Abstract
Proper placement of the division apparatus in Escherichia coli requires pole-to-pole oscillation of the MinC division inhibitor. MinC dynamics involves a membrane association-dissociation cycle that is driven by the activities of the MinD ATPase and the MinE topological specificity factor, which themselves undergo coupled oscillatory localization cycles. To understand the biochemical mechanisms underlying Min protein dynamics, we studied the interactions of purified Min proteins with phospholipid vesicles and the role of ATP in these interactions. We show that (i) the ATP-bound form of MinD (MinD.ATP) readily associates with phospholipid vesicles in the presence of Mg2+, whereas the ADP-bound form (MinD.ADP) does not; (ii) MinD.ATP binds membrane in a self-enhancing fashion; (iii) both MinC and MinE can be recruited to MinD.ATP-decorated vesicles; (iv) MinE stimulates dissociation of MinD.ATP from the membrane in a process requiring hydrolysis of the nucleotide; and (v) MinE stimulates dissociation of MinC from MinD.ATP-membrane complexes, even when ATP hydrolysis is blocked. The results support and extend recent work by Z. Hu et al. (Z. Hu, E. P. Gogol, and J. Lutkenhaus, Proc. Natl. Acad. Sci. USA 99:6761-6766, 2002) and support models of protein oscillation wherein MinE induces Min protein dynamics by stimulating the conversion of the membrane-bound form of MinD (MinD.ATP) to the cytoplasmic form (MinD.ADP). The results also indicate that MinE-stimulated dissociation of MinC from the MinC-MinD.ATP-membrane complex can, and may, occur prior to hydrolysis of the nucleotide.
Cytokinesis in bacteria is mediated by the septal ring, an envelope-associated organelle which is present at the site of cell wall constriction (26, 37). Formation of the organelle in Escherichia coli involves the ordered assembly of at least 10 essential division proteins at the prospective site of division. Assembly is thought to initiate with polymerization of the FtsZ protein and decoration of these polymers with FtsA and ZipA, leading to the formation of an intermediate membrane-associated structure (Z ring) to which the remaining components are recruited in a specific order (2, 3, 12, 32). Equipartitioning of cell components during division requires that Z ring formation be accurately confined to a narrow zone around midcell. Positioning of the Z ring in E. coli is primarily controlled through prevention of its assembly at membrane sites that lie off-center (26). Two negative-regulatory systems are known. The dominant and best-understood determinant is the Min system, which relies on the activities of the minB operon products: MinC, MinD, and MinE (9). The other is the location of the nucleoid(s), in that Z rings preferentially form at sites that are relatively free of nucleoid material (30, 45). This nucleoid occlusion phenomenon indicates that the presence of a nucleoid somehow interferes with Z ring assembly in its close vicinity. Nucleoid occlusion and the Min system act independently, and the effects of nucleoid occlusion on placement of the Z ring are largely redundant with respect to the restrictions imposed by the Min system (4, 40, 41).
The MinC protein consists of 231 residues which form two domains of roughly equal size (5, 9, 18). The N-terminal domain (ZMinC) inhibits FtsZ polymerization in vitro and Z ring assembly in vivo (18, 21, 23, 31). The C-terminal (DMinC) domain mediates homodimerization, as well as binding to the MinD protein in yeast two-hybrid assays (5, 18, 23, 43). The location of the Z ring is determined by the cellular distribution of MinC. Interestingly, this division inhibitor undergoes a highly dynamic and rapid localization cycle. During this cycle, the protein alternately accumulates on the membrane at either cell half every other 45 to 50 s (20, 34). A consequence of this pole-to-pole oscillation is that the average interval during which a membrane site is occupied by MinC increases with its distance from the cell center. We proposed that this time-averaged concentration differential effectively forces proper positioning of the Z ring to midcell because the level of interference with FtsZ polymerization by MinC at this site should be minimal (13). The feasibility of such a mechanism is supported by computer simulations of theoretical models that mimic Min protein dynamics in silico (16, 25, 29).
