Abstract
Aer, the Escherichia coli receptor for behavioral responses to oxygen (aerotaxis), energy, and redox potential, contains a PAS sensory-input domain. Within the PAS superfamily, the N-terminal segment (N-cap) is poorly conserved and its role is not well understood. We investigated the role of the N-cap (residues 1 to 19) in the Aer PAS domain by missense and truncation mutagenesis. Aer-PAS N-cap truncations and an Aer-M21P substitution resulted in low cellular levels of the mutant proteins, suggesting that the N-terminal region was important for stabilizing the structure of the PAS domain. The junction of the N-cap and PAS core was critical for signaling in Aer. Mutations and truncations in the sequence encoding residues 15 to 21 introduced a range of phenotypes, including defects in FAD binding, constant tumbling motility, and an inverse response in which E. coli cells migrated away from oxygen concentrations to which they are normally attracted. The proximity of two N-cap regions in an Aer dimer was assessed in vivo by oxidatively cross-linking serial cysteine substitutions. Cross-linking of several cysteine replacements at 23°C was attenuated at 10°C, indicating contact was not at a stable dimer interface but required lateral mobility. We observed large multimers of Aer when we combined cross-linking of N-cap residues with a cysteine replacement that cross-links exclusively at the Aer dimer interface. This suggests that the PAS N-cap faces outwards in a dimer and that PAS-PAS contacts can occur between adjacent dimers.
Motile bacteria migrate to nutrient-rich environments by suppressing directional changes (tumbling) when swimming toward a favorable environment. The receptor responsible for oxygen sensing in Escherichia coli, Aer, is a membrane-bound, homodimeric flavoprotein that is believed to respond to the redox state of the electron transport chain and signal this status to the two-component chemotaxis cascade that ultimately controls flagellar rotation (6, 48). Each monomer of Aer contains a cytosolic PAS domain that is connected by an F1 domain to a membrane anchor consisting of two transmembrane sequences and a short periplasmic loop (3). The second transmembrane sequence is linked to the signaling region of the protein by a HAMP domain (4, 37). The PAS (an acronym of Per-Arnt-Sim) domain is of particular interest because it is a cytoplasmic sensor and a member of the PAS superfamily of sensors found in all kingdoms of life (10, 53, 62; http://smart.embl-heidelberg.de/). PAS domains are sensory input modules that sense light, oxygen, redox potential, energy, and voltage, bind small ligands, and participate in protein-protein interactions (53).
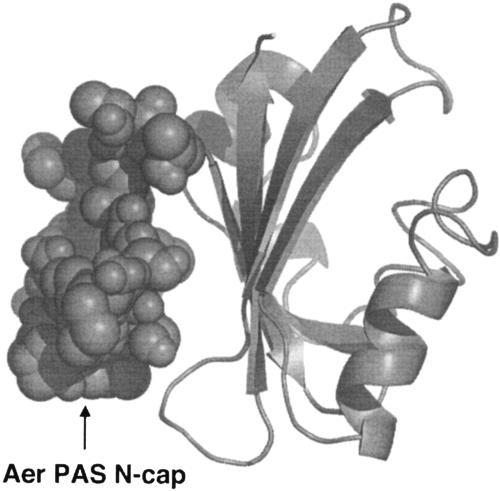
Although sequence identity is low in the PAS superfamily (53), the three-dimensional structures of PAS domains are highly conserved (60), suggesting that common mechanisms may be used for signaling. Three-dimensional crystal structures have been resolved for one or more of the PAS domains from the Ectothiorhodospira halophila blue light receptor chromophore 4-hydroxycinnamoyl (PYP) (7, 46), the related Ppr-PYP domain from Rhodospirillum centenum (47), the human HERG voltage-dependent potassium channel (41), the Phy3 LOV2 domain from Adiantum (11), the oxygen-receptor FixL from Bradyrhizobium japonicum (19-21) and Rhizobium meliloti (40), the phosphodiesterase Ec DOS from E. coli (33), and the Drosophila PERIOD protein (58). In addition, the structures of the PAS domains from human PAS kinase (2) and the transcription factor HIF-2α (15) have been determined by nuclear magnetic resonance. The common structural features of these resolved PAS domains, as well as those predicted for other PAS domains, include (i) an N-terminal cap, (ii) a PAS core, (iii) a helical linker that connects the PAS core to the β-scaffold, and (iv) the β-scaffold (Fig. 1). The overall architecture has been described as a left-handed glove that can enclose a cofactor (60). The specificity of a PAS domain for a sensory input signal is determined partly by its associated cofactor. Currently identified PAS cofactors include the PYP, heme (FixL, Ec DOS), FMN (LOV2), and FAD (Aer, NifL) (6, 11, 19-21, 24, 33, 40, 46, 48, 50). Some PAS domains also appear to function without a cofactor (HERG, HIF-2α, and PERIOD). Signal transduction is presumably initiated by a change in the state of the cofactor that alters local interactions in the binding cleft of the PAS domain. These changes are transmitted to, and amplified in, other regions of the protein (for an example, see reference 18).
FIG. 1.
Space-filled model of the Aer-PAS N-cap (residues 1 to 19) mapped onto the ribbon backbone of the Aer-PAS domain (49).
This laboratory has undertaken investigations of the sensory transduction pathway used by the PAS domain of the Aer protein. Residues important for FAD binding, signaling, and structural integrity have been identified (23, 49, 57). A model for FAD sensing was proposed in which the fully oxidized (FAD) and fully reduced (FADH2) forms of FAD activate CheA autophosphorylation and initiate a clockwise (CW) signal (tumbling) (49). The semiquinone form of FAD (FADH·) is thought to inhibit CheA autophosphorylation, resulting in a counterclockwise (CCW) rotational signal that suppresses tumbling (49).
To date, the N-terminal cap (N-cap) region of the Aer-PAS domain (residues 1 to 19) (Fig. 1), as well as the N-cap regions of other PAS domains, has not been extensively explored. The N-cap is the least conserved region among PAS domains, and early alignments of PAS domains omitted the N-cap sequence because of its variability (53). The three-dimensional structure of the PYP PAS domain is similar in the presence (7, 46) or absence (55) of its N-cap, although the PYP N-cap plays a role in signaling (9, 22), as do the PAS N-cap regions from HERG (41), Ec DOS (33), and Streptococcus pneumoniae MicB (14). There is, however, no current consensus on a specific role for the N-cap region from different PAS proteins. The purpose of the present study was to investigate the role of the PAS N-cap in Aer function. We demonstrate that the Aer-PAS N-cap is important for signaling and projects outward, away from the Aer dimer interface.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmids containing mutated aer genes were expressed in strain BT3388 (Δaer::erm Δtsr-7021 Δtar-tap-5201 trg::Tn10) (59), BT3312 (Δaer-1 Δtsr-7028I) (49), or BT3400 (Δaer-1 Δtsr-7028 recA::catI) (56). These strains were derived from E. coli RP437, which is the wild type (WT) for chemotaxis (45). Bacteria were grown at 30°C in Luria-Bertani medium (13) supplemented with 0.5 μg ml−1 thiamine.
