Abstract
The group A streptococcus (GAS), or Streptococcus pyogenes, is a strict human pathogen of medical significance, causing infections ranging from pharyngitis (strep throat) to necrotizing fasciitis (flesh-eating disease). Several virulence genes that encode factors important for colonization, internalization, and immune evasion are under the control of the multiple gene regulator of the GAS, or Mga. Mga functions as a DNA-binding protein that interacts with sites both proximal (Pemm and PscpA) and distal (PsclA) to the start of transcription for the genes that it regulates. The genes encoding serum opacity factor, sof, and a novel fibronectin-binding protein, sfbX, are cotranscribed and represent two uncharacterized Mga-regulated virulence genes in the GAS. Analysis of the promoter region of sof-sfbX identified a putative Mga-binding site 278 bp upstream of the regulated start of transcription as determined by primer extension. Electrophoretic mobility shift assays demonstrated that Mga is able to bind specifically to the single distal site in a fashion similar to the previously characterized PsclA. In order to better understand the events that take place at this and other Mga-regulated promoters, an in vitro transcription assay was established. Using this assay, we showed that Mga is sufficient to activate transcription in vitro for Mga-regulated promoters containing both proximal (Pemm) and distal (PsclA and Psof-sfbX) binding sites. These results indicate that additional factors are not required for Mga-specific activation at diverse promoters in vitro, although they do not rule out the potential influence of other components on the Mga virulence regulon in vivo.
Differential control of gene expression in bacteria is an important adaptive response to various environments and is often mediated by transcription factors that either activate or repress initiation of transcription. In the case of activators, the factors are thought to increase transcription initiation by recruiting RNA polymerase (RNAP) to the promoter via direct protein-protein interactions (10). Such interactions between RNAP and the activator have been shown to occur with the carboxyl terminus of the α subunit for class I transcription factors or with the σ subunit for class II transcription factors. Class I activators typically bind to DNA just upstream of the −35 region of the promoters that they regulate in order to facilitate their interaction with the α subunits. Class II activator binding sites, on the other hand, tend to overlap the −35 region while still promoting transcriptional activation (10). Less common, however, are transcription factors capable of activating transcription from a single or multiple distant sites, especially when the Escherichia coli housekeeping σ70 is employed by the RNAP (6). In fact, the majority of transcription factors that activate from far upstream sites are required to initiate σ54-dependent promoters, such as the prototypical regulator NtrC. Despite their binding distal to the −35 region, they are still considered a class II transcription factor (10).
Transcriptional regulators play a vital role in the pathogenesis of the group A streptococcus (GAS), or Streptococcus pyogenes. The GAS is a strict human pathogen that is capable of causing disease in varied locations throughout the body, including the skin (impetigo), the respiratory tract (pharyngitis), and deeper tissues (necrotizing fasciitis). Unlike many other prokaryotes, the ability of the GAS to coordinately regulate expression of genes does not depend on alternative sigma factors, since the GAS has only one that is thought to control transcription of a limited number of genes (21). Rather, the organism is able to respond to environmental cues by relaying information to a variety of transcriptional regulators.
One such regulator is the multiple gene regulator of the GAS, or Mga. Mga is responsible for activating the expression of several surface-associated or secreted factors that enable the bacterium to adhere to epithelial surfaces, invade cells, and evade the immune system. These factors include the serum opacity factor (OF), M protein, and the streptococcal collagen-like protein (SclA) (4, 15, 20, 23, 24). Mutations in mga and Mga-regulated genes result in strains that are significantly attenuated for virulence in a variety of animal models (7, 12, 13, 15, 25). Furthermore, a recent longitudinal study of pharyngitis in macaques found that the Mga regulon was highly expressed in vivo during the acute phase of infection (30). Mga is known to bind to the promoters of the genes that it activates (1, 16, 19), but its mechanism of action in transcriptional activation remains to be determined.
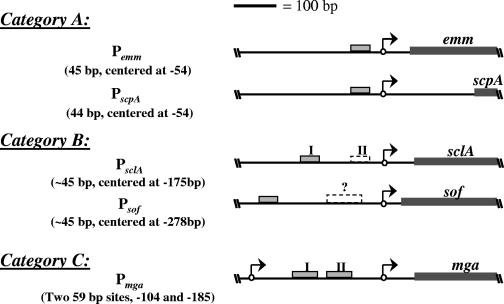
Since the Mga-binding sites in the emm (M protein) and scpA (C5a peptidase) promoters are located proximal to the start of transcription (16), it is thought that Mga acts like a class I transcription factor to activate gene expression by binding to the α subunit of the RNAP. However, it was recently discovered that Mga is capable of activating transcription from a distal binding site in the sclA promoter (1). Thus, Mga-regulated promoters were grouped on the basis of the number and location of Mga-binding sites: a single proximal site (category A; emm and scpA), a single distal site (category B; sclA), and multiple distal sites (category C; mga) (19). The differences in these promoters suggest that Mga may be capable of activating transcription by multiple mechanisms. In this study, a second category B Mga-regulated promoter is identified (sof-sfbX), and Mga is demonstrated to be sufficient to activate both category A and B promoters in vitro.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains used in this study are listed in Table 1. The GAS was cultured in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY), and growth was followed by optical density using a Klett-Summerson photoelectric colorimeter with the A filter. E. coli was grown in Luria-Bertani broth (LB). Antibiotics were used at the following concentrations: ampicillin at 50 to 100 μg/ml for E. coli, rifampin at 150 μg/ml for E. coli, and spectinomycin at 100 μg/ml for the GAS.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| GAS | ||
| AL168str-r | M22, OF+, streptomycin-resistant derivative of AL168 | 28 |
| nr4 | M22, Tn916 mutant of AL168str-r in mga | 28 |
| JRS4 | M6, streptomycin-resistant derivative of D471 | 26 |
| JRS4-PolHis (12Z05A) | M6, derivative of JRS4 with chromosomal His-tagged β′ subunit of RNA polymerase | 21 |
| JRS519 | M6, derivative of JRS4 (mga-10) | 18 |
| SF370 | M1, sequenced strain | 27 |
| AP4 | M4, OF+ | 14 |
| E. coli | ||
| DH5α | hsdR17 recA1 gyrA endA1 relA1 | 9 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pBluescript II KS(−) | ColE1 ori AmprlacZα | Stratagene |
| pCD2 | PT7-σA (Bacillus subtilis) | 5 |
| pKSM164 | Pmga-mga-his | 1 |
| pKSM170 | PT7-mga-his | 1 |
| pKSM415 | PrpsL, 3′ HindIII site | This study |
| pKSM416 | Pemm, 3′ EcoRI site | This study |
| pKSM417 | PsclA (sequence downstream of Mga-binding site), 3′ XhoI site | 1 |
| pKSM418 | PsclA (ΔMga-binding site), 3′ XhoI site | 1 |
| pKSM419 | PsclA (full length), 3′ XhoI site | 1 |
| pKSM420 | Psof, 3′ BamHI site | This study |
DNA manipulations.