MinC oscillation is driven by MinD and MinE. In fact, MinC itself plays no discernible role in the mechanism of oscillation (20, 34). Rather, the cellular location of MinC is directly dictated by that of MinD, which undergoes a similar oscillatory localization cycle (36, 38). MinD is an ATPase that associates with the cytoplasmic membrane in a peripheral manner (7, 36, 38). In addition, MinD interacts with itself, as well as with both MinC and MinE in yeast two-hybrid assays (15, 18, 22, 23, 27, 42). Consistent with these interactions, MinD is required to recruit both MinC and MinE to the membrane in vivo (19, 22, 34-36). Different lines of evidence indicate that this recruitment is essential for the functions of all three proteins. In MinD− cells, MinC remains in the cytoplasm and fails to inhibit Z ring assembly (20, 34). As a result, cells show the classical minicell phenotype (Min−) in which they frequently divide asymmetrically near either one of the cell poles (1, 9, 44).
Recruitment of MinE is required for the pole-to-pole oscillation of MinD and, hence, that of MinC (36, 38). In the absence of MinE, MinD still recruits MinC, but both proteins accumulate along the entire membrane and fail to display any obvious bulk movement. This static distribution of MinC results in a block of Z ring assembly at all membrane sites and, therefore, the formation of long nonseptate filamentous cells (Sep−) (9, 20, 34). The 88-residue MinE peptide contains two domains. The N-terminal domain (DMinE, residues 1 to 32) is both required and sufficient for interaction with MinD, as well as to counteract MinC/MinD-mediated division inhibition (22, 33, 35, 36, 47). The C-terminal domain (TSMinE) mediates homodimerization of MinE. Mutations in this domain give rise to a Min− phenotype, implying that it is involved in restricting suppression of MinC/MinD action to the cell center (24, 33, 39, 46, 47).
MinE undergoes a localization cycle that appears to be intimately coupled to that of MinD (11, 13, 39). At the start of a cycle, MinD has assembled on the membrane appearing as a “test tube” that covers the membrane from one cell pole (pole 1) up to approximately midcell. A portion of the MinE protein decorates the whole tube, but the bulk appears to accumulate at its “rim” in the shape of a ring (the E ring). The rim of the tube then retracts toward pole 1, and the E ring moves with it. As this happens, a portion of MinD becomes cytoplasmic, suggesting that molecules are released from the tube into the cytoplasm. When the rim reaches the pole, the associated E ring dissipates and a substantial portion of MinE appears to become cytoplasmic as well. Meanwhile, a new MinD “tube” assembles on the membrane at the opposite cell end. A new E ring forms on the rim of this new tube, and both move to pole 2. This is followed by reassembly of MinD at pole 1 again, etc. Importantly, MinE remains dispersed in the cytoplasm of MinD− cells (35), implying that its dynamic localization pattern is strictly dependent on that of MinD. Given that MinD fails to oscillate in the absence of MinE (36, 38), this further implies that the dynamic behaviors of the two proteins are mutually dependent phenomena.
The dynamics of the Min proteins have raised the question of how the interactions between MinD, ATP, membrane, and MinE culminate in the observed oscillating behaviors. On the basis of the localization patterns of the MinD and MinE proteins, we proposed a model in which MinE has a high affinity for membrane-associated MinD and, once accumulated to a sufficient extent, stimulates the dissociation of MinD from the membrane, eventually leading to its own release from the membrane as well (13). Support for this idea has come from the demonstration that MinD's ATPase activity can be stimulated by MinE in vitro, provided phospholipid vesicles are also supplied (19). In addition, computer modeling has shown that MinD and MinE dynamics can be realistically simulated by using a minimal set of assumptions (16, 25, 29). An important property of the underlying models is that dynamic patterns are self-generating and do not require any fixed topological markers. The assumption that MinE forces the release of MinD from the membrane, however, is crucial to MinD/MinE oscillation in each of the models (16, 25, 29).