The pGH1 and pMB1 plasmids were used as templates to mutate the aer gene. The pGH1 plasmid is a pTrc99A derivative expressing WT Aer under the control of an IPTG (isopropyl-β-D-thiogalactopyranoside)-inducible ptrc promoter (48). The pMB1 plasmid was constructed by removing the 1.17-kb SmaI-SalI fragment from pGH1 (encompassing Aer residues 116 to 506) and replacing it with the SmaI-SalI fragment from a construct encoding nondisruptive replacements for the three native cysteines of Aer (C193S, C203A, and C253A) (38). The pDS7 plasmid expresses WT Aer from pACYC184 (8), using a tightly regulated sodium salicylate-inducible promoter (pnahG) (56). This construct contains a p15A replication origin, allowing coexpression of genes with pTrc99A-derived plasmids.
Mutagenesis.
Codons 5 to 21 of the aer gene were mutated to all possible codons in pGH1, using PfuTurbo (Stratagene, La Jolla, Calif.) and degenerate primers that were created using an equimolar mix of all four nucleotides at each codon. The Aer-PAS substitution S28G was introduced into several mutant Aer proteins by site-directed mutagenesis of the aer gene, using specific primers (57). We introduced cysteine codons in place of aer codons 4 to 22 in pMB1, using PfuTurbo and primers specific for each cysteine replacement. The primers were constructed as described in the QuikChange site-directed mutagenesis kit (Stratagene). An additional cysteine replacement, A184C (located in the membrane region of Aer), was introduced into the constructs expressing Aer-H4C and Aer-P5C and into pMB1 by the same method.
Four N-terminal Aer truncations, with deletions of residues 1 through 6, 1 through 14, 1 through 19, or 1 through 24, were created by PCR of pGH1, using primers complementary to the NcoI site at pTrc99A nucleotide 265 (Aer7NcoIF, Aer15NcoIF, Aer20NcoIF, and Aer25NcoIF), each paired with a primer containing an engineered SalI site (Aer506SalIR). The AerNcoIF primers incorporated a start codon, followed by the codon for either residue 7, 15, 20, or 25, while the Aer506SalIR primer included residue 506, followed by the normal stop codon of Aer. It was necessary to add a glycine codon (ggc) before codon 20 in the Aer20NcoIF primer, since position one of the triplet had to be guanine. PCR products were digested with NcoI and SalI and ligated to the 4.14-kb NcoI-SalI fragment of pTrc99A. The expression of full-length and truncated Aer proteins was verified by chemiluminescent Western blotting, using anti-Aer2-166 antisera (49). The expected mutations and/or truncations were confirmed for all constructs by DNA sequencing.
Phenotypic screening.
Mutant Aer proteins that did not support aerotaxis were identified by their effects on the behavior of BT3388 cells on tryptone semisoft agar containing 50 μg ml−1 ampicillin and 20 μM to 1 mM IPTG and on BT3312 cells on 30 mM succinate semisoft agar containing 50 μg ml−1 ampicillin and 0 to 1 mM IPTG. The response of BT3388 and BT3312 mutants to oxygen in a gas perfusion chamber was examined after induction with 200 μM IPTG or with 1 mM IPTG if no response was observed, as described previously (48). The induction levels required for an aerotactic response in the gas perfusion chamber were higher than those required for swarming. This may reflect the larger stimulus that is required for the temporal assay of aerotaxis. Phenotypes were determined by evaluating swimming patterns with video microscopy.
Dominance testing.
To test for dominant behavior, mutant aer constructs were cotransformed with pDS7 into BT3400 cells. Cotransformants were selected on Luria-Bertani agar containing 100 μg ml−1 ampicillin and 15 μg ml−1 tetracycline and transferred to 30 mM succinate semisoft agar containing 50 μg ml−1 ampicillin and 7.5 μg ml−1 tetracycline. Sodium salicylate (0.5 μM or 1 μM) and IPTG (between 0 and 0.6 mM) were also included in the semisoft agar in a series of titrations to vary plasmid induction levels. Induction levels that produced approximately 1:1 expression ratios were determined by Western blotting. Relevant mutant constructs were also transformed into BT3400 and tested individually for their aerotactic ability.
Measurement of total cellular FAD.
Total cellular FAD was extracted from BT3312 cells and measured by the modifications of Keppler et al. (29) and Hinkkanen and Decker (25), respectively. Cells (4 ml) from late log phase were harvested, resuspended in 300 μl of ice-cold 0.6 N perchloric acid to extract cellular FAD, and centrifuged at 5,000 × g for 10 min. The supernatant was transferred to a tube containing 42 mg of sodium bicarbonate. This brought the pH to approximately 8.2, precipitated the perchlorate, and left the FAD in the soluble fraction. After centrifugation at 5,000 × g for 10 min, the supernatant was removed and stored in the dark at −20°C until assayed. Stock solutions of horseradish peroxidase (0.8 mg · ml−1; Sigma, St. Louis, Mo.), d-alanine (1.12 M; Sigma), and apo-d-amino acid oxidase (apo-DAAO; 1 mg · ml−1; Calzyme, San Luis Obispo, Calif.) were prepared in 0.1 M sodium phosphate (pH 7.0). Luminol (3.75 mM; Sigma) was dissolved by vortexing in 0.1 M sodium bicarbonate-0.1 M NaCl (pH 9.2) (reaction buffer). For FAD determination, 3 μl of cell extract was added to 750 μl of a prereaction mixture containing 23 mM d-alanine, 7.5 μg horseradish peroxidase, and 25 μM luminol. Triplicate 200-μl aliquots for each sample were transferred to a 96-well plate, and the reaction was initiated by 5 μl of 50 μg ml−1 (250 ng per well) apo-DAAO. Luminescence was counted for 0.1 s every 60 s to 90 s (depending on the number of wells being counted) for 60 min in a Biolumat luminometer (Berthold, Germany) at 37°C. Peak luminometric readings (approximately 27 min) were plotted on a standard curve of FAD standards ranging from 20 fmol to 6,000 fmol. Ratios of the FAD levels from cells induced with 1 mM IPTG to those measured in uninduced samples were used to assess FAD binding.