Plasmid DNA was isolated from E. coli using either the Wizard Miniprep system (Promega) or Maxi/Midi prep purification systems (QIAGEN). DNA fragments were isolated from agarose gels using the QIAquick gel extraction kit (QIAGEN). Chromosomal DNA from the GAS was isolated using the FastDNA kit and a FastPrep cell disruptor (Bio101, Inc.). PCR for cloning and promoter probes was performed using Platinum Pfx high-fidelity DNA polymerase (Invitrogen), and reactions were purified using the QIAquick PCR purification system (QIAGEN). PCR for diagnostic assays was performed using Taq DNA polymerase (New England Biolabs). DNA sequencing was performed either using the Excel II cycle sequencing kit (Epicenter, Inc.) or through the automated sequencing core facility in the McDermott Center at University of Texas Southwestern Medical Center.
Expression and purification of Mga-His protein from E. coli.
Mga-His protein was purified from E. coli as previously described (1). Briefly, cultures of E. coli BL21(DE3) containing pKSM170 (PT7-mga-his) were grown to an optical density at 600 nm (OD600) of 0.6 at 30°C in LB plus ampicillin (50 μg/ml), and expression of the protein was induced for 1.5 h by addition of 1 mM IPTG (isopropyl-β-d-galactopyranoside). Cells were lysed by two passages through a prechilled French pressure cell, and Mga-His was purified over a Ni-nitrilotriacetic acid (Ni-NTA) resin column (QIAGEN) under native conditions. Protein concentrations were determined using the Bio-Rad protein assay kit. Integrity of the purified protein was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis as described below. Mga-His purified for the purposes of in vitro transcription was dialyzed twice against 1 liter of transcription buffer minus bovine serum albumin (33 mM Tris acetate, pH 8, 10 mM magnesium acetate, and 0.5 mM dithiothreitol) for 4 h to overnight prior to storage at −80°C.
Western blot analysis.
Anti-Mga-His immunoblots were performed as described previously (17). Briefly, purified Mga-His proteins were separated on SDS-10% PAGE. Proteins were transferred to nitrocellulose membranes and reacted with the anti-His monoclonal antisera (Novagen) at a 1:2,000 dilution for 2 h at room temperature. Blots were reacted with horseradish peroxidase-conjugated anti-mouse immunoglobulin G secondary antibody (Chemicon Int.), developed using the Western Lighting chemiluminescence system (Perkin-Elmer), and visualized on autoradiography film (Kodak).
Electrophoretic mobility shift assays (EMSAs).
Promoter probes were generated by PCR amplification using either serotype M4 AP4 (OF+), serotype M1 SF370 (OF−), or serotype M6 strain JRS4 (OF−) chromosomal DNA and the relevant primer pairs (Psof, Pemm, PscpA, Pmga, PsclA, and nonspecific) listed in Table 2. PCR fragments were end labeled with [γ32P]ATP using T4 polynucleotide kinase (New England Biolabs). Labeled fragments were excised from a 5% polyacrylamide gel, extracted by crush and soak elution, and purified using the QIAquick PCR purification system (QIAGEN).
TABLE 2.
PCR primers used in this study
| Target | Primer | Sequence (5′-3′) | Reference |
|---|---|---|---|
| Nonspecific | rpsL-1 | GAATGTAGATGCCTACAATTAACCA | 17 |
| rpsL-2 | GTGCGCCACGAACGATATG | 17 | |
| Pemm | Pemm-L1 | GCATGGATCCCATCGCAAAGAGCTTA | 17 |
| Pemm-R1 | GCGGCTCGAGTAGTGTCTATTCGTGTTATT | 17 | |
| Pemm1 | Pemm1-L1 | AAGCGATTATTGACAAGCTC | 1 |
| Pemm1-R1 | AGTCAAAGCTACCGCTACTG | 1 | |
| Pmga | OYL-25 | TACCATAAAATACCTTTC | 17 |
| OYR-25 | GGTTGTACCATAACAGTC | 17 | |
| PrpsL | GAS-rpsL1 | TGTCTAAAATCACATCTTCGA | This study |
| GAS-rpsL7 | TTACGTACCAACTGGTTAATT | This study | |
| PsclA | PsclA-L2 | TGGTAATCGTAATTGTCTGC | 1 |
| PsclA-R | ATGGTGCTTTGATGTCAACA | 1 | |
| PscpA | C5-L1 | AAGAATGAGATTAAGGAGGTCACA | 16 |
| C5-R1 | GCGCAATGGCAAGTTTGTC | 16 | |
| Psof | Psof-L3 | TTTGGTCTCAGACGGCGCCA | This study |
| Psof-L4 | AATCAAGGTCACAAACTTTC | This study | |
| Psof-L5 | AATCGTCTGATAAGCTCTTG | This study | |
| Psof-L6 | CACTCAGTTGCCTCCAAGCT | This study | |
| Psof-R1 | ACCTCCTGTCCTAAAACTGT | This study | |
| Psof-R2 | GAAAAAAGGGGTTGCCGCGA | This study | |
| Psof-R3 | GTCAATGTTTCGTTAACCTT | This study | |
| Psof-R4 | GGAAAGTTTGTGACCTTGAT | This study | |
| Psof-R5 | CGTCTGAGACCAAAGCGTTA | This study | |
| Psof-R6 | GCTGCGCAAGGAGAAGCTCA | This study |
EMSA was performed as described previously (16). Briefly, a constant amount of labeled promoter probe DNA (ca. 1 to 5 ng) and increasing amounts of purified Mga-His (1 to 2 μg) were used in each reaction mixture. Competition assays were performed by addition of 750 ng unlabeled promoter probes to binding reaction mixtures. Reaction mixtures were run on a 5% polyacrylamide gel at 4°C. The gel was dried under vacuum at 80°C for 1 h and exposed overnight to a phosphorimaging screen. The phosphor screen was scanned using a Storm820 (Amersham Biosciences), and resulting data were analyzed with the ImageQuant analysis software (version 5.2).