Here we explored the interactions between the Min proteins and phospholipid vesicles in vitro. Using vesicle sedimentation assays, we found that, in contrast to the ADP-bound form of MinD, the ATP-bound form readily associates with phospholipid vesicles in a self-enhancing manner, as well as recruits MinC and MinE to these vesicles.
We further found that MinE stimulates dissociation of both MinD and MinC from the vesicles. MinD became resistant to MinE-stimulated dissociation when ATP was substituted with ATPγS, indicating that release of MinD from the membrane requires hydrolysis of the nucleotide. In contrast, MinE stimulated the release of MinC from MinD-decorated vesicles, even in the presence of ATPγS. In vitro binding of MinD.ATP to phospholipid vesicles and the ability of MinE to stimulate membrane release of MinD was recently also reported by Hu et al. (17). The present work supports and extends these results. Both studies show that MinD can bind membrane by a direct interaction with phospholipids and support models wherein MinE contributes to Min protein dynamics by stimulating the conversion of the membrane-bound form of MinD (MinD.ATP) to the cytoplasmic form (MinD.ADP). In addition, our finding that MinE stimulates the release of MinC from MinD.ATPγS-decorated vesicles suggests that, in vivo, dissociation of MinC from the MinC-MinD.ATP-membrane complex may occur prior to hydrolysis of the nucleotide.
MATERIALS AND METHODS
Strains and plasmids.
Strains PB114 (dadR trpE trpA tna ΔminCDE::aph) (9) and LL1 (PB114, lon::Tn10) (23) were described before. Strains BL21(λDE3) [ompT rB− mB− (PlacUV5::T7gene1)] and BL21(λDE3)/plysS were purchased from Novagen, and ER2566 [fhuA2 (lon) ompT lacZ::T7gene1 gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr73::miniTn10)2 R(zgb-210::Tn10)1 (Tets) endA1 (dcm)] was from New England Biolabs.
Strains JE9(λDE3), AS1(λDE3)/plysS, and LL2 were obtained by P1-mediated transduction of ΔminCDE::aph from PB114 to BL21(λDE3), BL21(λDE3)/plysS, and ER2566, respectively.
Plasmids pZC100 (47), pDR112 and pDR139 (35), pDR119 and pDR122 (36), pDR175 (34), pCH49 (14), and pDB389, pLL6, pLL18, and pLL19 (23) have been described previously. pET21a, pET21b, plysS, and plysS-RARE were obtained from Novagen.
Plasmid pDR192 (PT7::minE-h) was obtained in a number of steps. A PCR with primers 5′-CAAGGAATTCATATGGCACGCATTATTGTTG-3′ and 5′-GGCTTACTCGAGCTCTTCTGCTTCCGG-3′ was used to amplify a minDE fragment. The product was treated with NdeI and XhoI (underlined), and the resulting 1,076-bp fragment was ligated to similarly treated pET21b. This yielded pDR31 (PT7::minD minE-h) encoding native MinD and MinE-H in which the last residue of MinE (K88) has been replaced with EHHHHHH. The 392-bp XmnI-StyI minE-h fragment of pDR31 was next flush ended by incubation with Klenow enzyme and nucleotides and inserted in pZC100 that had been linearized with BamHI and flush ended as well. This yielded pDR42 (aadA::minE-h), which carries minE-h in the same orientation as the aadA gene of the vector. Finally, pDR192 was prepared by replacing the 117-bp XbaI-XhoI fragment of pET21b with the 301-bp XbaI-XhoI fragment of pDR42, thereby placing minE-h downstream of PT7.
