Abstract
Two new insertion sequences, ISCce1 and ISCce2, were found to be inserted into the cipC gene of spontaneous mutants of Clostridium cellulolyticum. In these insertional mutants, the cipC gene was disrupted either by ISCce1 alone or by both ISCce1 and ISCce2. ISCce1 is 1,292 bp long and has one open reading frame. The open reading frame encodes a putative 348-amino-acid protein with significant levels of identity with putative proteins having unknown functions and with some transposases belonging to the IS481 and IS3 families. Imperfect 23-bp inverted repeats were found near the extremities of ISCce1. ISCce2 is 1,359 bp long, carries one open reading frame, and has imperfect 35-bp inverted repeats at its termini. The open reading frame encodes a putative 398-amino-acid protein. This protein shows significant levels of identity with transposases belonging to the IS256 family. Upon transposition, both ISCce1 and ISCce2 generate 8-bp direct repeats of the target sequence, but no consensus sequences could be identified at either insertion site. ISCce1 is copied at least 20 times in the genome, as assessed by Southern blot analysis. ISCce2 was found to be mostly inserted into ISCce1. In addition, as neither of the elements was detected in seven other Clostridium species, we concluded that they may be specific to the C. cellulolyticum strain used.
Insertion sequences (ISs) are small mobile genetic elements that are between 0.7 and 3.5 kb long and are found in the genomes of numerous bacteria. They contain only genes involved in their transposition. They usually have inverted repeats (IRs) at their termini and duplicate a sequence consisting of several base pairs at the target site upon transposition. Based on the homology between their transposase sequences and common structural features, these sequences have been classified in various IS families (for reviews see references 8 and 32). Insertion of an IS element can cause gene disruption or activation due to the creation or insertion of upstream promoters, and this contributes significantly to the plasticity of the host cell genome. Mobile elements are commonly associated with the virulence functions of many pathogens, such as Escherichia coli (11), Vibrio cholerae (43), Yersinia pestis (15), and Clostridium perfringens (6). IS elements are frequently used as markers in restriction fragment length polymorphism studies for epidemiological purposes, like those performed with Salmonella enterica serovar Typhimurium (IS200) (42) and Mycobacterium tuberculosis (IS6110) (27). In addition, these sequences are valuable tools for identifying relevant genes and the functions in which they are involved.
Only a few IS elements have been described so far in clostridia; four have been reported in Clostridium perfringens (6), one has been reported in Clostridium beijerinckii NCIMB 8052 (30), and one has been reported in the cellulolytic bacterium Clostridium thermocellum (39). Clostridium cellulolyticum is a mesophilic anaerobic cellulolytic bacterium which secretes enzymatic complexes called cellulosomes (5, 17). These complexes are composed of several enzymes, most of which are cellulases (Cel proteins); these enzymes are anchored to a large scaffolding protein (160 kDa) that lacks catalytic activity, designated CipC (17, 36). Many of the cel genes form a large cluster spanning 24 kb beginning with the cipC gene (3, 38). Functional studies of the cellulosomes have been restricted so far to biochemical studies of recombinant subunits overproduced in E. coli (4, 16, 19, 38). Gene transfer techniques were recently developed for C. cellulolyticum (25, 44) and were used to modify its fermentation pathways (22). However, no description of a mutagenic system allowing random or targeted mutagenesis has been described so far for this bacterium. Naturally occurring ISs would therefore be valuable tools for developing a transposon-based mutagenesis system.
In this paper, we describe two different IS elements which were found in the cipC gene of various isolated clones of C. cellulolyticum ATCC 35319. The features of these sequences, which are designated ISCce1 and ISCce2, are described below, and their membership in various IS families is discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Table 1 lists all the bacterial strains and plasmids used in this study. E. coli DH5α was used as the recipient strain for the recombinant plasmids (derivatives of pUC18, pUC19, or pGEM-T-Easy). It was grown at 37°C in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) (23).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source (reference) |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F−endA1 hsdR17(rK− mK+) supE44 thi-1 λ−gyrA96 relA1 Δ(lacZYA argF)U169 (φ80 lacZΔM15) recA1 | Roche Diagnostics (23) |
| Clostridium cellulolyticum ATCC 35319 | Wild type | E. Petitdemange (37) |
| cipCMut1 | cipC::ISCce1::ISCce2 | This study |
| cipCMut2 | cipC::ISCce1 | This study |
| Clostridium papyrosolvens DSM 2782 | Wild type | DSM (31)a |
| Clostridium termitidis ATCC 51486 | Wild type | J. L. Caillol/(24) |
| Clostridium cellobioparum DSM 1351 | Wild type | DSM (10) |
| Clostridium thermocellum DSM 1237 | Wild type | DSM (29) |
| Clostridium saccharobutylicum ATCC 860 | Wild type | A. L. Contreras (26) |
| Clostridium acetobutylicum ATCC 328 | Wild type | P. Soucaille (41) |
| Plasmids | ||
| pUC18 | Cloning vector; Apr | Appligene Oncor |
| pUC19 | Cloning vector; Apr | Appligene Oncor |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pH62 | 3.8-kb PvuII fragment containing cipC::ISCce1::ISCce2, cloned into pUC18 | This study |
| pS29 | 2-kb SspI fragment containing part of ISCce1::ISCce2, cloned into pUC19 | This study |
DSM, Deutsche Sammlung von Mikroorganismen.
C. cellulolyticum ATCC 35319 and mutant strains cipCMut1 and cipCMut2 were grown anaerobically at 32°C on basal medium (20) supplemented with either cellobiose (2 g/liter; Sigma-Aldrich) or MN300 cellulose (5 g/liter; Serva) as the carbon and energy source. Colonies of the mutant strains were isolated on solid medium (basal medium supplemented with 2 g of cellobiose per liter and 15 g of agar per liter) under the anaerobic atmosphere in a glove box (N2-H2, 95:5 [vol/vol]). Plates were incubated in anaerobic jars under 2 × 105 Pa of an N2-CO2 atmosphere (80:20, vol/vol).
The other Clostridium strains were grown as previously described (10, 24, 26, 29, 31, 41).
DNA manipulations.