Primer extension analysis.
Total RNA was extracted from samples in late exponential phase as described previously (17) using the FastRNA kit and a FastPrep cell disruptor (Bio101, Inc.). A primer extension was performed on 5 μg of total RNA from GAS strains AL168str-r and nr4 as described previously (19) using the primer Psof-R1 (Table 2). The primer extension product was run on a 6% denaturing polyacrylamide gel (Amresco), and the gel was processed as described above for EMSAs. Sequence was generated using labeled Psof-R1 primer on a PCR product amplified from AL168str-r genomic DNA (Table 2; Psof-L5 and Psof-R1) as described above.
Construction of in vitro transcription plasmids.
A 421-bp fragment of PrpsL was PCR amplified from the M1 strain SF370 by using the primers GAS-rpsL1 and GAS-rpsL7 (Table 2) and ligated into the EcoRV-digested pBluescript II KS(−) (Stratagene) to form pKSM415 (Table 1). A 420-bp fragment of Pemm containing the Mga-binding site was PCR amplified from SF370 by using the primers Pemm1-L1 and Pemm1-R1 (Table 2) and ligated into the EcoRV-digested pBluescript II KS(−) to form pKSM416 (Table 1). A 692-bp fragment of Psof containing the Mga-binding site was PCR amplified from the M4 strain AP4 by using the primers Psof-L5 and Psof-R1 (Table 2) and ligated into the EcoRV-digested pBluescript II KS(−) to form pKSM420 (Table 1).
Expression and purification of Mga-His protein from the GAS.
Mga-His protein was purified from the GAS as previously described (1). Briefly, cultures of JRS519 carrying pKSM164 (Pmga-mga-his) were grown to late logarithmic phase at 37°C in THY plus spectinomycin. Cells (20 50-ml aliquots) were lysed using five 9-s pulses at 4°C on a FastPrep cell disruptor (Bio101, Inc.), and Mga-His was purified over a Ni-NTA resin column (QIAGEN) under native conditions. Protein concentrations were determined using the Bio-Rad protein assay kit, and the integrity of the purified protein was assessed by SDS-PAGE and Western blot analysis as described above. Mga-His purified for the purposes of in vitro transcription was dialyzed twice against 1 liter of transcription buffer minus bovine serum albumin (33 mM Tris acetate, pH 8, 10 mM magnesium acetate, and 0.5 mM dithiothreitol) for 4 h to overnight prior to storage at −80°C.
Expression and purification of core RNA polymerase from the GAS.
Core RNA polymerase was purified from the GAS as previously described (21) with modifications. Briefly, cultures of JRS4-PolHis were grown to an OD600 of 0.8 at 37°C in THY plus spectinomycin. Cells (20 50-ml aliquots) were lysed using five 9-s pulses at 4°C on a FastPrep cell disruptor (Bio101, Inc.), and RNA polymerase was purified over a Ni-NTA resin column (QIAGEN) under native conditions. Fractions containing the polymerase were then passed over a HiTrap Q Sepharose FF anion exchange column (Amersham Biosciences) and eluted with a linear salt gradient. Protein concentrations were determined using the Bio-Rad protein assay kit, and the integrity of the purified protein complex was assessed by SDS-PAGE.
Expression and purification of σA of Bacillus subtilis from E. coli.
σA was purified from E. coli as previously described (3). Briefly, E. coli BL21(DE3) carrying pCD2 (5) was grown to an OD600 of 0.8 at 37°C in LB plus ampicillin (100 μg/ml), and expression of the protein was induced for 0.5 h by addition of 1 mM IPTG. Rifampin was added at 150 μg/ml, and the cells were grown for another 3.5 h. Cells were lysed by two passages through a prechilled French pressure cell, and sodium deoxycholate was added to solubilize the cell membranes. Inclusion bodies containing σA were pelleted, washed, and solubilized with 0.4% Sarkosyl, and the protein was then refolded by dialysis. The protein was loaded onto a HiTrap Q Sepharose FF anion exchange column (Amersham Biosciences) and eluted by a linear salt gradient. Protein concentrations were determined using the Bio-Rad protein assay kit, and the integrity of the purified protein was assessed by SDS-PAGE.
In vitro transcription analysis.
In vitro transcription reactions were performed as previously described (31). Briefly, core RNA polymerase, σA, and Mga were incubated together on ice for 10 min. Transcription buffer, RNaseOUT (Invitrogen), linearized plasmid template, and distilled H2O were added to a final volume of 40 μl, and each mixture was incubated at 37°C for 10 min. The reaction was started by the addition of ribonucleotides (including [α32P]CTP) and incubated at 37°C for 2 min. Heparin (10 μg) was added to inhibit reinitiation of transcription, and each mixture was incubated at 37°C for 5 min. The reaction products were then chased with 1 μl of 100 mM cold CTP and incubated at 37°C for 5 min. Products were ethanol precipitated, resuspended in formamide gel loading dye, and run on a 6% denaturing polyacrylamide gel (Amresco), and gels were processed as described above.