For pDR193 [PT7::minE(34-87)-h], a partial minE fragment was amplified with the primers 5′-CGTCTACATATGGCAGAACCGCATTATCTGCCG-3′ and 5′-TTACCGAGCTCTTATTTCAGCTCTTCTGCTTCCGG-3′. The product was digested with NdeI and SacI (underlined), and the resulting 177-bp fragment was ligated to similarly digested pET21a, yielding pCH81 [PT7::minE(34-88)]. The 3,062-bp SapI fragment of pDR192 was then replaced with the 2,965-bp SapI fragment of pCH81, yielding pDR193. Plasmid pDR193 is identical to pDR192 except that it encodes a derivative of MinE-H in which residues 2 to 33 are absent.
For pLL56 (PT7::h-minD), the 821-bp NdeI-HindIII minD fragment of pDR119 was inserted into similarly treated pDB389. This resulted in pDB394 (PT7::stII-minD), encoding MinD fused to the Streptag II peptide (Genosys). The oligonucleotides 5′-CATGCGTGGTTCTCACCACCACCACCACCA-3′ (sense) and 5′-TATGGTGGTGGTGGTGGTGAGAACCACG-3′ (antisense) were annealed, and the resulting fragment was used to replace the 45-bp NcoI-NdeI stII fragment of pDB394, yielding pLL56. This plasmid encodes H-MinD in which the peptide MRGSHHHHHH (H) is fused to the N terminus of MinD.
Plasmid pLL30 [PT7::gfp-t-minC(5-231)-h] was obtained by replacing the 326-bp XbaI-AgeI fragment of pLL6 with the 977-bp XbaI-AgeI fragment of pLL18. The plasmid encodes a functional MinC derivative in which residues 1 to 4 have been replaced with Gfpmut2 (6) and the linker peptide ASMTGGQQMGRIP (T), and the peptide LEHHHHHH is fused to the C-terminal residue.
To construct pLL61 (Plac::h-minD minE), the 1,008-bp XbaI-PacI fragment of pDR122 was replaced with the 294-bp XbaI-PacI fragment of pLL56.
Plasmid pLL67 (Plac::h-minD minE-h) was obtained in a number of steps. The 1,790-bp BstEII-XhoI fragment of pCH49 was replaced with the 1734-bp BstEII-XhoI fragment of pDR139, yielding pDR168 [Plac::minD minE(1-33)-h]. Next, the 2,539-bp PacI-SacI fragment of pDR119 was replaced with the 2,757-bp PacI-SacI fragment of pDR168, resulting in pDR170 [Plac::gfp-t-minD minE(1-33)-h]. Plasmid pCH157 (Plac::gfp-t-minD minE-h) was then created by replacing the 692-bp PacI-XhoI fragment of pDR170 with the 851-bp PacI-XhoI fragment of pDR112. Finally, pLL67 was prepared by replacing the 1,008-bp XbaI-PacI fragment of pCH157 with the 294-bp XbaI-PacI fragment of pLL61.
Functionality of tagged proteins in vivo.
Strain LL1/pLL19 [ΔminCDE::aph lon::Tn10/cI857(ts)PλR::gfp-t-minC(5-231)-h], carrying either pLL61 (Plac::h-minD minE) or pLL67 (Plac::h-minD minE-h), was grown at 37°C in M9 medium supplemented with l-tryptophan (50 μg/ml), Casamino Acids (0.2%), maltose (0.2%), ampicillin (50 μg/ml), and either glucose or IPTG (isopropyl-β-d-thiogalactopyranoside) as indicated. Cells were imaged and analyzed as described before (23, 34).
Purification of proteins.