Chromosomal DNA was obtained from the various Clostridium strains by using a genomic DNA purification kit (Promega). DNA from Clostridium cellulovorans was a generous gift from R. H. Doi (University of California, Davis). Large-scale plasmid purification from E. coli and small-scale plasmid purification from E. coli were performed by using kits from Qiagen and Promega. Restriction enzymes and DNA-modifying enzymes were purchased from Promega and Roche Applied Science and were used as recommended by the manufacturers. DNA sequencing was performed by Genome Express (Grenoble, France).
Primers and probes.
Primers were purchased from MWGAG-Biotech (Courtaboeuf, France) (Table 2). Primers c1 and c2 were used to amplify sequences that disrupt the cipC gene in the cipCMut1 and the cipCMut2 strains. Primers A, B, C, D, E, F, G, and H were used in inverse PCR experiments to analyze insertion sites of the two IS elements (see below and Fig. 2). The various primers were also used for sequencing ISCce1 and ISCce2.
TABLE 2.
Primer sequences
| Primer | Sequence (5′ to 3′) |
|---|---|
| c1 | ACATTCAAATTAAAGAGTGTAGCGG |
| c2 | AGAGTTACATTTACATTTGCAGGAA |
| A | CAAAACTTAATATTTCCCAGAGG |
| B | CGCTCGCATATACCGTTTGTT |
| C | GGTAACTTAATTCTACCAAATGACTG |
| D | CCTGGTTAGGAGTAGTGTTGAGGA |
| E | TAGTCCGTCCGCGAATCTCC |
| F | GTACACAATGGGGCAGCAAGAA |
| G | CTGATTGAGTTGGCCGAATAT |
| H | TGTGCTGTCATAATTGAAATCTCC |
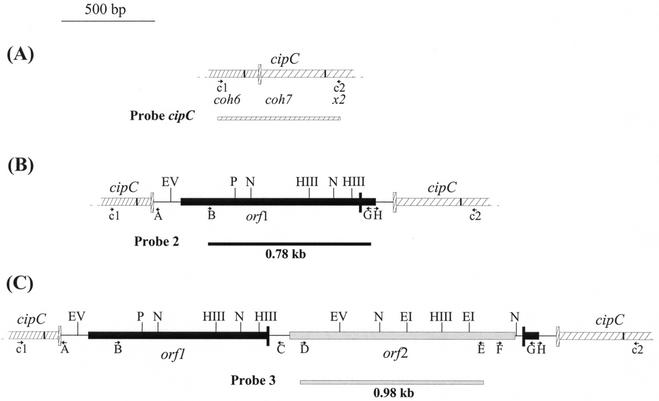
FIG. 2.
Maps of the cipC gene in the wild-type strain (A) and mutant strains cipCMut2 (B) and cipCMut1 (C). orf1 (solid box) and orf2 (gray box) encode the putative transposases of ISCce1 and ISCce2, respectively. The vertical boxes represent insertion sites of ISCce1 in the cipC gene (cross-hatched box) and of ISCce2 in ISCce1 (solid box). The positions of primers A, B, C, D, E, F, G, H, c1, and c2 are indicated by arrows. Probe 2 and probe 3 are internal probes of ISCce1 and ISCce2, respectively. Restriction sites: EV, EcoRV; P, PstI; N, NdeI; HIII, HindIII; EI, EcoRI.
Probe 1 was obtained by PCR performed with primers M13-upward and M13-downward by using the pS29 plasmid (Table 1) as the template (Fig. 1). Primers G and B and primers D and E were used to synthesize probes 2 and 3, respectively. The cipC probe was synthesized by using primers c1 and c2 with wild-type DNA.
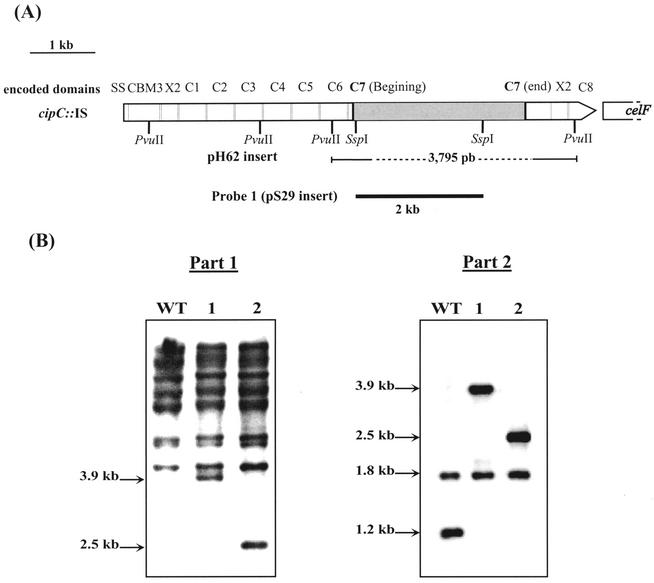
FIG. 1.
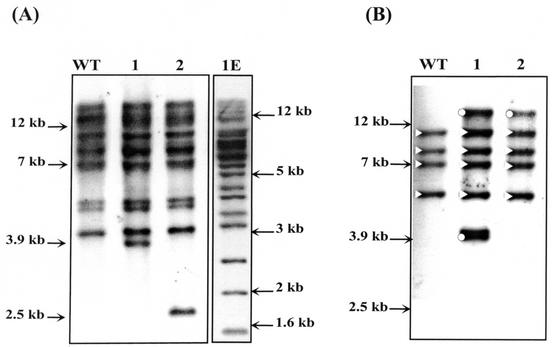
Discovery of insertion elements in C. cellulolyticum. (A) Map of the cipC gene disrupted by an insertion element (shaded box). The encoded domains are indicated above the gene (SS, signal sequence; CBM3, carbohydrate binding module of family 3; X2, unknown function module of family 2; C1 to C8, cohesin modules). (B) Southern blot analysis of PvuII-digested genomic DNA from various strains. The blot was probed with PCR digoxigenin-labeled probe 1 (part 1) and with the cipC probe (part 2). Lane WT, wild type; lane 1, cipCMut1; lane 2, cipCMut2. Sizes (in kilobase pairs) are indicated on the left.
Southern blot analysis.