RESULTS
The sof-sfbX promoter contains a single distal Mga-binding site.
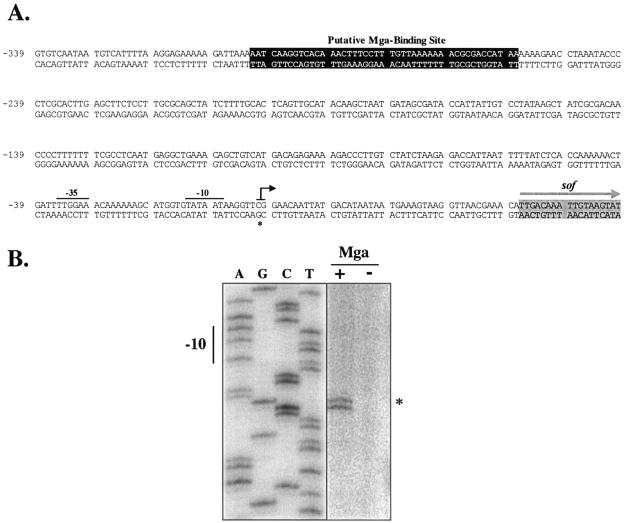
Although expression of the cotranscribed sof, encoding serum opacity factor, and sfbX, encoding a fibronectin-binding protein, are Mga dependent (20, 22), a direct role for Mga in this process has yet to be established. Using sequence from the chromosomal locus of sof-sfbX in a serotype M12 strain of the GAS (11), a region homologous to the Mga-binding site consensus (16) was found approximately 278 bp upstream of a predicted start of transcription that was identified on the basis of its similarity to conserved regions of the E. coli σ70 promoter (Fig. 1A). A comparison of the sof-sfbX promoter regions from this M12 strain, a recently sequenced OF+ M28 strain (8), and the sequence of an OF+ M22 strain available in the laboratory determined that the putative Mga-binding site and its distance to the predicted start of transcription were highly conserved among several OF+ strains. The location of the Mga-binding site is reminiscent of that found in the promoter of sclA (1), suggesting that Mga may function to activate transcription of these two promoters by a similar mechanism.
FIG. 1.
Identification of an Mga-regulated transcriptional start site for sof-sfbX. (A) Sequence of sof-sfbX promoter region from serotype M22 indicating the transcriptional start site (asterisk and black arrow), −10 and −35 regions (solid bars), the putative Mga-binding site (darkened box), and the sof start of translation (gray arrow). The sequence is numbered using the Psof-sfbX transcriptional start site as +1. (B) Primer extension analysis was performed on total RNA isolated from serotype M22 AL168str-r (Mga+) and nr4 (Mga−) using the radiolabeled antisense primer Psof-R1 (Table 2) as described in Materials and Methods. The start of transcription for sof-sfbX (asterisk) and its corresponding −10 hexamer are shown.
Identification of the Mga-regulated transcriptional start site of sof-sfbX.
In order to confirm the location of the Mga-binding site in the sof-sfbX promoter, its transcriptional start site was mapped by a primer extension analysis using total RNA from the OF+ M22 strain AL168str-r and its Mga− derivative nr4. Using the antisense primer Psof-R1 (Table 2) that is located at the 5′ end of sof-sfbX in the extension reaction mixture, a doublet product was clearly observed in the Mga+ strain AL168str-r that was noticeably absent from the Mga− strain nr4 (Fig. 1B). A repeat of the primer extension with a second antisense primer Psof-R5 (data not shown) also showed a doublet at this location and confirmed it as the true Mga-regulated transcriptional start site. Notably, the −10 and −35 sequences (Fig. 1A) of this site are homologous to established E. coli consensus sequences (six/six nucleotides and four/six nucleotides, respectively).
Specific binding of Mga-His to Psof in vitro.
A series of electrophoretic mobility shift assays were performed to determine whether Mga binds in vitro to the putative binding site in the sof-sfbX promoter. Increasing concentrations of Mga-His were incubated with overlapping promoter probes that were PCR amplified from the OF+ serotype M4 strain AP4. Initially, an M4 strain was chosen for EMSA analysis to allow use of purified M4 Mga-His, which was available in the laboratory. However, because of technical difficulties in purifying M4 Mga-His to sufficient levels for EMSA analysis, the established M6 Mga-His (1, 29) was used in future experiments. The ability of M6 Mga-His to bind to Psof was found to be similar to that of M4 Mga-His (data not shown), validating its use.
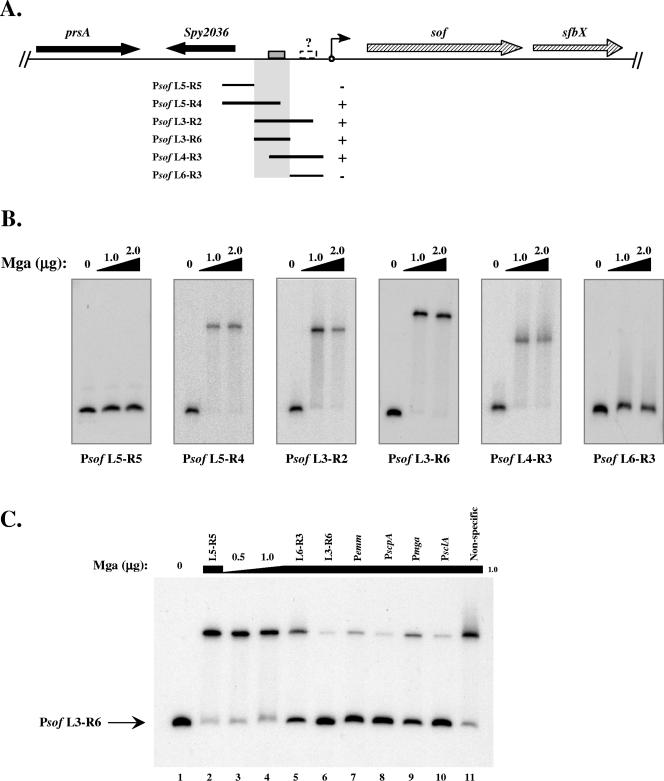
A summary of the probes used and the ability of Mga to bind those probes are shown in Fig. 2A. The promoter fragments that shifted upon addition of protein helped to delineate an Mga-binding region (highlighted), which included the Mga-binding site predicted by the consensus (gray box in Fig. 2A). However, promoter probes not including this region did not shift upon incubation with Mga (Fig. 2B, L5-R5 and L6-R3). Conversely, promoter probes that shared sequence with the highlighted region shifted upon incubation with Mga (Fig. 2B, L5-R4, L3-R2, L3-R6, and L4-R3). Interestingly, the Psof L6-R3 promoter probe appeared to shift slightly (smearing above the unbound probe) in these assays (Fig. 2B, L6-R3), suggesting that, like PsclA, a second low-affinity Mga-binding site (dashed box with a question mark) may be present proximal to the start of transcription. Unlike PsclA, however, this Mga-binding site was not detected by homology to the consensus Mga-binding sequence.
