| yflT | Unknown | 0.35 | 1.10−4 | + |
| yheK | Putative regulator of nhaC (Na+/H+ antiporter) | 0.48 | 1.10−4 | Probable |
| yjbC | Unknown | 0.54 | 2.10−3 | |
| ykzA | Similar to organic hydroperoxide resistance protein | 0.35 | 1.10−4 | + |
| yoxC | Protein containing a divergent version of the methyl-accepting chemotaxis-like domain | 0.55 | 1.10−4 | + |
| yqgZ | Unknown | 0.29 | 1.10−4 | + |
| ytxG | Similar to general stress protein | 0.60 | 4.10−3 | + |
| ytxH | Similar to general stress protein | 0.44 | 1.10−4 | |
| ytxJ | Similar to general stress protein | 0.47 | 2.10−3 | |
| yvaA | Predicted dehydrogenase (MviM); oxidoreductase-like (GFO-IDH-MocA) | 0.43 | 2.10−3 | Probable |
| yvrE | Similar to senescence marker protein 30 | 0.56 | 9.10−3 | + |
| ywiE | Similar to cardiolipin synthetase | 0.51 | 1.10−4 | Probable |
| ywjA | ABC transporter (ATP-binding protein) | 0.57 | 3.10−2 | |
| ywjC | Unknown | 0.54 | 1.10−4 | Probable |
| ywmG | Unknown | 0.30 | 1.10−3 | + |
| ywzA | Unknown | 0.44 | 4.10−4 | Probable |
| yxcC | Putative sugar transporter | 0.48 | 1.10−4 | + |
The results obtained are representative of eight hybridizations from four independent cultures. The data sets generated were loaded into the GenoScript Database (http://genodb.pasteur.fr).
Several genes related to stress response were expressed at lower levels in the ΔyrzC mutant as compared to the wild-type strain (Table 3). This included the heat shock-related chaperone (DnaK and GrpE), the LonA protease, and the cell division protein FtsH. Two proteins probably involved in the oxidative stress response were found: a protein similar to thioredoxin reductases (yumC) and a member of the PerR regulon of unknown function (yoeB) (15). Likewise, several members (30 genes) of the σB regulon (38, 40) were also expressed at lower levels in the mutant. Among them, we found the rsbVW-sigB-rsbX operon, which encodes sigB itself and proteins involved in σB signaling networks.
Surprisingly, the expression of a large set of genes usually associated with transition between aerobic and anaerobic conditions (34, 57) differed between a ΔyrzC mutant and a wild-type strain. The expression of several genes decreased during anaerobic growth conditions as well as in a ΔyrzC background. A repression of Krebs cycle genes was generally observed during oxygen limitation (36). Similarly, the level of expression of Krebs cycle genes as well as pycA encoding the pyruvate carboxylase, which provides the Krebs cycle with oxaloacetate, was 1.7- to 2.8-fold reduced in the ΔyrzC mutant. The level of expression of the yjlCD operon encoding an unknown protein and an NADH dehydrogenase and the yvfV gene encoding a probable [Fe-S] protein also decreased in a ΔyrzC mutant. In contrast, several genes induced during a shift to anaerobiosis were derepressed in a ΔyrzC background. They correspond to hemHY, involved in heme biosynthesis; csn, encoding a chitosanase; yxaK, encoding a serine/threonine protein kinase; and two genes of unknown function (yvaX and ywfI). Moreover, an induction of the expression level of genes related to subtilosin (sbo-albA) and sublancin 168 lantibiotic production was also observed.