DNAs from Clostridium strains were cut with PvuII or EcoRI and separated by electrophoresis in a 0.7% agarose gel. DNA fragments were transferred by Southern blotting onto a nylon membrane (Roche Applied Science) and hybridized to the PCR-generated digoxigenin-labeled probe at 68°C (in the case of C. cellulolyticum DNA) and at 68 or 55°C (in the case of heterologous Clostridium DNAs). Targets were detected by chemiluminescence by using a DIG luminescent detection kit (Roche Applied Science). The probe was removed after each experiment by incubating the blot twice for 20 min in a 0.2 M NaOH-0.1% sodium dodecyl sulfate solution at 37°C in order to hybridize one blot successively with many probes.
Inverse PCR.
DNA sequences flanking the IS elements in the genome of C. cellulolyticum were amplified by inverse PCR (34). Total chromosomal DNA of the cipCMut1 strain was digested by a restriction enzyme cutting the IS element once near the unknown sequence. To determine the sequences flanking ISCce1 at its left junction, DNA was digested with PstI or NdeI (Fig. 2C). The resulting fragments were ligated and used as templates for PCR amplification with divergent primers A and B. The inverse PCR products were then purified by using a Qiaex II gel purification kit (Qiagen) and were ligated to linearized pGEM-T-Easy vector. Ligation mixtures were used to transform competent E. coli DH5α cells. Ampicillin-resistant colonies were isolated. Plasmid DNA was purified and subjected to restriction analysis. Depending on the orientation of the insert, the T7 or SP6 primer was used to sequence the junction. The same protocol was used to determine the right junctions of ISCce1, but in this case the DNA was digested with NdeI and the PCR was carried out with primers G and H (Fig. 2C). In order to find the right junctions of combined ISs, the DNA was digested with EcoRI or HindIII, and the PCR was performed with primers E and H (Fig. 2C). Fragments flanking ISCce2 at its left junctions were synthesized with primers C and D from ligated EcoRV DNA fragments. Right junctions of ISCce2 were analyzed from inverse PCR products obtained with primers E and F by using ligated EcoRI or HindIII fragments as the templates.
Computer analysis.
Nucleotide sequences were analyzed with the DNASIS program, version 2.1. The BLAST program (1) was used for a homology search of the nucleotide and protein sequences in the GenBank and IS (www-is.biotoul.fr) databases. The DNA binding motifs in the proteins were predicted by using the Helix-Turn-Helix program (13). Multiple-sequence alignments, obtained with ClustalW, version 1.7 (45), were used to construct phylogenetic trees with Phylo_win (18).
Nucleotide sequence accession numbers.
The nucleotide sequences of the IS elements described here, ISCce1 and ISCce2, have been deposited in the GenBank database under accession numbers AY130778 and AY130779, respectively.
RESULTS
Discovery of insertion elements in C. cellulolyticum.
A pUC18-PvuII genomic DNA library of C. cellulolyticum was previously constructed in E. coli DH5α and screened by colony hybridization with a 285-bp probe complementary to the 3′ end of cipC (35). The 3.8-kb PvuII fragment inserted into the pH62 recombinant plasmid of one of the selected clones was found to contain an internal part of the cipC gene interrupted by a 2,659-bp sequence. This sequence contained two open reading frames (ORFs) encoding proteins which showed significant levels of identity with transposases. The cipC gene was disrupted at the beginning of the sequence encoding cohesin 7 of the scaffolding protein CipC (Fig. 1A).
A liquid culture of the strain used to construct the DNA library was plated onto solid medium. DNA was extracted from 17 isolated colonies, digested with PvuII, and subjected to a Southern blot analysis by using probe 1 (Fig. 1A). Based on comparisons between the various patterns obtained, three major groups were distinguished. The patterns of the first group were comparable to the pattern obtained with the DNA purified from the reference strain (ATCC 35319). Probe 1 hybridized with many fragments (Fig. 1B, part 1, lane WT), indicating that many copies of this DNA sequence were inserted at various loci on the chromosome. In the second group, an additional fragment was detected in the DNA; this fragment was 3.9 kb long (a representative example is shown in Fig. 1B, part 1, lane 1). In the third group, the additional fragment was 2.5 kb long (Fig. 1B, part 1, lane 2). When the cipC probe was used, 1.2- and 1.8-kb fragments were detected in the DNA of the wild-type strain (Fig. 1B, part 2); the probe hybridized with two PvuII fragments of the cipC gene. The 1.8-kb fragment was also detected in lanes containing DNA from strains 1 and 2, but the 1.2-kb fragment was not detected. Instead of the latter fragment, 3.9- and 2.5-kb fragments were detected in strains 1 and 2, respectively (Fig. 1B, part 2), which indicated that the cipC gene had been disrupted in these two strains (which were designated cipCMut1 and cipCMut2).
Structural analysis of the ISs.
Genomic DNAs from strains cipCMut1 and cipCMut2 were used to amplify the sequences inserted into cipC. The PCR fragments were synthesized by using primers c1 and c2 designed from the cohesin 6- and C-terminal X2 module-encoding sequences, respectively (Fig. 2). The sequences were analyzed after cloning into the pGEM-T-Easy vector by using primers c1, c2, B, C, D, and E (Fig. 2B and C). A 2,659-bp sequence was inserted into cipC in cipCMut1 DNA; this sequence was identical to the copy found in the PvuII fragment of pH62 and inserted at the same place (Fig. 1A and 2C). Another insertion element, which was 1,292 bp long, was found in the same place in cipCMut2 DNA. This element corresponds to the 2,659-bp sequence with its internal part deleted (Fig. 2B).