FIG. 2.
Identification of a specific Mga-binding site in the sof-sfbX promoter region. The ability of M6 Mga-His to bind to DNA within the sof-sfbX promoter was assessed by an EMSA. (A) Region surrounding sof-sfbX (hatched arrows) in the GAS genome, including a protein export protein (prsA) and a hypothetical cytosolic protein (Spy2036 in the SF370 M1 genome) (black arrows). Overlapping promoter probes in relation to the transcriptional start site of sof-sfbX (circle with arrow) are shown by the black lines. The ability of Mga-His to bind to the probes is indicated by both a plus sign and shaded region. The putative Mga-binding site (gray box) and a possible second low-affinity binding site (dashed box) are denoted on the chromosomal diagram. (B) EMSAs on Psof promoter probes (Table 2). Constant amounts (1 to 2 ng) of labeled promoter probes were incubated with increasing amounts (1.0 to 2.0 μg) of Mga-His for 15 min at 16°C prior to separation on a 5% polyacrylamide gel. (C) Radiolabeled Psof L3-R6 probe was assayed by an EMSA following incubation with 0, 0.5, and 1.0 μg Mga-His (lanes 1, 3, and 4). To the remaining binding reaction mixtures (lanes 2 and 5 to 11), 1.0 μg Mga-His and a constant amount of unlabeled competitor probe (750 ng) were added corresponding to Psof L5-R5, Psof L6-R3, Psof L3-R6, Pemm, PscpA, Pmga, PsclA, and a nonspecific rpsL fragment using the PCR primer pairs listed in Table 2.
Binding to these probes was shown to be specific by an EMSA using the Psof L3-R6 promoter probe with several other unlabeled competitor probes. Psof L3-R6 shifted when incubated with Mga alone (Fig. 2C, lanes 3 and 4); however, the bound species was almost completely chased back upon addition of cold L3-R6 probe (Fig. 2C, lane 6). The same effect was observed when the promoters of emm, scpA, mga, and sclA, to which Mga is known to bind, were used as competitors (Fig. 2C, lanes 7 to 10). On the other hand, the addition of the L5-R5 and nonspecific DNA promoter probes had no detectable effect on the ability of Mga to bind to the L3-R6 probe (Fig. 2C, lanes 2, 5, and 11). Yet, as observed above, the L6-R3 probe appeared to compete slightly with binding to the labeled Psof L3-R6 promoter probe, confirming the possibility of a second Mga-binding site in the promoter (Fig. 2C, lane 5). Nevertheless, the results of this experiment demonstrate that Mga is capable of binding specifically to the sof-sfbX promoter region in vitro.
Mga is sufficient to activate transcription of a category A promoter, Pemm, in vitro.
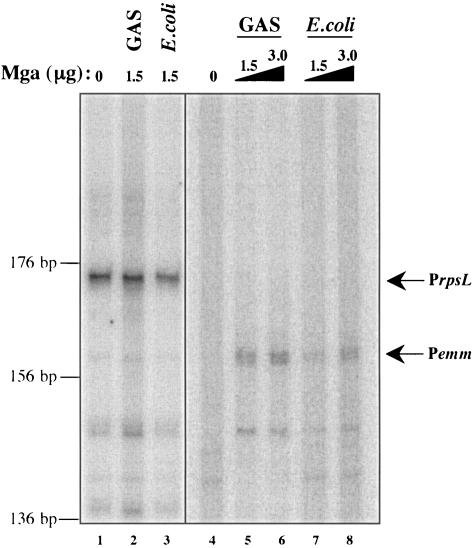
The identification of multiple categories of Mga-regulated promoters warrants an investigation into how Mga functions to activate transcription. In order to better understand the events that take place at Mga-regulated promoters, an in vitro transcription assay was adapted from one developed for the study of the secondary sigma factor (σX) in the GAS (21). Basically, transcription was allowed to occur by incubating core RNA polymerase purified directly from the GAS with purified recombinant Bacillus subtilis housekeeping sigma factor σA, a linearized plasmid DNA template containing a promoter with approximately 100 bp of downstream sequence, and radiolabeled ribonucleotides. Products were precipitated and separated on a denaturing polyacrylamide gel. When the housekeeping gene rpsL and its promoter were tested in this system, a band of the expected size was identified, and transcription levels were found to depend on the amount of RNA polymerase added to the reaction mixture (Fig. 3, lane 1, and data not shown).
FIG. 3.
In vitro transcription of the category A Mga-regulated gene emm. Transcription of emm was tested in an in vitro transcription assay with increasing amounts of GAS- and E. coli-purified M6 Mga-His (0, 1.5, and 3.0 μg, lanes 4 to 8) as described in Materials and Methods. DNA template (pKSM416 digested with EcoRI) was incubated with purified σA, core RNA polymerase, Mga (where indicated), and a radiolabeled ribonucleotide mixture. Reaction products were ethanol precipitated and separated on a 6% denaturing polyacrylamide gel. Transcription of the Mga-independent rpsL gene was used as a control in the assay. In vitro transcription was performed on rpsL as described above in the absence and presence of Mga purified from the GAS and from E. coli (lanes 1 to 3). Bands representing the transcribed products of the expected size are indicated by arrows and the names of the promoters. The sizes of the transcripts (in base pairs) were estimated on the basis of the migration of a DNA sequencing ladder.