DISCUSSION
In this article, we have shown that YrzC is a master regulator of sulfur metabolism in Bacillus subtilis. Two different classes of genes involved or potentially involved in sulfur metabolism are differently expressed in the wild-type strain and in a ΔyrzC background (Table 2) (5). The first set corresponds to genes belonging to the S-box regulon. Their expression is higher with sulfate or cysteine than with methionine (1) and decreases in the ΔyrzC mutant. The second class contains all genes (cysK, tcyP, ytlI, and ydbM genes as well as the ssu, yxeK, ytmI, and yrrT operons) previously identified by transcriptome or molecular genetic analysis as repressed during growth of the wild-type strain in the presence of sulfate or cysteine (1, 55). Considering this second set of genes, YrzC appears as a repressor controlling several pathways leading to cysteine formation, including the OAS-thiol-lyase (CysK), l-cystine transporters (TcyP and TcyJKLMN), sulfonate assimilation (SsuABCD), and the methionine-to-cysteine conversion involving YrhA and YrhB (1, 4, 55, 56; unpublished results). We thus propose to rename YrzC CymR, for “cysteine metabolism repressor.” Several data strongly support the key role of CymR in the control of cysteine availability in the cell. Indeed, the growth of a ΔcymR mutant was considerably affected in the presence of cystine (Fig. 2A). Derepression of several cystine transporters as well as pathways leading to the intracellular production of cysteine in this mutant probably resulted in cysteine accumulation in the cell to a toxic level (12, 17). In contrast, reduced growth of strain BSIP1807 overexpressing cymR was observed in the presence of methionine and xylose. This was due to cysteine depletion since the addition of cystine restored BSIP1807 growth (Fig. 2B). The repression of the methionine-to-cysteine conversion pathway (yrhAB), as confirmed by measurement of the expression of an yrrT′-lacZ fusion (data not shown), might account for this growth defect.
CymR represses the expression of the cysK, tcyP, ytlI, and ydbM genes and the ssu, yxeK, ytmI, and yrrT operons. CymR-dependent binding to the promoter regions of these genes was demonstrated using gel shift experiments, except in the case of ytmI, which is indirectly controlled by CymR via YtlI (5). However, since crude extracts were used in these experiments, we cannot completely rule out that E. coli proteins or metabolites may trigger CymR binding. Deletions and point mutations in the promoter region of several targets of CymR combined with in silico analysis allowed us to propose a 27-bp CymR binding motif (Fig. 8). In a DNase footprint experiment, the protected region in the ytlI promoter contains this consensus sequence (Fig. 4). The CymR-dependent binding to the ytlI promoter region in gel mobility shift assays was also completely abolished after deletion of the proposed target sequence and less efficient when a point mutation was introduced in the motif (Fig. 3C and D). In agreement with these data, total or partial deletions of this conserved sequence in the promoter region of ytlI, yrrT, and yxeK (Fig. 6 and 7) (5) led to constitutive expression. Likewise, point mutations, distributed throughout this motif, obtained either by random or site-directed mutagenesis, resulted in the complete or partial loss of sulfate-dependent repression (5, 51; this study). This conserved sequence is not found upstream of the S-box genes, whose expression decreased in a ΔcymR mutant, suggesting an indirect effect of CymR on part of the S-box regulon. CymR belongs to the Rrf2 family of transcriptional regulators (2, 18, 44, 58). Only four members have been characterized, and a potential binding sequence has been identified for only one of them, RirA. The position of the CymR (Fig. 8) and RirA (58) binding sites relative to the transcriptional start site differs from one gene to another. They either overlap the −35 box or are located downstream of the promoters. As for CymR, the RirA binding sequence is rather poorly conserved (58), a property often found with pleiotropic regulators.
Physiological data suggest a role of OAS in the signaling pathway controlling expression of ytlI encoding the activator of the ytmI operon and of the ssu and cysH operons involved in sulfonate or sulfate assimilation (5, 25, 54). The addition of OAS to the culture medium led to partial derepression of these genes. Moreover, constitutive expression of the ssu operon and the ytlI gene has been observed in a cysK mutant, which could accumulate OAS (5, 54). This compound is synthesized by the serine transacetylase, CysE. In B. subtilis, cysE expression is regulated by transcription antitermination at a cysteine-specific T-box and is thus induced under conditions of cysteine starvation (8). The level of OAS could be inversely correlated with the level of cysteine in the cell. Interestingly, the addition of OAS in gel shift experiments resulted in the release of DNA from the CymR-DNA complex for all of the targets of CymR tested (Fig. 3 and 4). OAS or its spontaneous derivative N-acetylserine modulates CymR-dependent binding. However, since crude extracts were used in these experiments, a direct or indirect effect could be proposed. A similar model of regulation has been previously proposed by Mansilla et al. (25) for the control of cysH expression, although the repressor, named CysR, has not been identified. We cannot completely exclude that CymR controls cysH, but we have not observed a derepression of cysH expression in a ΔcymR mutant grown with sulfate in transcriptome experiments (Table 2) or with a cysH′-lacZ fusion (data not shown).