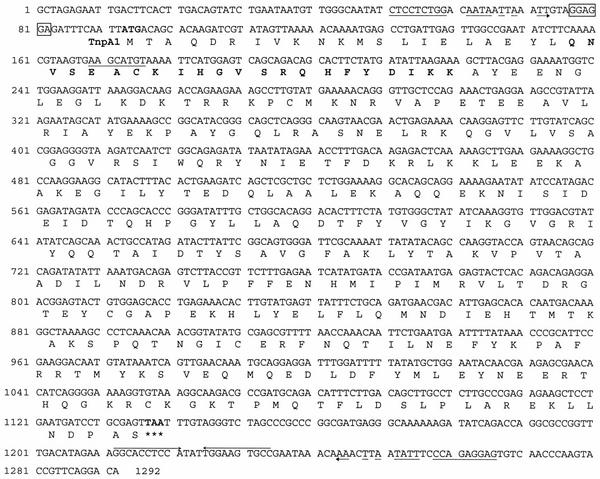
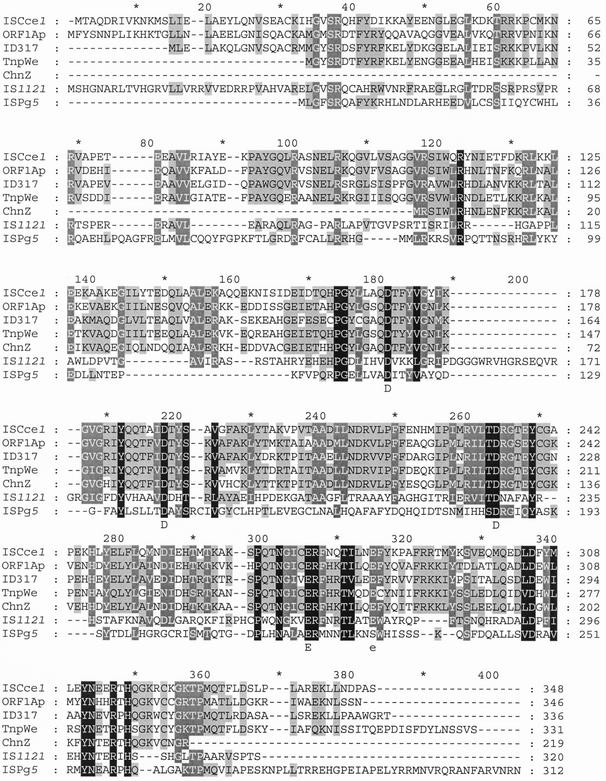
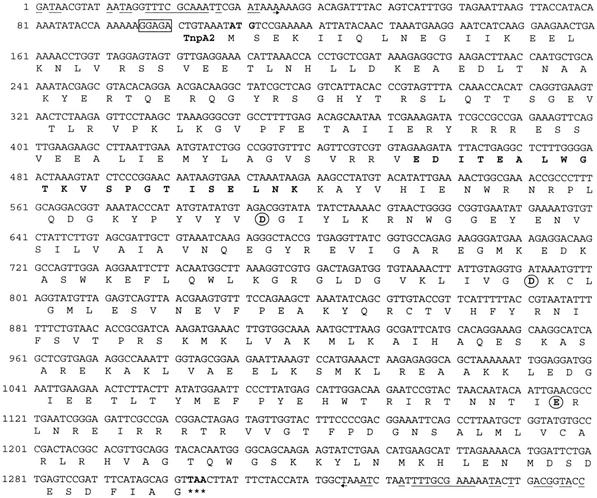
The 1,292-bp DNA sequence contained only one ORF (orf1), which was 1,047 bp long and spanned almost the entire element (Fig. 3); it was flanked by 23-bp IRs with six mismatches. The left and right IRs were found at 50 and 26 bp of the extremities, respectively. Many characteristics typical of an IS were observed: (i) insertion of the 1,292-bp sequence yielded an 8-bp direct repeat (DR) footprint in the target sequence and (ii) the large ORF encoded a 348-amino-acid protein (40.2 kDa), designated TnpA1, which exhibited significant levels of identity with a hypothetical protein (designated ORF1Ap [see below]) from Actinobacillus pleuropneumoniae (57%) (2), with a putative transposase (TnpWe) from a Wolbachia endosymbiont of Drosophila simulans (57%) (accession number AAK69114), and with the putative proteins ID317 (55%) (21) and ChnZ (62%) (9) from Bradyrhizobium japonicum and Acinetobacter sp. strain SE19, respectively. It also exhibited some identity with the transposases encoded by many ISs belonging to the IS481 family (20% to 38%) and with one IS (ISPg5 [7]) belonging to the IS3 family (28%) (8). A multiple alignment of some of these proteins (Fig. 4) enlightened many stretches of conserved amino acids, including three aspartic residues and two glutamic residues. Three of these amino acids might constitute the DDE catalytic triad (Fig. 4). Protein structure predictions suggested that an α-helix-turn-helix (HTH) DNA binding motif was present at the N terminus of TnpA1 (Fig. 3). Based on all these criteria, the element was designated ISCce1, although it does not have any canonical IRs at its extremities.
FIG. 3.
Nucleotide sequence of ISCce1 and predicted amino acid sequence of transposase TnpA1. The putative ribosome binding site sequence is enclosed in a box. The ORF encoding transposase TnpA1 starts with an ATG codon at position 93 (boldface type) and ends with a TAA stop codon at position 1137 (asterisks). The deduced amino acid sequence is indicated under the corresponding nucleotide sequence. The putative ribosome binding site sequence is boxed. A palindromic sequence is overlined with arrows. The 8-bp duplicated sequence at the insertion site of ISCce2 into ISCce1 is underlined. Imperfect terminal IRs are indicated by incomplete arrows, with mismatches indicated by interruptions. The potential HTH DNA binding motif in the TnpA1 amino acid sequence is indicated by boldface type.
FIG. 4.
Alignment of TnpA1(from ISCce1) with ORF1Ap (A. pleuropneumoniae), ID317 (B. japonicum), TnpWe (Wolbachia endosymbiont of D. simulans), ChnZ (Acinetobacter sp. strain SE19), and proteins encoded by IS1121 (Clavibacter michiganensis) and ISPg5 (Porphyromonas gingivalis). A black background indicates identical amino acids, a dark gray background indicates very similar amino acids, and a light gray background indicates weakly similar amino acids. Conserved aspartic acid (D) and glutamic acid (E) residues are indicated below the alignment.
The 1,359-bp DNA sequence that was found in the large 2,659-bp element and was inserted into ISCce1 was designated ISCce2. The sequence surrounding a large ORF (orf2; length, 1,195 bp) was found to fulfill all the criteria required for an IS: an 8-nucleotide target sequence was found to be duplicated on both sides, and the extremities of the IS contained 35-bp imperfect IRs with 13 mismatches (Fig. 5). The 398-amino-acid protein (45.9 kDa) encoded by orf2 exhibited significant levels of sequence identity (20 to 30%) with the transposases encoded by ISs belonging to the IS256 family (8). This protein, designated TnpA2, also contains a potential HTH DNA binding motif and the DDE triad of the catalytic domain (Fig. 5).
FIG. 5.