Purified proteins in the present study carried His6 tags (H). Gfp-T-MinC(5-231)-H is referred to as Gfp-MinC-H, MinE(1-87)-H is referred to as MinE-H, and MinE(34-87)-H is referred to as *MinE-H. Proteins were purified after overexpression in strains JE9(λDE3)/pLL30 (Gfp-MinC-H), LL2/plysS-RARE/pLL56 (H-MinD), or AS1(λDE3)/plysS carrying either pDR192 (MinE-H) or pDR193 (*MinE-H). Cells were grown overnight in Luria-Bertani (LB) medium with ampicillin (50 μg/ml), glucose (0.1%) and, when appropriate, chloramphenicol (25 μg/ml). Cultures were diluted 100-fold in LB medium with ampicillin (50 μg/ml) and 0.04% glucose, and cells were grown at 30°C (Gfp-MinC-H) or 37°C (other proteins) to an optical density at 600 nm (OD600) of 0.2 to 0.7. IPTG was added to 840 μM, and growth was continued for an additional 1 to 5 h. Cells were harvested by centrifugation, washed once in saline, resuspended in 1/100 volume of buffer (see below), frozen quickly in a dry ice-acetone bath, and stored at −80°C.
For MinE-H and *MinE-H, frozen cell suspensions in NiCS(50/5) buffer (20 mM Tris-Cl, 10% glycerol, 50 mM NaCl, 5 mM imidazole; pH 8.0) were quickly thawed at 37°C and subjected to two more freeze-thaw cycles. The homogenate was sonicated briefly to reduce viscosity and clarified by centrifugation at 200,000 × g for 3 h at 5°C. The supernatant was applied to a column of Fast Flow Chelating Sepharose (Pharmacia) that had been charged with NiCl2 and equilibrated in NiCS(50/5). The column was washed with NiCS(500/5) [same as NiCS(50/5), but containing 500 mM NaCl]. Protein was eluted with NiCS(500/150) [same as NiCS(500/5), but containing 150 mM imidazole], and EDTA was added to 5 mM. Protein was further fractionated on a Mono-Q column by using a linear gradient of NaCl (20 to 1,000 mM) in 20 mM Tris-Cl (pH 8.0)-20 mM NaCl-2 mM EDTA. Peak fractions were pooled and dialyzed against MinE storage buffer (50 mM HEPES-KOH, 0.1 mM EDTA, 10% glycerol; pH 7.2).
For Gfp-MinC-H, cell suspensions in NiCSk(70/50) buffer (20 mM Tris-Cl, 10% glycerol, 70 mM KCl, 50 mM imidazole; pH 8.0) were thawed and broken by three passages through a French pressure cell at 10,000 lb/in2. The homogenate was clarified as described above, and the supernatant was applied to a chelating Sepharose column that had been charged with NiCl2 and equilibrated in NiCSk(70/50). The column was washed with NiCSk(500/50) [same as NiCSk(70/50), but containing 500 mM KCl], and protein was eluted with NiCSk(500/500) [same as NiCSk(500/50), but containing 500 mM imidazole]. EDTA was added to 5 mM, and desired fractions were pooled and dialyzed against MinC storage buffer (50 mM HEPES-KOH, 150 mM KCl, 0.1 mM EDTA, 10% glycerol; pH 7.2).
For H-MinD, cells were resuspended in NiNTA(300/10) buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole; pH 8.0) supplemented with 50 μM ADP and 1 mM MgCl2. Frozen suspensions were quickly thawed at 37°C and subjected to three more freeze-thaw cycles. The homogenate (10 ml) was sonicated briefly, and one tablet of Complete, Mini, EDTA-free protease inhibitor cocktail (Boehringer Mannheim) was added before clarification by centrifugation as described above. The supernatant was applied to a column of NiNTA agarose (Qiagen) that had been equilibrated in NiNTA(300/10) containing 50 μM ADP. The column was washed with 50 μM ADP in NiNTA(300/20) [same as NiNTA(300/10), but containing 20 mM imidazole], and protein was eluted with NiNTA(300/100) [same as NiNTA(300/10), but containing 100 mM imidazole] containing 50 μM ADP. EDTA was added to 5 mM, and pooled fractions were dialyzed against MinD storage buffer (50 mM HEPES-KOH, 150 mM KCl, 0.1 mM EDTA, 10% glycerol; pH 8.0).