Since the in vitro transcription assay produced a transcript with the constitutive rpsL promoter, the well-studied emm promoter was chosen to test the necessity and sufficiency of Mga to regulate transcription. Mga-His was purified from soluble lysates of both the GAS and E. coli, and each was added to a Pemm in vitro transcription assay in increasing amounts. As expected, a transcribed product was absent when Mga was not added to the reaction mixture (Fig. 3, lane 4), while a product of the correct size was observed when Mga purified either from the GAS or from E. coli was added (Fig. 3, lanes 5 to 8). Interestingly, the product of emm transcription appeared to be a doublet, suggesting that transcription initiates, in vitro, from two adjacent sites.
Although not a quantitative assay, repeated in vitro experiments showed Mga to have increased transcription efficiency when it was purified from the GAS compared to when it was purified from E. coli (Fig. 3, lanes 5 and 6 versus lanes 7 and 8). For a control in these assays, Mga was added to an rpsL reaction mixture, which showed that its stimulatory effect on transcription was specific only to the emm promoter (Fig. 3, lanes 1 to 3). The results of this experiment indicate that Mga is necessary and sufficient to activate transcription at the category A emm promoter, but its activity is enhanced when it is expressed in the GAS.
Mga is sufficient to activate transcription of the category B promoters, PsclA and Psof-sfbX, in vitro.
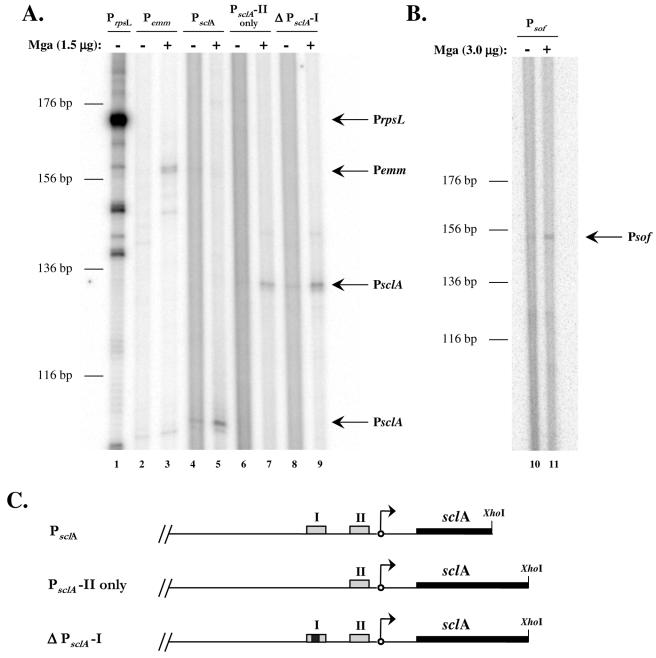
Similar in vitro transcription experiments were devised for the category B promoters, PsclA and Psof-sfbX, to determine whether activation from a distal site requires additional factors. As expected, transcription initiated from the full-length promoters of both sclA and sof-sfbX was dependent on the presence of Mga (Fig. 4A, lanes 4 and 5, and Fig. 4B, lanes 10 and 11, respectively). Likewise, expression of the control promoters, PrpsL and Pemm, followed expected patterns in this experiment (Fig. 4A, lanes 1 to 3).
FIG. 4.
In vitro transcription of the category B Mga-regulated genes sclA and sof-sfbX. (A and B) Transcription of the sclA and sof-sfbX genes were tested in an in vitro transcription assay in the absence (−) or presence (+) of 1.5 μg of GAS-purified M6 Mga-His as described in Materials and Methods. Transcription of the Mga-independent rpsL gene and the Mga-dependent emm gene were used as controls in the assay. DNA templates (plasmid and promoter) were as follows: pKSM415, PrpsL (lane 1); pKSM416, Pemm (lanes 2 and 3); pKSM419, PsclA (lanes 4 and 5); pKSM417, PsclA-II only (lanes 6 and 7); pKSM418, ΔPsclA-I (lanes 8 and 9); and pKSM420, Psof-sfbX (lanes 10 and 11). DNA templates were incubated with purified σA, core RNA polymerase, Mga (where indicated), and a radiolabeled ribonucleotide mixture. Reaction products were ethanol precipitated and separated on a 6% denaturing polyacrylamide gel. Bands representing the transcribed products of the expected size are indicated by arrows and the name of the promoter. The sizes of the transcripts (in base pairs) were estimated on the basis of the migration of a DNA sequencing ladder. (C) Schematic of PsclA constructs including Mga-binding sites (gray boxes), deleted Mga-binding site (gray and black box), transcriptional start site (arrows), sclA (thick black lines), and location of restriction enzyme cut sites (XhoI).
Previous studies of the sclA promoter showed it to have more than one Mga-binding site by sequence analysis, while Mga was able to bind to only one of those sites (PsclA-I) by an EMSA (1). Consequently, multiple constructs of the promoter were tested in the in vitro transcription system to further examine the necessity of each site. Surprisingly, Mga was still capable of activating transcription at PsclA in the absence of PsclA-I, whether by a deletion of the site or by elimination of all sequences upstream of the proximal Mga-binding site (PsclA-II) (Fig. 4A, lanes 6 to 9, and Fig. 4C). This result indicates that, unlike in vivo experiments (1), the presence of PsclA-II is sufficient for Mga to activate transcription of sclA in vitro.
DISCUSSION
Categories of Mga-regulated promoters.