Nucleotide sequence of ISCce2 and predicted amino acid sequence of the transposase TnpA2. The ORF encoding transposase TnpA2 starts with an ATG codon at position 109 (boldface type) and ends with a TAA stop codon at position 1303 (asterisks). The putative ribosome binding site sequence is enclosed in a box. The potential HTH DNA binding motif and the potential DDE catalytic triad motif in the TnpA2 amino acid sequence are indicated by boldface type and by circled residues, respectively. Terminal IRs are indicated by incomplete arrows, with mismatches indicated by interruptions.
tnpA1 and tnpA2 are preceded by purine-rich sequences indicative of potential ribosome binding sites (Fig. 3 and 5). The G+C contents of ISCce1 and ISCce2 were 42 and 40%, respectively; these values are similar to the G+C content of the C. cellulolyticum genome (40%). The genetic code usage in tnpA1 and tnpA2 corresponds to the usage which was previously observed in a set of functional C. cellulolyticum genes (unpublished data).
Insertion sites of ISCce1 and ISCce2 in the genome of C. cellulolyticum.
To determine the sequences flanking ISCce1 in the genome of C. cellulolyticum, chromosomal DNA of the cipCMut1 strain was digested with NdeI, EcoRI, HindIII, or PstI. Ligated fragments were used as templates for inverse PCR performed with primers A and B for the left junctions and primers G and H or primers E and H for the right junctions (Fig. 2C). PCR products were cloned into the pGEM-T-Easy vector and then analyzed by sequencing. Fourteen plasmids harboring fragments different from the cipC gene and disrupted by ISCce1 were obtained. Three of the ISCce1 copies were combined with an ISCce2 copy. The ISCce1 target sequences found were AT rich, but no consensus sequence could be identified (Table 3).
TABLE 3.
Frequency of base occurrence at each position of the ISCce1 insertion sites
| Base | Frequency of occurrence (%) at target site positiona:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| A | 80.0 | 40.0 | 26.6 | 0 | 46.6 | 80.0 | 26.6 | 6.6 |
| T | 6.7 | 26.6 | 60.0 | 100 | 40.0 | 13.4 | 53.4 | 20.0 |
| C | 6.7 | 20.0 | 13.4 | 0 | 6.7 | 6.6 | 6.6 | 53.4 |
| G | 6.6 | 13.4 | 0 | 0 | 6.7 | 0 | 13.4 | 20.0 |
The frequency of occurrence of a base was calculated for each position at 15 transposition sites in the chromosome. Boldface type indicates overrepresented bases.
To determine the insertion sites of ISCce2, EcoRI-, HindIII-, or EcoRV-digested and ligated DNA was used in inverse PCR performed with primers C and D to amplify the left junctions and with primers E and F to amplify the right junctions (Fig. 2C). Only four different recombinant plasmids harboring ISCce2 junctions were obtained. In one of them, ISCce2 was inserted into an ISCce1 copy. This combined IS might be one of those found when we searched for ISCce1 junctions (see above). No consensus target sequence was identified from the four insertion sites obtained, ACATGCTT (in ISCce1), CATAATAA, CAGCACTT, and GCTTTTAT.
In other respects, partial sequences determined for various copies of each IS were found to be exactly identical to the sequences of the copies initially found in the cipC gene (data not shown). Furthermore, the noncanonical location of IRs, which were found near the end of ISCce1, was confirmed by looking at other copies cloned from genomic DNA.
Close physical link between ISCce1 and ISCce2.
ISCce2 was initially found within ISCce1 in the cipCMut1 strain. In addition, the sequence analysis of the IS junctions showed that at least four of the seven ISCce2 copies found in this strain were inserted into ISCce1. In order to examine this unusual association in another way, the Southern blot which was previously probed with a combined IS element was reprobed with probe 2 to detect only ISCce1 (Fig. 2B) and then with probe 3 to detect only ISCce2 (Fig. 2C). The four bands detected in the wild-type lane with probe 3 could be superimposed on the bands revealed with probe 2 (Fig. 6B). Furthermore, the pattern obtained with the cipCMut1 DNA (when probe 3 was used) contained three bands which were absent in the wild-type DNA lane (Fig. 6B); two of these bands could be superimposed with those detected in the same lane with probe 2 (Fig. 6A). These findings suggest that many PvuII fragments contain both IS elements, although the possibility that some of the common bands may have resulted from comigration of two different fragments, each containing one of the two ISs, cannot be ruled out. Nevertheless, of the seven ISCce2 copies studied, at least four were found to be inserted into ISCce1 (see above). Taken together, these results strongly suggest that ISCce1 is a hot spot for the transposition of ISCce2.
FIG. 6.
Distribution and association of ISCce1 and ISCce2 in C. cellulolyticum: Southern blot analysis of PvuII-digested genomic DNAs of the wild type (lane WT), cipCMut1 (lane 1), and cipCMut2 (lane 2) and of EcoRI-digested cipCMut1 DNA (lane 1E). Blots were hybridized with the ISCce1 probe (A) and with the ISCce2 probe (B). Superimposable bands detected in both hybridization experiments for all strains are indicated in panel B by arrowheads. The bands indicated by circles are superimposable bands obtained only with the mutant strains. Sizes (in kilobase pairs) are indicated on the left and on the right.
ISCce1 and ISCce2 distribution among Clostridium species.
Southern blotting of the PvuII-digested DNAs from C. cellulolyticum strains probed with ISCce1 revealed between 10 and 14 bands. When the DNAs were probed with ISCce2, four to seven bands were detected, depending on the strain (Fig. 6). To determine the number of ISCce1 copies more exactly, an EcoRI digestion was performed. Up to 20 bands were detected, leading to the conclusion that there were about 20 copies of ISCce1 (Fig. 6A, lane 1E).