Cysteine cross-linking.
Cysteine cross-linking was performed for 10 min at 23°C in intact BT3388 cells induced with 50 μM IPTG, using the oxidant copper phenanthroline as described previously (26, 38). Western blots of cross-linked Aer products were quantified on an Alpha Innotech digital imaging system. Rates of cross-linking at 23°C were determined in time courses performed over 15 min. Individual cysteine mutants that produced the most-substantial dimer formation were also cross-linked for 20 min at 10°C to differentiate stable from transient interactions. Aer proteins with a cysteine replacement in the PAS domain and an A184C substitution were cross-linked in BT3312 cells at 23°C for 30 min to differentiate cross-linking within dimers from cross-linking between adjacent dimers.
RESULTS
Mutagenesis of the Aer-PAS N-cap.
We studied the behavior of Aer mutants generated by site-directed mutagenesis of codons 5 to 21 of the aer gene, which encode most of the N-cap (residues 1 to 19) and two residues from the PAS core (20 and 21). This region was mutated to all possible codons using degenerate primers, each containing a codon synthesized from an equimolar mix of all four nucleotides. Plasmids containing mutated aer genes were introduced into a receptorless E. coli strain (BT3388) that lacked all five chemoreceptors (Aer, Tsr, Tar, Tap, and Trg). Transformants were screened for a loss of aerotactic function on tryptone semisoft agar containing 20 μM IPTG. Mutants with an abnormal phenotype on semisoft agar were then tested for the presence of full-length Aer protein by Western blotting. As a result of the selection procedure used, we did not obtain a mutation frequency; however, 300 colonies, on average, were screened for each mutated codon. From these analyses, we identified 19 unique Aer mutants that were nonaerotactic and which formed cylinder-shaped colonies lacking an aerotactic ring at their edges (Fig. 2 and 3A). These mutants remained nonaerotactic when IPTG concentrations were varied from 20 μM to 1 mM in swarm plates, differentiating true null aerotaxis phenotypes from simple deficiencies in Aer expression.
FIG. 2.
Domain organization of the Aer protein showing residue substitutions, N-terminal deletions, and associated phenotypes. All missense mutations, except for T12C, were identified by mutagenesis of pGH1. Aer-T12C was identified during cysteine substitution experiments. Mutant Aer proteins that were phenotypically rescued in the Δaer Δtsr strain BT3312 are indicated in roman type, while mutant proteins that were not phenotypically rescued are underlined and italicized. An asterisk (Aer[20-506]*) indicates that a glycine codon was added before codon 20 in this construct, as explained in Materials and Methods. The phenotypes listed indicate behavior in a temporal oxygen gradient for the receptorless strain BT3388, expressing the mutant Aer proteins. Abbreviations: TM, transmembrane domain; PS, proximal signaling region; HCD, highly conserved domain.
FIG. 3.
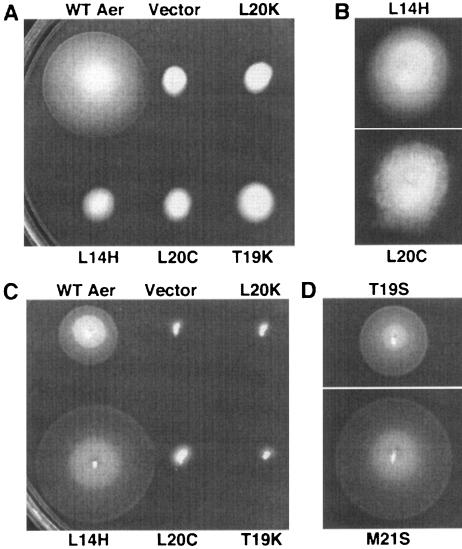
Aerotaxis elicited by Aer missense mutants in the receptorless E. coli strain BT3388 (A and B) or the Δaer Δtsr strain BT3312 (C and D). (A and B) Taxis on tryptone semisoft agar by Aer mutants in the receptorless strain induced with 20 μM IPTG after incubation at 30°C for 14 h (A), or 19 h (B). (A) Examples of Aer missense mutants demonstrating the different phenotypes observed. Upper row: WT Aer, plasmid vector pTrc99A, Aer-L20K (smooth swimming). Bottom row: Aer-L14H (increased tumbling frequency), Aer-L20C (constant tumbling), Aer-T19K (inverse response). (B) Enlargements showing details of the fuzzy, cotton ball edges produced by the constant tumbling of Aer-L20C in comparison with the more-regular edges of other nonaerotactic mutants such as Aer-L14H. (C and D) Aerotaxis on succinate semisoft agar by Aer mutants in the Δaer Δtsr strain after incubation at 30°C for 17 h. (C) Aer missense mutants from panel A. (D) Aer-T19S and Aer-M21S.
In a temporal assay, receptorless E. coli expressing WT Aer from pGH1 swam smoothly (CCW rotation) in response to an oxygen increase and tumbled (CW rotation) in response to an oxygen decrease. Control cells transformed with a vector alone (pTrc99A) retained a constant smooth swimming (CCW rotation) phenotype in response to oxygen increases and decreases. The 19 Aer mutants displayed four distinct aerotaxis phenotypes in a temporal oxygen gradient: (i) smooth swimming (no response), (ii) constant tumbling (signal locked on CW rotation), (iii) increased tumbling frequency (CW biased), and (iv) inverse response (bacteria were repelled by oxygen instead of attracted to it; i.e., a CCW signal elicited a CW response) (Fig. 2). Cells expressing mutant proteins that caused increased tumbling frequency responded correctly to an increase or decrease in oxygen, demonstrating signal processing by those mutant proteins. Inverse mutants tumbled in response to an oxygen increase and under steady-state aerobic conditions but swam smoothly in response to an oxygen decrease. Mutants designated as constantly tumbling, on the other hand, remained tumbling in response to an oxygen increase or decrease and under steady-state aerobic conditions. They produced nonaerotactic swarms with fuzzy, “cotton ball” edges, unlike the more-even edges of other nonaerotactic mutants (see example in Fig. 3B).