Considering the set of controlled genes, the B. subtilis CymR regulator can be seen as a functional equivalent of CysB from E. coli (19, 20). However, the sulfate assimilation pathway is regulated by CysB but not by CymR under the conditions tested (Table 2). In B. subtilis, the expression of the cysJI operon, encoding the sulfite reductase, is positively regulated by the CysL activator (11). These two organisms have retained different strategies of regulation: an LysR-type activator, CysB, in E. coli and an Rrf2-type repressor, CymR, in B. subtilis. Likewise, the expression of the methionine biosynthesis pathway in these microorganisms is controlled by two different mechanisms of regulation: a repressor, MetJ, in E. coli and the S-box transcriptional antitermination system in B. subtilis (9, 10, 29). In L. lactis, a unique LysR-type activator, CmbR (FhuR), controls most of the genes of the cysteine and methionine biosynthesis pathways (52). OAS increases CmbR binding affinity to their targets. Similarly, the McbR repressor (TetR type) of Corynebacterium glutamicum modulated by the effector S-adenosylhomocysteine (Fig. 1) controls the transcription of genes involved in sulfate or sulfonate assimilation as well as cysteine and methionine synthesis (41). As previously proposed for methionine metabolism (42), a large diversity of molecular mechanisms participates in the fine-tuning regulation of cysteine metabolism in bacteria.
Previous studies have established the existence of links between sulfur metabolism and oxidative stress. A higher expression of several genes involved in sulfur metabolism is observed in a strain depleted of thioredoxin A (48) and in response to superoxide and disulfide stresses (23, 31). In particular, most of the sulfur metabolism genes repressed by CymR are induced under these conditions. Cysteine itself has been proposed to be involved in the oxidative stress response in the pathogen Staphylococcus aureus (24). An S. aureus cysM-knockout mutant, inactivated for the OAS-thiol-lyase, is more sensitive to oxidative and disulfide stresses. This increased sensitivity could be related to a depletion of the cysteine pool (24). As proposed by Leichert et al. (23), the possible involvement of cysteine in the oxidative stress response in B. subtilis could explain the induction of cysteine biosynthesis pathways during disulfide stress. Free cysteine could be involved in thiol homeostasis within the cell by playing a role as a low-molecular-weight thiol antioxidant similar to that of glutathione in other organisms. B. subtilis lacks the glutathione biosynthesis pathway (21), and this compound is not detected in this bacterium (37). In addition, a protein sharing only weak similarities with glutaredoxin (YtnI) is present in B. subtilis but no glutaredoxin reductase is found. The sulfur-containing compound involved in the thiol homeostasis in B. subtilis remains to be identified. Interestingly, while the sulfur metabolism genes repressed by CymR are induced by oxidative stress, the inactivation of cymR does not seem to modulate the expression of genes specific to the oxidative stress response. Only two genes specifically induced in response to oxidative stress (15, 31) are less expressed in a ΔcymR mutant: yoeB, a member of the PerR regulon; and yumC, encoding a protein similar to thioredoxin reductase. Nevertheless, a remarkable consequence of the cymR gene deletion is the lower expression of several genes of the σB regulon (Table 3) (38, 39). An increased expression of σB-dependent genes is observed after peroxide and/or paraquat treatment (15, 31). The effect of a cymR gene disruption on the σB regulon is thus opposite to that observed under conditions of oxidative stress. The signaling pathway leading to down-regulation of the σB regulon in a ΔcymR mutant is still unknown but may involve either the energetic or the physical branch of the σB complex signal transduction network (14, 40). The expression of several genes generally affected by changes in oxygen availability also varied in a ΔcymR mutant compared to that observed in the wild-type strain (Table 4), including genes related to the Krebs cycle, fermentation pathways, and electron transport, as well as antibiotic production (13, 33, 35, 36, 57). As we did not identify a CymR binding site in the promoter region of these genes, the role of CymR in this regulation is most probably indirect via ResD/E, FNR, or a different regulator (35). In a ΔcymR background, the derepression of several pathways involved in cysteine formation and normally repressed in the presence of sulfate may have led to a higher cysteine pool. This could result in an altered thiol homeostasis and consequently a more reduced state of the cell, which may influence both the genes of anaerobiosis and the σB regulon.
In conclusion, this article gives new insights into the regulatory network controlling sulfur metabolism in B. subtilis with the identification of a master regulator, CymR. CymR-like repressors are found in Bacillus, in Listeria, and in Staphylococcus, suggesting common mechanisms of regulation of cysteine metabolism in these bacteria. This work also supports connections between the sulfur metabolism and the redox state of the cell, although several questions remain to be answered in this respect. In particular, the putative contribution of cysteine to cellular thiol homeostasis and the signaling pathway involved in the derepression of the CymR-regulated genes involved in sulfur metabolism in response to oxidative stress deserve further investigation.