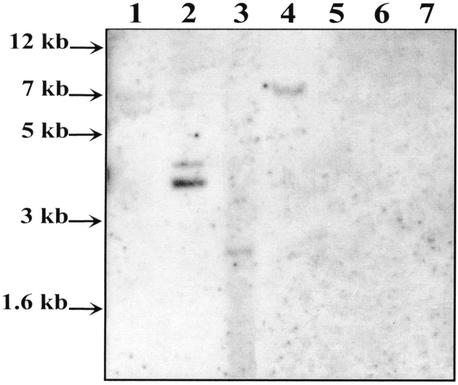
To determine whether ISCce1 and ISCce2 were present in other clostridia, DNAs extracted from some selected stains were digested by PvuII, electrophoresed, and hybridized with probe 2 or probe 3. No fragments homologous to ISCce1 were detected in any of the strains tested with probe 2 in hybridization experiments carried out at 68 or 55°C (data not shown). However, probe 3 hybridized with one or two DNA fragments from Clostridium cellobioparum, Clostridium papyrosolvens, Clostridium termitidis, and C. cellulovorans in the experiments carried out at 55°C (Fig. 7). These fragments may therefore have low levels of sequence similarity with ISCce2.
FIG. 7.
Distribution of ISCce2 in Clostridium strains: Southern blotting of PvuII-digested DNAs of clostridia hybridized with probe 3 at 55°C. Lane 1, C. cellobioparum; lane 2, C. papyrosolvens; lane 3, C. termitidis; lane 4, C. cellulovorans; lane 5, C. saccharobutylicum; lane 6, C. thermocellum; lane 7, C. acetobutylicum.
Our results indicate that ISCce1 and ISCce2 might be ISs specific to C. cellulolyticum.
DISCUSSION
In this paper, we report the discovery and characterization, in the cellulolytic bacterium C. cellulolyticum, of novel 1,292- and 1,359-bp IS elements, designated ISCce1 and ISCce2, respectively. These two elements were found to be frequently associated. In at least four cases, ISCce2 was found to be located within ISCce1 itself; this situation is comparable to that reported for two ISs in Pseudomonas syringae (40) and two other ISs in Sinorhizobium meliloti (28).
The nucleotide sequence of ISCce1 has only one long ORF, which codes for a putative transposase designated TnpA1. The deduced amino acid sequence showed significant levels of identity with proteins ORF1Ap (2), ID317 (21), TnpWe (accession number AAK69114), and ChnZ (9) and with some transposases encoded by ISs belonging to the IS481 family (33) and by ISPg5 belonging to the IS3 family (8) (Fig. 4).
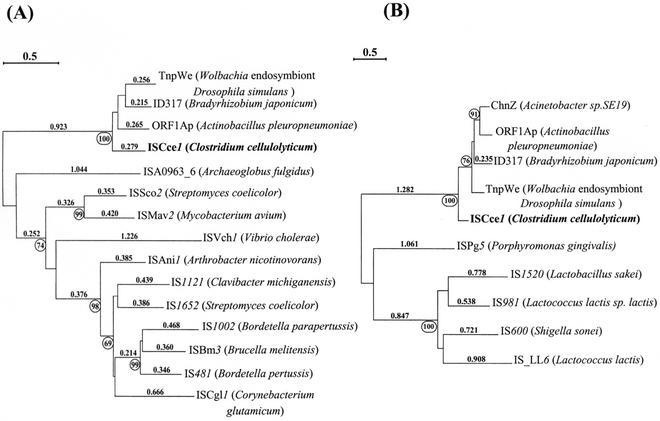
To investigate the evolutionary relationships between TnpA1 and these proteins, phylogenetic trees were drawn. The Phylo_Win program (18) was applied to the multiple-sequence alignment of TnpA1, ORF1Ap, ID317, TnpWe, and 11 transposases encoded by elements belonging to the IS481 family (8), which was obtained with the program CLUSTAL W (45). The resulting tree shows that TnpA1 (ISCce1), ORF1Ap, ID317, and TnpWe may constitute a group (Fig. 8A). This group is separated from the 11 members of the IS481 family, and the existence of a relationship between the two groups was not confirmed by a high bootstrap confidence level. A similar analysis was carried out with TnpA1 (ISCce1), ORF1Ap, ID317, TnpWe, ChnZ, and six transposases encoded by ISs belonging to the IS3 family (all of which belong to the IS3 group) (8). As ChnZ is only 219 amino acids long, the tree was generated from part of the multiple-sequence alignment. As described above, TnpA1 (ISCce1), ORF1Ap, ID317, TnpWe, and ChnZ constitute a group separated from the group formed by the IS3 family members (Fig. 8B). Again, the relationship between the ISCce1 group and the IS3 group was not confirmed by a high bootstrap confidence level. Like TnpA1, the other proteins in the ISCce1 group also show some homology with the transposases of the IS481 family and with some members of the IS3 family (data not shown).
FIG. 8.
Phylogenetic trees showing relationships between ISCce1 and members of the IS481 and IS3 families. The trees were constructed from a multiple-sequence alignment (ClustalW) (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html) of transposases of ISs and proteins showing identity with TnpA1 (ISCce1) by using the neighbor-joining method (Phylo_Win) (http://biom1.univ-lyon1.fr/software/phylowin.html). The circled numbers are the percentages of support (bootstrap values) for individual nodes in the tree obtained by performing 100 replicate searches. Only values higher than 65% are indicated. A percentage of accepted mutation distance is indicated above each clade. (A) Tree constructed from the entire multiple-sequence alignment of TnpA1 (ISCce1), 11 members of the IS481 family, and other homologous proteins. The accession numbers for the members of the IS481 family used are as follows: ISA0963_6, AE000986; ISSco2, AL10949; ISMav2, AF286339; ISVch1, AF034434; ISAni1, X97015; IS1121, AF079817; IS1652, AL109949; IS1002, Z54268; ISBm3, AF047478; IS481, M22031; and ISCgl1, U85507. The accession numbers for the other homologous proteins used are as follows: TnpWe, AAK69114; ID317, AAG60838; and ORF1Ap, S27482. (B) Tree constructed from part of the multiple-sequence alignment of TnpA1, five members of the IS3 family, and other homologous proteins. The accession numbers for the members of the IS3 family used are as follows: ISPg5, AF224744; IS1520, AJ250598; IS981, M33933; IS600, X05992; and IS_LL6, U23813. The accession numbers for the other homologous proteins used (TnpWe, ID317 and ORF1Ap) are as described above; in addition, ChnZ (accession number AAG10024) was used.
orf1Ap from A. pleuropneumoniae, like ISCce1 and ISCce2, was found when spontaneous mutants of the strain were characterized (2). This sequence is flanked by 26-bp IRs with four mismatches. Because the published sequence resulted from a recombination event, it was impossible to identify eventual DRs at the extremities of the sequence. The sequences flanking chnZ and id317, available in the GenBank database, were analyzed. Only id317 was flanked by 43-bp imperfect IRs (with 13 mismatches), but no DRs were found near these IRs. The absence of DRs might be due to genomic rearrangements, as in the case of the orf1Ap from A. pleuropneumoniae. No data are available on IRs and DRs in the flanking sequences of the region encoding TnpWe.