Plasmids encoding mutant Aer proteins that elicited aberrant phenotypes in the receptorless strain (BT3388) were transformed into a Δaer Δtsr strain (BT3312), which expresses the Tar, Trg, and Tap chemoreceptors. On 30 mM succinate semisoft agar without induction, 12 of the 19 mutant proteins were phenotypically rescued in the Δaer Δtsr strain (Fig. 2 and 3C and D). The majority of these were aerotactic superswarmers, with a swarm diameter at least 1.3-fold larger than that generated by pGH1 in the Δaer Δtsr strain (Fig. 3D, lower panel). The mechanism for superswarming is unknown, although Aer expression levels can affect swarm size. For instance, when WT Aer is expressed from pGH1 to concentrations above the uninduced “leaky” expression level, there is a decrease in the size of swarms. However, when superswarming mutants were coexpressed with WT Aer from a compatible plasmid, the colony swarm size still increased, indicating that the substitution in the Aer protein, and not the expression level, caused the phenotype (data not shown).
Seven of the mutant Aer proteins did not resurrect aerotaxis in the Δaer Δtsr strain (Table 1 and Fig. 3C), suggesting that the associated mutations caused more-serious defects than the mutations that were rescued in this strain. One of these Aer proteins had a conservative substitution (L20C), a second contained a hydrophobic to polar substitution (L20T), while the other five mutants were the result of charged amino acid or proline substitutions (L20K, M21D, M21P, T19K, and T19R). The Aer-M21P construct expressed only 20% the amount of WT Aer protein (as expressed from pGH1), while the other mutant proteins expressed 50% to 90% as well as the WT (Table 1). When expression of these mutant Aer proteins was induced with IPTG concentrations between 20 μM and 1 mM, none supported aerotaxis on succinate semisoft agar. The individual phenotype orchestrated by each of the mutants in a temporal oxygen gradient was identical in both the Δaer Δtsr strain and the receptorless strain. In addition, Aer-L20K, Aer-M21D, and Aer-M21P remained nonfunctional in temporal assays when they were induced with 1 mM IPTG.
TABLE 1.
Characteristics of mutant Aer proteins in the Δaer Δtsr strain BT3312
| Aer mutant | Phenotypea | Aer stabilityb | Dominancec | FAD ratio (induced: uninduced)d |
|---|---|---|---|---|
| L20K | No response (smooth) | +/− | Dominant | 1.04 ± 0.09 |
| M21D | No response (smooth) | +/− | Partially dominant | 1.01 ± 0.06 |
| M21P | No response (smooth) | − | Partially dominant | 0.92 ± 0.08 |
| L20C | Constantly tumbling | + | Recessive | 1.66 ± 0.16 |
| T19K | Inverse response | +/− | Recessive | 1.19 ± 0.15 |
| T19R | Inverse response | +/− | Recessive | 1.14 ± 0.11 |
| L20T | Inverse response | +/− | Recessive | 1.06 ± 0.15 |
Phenotypes were determined in a temporal oxygen gradient after induction with 200 μM IPTG.
Stability was estimated from the cellular concentration of mutant Aer proteins relative to the concentration of pGH1-expressed WT Aer protein. −, unstable, 20% or less of the amount of pGH1-expressed Aer; +/−, intermediate stability, 50% to 75% of the amount of pGH1-expressed Aer; +, stable, 76% to 100% of the amount of pGH1-expressed Aer.
Phenotypic dominance was tested by coexpressing the mutant Aer proteins with WT Aer in a recA derivative of BT3312 (BT3400), as described in the text.
Ratios of FAD concentrations in whole cells before and after induction of Aer with 1 mM IPTG. Averages and standard deviations were derived from the results of 3 to 5 independent experiments. The ratio for WT Aer (as expressed from pGH1) was 1.64 ± 0.09, compared to 0.99 ± 0.05 for the plasmid vector pTrc99A. For the various Aer mutants, a ratio of less than 1.2 indicates that the mutant Aer protein did not bind FAD.
To determine whether the seven Aer missense mutants affected interactions within dimers or larger signaling units, we tested for phenotypic dominance by coexpressing the mutant proteins with WT Aer, where WT expression was induced from a tightly regulated pnahG promoter in pDS7. We introduced plasmids into a recA derivative of the Δaer Δtsr strain (BT3400) and determined the concentrations of sodium salicylate (WT Aer) and IPTG (Aer mutants) that induced a WT:mutant expression ratio of approximately 1:1 for each combination. Under these conditions, the Aer-L20K allele was dominant and spoiled aerotaxis signaling by WT Aer. Aer-M21D and Aer-M21P were partially dominant (Table 1): in both cases the colonies swarmed, but an aerotactic ring was absent. In comparison with the null aerotaxis mutants, the CW-locked and inverse Aer mutants were recessive alleles and had no effect on aerotaxis directed by WT Aer proteins (Table 1).
Truncation of the Aer-PAS N-cap.
We examined the ability of Aer mutants with N-terminal truncations to support aerotaxis. Three of the truncations lacked part or all of the PAS N-cap; the fourth truncation lacked the entire N-cap and five PAS core residues (Fig. 2). The four truncated Aer proteins were significantly unstable; steady-state cellular levels ranged from 5% to 20% of pGH1-expressed WT Aer, suggesting that the N-cap might stabilize the structure of the Aer PAS domain. When the truncated Aer mutants were expressed in the receptorless strain, only Aer[7-506] supported aerotaxis on tryptone semisoft agar, although induction with 100 μM IPTG was required. The other truncation mutants did not support aerotaxis when expressed in the presence of IPTG concentrations up to 1 mM. When expressed in Δaer Δtsr cells on succinate semisoft agar, Aer[7-506] and Aer[15-506] supported aerotactic superswarming, while cells with the larger truncations (Aer[20-506] and Aer[25-506]) remained nonaerotactic at all levels of IPTG induction (up to 1 mM).
When expressing Aer[25-506], neither strain showed a response to a temporal oxygen gradient. Similarly, when Aer[20-506] was expressed, there was no response to a temporal oxygen gradient in the receptorless strain, but there was a weak inverse response in the Δaer Δtsr strain. This response consisted of a weak, smooth-swimming response when oxygen was decreased and increased tumbling (although not constant tumbling) under steady-state aerobic conditions. This phenotypic difference between strains may reflect the smooth-swimming bias of the receptorless strain. Unlike Aer[20-506], Aer[15-506] caused a strong inverse response in the receptorless strain, while Aer[7-506] allowed cells to respond normally to an oxygen increase or decrease but caused increased tumbling under steady-state aerobic conditions. This bias did not prevent aerotaxis directed by Aer[7-506] on tryptone swarm plates under the conditions used for these assays (200 μM IPTG).