Supplementary Material
Acknowledgments
We are grateful to J. Fert, M. F. Hullo, E. Brito, and L. Maranghi for their technical assistance during this work and to M. A. Dillies for the design of the transcriptome experiment. We are also grateful to C. Laurent and S. Moreira for their help for statistical analysis and use of the Genoscript database. We thank G. Rapoport and U. Mechold for critical reading of the manuscript.
Sergine Even is an “ATER” from the Université Paris 7. This research was supported by grants from the ANR [Dynamocell (NT05-2-44860)], the “Ministère de l'Education Nationale de la Recherche et de la Technologie,” the “Centre National de la Recherche Scientifique” (URA 2171), the “Institut Pasteur,” and the “Université Paris 7.”
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaumont, H. J. E., S. I. Lens, W. N. M. Reijnders, H. V. Westerhoff, and R. J. M. van Spanning. 2004. Expression of nitrite reductase in Nitrosomonas europaea involves NsrR, a novel nitrite-sensitive transcription repressor. Mol. Microbiol. 54:148-158. [DOI] [PubMed] [Google Scholar]
- 3.Berndt, C., C. H. Lillig, M. Wollenberg, E. Bill, M. C. Mansilla, D. de Mendoza, A. Seidler, and J. D. Schwenn. 2004. Characterization and reconstitution of a 4Fe-4S adenylyl sulfate/phosphoadenylyl sulfate reductase from Bacillus subtilis. J. Biol. Chem. 279:7850-7855. [DOI] [PubMed] [Google Scholar]
- 4.Burguiere, P., S. Auger, M.-F. Hullo, A. Danchin, and I. Martin-Verstraete. 2004. Three different systems participate in l-cystine uptake in Bacillus subtilis. J. Bacteriol. 186:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burguière, P., J. Fert, I. Guillouard, S. Auger, A. Danchin, and I. Martin-Verstraete. 2005. Regulation of the Bacillus subtilis ytmI operon, involved in sulfur metabolism. J. Bacteriol. 187:6019-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppee, J. Y., S. Auger, E. Turlin, A. Sekowska, J. P. Le Caer, V. Labas, V. Vagner, A. Danchin, and I. Martin-Verstraete. 2001. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 147:1631-1640. [DOI] [PubMed] [Google Scholar]
- 7.Erwin, K. N., S. Nakano, and P. Zuber. 2005. Sulfate-dependent repression of genes that function in organosulfur metabolism in Bacillus subtilis requires Spx. J. Bacteriol. 187:4042-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagnon, Y., R. Breton, H. Putzer, M. Pelchat, M. Grunberg-Manago, and J. Lapointe. 1994. Clustering and co-transcription of the Bacillus subtilis genes encoding the aminoacyl-tRNA synthetases specific for glutamate and for cysteine and the first enzyme for cysteine biosynthesis. J. Biol. Chem. 269:7473-7482. [PubMed] [Google Scholar]
- 9.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 10.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 11.Guillouard, I., S. Auger, M.-F. Hullo, F. Chetouani, A. Danchin, and I. Martin-Verstraete. 2002. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 184:4681-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, C. L. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J. Bacteriol. 145:1031-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Härtig, E., H. Geng, A. Hartmann, A. Hubacek, R. Munch, R. W. Ye, D. Jahn, and M. M. Nakano. 2004. Bacillus subtilis ResD induces expression of the potential regulatory genes yclJK upon oxygen limitation. J. Bacteriol. 186:6477-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 15.Helmann, J. D., M. F. W. Wu, A. Gaballa, P. A. Kobel, M. M. Morshedi, P. Fawcett, and C. Paddon. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hullo, M. F., S. Auger, E. Dassa, A. Danchin, and I. Martin-Verstraete. 2004. The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, d- and l-methionine. Res. Microbiol. 155:80-86. [DOI] [PubMed] [Google Scholar]
- 17.Kari, C., Z. Nagy, P. Kovacs, and F. Hernadi. 1971. Mechanism of the growth inhibitory effect of cysteine on Escherichia coli. J. Gen. Microbiol. 68:349-356. [DOI] [PubMed] [Google Scholar]
- 18.Keon, R. G., R. D. Fu, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 19.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 20.Kredich, N. M. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747-2753. [DOI] [PubMed] [Google Scholar]