These results and those of the phylogenetic analysis suggest that the unknown protein from A. pleuropneumoniae (2) (ORF1Ap), protein ID317 from B. japonicum (21), ChnZ from Acinetobacter sp. strain SE19 (9), and the putative transposase from a Wolbachia endosymbiont of D. simulans (accession number AAK96114) might be encoded by complete or truncated ISs. These sequences, along with ISCce1, might form a new group of ISs, which is probably related to the IS481 and the IS3 families. In this group, ISs would (i) exhibit IRs, but not at the extremities of the element; (ii) generate the formation of DRs in the target upon transposition, although this feature could be identified only for ISCce1; and (iii) contain only one ORF encoding a putative transposase. The strict conservation of several D and E residues strongly suggests that the catalytic mechanism of these transposases involves a DDE triad.
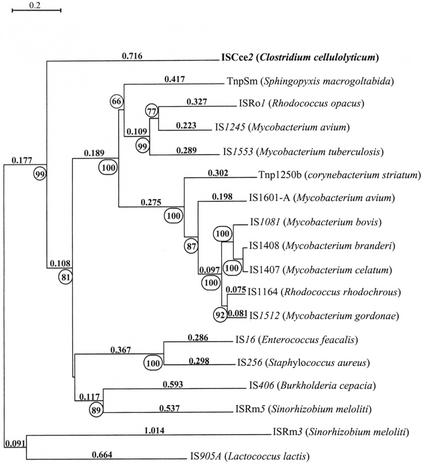
The nucleotide sequence of ISCce2 has one large ORF (tnpA2) that putatively encodes a transposase (TnpA2). This protein has significant levels of identity with many transposases belonging to the IS256 family. A phylogenetic tree was generated for 18 IS elements belonging to the IS256 family or showing some identity with members of this family (Fig. 9). This tree shows that ISCce2 might be a member of the IS256 family, but as it is located on a separate clade of the tree, it does not have any close relatives belonging to this family. ISCce2 has all the features of the IS256 family (8): (i) it has IRs at its extremities (35 bp in ISCce2); (ii) it duplicates an 8-bp target site sequence upon transposition; (iii) TnpA2 has a DDE motif that has extended regions similar to that of the transposases of the IS256 family; and (iv) TnpA2 exhibits similarities with the putative MurA gene product of the autonomous mutator element of Zea mays, MuDR (14).
FIG. 9.
Phylogenetic tree of some members of the IS256 family. This tree was constructed from a multiple-sequence alignment (ClustalW) (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html) of transposases of ISs and proteins showing identity with TnpA2 (ISCce2) by using the neighbor-joining method (Phylo_Win) (http://biom1.univ-lyon1.fr/software/phylowin.html). The circled numbers are the percentages of support (bootstrap values) for individual nodes on the tree obtained by performing 100 replicate searches. Only values higher than 65% are indicated. A percentage of accepted mutation distance is indicated above each clade. The accession numbers for the proteins and ISs are as follows: TnpSm, BAB07803; ISRo1, U70364; IS1245, L33879; IS1553I, NP_338287; Tnp1250b, AF024666; IS1601-A, AAD44203; IS1081, X61270; IS1408, U62766; IS1407, X97307; IS1164, D67027; IS1512, U95314; IS16, U35366; IS256, M18086; IS406, M83145; ISRm5, U08627; ISRm3, M60971; and IS905A, L20851.
ISCce1 and ISCce2 seem to be specific to C. cellulolyticum. Indeed, they were not found in any of the species that are phylogenetically closely related to C. cellulolyticum, such as C. papyrosolvens, C. cellobioparum, and C. termitidis, or in the distantly related species, such as C. cellulovorans, Clostridium saccharobutylicum, C. thermocellum, and Clostridium acetobutylicum (12).
Since ISCce1 and ISCce2 were isolated after they were inserted into the cipC gene, they are therefore transpositionally active. The use of these ISs for construction of mutagenic tools is interesting. Such tools should allow identification of new relevant genes involved in cellulolysis.
Acknowledgments
We thank R. H. Doi for kindly providing C. cellulovorans genomic DNA and G. Fichant for her help with the Phylo_Win program. We are grateful to O. Valette for her expert technical assistance and to J. Blanc for revising the English in the manuscript. We thank A. Bélaich, H. P. Fièrobe, and S. Pagés for helpful discussions.