FAD binding by the mutant Aer proteins.
The level of FAD bound to Aer was determined by measuring total cellular FAD extracted from Δaer Δtsr cells before and after overexpression of Aer. FAD levels were quantified by the ability of extracts to resurrect apo-DAAO activity (25, 29). There were several advantages of this method over the one described previously (5, 49). The new protocol did not require fractionation or extraction of membranes, there was minimal loss of FAD during sample preparation, and femtomol sensitivity eliminated the requirement for large culture volumes. However, the method did require Aer to increase total cellular FAD enough above background levels to be resolved. Since a single Δaer Δtsr cell expressed approximately 30,000 copies of FAD (55 μM; data not shown), a 20% rise in total FAD required some 6,000 copies of Aer monomers to be expressed, assuming 1:1 stoichiometry. Using this method, six of the seven proteins that contained missense mutations and that did not support aerotaxis in Δaer Δtsr cells did not appear to bind FAD compared to the WT Aer and vector controls (Table 1). The FAD-binding defect of Aer-L20K, Aer-M21D, and Aer-M21P explains their lack of response in temporal assays. However, conclusions cannot be drawn for Aer-M21P, since the cellular concentration of this Aer mutant was very low.
The mutant Aer proteins associated with an inverse temporal response, Aer-T19K, Aer-T19R, and Aer-L20T, likewise appeared to have defective FAD binding (Table 1). These mutant proteins were present at no lower than 50% the concentration of WT Aer as expressed from pGH1. This should have yielded induced to uninduced ratios of at least 1.3 had they bound FAD with affinity similar to that of WT Aer (induced to uninduced ratio, 1.6). Since Aer apparently requires FAD to switch between CW- and CCW-signaling states, it is likely the mutant Aer proteins that elicit an inverse response bind FAD. However, they may have a lower FAD-binding affinity that resulted in less FAD sequestered and therefore lower total FAD concentrations in the cell. The constantly tumbling mutant Aer-L20C, which bound FAD to an extent similar to that of WT Aer, is assumed to have a normal affinity for FAD (Table 1).
Second-site suppressor analysis.
The Aer-S28G nonspecific suppressor functionally resurrects FAD binding and aerotaxis to many Aer-F1 and Aer-HAMP mutants (57; M. Kang, M. S. Johnson, and B. L. Taylor, unpublished data). When the S28G missense mutation was introduced into the Aer-L20K, Aer-M21D, and Aer-M21P constructs, it did not resurrect function. This suggests that the type of conformational changes induced by substitutions in Aer residues 20 and 21 are different from the type of conformational changes induced by F1 or HAMP mutants.
Cross-linking the Aer-PAS N-cap to determine orientation and proximity.
Codons 4 to 22 of the aer gene were serially mutated to encode cysteine in a construct that encoded substitutions at the three native cysteine sites of Aer (C193S, C203A, and C253A [pMB1]). The single-cysteine replacements did not affect aerotaxis, with the exception of Aer-T12C and Aer-L20C, which were nonaerotactic in the receptorless strain (Fig. 2) and were therefore excluded from the cross-linking experiments.
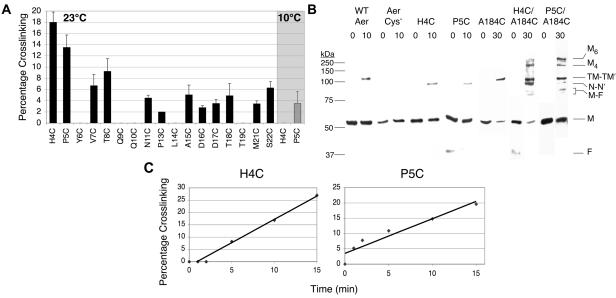
We investigated the orientation of the N-terminal regions from two PAS domains in an Aer dimer by attempting to cross-link cysteine replacements. Cross-linking was assessed by disulfide formation after whole cells were treated with the oxidant copper phenanthroline. Overall, the extent of cross-linking at 23°C was low, although 12 of 17 residues formed disulfide bonds, indicating that the N terminus of two monomers can collide (Fig. 4). The highest level of cross-linking was observed for Aer-H4C and Aer-P5C (18% and 13.5% after 10 min, respectively) (Fig. 4A). At 10°C, Aer-H4C did not cross-link and P5C showed a significant reduction in cross-linking (Fig. 4A). In contrast, Aer-A184C, which cross-links exclusively at the dimer interface in the membrane region, formed extensive disulfide bonds at 23°C (3) and at 10°C (49% after 20 min). Since 10°C is below the lipid phase transition temperature of E. coli membranes (18°C) (44), the higher level of cross-linking observed at 23°C for Aer-H4C and Aer-P5C suggests that N-cap cross-linking depends upon random collisions from lateral movement in the membrane bilayer rather than stable structural proximity. The remaining N-cap residues did not cross-link, or they cross-linked weakly at 23°C (<10% after 10 min). Cross-linked N-cap dimers migrated during sodium dodecyl sulfate-polyacrylamide gel electrophoresis, with an apparent size approximately 10-kDa smaller than cross-linked WT Aer, which cross-links at C253 and C203 (38) (Fig. 4B).
FIG. 4.
In vivo disulfide bond formation between N-cap residues of cognate Aer-PAS domains. Disulfide bond formation was determined after exposing intact cells to copper phenanthroline as described in the text. (A) Percentage of specified cysteine replacements that formed disulfide bonds in the receptorless strain after 10 min at 23°C (black bars), or after 20 min at 10°C (shaded region). Averages were derived from the results of 2 to 4 independent experiments, and error bars indicate the standard deviations from the means. (B) Western blots of cross-linked Aer proteins. Cellular proteins were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Aer was visualized by chemiluminescent Western blotting, using anti-Aer2-166 antisera. Cross-linked N-cap dimers (N-N′) were approximately 10-kDa smaller than cross-linked WT Aer (which cross-links at C253 and C203) or Aer-A184C (TM-TM′). The double-cysteine mutants Aer-H4C/A184C and Aer-P5C/A184C cross-linked to form higher-order products, which may represent tetramers (M4) and hexamers (M6) of Aer. A 37-kDa proteolytic fragment of Aer (F) was often present (38); this fragment dimerized with a full-length Aer monomer (M) to yield two faint bands (M-F) generated from single disulfide bonds at A184C or an N-Cap cysteine. Protein standards are on the left (Precision Plus, Bio-Rad). (C) Rates of Aer-H4C and Aer-P5C cross-linking over 15 min. After determining that a 10-min exposure to oxidant was in the linear range for disulfide formation, we subsequently used 10 min for fixed-time assays at 23°C.