- 21.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 22.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leichert, L. I. O., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lithgow, J. K., E. J. Hayhurst, G. Cohen, Y. Aharonowitz, and S. J. Foster. 2004. Role of a cysteine synthase in Staphylococcus aureus. J. Bacteriol. 186:1579-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansilla, M. C., D. Albanesi, and D. de Mendoza. 2000. Transcriptional control of the sulfur-regulated cysH operon, containing genes involved in l-cysteine biosynthesis in Bacillus subtilis. J. Bacteriol. 182:5885-5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansilla, M. C., and D. de Mendoza. 2000. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology 146:815-821. [DOI] [PubMed] [Google Scholar]
- 27.Mansilla, M. C., and D. de Mendoza. 1997. l-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J. Bacteriol. 179:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDaniel, B. A. M., F. J. Grundy, I. Artsimovitch, and T. M. Henkin. 2003. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 32.Nair, S., I. Derre, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 33.Nakano, M. M., G. L. Zheng, and P. Zuber. 2000. Dual control of sbo-alb operon expression by the Spo0 and ResDE systems of signal transduction under anaerobic conditions in Bacillus subtilis. J. Bacteriol. 182:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 35.Nakano, M. M., and P. Zuber. 2002. Anaerobiosis, p. 393-404. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 36.Nakano, M. M., P. Zuber, and A. L. Sonenshein. 1998. Anaerobic regulation of Bacillus subtilis Krebs cycle genes. J. Bacteriol. 180:3304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 40.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 41.Rey, D. A., S. S. Nentwich, D. J. Koch, C. Ruckert, A. Puhler, A. Tauch, and J. Kalinowski. 2005. The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol. Microbiol. 56:871-887. [DOI] [PubMed] [Google Scholar]
- 42.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2004. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 32:3340-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Schwartz, C. J., J. L. Giel, T. Patschkowski, C. Luther, F. J. Ruzicka, H. Beinert, and P. J. Kiley. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. USA 98:14895-14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekowska, A., and A. Danchin. 1999. Identification of yrrU as the methylthioadenosine nucleosidase gene in Bacillus subtilis. DNA Res. 6:255-264. [DOI] [PubMed] [Google Scholar]
- 46.Sekowska, A., and A. Danchin. 25 April 2002, posting date. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol. 2:8. [Online.] doi: 10.1186/1471-2180-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekowska, A., S. Robin, J. J. Daudin, A. Henaut, and A. Danchin. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smits, W. K., J.-Y. F. Dubois, S. Bron, J. M. van Dijl, and O. P. Kuipers. 2005. Tricksy business: transcriptome analysis reveals the involvement of thioredoxin A in redox homeostasis, oxidative stress, sulfur metabolism, and cellular differentiation in Bacillus subtilis. J. Bacteriol. 187:3921-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soda, K. 1987. Microbial sulfur amino acids: an overview. Methods Enzymol. 143:453-459. [DOI] [PubMed] [Google Scholar]
- 50.Solovieva, I. M., R. A. Kreneva, D. J. Leak, and D. A. Perumov. 1999. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology 145:67-73. [DOI] [PubMed] [Google Scholar]
- 51.Solovieva, I. M., R. A. Kreneva, L. E. Lopes, and D. A. Perumov. 2005. The riboflavin kinase encoding gene ribR of Bacillus subtilis is a part of a 10 kb operon, which is negatively regulated by the yrzC gene product. FEMS Microbiol. Lett. 243:51-58. [DOI] [PubMed] [Google Scholar]
- 52.Sperandio, B., P. Polard, D. S. Ehrlich, P. Renault, and E. Guédon. 2005. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187:3762-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stülke, J., I. Martin-Verstraete, M. Zagorec, M. Rose, A. Klier, and G. Rapoport. 1997. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 25:65-78. [DOI] [PubMed] [Google Scholar]
- 54.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Expression of the Bacillus subtilis sulphonate-sulphur utilization genes is regulated at the levels of transcription initiation and termination. Mol. Microbiol. 39:1356-1365. [PubMed] [Google Scholar]
- 55.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol. Lett. 201:29-35. [DOI] [PubMed] [Google Scholar]
- 56.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 57.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeoman, K. H., A. R. J. Curson, J. D. Todd, G. Sawers, and A. W. B. Johnston. 2004. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology 150:4065-4074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.