We acknowledge the financial support received from the Centre National de la Recherche Scientifique and Université de Provence, from Conseil Général des Bouches du Rhône, and from Région Provence-Alpes-Côtes d'Azur. H. Maamar received a fellowship from the Tunisian Government.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, C., A. A. Potter, and G. F. Gerlach. 1991. Isolation and molecular characterization of spontaneously occurring cytolysin-negative mutants of Actinobacillus pleuropneumoniae serotype 7. Infect. Immun. 59:4110-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J. P. Belaich.1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 4.Belaich, A., G. Parsiegla, L. Gal, C. Villard, R. Haser, and J. P. Belaich. 2002. Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 184:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belaich, J. P., C. Tardif, A. Belaich, and C. Gaudin. 1997. The cellulolytic system of Clostridium cellulolyticum. J. Biotechnol. 57:3-14. [DOI] [PubMed] [Google Scholar]
- 6.Brynestad, S., B. Synstad, and P. E. Granum. 1997. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 143:2109-2115. [DOI] [PubMed] [Google Scholar]
- 7.Califano, J. V., T. Kitten, J. P. Lewis, F. L. Macrina, R. D. Fleischmann, C. M. Fraser, M. J. Duncan, and F. E. Dewhirst. 2000. Characterization of Porphyromonas gingivalis insertion sequence-like element ISPg5. Infect. Immun. 68:5247-5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler, M., and J. Mahillon. 2002. Insertion sequences, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 9.Cheng, Q., S. M. Thomas, K. Kostichka, J. R. Valentine, and V. Nagarajan. 2000. Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 by in vitro transposition. J. Bacteriol. 182:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, K. T. 1976. Inhibitory effects of H2 on growth of Clostridium cellobioparum. Appl. Environ. Microbiol. 31:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, C. M., and D. M. Gutman. 1992. Insertional inactivation of an Escherichia coli urease gene by IS3411. J. Bacteriol. 174:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 13.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, J. A., M. I. Benito, and V. Walbot. 1994. Sequence similarity of putative transposases links the maize mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22:2634-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fetherston, J. D., and R. D. Perry. 1994. The pigmentation locus of Yersinia pestis KIM6+ is flanked by an insertion sequence and includes the structural genes for pesticin sensitivity and HMWP2. Mol. Microbiol. 13:697-708. [DOI] [PubMed] [Google Scholar]
- 16.Gal, L., C. Gaudin, A. Belaich, S. Pages, C. Tardif, and J. P. Belaich. 1997. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J. Bacteriol. 179:6595-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gal, L., S. Pages, C. Gaudin, A. Belaich, C. Reverbel-Leroy, C. Tardif, and J. P. Belaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galtier, N., M. Gouy, and C. Gautier. 1996. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Applic. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 19.Gaudin, C., A. Belaich, S. Champ, and J. P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giallo, J., C. Gaudin, J. P. Belaich, E. Petitdemange, and F. Caillet-Mangin. 1983. Metabolism of glucose and cellobiose by cellulolytic mesophilic Clostridium sp. strain H10. Appl. Environ. Microbiol. 45:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottfert, M., S. Rothlisberger, C. Kundig, C. Beck, R. Marty, and H. Hennecke. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J. Bacteriol. 183:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guedon, E., M. Desvaux, and H. Petitdemange. 2002. Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering. Appl. Environ. Microbiol. 68:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning, a practical approach, vol. 1. IRL Press, Oxford, United Kingdom.
- 24.Hethener, P., A. Brauman, and J. L. Garcia. 1992. Clostridium termitidis sp. nov., a cellulolytic bacterium from the gut of the wood-feeding termite, Nasutitermes lujae. Syst. Appl. Microbiol. 15:52-58. [Google Scholar]
- 25.Jennert, K. C., C. Tardif, D. I. Young, and M. Young. 2000. Gene transfer to Clostridium cellulolyticum ATCC 35319. Microbiology 12:3071-3080. [DOI] [PubMed] [Google Scholar]
- 26.Keis, S., R. Shaheen, and D. T. Jones. 2001. Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int. J. Syst. Evol. Microbiol. 51:2095-2103. [DOI] [PubMed] [Google Scholar]
- 27.Kivi, M., X. Liu, S. Raychaudhuri, R. B. Altman, and P. M. Small. 2002. Determining the genomic locations of repetitive DNA sequences with a whole-genome microarray: IS6110 in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:2192-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laberge, S., A. T. Middleton, and R. Wheatcroft. 1995. Characterization, nucleotide sequence, and conserved genomic locations of insertion sequence ISRm5 in Rhizobium meliloti. J. Bacteriol. 177:3133-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liyanage, H., P. Holcroft, V. J. Evans, S. Keis, S. R. Wilkinson, E. R. Kashket, and M. Young. 2000. A new insertion sequence, ISCb1, from Clostridium beijernickii NCIMB 8052. J. Mol. Microbiol. Biotechnol. 2:107-113. [PubMed] [Google Scholar]
- 31.Madden, R. H., M. J. Bryder, and N. J. Poole. 1982. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium papyrosolvens sp. nov. Int. J. Syst. Bacteriol. 32:87-91. [Google Scholar]
- 32.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPheat, W. L., and T. McNally. 1987. Isolation of a repeated DNA sequence from Bordetella pertussis. J. Gen. Microbiol. 133:323-330. [DOI] [PubMed] [Google Scholar]
- 34.Ochman, H., M. M. Medhora, D. Garza, and D. L. Hartl. 1990. Amplification of flanking sequences by inverse PCR, p. 219-227. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. Academic Press, Inc., New York, N.Y.
- 35.Pages, S., A. Belaich, C. Tardif, C. Reverbel-Leroy, C. Gaudin, and J. P. Belaich. 1996. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 178:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pages, S., A. Belaich, H. P. Fierobe, C. Tardif, C. Gaudin, and J. P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petitdemange, E., F. Caillet, J. Giallo, and C. Gaudin. 1984. Clostridium cellulolyticum sp. nov., a cellulolytic, mesophilic species from decayed grass. Int. J. Syst. Bacteriol. 34:155-159. [Google Scholar]
- 38.Reverbel-Leroy, C., A. Belaich, A. Bernadac, C. Gaudin, J. P. Belaich, and C. Tardif. 1996. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology 142:1013-1023. [DOI] [PubMed] [Google Scholar]
- 39.Snedecor, B., E. Chen, and R. F. Gomez. 1983. In Proceedings of the IVth International Symposium on the Genetics of Industrial Microorganisms, p. 356-360.
- 40.Soby, S., B. Kirkpatrick, and T. Kosuge. 1993. Characterization of an insertion sequence (IS53) located within IS51 on the iaa-containing plasmid of Pseudomonas syringae pv. savastanoi. Plasmid 29:135-141. [DOI] [PubMed] [Google Scholar]
- 41.Soucaille, P., and G. Goma. 1986. Acetonobutylic fermentation by Clostridium acetobutylicum ATCC 824: autobacteriocin production, properties, and effects. Curr. Microbiol. 13:163-169. [Google Scholar]
- 42.Stanley, J., N. Baquar, and E. J. Threlfall. 1993. Genotypes and phylogenetic relationships of Salmonella typhimurium are defined by molecular fingerprinting of IS200 and 16S rrn loci. J. Gen. Microbiol. 139:1133-1140. [DOI] [PubMed] [Google Scholar]
- 43.Stroeher, U. H., K. E. Jedani, B. K. Dredge, R. Morona, M. H. Brown, L. E. Karageorgos, M. J. Albert, and P. A. Manning. 1995. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc. Natl. Acad. Sci. USA 92:10374-10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tardif, C., H. Maamar, M. Balfin, and J. P. Belaich. 2001. Electrotransformation studies in Clostridium cellulolyticum. J. Ind. Microbiol. Biotechnol. 27:271-274. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]