Cross-linking to differentiate interactions within and between dimers.
To determine whether cross-linking of the Aer N-cap occurs within or between Aer dimers, Aer-A184C was engineered into Aer-H4C and Aer-P5C. Evidence that Aer-A184C cross-links exclusively within Aer dimers at the dimer interface and not between adjacent dimers was derived from experiments in which A184C was paired with other cysteine substitutions known to form intra- and interdimeric disulfide bonds. Dimers formed exclusively when A184C was paired with cysteines that form intradimer cross-links, but higher-order multimers were present when paired with cysteines known to form interdimer cross-links (D. Amin, B. L. Taylor, and M. S. Johnson, unpublished data).
Both Aer-H4C/A184C and Aer-P5C/A184C were aerotactic in Δaer Δtsr cells, although Aer-H4C/A184C was nonaerotactic in the receptorless strain. Disulfide formation was assayed in Δaer Δtsr cells expressing these proteins. In this case, cells were incubated with copper phenanthroline for 30 min, rather than 10 min, to enhance the formation of higher-order oligomers. If Aer-H4C or Aer-P5C cross-linked exclusively within a dimer, the highest complexes formed upon oxidation would be dimers. Multimers of Aer would be possible only if these N-cap cysteines could form disulfide bonds with neighboring dimers. Aer-H4C and Aer-P5C each cross-linked to form approximately 11% more dimers at 30 min than at 10 min (29 ± 4.9% and 25 ± 0.99% dimers, respectively; compare with 10-min values in Fig. 4A), but neither formed larger multimers. Similarly, Aer-A184C cross-linked strongly to form a dimer at 30 min (53.5 ± 5%), but did not form larger multimers (Fig. 4B). On the other hand, cross-linking of Aer-H4C/A184C or Aer-P5C/A184C under the same conditions produced bands on Western blots corresponding to dimers and larger multimers that may correspond to tetramers and hexamers (Fig. 4B). Because A184C cross-links only within a dimer, this indicates that Aer N-cap cross-linking can occur between sets of adjacent dimers.
DISCUSSION
Function of the Aer-PAS N-cap.
In this study, we examined the function of Aer proteins containing substitutions or deletions in the N-cap of the PAS domain. The findings suggest that the N-cap is important for stabilizing the structure of the PAS domain, perhaps by protecting the Aer-PAS core from degradation. PAS N-cap truncations and an M21P substitution (in full-length Aer) resulted in levels of mutant Aer proteins that were less than 20% of pGH1-expressed WT Aer. Other substitutions in the N-cap had less severe effects on Aer stability (Table 1).
The most common phenotype caused by missense mutations in the N-cap region was frequent or constant tumbling. This resulted from an increased CW bias of the flagellar motors (39). Other aerotaxis phenotypes observed included null and inverse behavior. The null aerotaxis phenotype was associated with a loss of FAD binding to mutant Aer proteins, suggesting that the N terminus of Aer stabilizes FAD binding to the PAS domain. The PAS N-cap regions from PYP and HERG play similarly important roles in signaling. When part of the PAS N-cap is removed from the PYP blue light receptor, the lifetime of the active form of PYP is markedly prolonged (27, 54). Removing the PAS N-cap from the HERG voltage-dependent potassium channel in humans severely affects the kinetics of channel deactivation and is indistinguishable from the results of deleting the entire HERG PAS domain (41). Aer N-cap truncations disrupted function in a graded manner. Aer[7-506] (Fig. 2) recovered function when overexpressed in the receptorless strain (BT3388). Aer[15-506] required the presence of other chemoreceptors to function, and truncations lacking the entire N-cap (Aer[20-506] and Aer[25-506] showed no function under any conditions tested.
Multiple and diverse mutations that convert aerotaxis and chemotaxis signaling from a normal phenotype to an inverse phenotype in which bacteria are repelled by ligands to which they are normally attracted have been reported (12, 28, 30, 42, 43). Models to explain these phenotypes have been proposed (28, 52). A suitable model that could account for the tumbling and inverted phenotypes orchestrated by the Aer-sensing (PAS) domain is a three-state model that was described previously (49). In this model, Aer normally cycles between the semiquinone form (FADH·; CCW rotation; CheA is not activated) under aerobic conditions and the hydroquinone form (FADH2; CW rotation; CheA is activated) under anaerobic conditions. The fully oxidized quinone form (FAD; CW rotation; CheA is activated) would be present under rare circumstances, such as during starvation (61) or at high oxygen concentrations (51). An inverted response could occur if a defect in Aer stabilized the quinone form (FAD; CW signal) under aerobic conditions and the semiquinone form (FADH·; CCW signal) under anoxic conditions. A constantly tumbling behavior could occur if the Aer defect stabilized the redox potential in either the fully oxidized or fully reduced states.
Fourteen of the Aer mutants that elicited a negative aerotaxis phenotype in the receptorless strain recovered aerotaxis when they were expressed in the Δaer Δtsr strain (BT3312), which retains the Tar, Trg, and Tap receptors (Fig. 2). The rescued mutants included all of those with an increased CW bias except L20C. Rescue probably resulted from bias correction in the Δaer Δtsr strain by the presence of Tar, which is present in much higher abundance than either Trg or Tap (17, 35). In this scenario, the CW bias of Aer increases the concentration of phospho-CheA, and therefore phospho-CheB methylesterase, which demethylates Tar, resulting in a concomitant decrease in CheA-P and a reduced CW bias of the flagellar motors. Of particular interest is the rescue of the inverse mutants, Aer-D17G, Aer-T19P, and Aer[15-506], by expression in the Δaer Δtsr strain. The mechanisms postulated to explain inverse responses do not account for rescue of an inverse mutation by correction of a signaling bias. However, high-abundance chemoreceptors are known to promote polar clustering of other low-abundance chemoreceptors (34, 36) (and likely Aer), so it is possible that clustering in some way stabilizes the native structure and function of Aer, despite defects in the N-cap region.