In this study, the sof-sfbX promoter was shown to have a single Mga-binding site distal to the start of transcription similar to the one found recently in PsclA (1). The architecture of this promoter lends further support to the idea that Mga-regulated promoters fall into multiple categories dependent on the number and location of the Mga-binding site(s) (1) (Fig. 5). Thus, Psof-sfbX represents a second category B promoter with the most distal Mga-binding site yet observed. Though category B promoters, with a single distal Mga-binding site, may have a second, low-affinity proximal Mga-binding site, the location and size of those sites are sufficiently different from those found in Pmga, validating their placement into a separate category (Fig. 5).
FIG. 5.
Categorization of biochemically characterized Mga-regulated promoters. Mga-regulated promoters fall into three categories on the basis of the number and location of Mga-binding sites (gray boxes) in relation to the transcriptional start sites (arrows). In category A, a single Mga-binding site exists proximal to the start of transcription. In category B, a single Mga-binding site exists distal to the start of transcription, while a second low-affinity Mga-binding site proximal to the start of transcription (dashed box) may be present. The single category C promoter, Pmga, has multiple binding sites that are larger than those in the other two categories (59 bp versus 45 bp) and are located between two transcriptional start sites. Position numbers are given in relation to the transcriptional start site, which is numbered +1. Dark gray boxes represent the translational coding region for each gene.
To date, five Mga-regulated promoters have been characterized biochemically (Fig. 5). All of the uncharacterized promoters in the Mga locus, including the promoters of sic and members of the emm gene superfamily, have predicted Mga-binding sites proximal to the start of transcription, suggesting that they would fall within the same category as Pemm and PscpA (category A). It is interesting to note, therefore, that the only two genes known to be regulated directly by Mga outside of this region have promoters that fall within a separate category (category B).
An Mga-binding site exists near the sof-sfbX insertion site.
The GAS is divided into two classes on the basis of the reactivity of the M protein to an antibody directed to the C repeat region of the M6 protein (2). Generally, the two classes also differ in their expression of the serum opacity factor, with class I strains lacking and class II strains possessing OF (2). An alignment of sequence from a class I serotype (M1) and two class II serotypes (M12 and M49) revealed that the region upstream of the sof-sfbX genetic locus in the class II serotypes was highly homologous to sequence approximately 10 kb upstream of emm in the class I genome (11). Presumably, the insertion or excision of sof-sfbX at this site occurred at about the same time that the M protein diverged into two classes (11).
Interestingly, the Mga-binding site in Psof-sfbX identified in this study falls near this insertion (or excision) region (Fig. 2A). In fact, an alignment of M1 sequence with the M12 and M28 (class II) sof-sfbX region revealed that the first 15 bp of the Mga-binding site is completely conserved among these strains but differs by 14 bp in the 3′ region of the site (data not shown). However, since the consensus Mga-binding site is not highly specific, both the class I and class II sites exhibit significant homology to the consensus. In fact, Mga-His has been found to bind to the class I site and regulate an open reading frame adjacent to the insertion region in an M1 strain (D. A. Ribardo and K. S. McIver, unpublished results). The presence of an Mga-binding site near a region of genetic exchange raises the question of whether the site existed prior to or was created as a result of the recombination event.
Mga is sufficient to activate both category A and category B promoters.
Little is known about how Mga functions to activate transcription. Mga is the most studied regulator in its family, making it difficult to speculate on its mechanism of action on the basis of the activity of a homologous regulator. However, DNase I protection assays with Mga have demonstrated that, unlike many other regulatory proteins, Mga does not bind to a region of DNA exhibiting dyad symmetry (16). This suggests that Mga may bind to DNA as a monomer. Further, Mga has been shown to bind to the two sites in its own promoter independently, leaving little evidence for cooperative binding in this case (19). Finally, although σ54-dependent promoters often require a regulator with a distal binding site similar in location to Mga at category B promoters, the GAS does not appear to possess a σ54 homolog. Therefore, it is unlikely that Mga activates transcription from a distal site in a similar fashion.
The results of the in vitro transcription experiments performed in this study help to resolve some of the issues raised concerning how Mga functions to activate transcription. First, the ability of Mga, as purified here, to direct transcription in the in vitro transcription assay reveals that it is capable of activating both category A and B promoters on its own. Furthermore, this activity does not depend on any modifications to the protein that are specific to the GAS, since Mga purified from E. coli is active in the assay. Although Mga purified from the GAS appears to show enhanced activity over protein purified from E. coli, this may simply be a consequence of purifying the proteins in buffers of slightly different compositions, since salts have been found to be inhibitory to the transcription reaction (data not shown). However, Mga may still be modified to repress activity under certain conditions, such as during the stationary phase of growth.
The sufficiency of Mga to activate transcription does not rule out the possibility that other factors may be necessary for maximal activity at Mga-regulated promoters. Given the prospect that Mga may bind to a low-affinity proximal site at both the sclA and sof-sfbX promoters, the consequence of that binding may be what is observed in the in vitro transcription reaction. If a DNA-bending protein were added to the reaction mixture, it is possible that activity at these promoters may be significantly enhanced. On the other hand, the DNA topology of the sclA and sof-sfbX promoters may be such that Mga is already in close proximity to the transcription machinery. Clearly, further experimentation is necessary to determine whether other factors may be involved in initiating transcription from these promoters.
The ability of Mga to activate transcription at PsclA in the absence of its distal Mga-binding site in the in vitro transcription assay was a surprising result, considering that this site was shown to be essential for activity of the promoter in vivo (1). The interpretation of these data is that the in vitro transcription assay is more sensitive in detecting Mga activity at category B promoters than both an EMSA and the β-glucuronidase (GusA) transcriptional reporter system. In fact, GusA has been observed to be a relatively insensitive reporter when promoters of low activity are tested, requiring long incubation times with the substrate before that activity is detected (A. C. Almengor and K. S. McIver, unpublished results). As mentioned above, the addition of other factors to facilitate the interaction of Mga with the RNAP may enhance the activity of the category B promoters in the in vitro transcription assay.