The mutant Aer proteins that did not support aerotaxis in either of the host strains used are assumed to have more-serious functional defects. There was a hot spot for these mutations at the junction of the N-cap and the PAS core, suggesting residues T19, L20, M21 (Fig. 2), and S22 (5) are critical for Aer structure and/or signaling. Aer proteins with substitutions at residues L20 or M21 affected FAD binding and demonstrated dominant behavior (Table 1), suggesting that the aberrant receptors disrupt both signaling and interactions with neighboring subunits.
Analysis of the Aer-PAS N-cap by oxidative cross-linking.
Although the Aer protein forms homodimers (38), the orientation of the Aer-PAS N-cap to the dimer interface had not been examined. We investigated this relationship by oxidative disulfide cross-linking of specific cysteine pairs that were engineered into the Aer protein (for a review of cross-linking, see reference 16). In addition, we examined the proximity of cognate N-cap residues as a possible means of determining the secondary structure, which is predicted to be a helix-loop-helix (49). At 23°C, the extent of cross-linking between cognate cysteine replacements showed periodicity, including local maxima corresponding to positions one and four of a heptad-repeating pattern (Fig. 4A). Although this suggests that the Aer-PAS N-cap might be helical, the low extent of cross-linking at 10°C makes any structural prediction premature. The crystal structures of other PAS domains showed that the N-cap is a helix-turn-helix (or helical lariat) in PYP (7, 46), helical in Ec DOS (33), and disordered in both FixL and HERG (20, 41). In the heme-binding E. coli DOS protein, the PAS N-cap region is an integral part of a dimer interface, with several residues providing dimer stabilization through hydrophobic interactions (33).
In Aer, H4C and P5C cross-linked substantially at 23°C but weakly at 10°C (Fig. 4A), which is below the lipid phase transition temperature of E. coli membranes (44). This indicates that these residues require lateral movement of the Aer membrane anchor to collide and are not in continuous contact, as would be expected if the Aer N-cap was at the dimer interface or at a contact domain between two dimers. We have recently shown that Aer dimers, like E. coli chemoreceptors (1, 31, 32), are organized as functional trimers of dimers (D. Amin, B. L. Taylor, and M. S. Johnson, unpublished data). In this study, a second cysteine replacement that cross-links exclusively within a dimer (A184C) was engineered into several Aer N-cap cysteine mutants. Larger multimers formed when either Aer-H4C/A184C or Aer-P5C/A184C was cross-linked, indicating the presence of cross-links at the dimer interface (A184C) as well as between dimers (H4C or P5C). Similar results were also observed when Aer-V260C (located in the proximal signaling region) was paired with these N-cap cysteine mutants (data not shown). These results suggest that the PAS N-cap faces outward, away from the dimer interface, and that the N-caps from two different dimers require lateral diffusion in the membrane bilayer to collide. Whether there is another face of the PAS domain that is involved in dimerization remains to be determined.
Acknowledgments
We are grateful to Divya Amin for providing the Aer-A184C construct and the results of unpublished work and to Maxwell Brandon for constructing pMB1. We thank Adanna James for constructing the Aer truncation mutants and carrying out initial truncation experiments and Nathan Abraham for technical assistance.
This work was supported by a grant from the National Institute of General Medical Sciences (GM29481) to B. L. Taylor.
REFERENCES
- 1.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amezcua, C., S. Harper, J. Rutter, and K. Gardner. 2002. Structure and interactions of PAS kinase N-terminal PAS domain. Model for intramolecular kinase regulation. Structure (Camb) 10:1349-1361. [DOI] [PubMed] [Google Scholar]
- 3.Amin, D. N., B. L. Taylor, and M. S. Johnson. 2006. Topology and boundaries of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 188:894-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 5.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borgstahl, G. E., D. R. Williams, and E. D. Getzoff. 1995. A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry 34:6278-6287. [DOI] [PubMed] [Google Scholar]
- 8.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, C. J., N. M. Derix, J. Hendriks, R. Boelens, K. J. Hellingwerf, and R. Kaptein. 2000. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in solution by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry 39:14392-14399. [DOI] [PubMed] [Google Scholar]
- 10.Crews, S. (ed.). 2003. PAS proteins: regulators and sensors of development and physiology. Kluwer Academic Publisher, Norwell, Mass.
- 11.Crosson, S., and K. Moffat. 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98:2995-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang, C. V., M. Niwano, J. Ryu, and B. L. Taylor. 1986. Inversion of aerotactic response in Escherichia coli deficient in cheB protein methylesterase. J. Bacteriol. 166:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 14.Echenique, J. R., and M. C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erbel, P. J., P. B. Card, O. Karakuzu, R. K. Bruick, and K. H. Gardner. 2003. Structural basis for PAS domain heterodimerization in the basic helix-loop-helix-PAS transcription factor hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA 100:15504-15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, X., J. W. Baumgartner, and G. L. Hazelbauer. 1997. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 179:6714-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genick, U. K., G. E. Borgstahl, K. Ng, Z. Ren, C. Pradervand, P. M. Burke, V. Srajer, T. Y. Teng, W. Schildkamp, D. E. McRee, K. Moffat, and E. D. Getzoff. 1997. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science 275:1471-1475. [DOI] [PubMed] [Google Scholar]
- 19.Gong, W., B. Hao, and M. K. Chan. 2000. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry 39:3955-3962. [DOI] [PubMed] [Google Scholar]
- 20.Gong, W., B. Hao, S. S. Mansy, G. Gonzalez, M. A. Gilles-Gonzalez, and M. K. Chan. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 95:15177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hao, B., C. Isaza, J. Arndt, M. Soltis, and M. K. Chan. 2002. Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 41:12952-12958. [DOI] [PubMed] [Google Scholar]
- 22.Harigai, M., Y. Imamoto, H. Kamikubo, Y. Yamazaki, and M. Kataoka. 2003. Role of an N-terminal loop in the secondary structural change of photoactive yellow protein. Biochemistry 42:13893-13900. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, S., S. Austin, T. Eydmann, T. Jones, and R. Dixon. 1996. Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc. Natl. Acad. Sci. USA 93:2143-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinkkanen, A., and K. Decker. 1983. Luminometric determination of FAD in subpicomole quantities. Anal. Biochem. 132:202-208. [DOI] [PubMed] [Google Scholar]
- 26.Hughson, A. G., and G. L. Hazelbauer. 1996. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide cross-linking in vivo. Proc. Natl. Acad. Sci. USA 93:11546-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamoto, Y., H. Kamikubo, M. Harigai, N. Shimizu, and M. Kataoka. 2002. Light-induced global conformational change of photoactive yellow protein in solution. Biochemistry 41:13595-13601. [DOI] [PubMed] [Google Scholar]
- 28.Jung, K. H., and J. L. Spudich. 1998. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J. Bacteriol. 180:2033-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keppler, D., J. Rudigier, and K. Decker. 1970. Enzymic determination of uracil nucleotides in tissues. Anal. Biochem. 38:105-114. [DOI] [PubMed] [Google Scholar]
- 30.Kihara, M., and R. M. Macnab. 1981. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J. Bacteriol. 145:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. H., W. Wang, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 99:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokawa, H., D. S. Lee, M. Watanabe, I. Sagami, B. Mikami, C. S. Raman, and T. Shimizu. 2004. A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor. J. Biol. Chem. 279:20186-20193. [DOI] [PubMed] [Google Scholar]
- 34.Li, M., and G. L. Hazelbauer. 2005. Adaptational assistance in clusters of bacterial chemoreceptors. Mol. Microbiol. 56:1617-1626. [DOI] [PubMed] [Google Scholar]
- 35.Li, M., and G. L. Hazelbauer. 2004. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 186:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lybarger, S. R., and J. R. Maddock. 2000. Differences in the polar clustering of the high- and low-abundance chemoreceptors of Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8057-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, Q., M. S. Johnson, and B. L. Taylor. 2005. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes flavin adenine dinucleotide-binding determinants to the AS-2 helix. J. Bacteriol. 187:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma, Q., F. Roy, S. Herrmann, B. L. Taylor, and M. S. Johnson. 2004. The Aer protein of Escherichia coli forms a homodimer independent of the signaling domain and FAD binding. J. Bacteriol. 186:7456-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macnab, R. M. 1977. Bacterial flagella rotating in bundles: a study in helical geometry. Proc. Natl. Acad. Sci. USA 74:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyatake, H., M. Mukai, S. Y. Park, S. Adachi, K. Tamura, H. Nakamura, K. Nakamura, T. Tsuchiya, T. Iizuka, and Y. Shiro. 2000. Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies. J. Mol. Biol. 301:415-431. [DOI] [PubMed] [Google Scholar]
- 41.Morais Cabral, J. H., A. Lee, S. L. Cohen, B. T. Chait, M. Li, and R. Mackinnon. 1998. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell 95:649-655. [DOI] [PubMed] [Google Scholar]
- 42.Muskavitch, M. A., E. N. Kort, M. S. Springer, M. F. Goy, and J. Adler. 1978. Attraction by repellents: an error in sensory information processing by bacterial mutants. Science 201:63-65. [DOI] [PubMed] [Google Scholar]
- 43.Nishiyama, S., T. Nara, M. Homma, Y. Imae, and I. Kawagishi. 1997. Thermosensing properties of mutant aspartate chemoreceptors with methyl-accepting sites replaced singly or multiply by alanine. J. Bacteriol. 179:6573-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overath, P., M. Brenner, T. Gulik-Krzywicki, E. Shechter, and L. Letellier. 1975. Lipid phase transitions in cytoplasmic and outer membranes of Escherichia coli. Biochim. Biophys. Acta 389:358-369. [DOI] [PubMed] [Google Scholar]
- 45.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pellequer, J. L., K. A. Wager-Smith, S. A. Kay, and E. D. Getzoff. 1998. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. USA 95:5884-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajagopal, S., and K. Moffat. 2003. Crystal structure of a photoactive yellow protein from a sensor histidine kinase: conformational variability and signal transduction. Proc. Natl. Acad. Sci. USA 100:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Repik, A., A. Rebbapragada, M. S. Johnson, J. O. Haznedar, I. B. Zhulin, and B. L. Taylor. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitz, R. A. 1997. NifL of Klebsiella pneumoniae carries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol. Lett. 157:313-318. [DOI] [PubMed] [Google Scholar]
- 51.Shioi, J., C. V. Dang, and B. L. Taylor. 1987. Oxygen as attractant and repellent in bacterial chemotaxis. J. Bacteriol. 169:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor, B. L., and M. S. Johnson. 1998. Rewiring a receptor: negative output from positive input. FEBS Lett. 425:377-381. [DOI] [PubMed] [Google Scholar]
- 53.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Horst, M. A., I. H. van Stokkum, W. Crielaard, and K. J. Hellingwerf. 2001. The role of the N-terminal domain of photoactive yellow protein in the transient partial unfolding during signalling state formation. FEBS Lett. 497:26-30. [DOI] [PubMed] [Google Scholar]
- 55.Vreede, J., M. A. van der Horst, K. J. Hellingwerf, W. Crielaard, and D. M. van Aalten. 2003. PAS domains. Common structure and common flexibility. J. Biol. Chem. 278:18434-18439. [DOI] [PubMed] [Google Scholar]
- 56.Watts, K. J., M. S. Johnson, and B. L. Taylor. 2006. Minimal requirements for oxygen sensing by the aerotaxis receptor Aer. Mol. Microbiol. 59:1317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watts, K. J., Q. Ma, M. S. Johnson, and B. L. Taylor. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J. Bacteriol. 186:7440-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yildiz, O., M. Doi, I. Yujnovsky, L. Cardone, A. Berndt, S. Hennig, S. Schulze, C. Urbanke, P. Sassone-Corsi, and E. Wolf. 2005. Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol. Cell. 17:69-82. [DOI] [PubMed] [Google Scholar]
- 59.Yu, H. S., J. H. Saw, S. Hou, R. W. Larsen, K. J. Watts, M. S. Johnson, M. A. Zimmer, G. W. Ordal, B. L. Taylor, and M. Alam. 2002. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol. Lett. 217:237-242. [DOI] [PubMed] [Google Scholar]
- 60.Zhong, X., B. Hao, and M. K. Chan. 2003. Structure of the PAS fold and signal transduction mechanisms, p. 1-16. In S. T. Crews (ed.), PAS proteins: regulators and sensors of development and physiology. Kluwer Academic Publishers, Boston, Mass.
- 61.Zhulin, I. B., E. H. Rowsell, M. S. Johnson, and B. L. Taylor. 1997. Glycerol elicits energy taxis of Escherichia coli and Salmonella typhimurium. J. Bacteriol. 179:3196-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhulin, I. B., B. L. Taylor, and R. Dixon. 1997. PAS domain S-boxes in archaea, bacteria and sensors for oxygen and redox. Trends Biochem. Sci. 22:331-333. [DOI] [PubMed] [Google Scholar]