The finding that Mga is capable of promoting expression of genes in the in vitro transcription assay is significant. This technique opens up the possibilities of what can be studied in the activation of Mga-regulated genes. For instance, the role of additional regulatory factors may be assessed simply by adding those factors to the in vitro reaction mixture. A number of other promoters, such as the autoregulated Pmga, may also be analyzed using this system. In vitro transcription would be especially useful in studying expression of mga, since it is thought to have a number of other regulatory factors that function to activate and repress its transcription. Finally, the interaction between Mga and the transcriptional machinery may be examined through the use of mutated components of the RNAP. Above all, the elucidation of the mechanism(s) by which Mga functions should aid in our understanding of the role of this important virulence gene regulator in the GAS.
Acknowledgments
We thank Charles Moran, June Scott, and Jason Opdyke for reagents and guidance in establishing the in vitro transcription system; Gunnar Lindahl and Lars-Olof Heden for strains; Traci Kinkel, Temekka Leday, and Cheryl Vahling for critical review of the manuscript; and Stacy Cantrell for technical assistance.
This work was supported by grants from the American Heart Association, Texas Affiliate (beginning-grant-in-aid 0060073Y to K.S.M.) and the National Institutes of Health (NIH AI-47928 to K.S.M.). A.C.A. is supported in part by an NIH/NIAID Molecular Microbiology training grant (5T32 AI07520) and an NIH/NIAID research supplement for underrepresented minorities (RSUM AI-47928-S).
REFERENCES
- 1.Almengor, A. C., and K. S. McIver. 2004. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J. Bacteriol. 186:7847-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessen, D., K. F. Jones, and V. A. Fischetti. 1989. Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. J. Exp. Med. 169:269-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgess, R. R. 1996. Purification of overproduced Escherichia coli RNA polymerase sigma factors by solubilizing inclusion bodies and refolding from Sarkosyl. Methods Enzymol. 273:145-149. [DOI] [PubMed] [Google Scholar]
- 4.Caparon, M. G., and J. R. Scott. 1987. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc. Natl. Acad. Sci. USA 84:8677-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, B. Y., and R. H. Doi. 1990. Overproduction, purification, and characterization of Bacillus subtilis RNA polymerase sigma A factor. J. Bacteriol. 172:3257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collado-Vides, J., B. Magasanik, and J. D. Gralla. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55:371-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courtney, H. S., D. L. Hasty, Y. Li, H. C. Chiang, J. L. Thacker, and J. B. Dale. 1999. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol. Microbiol. 32:89-98. [DOI] [PubMed] [Google Scholar]
- 8.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. Lefebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group A streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192:760-770. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan, D., and M. Meselson. 1983. Plasmid screening at high colony density. Methods Enzymol. 100:333-342. [DOI] [PubMed] [Google Scholar]
- 10.Ishihama, A. 1993. Protein-protein communication within the transcription apparatus. J. Bacteriol. 175:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeng, A., V. Sakota, Z. Li, V. Datta, B. Beall, and V. Nizet. 2003. Molecular genetic analysis of a group A streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 185:1208-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji, Y., N. Schnitzler, E. DeMaster, and P. Cleary. 1998. Impact of M49, Mrp, Enn, and C5a peptidase proteins on colonization of the mouse oral mucosa by Streptococcus pyogenes. Infect. Immun. 66:5399-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kihlberg, B. M., J. Cooney, M. G. Caparon, A. Olsen, and L. Bjorck. 1995. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 19:299-315. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl, G. 1989. Cell surface proteins of a group A streptococcus type M4: the IgA receptor and a receptor related to M proteins are coded for by closely linked genes. Mol. Gen. Genet. 216:372-379. [DOI] [PubMed] [Google Scholar]
- 15.Lukomski, S., K. Nakashima, I. Abdi, V. J. Cipriano, R. M. Ireland, S. D. Reid, G. G. Adams, and J. M. Musser. 2000. Identification and characterization of the scl gene encoding a group A streptococcus extracellular protein virulence factor with similarity to human collagen. Infect. Immun. 68:6542-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIver, K. S., A. S. Heath, B. D. Green, and J. R. Scott. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J. Bacteriol. 177:6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1602. [DOI] [PubMed] [Google Scholar]
- 18.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIver, K. S., A. S. Thurman, and J. R. Scott. 1999. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J. Bacteriol. 181:5373-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLandsborough, L. A., and P. P. Cleary. 1995. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of ScpA, FcRA, OF, and M protein. FEMS Microbiol. Lett. 128:45-51. [DOI] [PubMed] [Google Scholar]
- 21.Opdyke, J. A., J. R. Scott, and C. P. Moran. 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495-502. [DOI] [PubMed] [Google Scholar]
- 22.Podbielski, A., M. Woischnik, B. Pohl, and K. H. Schmidt. 1996. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med. Microbiol. Immunol. 185:171-181. [DOI] [PubMed] [Google Scholar]
- 23.Rasmussen, R., A. Eden, and L. Bjorck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robbins, J. C., J. G. Spanier, S. J. Jones, W. J. Simpson, and P. P. Cleary. 1987. Streptococcus pyogenes type 12 M protein gene regulation by upstream sequences. J. Bacteriol. 169:5633-5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt, K. H., E. Gunther, and H. S. Courtney. 1996. Expression of both M protein and hyaluronic acid capsule by group A streptococcal strains results in a high virulence for chicken embryos. Med. Microbiol. Immunol. 184:169-173. [DOI] [PubMed] [Google Scholar]
- 26.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suvorov, A. N., and J. J. Ferretti. 1996. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J. Bacteriol. 178:5546-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thern, A., M. Wastfelt, and G. Lindahl. 1998. Expression of two different antiphagocytic M proteins by Streptococcus pyogenes of the OF+ lineage. J. Immunol. 160:860-869. [PubMed] [Google Scholar]
- 29.Vahling, C. M., and K. S. McIver. 2006. Domains required for transcriptional activation show conservation in the Mga family of virulence gene regulators. J. Bacteriol. 188:863-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wade, K. H., G. Schyns, J. A. Opdyke, and C. P. Moran, Jr. 1999. A region of σK involved in promoter activation by GerE in Bacillus subtilis. J. Bacteriol. 181:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]