Purified proteins (0.4 to 5.0 mg/ml) were stored in small aliquots at −80°C. Protein concentrations were determined by the noninterfering protein assay (Genotech) according to the manufacturer's instructions, with bovine serum albumin as a standard.
Preparation of phospholipid vesicles.
Total E. coli phospholipids and headgroup-labeled lissamine rhodamine B phosphatidylethanolamine (both supplied in chloroform by Avanti Polar Lipids, Inc.) were mixed at a ratio of 200:1 (wt/wt) before evaporation of the solvent under a stream of air. The lipid film was hydrated by the addition of reaction buffer (RB; 25 mM Tris-Cl, 50 mM KCl; pH 7.5) to yield 10 mg of phospholipid/ml, followed by incubation of the mixture at 65°C for 2 h. The mixture was passed in and out of a microtip 10-fold by aid of a micropipette, resulting in a suspension of uni- and multilamellar vesicles ranging in diameter from 0.2 to 5.0 μm, as judged by both light and electron microscopy (not shown).
Vesicle sedimentation assays.
Unless specified otherwise, purified proteins were added to a suspension containing phospholipid vesicles and nucleotide in RB. Mixtures were placed at 30°C, and reactions were started by addition of MgCl2 to 1 mM. Reaction volumes were kept at 20 to 30 μl. After the specified times, vesicles were sedimented by centrifugation at 16,000 × g at room temperature for 1 or 25 min. The supernatant of each mixture was collected, and the pellet was resuspended in RB to the original volume.
The amounts of phospholipid in the supernatant (S) and pellet (P) fractions were determined by fluorometry. Aliquots of fractions were diluted 30-fold in RB, and rhodamine fluorescence was measured by using a Fluoromax-3 fluorometer (Jobin Yvon Horiba) with slits at 2 nm and excitation and emission filters set at 570 and 589 nm, respectively. Where relevant, the amounts of Gfp-MinC-H were measured in the same fashion except that excitation and emission filters were set at 467 and 506 nm, respectively. Amounts of H-MinD, MinE-H, and *MinE-H were determined by quantitative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, gels were incubated overnight in SYPRO Ruby Protein Gel Stain (Molecular Probes) and washed in 10% methanol-7% acetic acid for 1 h. Fluorescent protein bands were visualized and quantitated by using a FluorS-Max Multi Imager and the accompanying Quantity One software (Bio-Rad). To improve accuracy, aliquots of the corresponding S and P fractions were loaded in adjacent lanes. The amounts of each protein loaded per pair of lanes (S+P) ranged from 7 to 200 ng, and measured bands contained from 1 to 160 ng of protein. For each measurement the intensities of both the minimal area containing a band and an equal area immediately above this band were determined, and the value of the latter was subtracted from the former. Fractions of sedimented phospholipid and proteins were calculated by dividing the amounts recovered in the pellet fraction by the sum of the amounts recovered in both fractions. Typically, more than 90% of the phospholipid was recovered in the pellet fraction, but in a few reactions recovery was in the 80 to 90% range. To compensate for incomplete sedimentation of vesicles, measured amounts of cosedimenting proteins were multiplied by a factor of 100/L, where L equals the percentage of phospholipid that was recovered in the pellet fraction. Note that this adjustment may not yield absolutely accurate values if nonpelleting (presumably small) vesicles bound protein with a different affinity than the pelleting ones. Also note that the adjustment procedure did not affect the conclusions of the study. Table 2 shows both measured and adjusted values. Subsequent tables and figures only show adjusted values.
TABLE 2.
ATP- and Mg2+-dependent association of H-MinD with phospholipid vesiclesa
| Nucleotide | % Content in pellet |
|||
|---|---|---|---|---|
| Without lipid, H-MinDb | With lipid |
|||
| Lipidb | H-MinDb | H-MinD | ||
| ATP | 13 | 93 | 51 | 55 |
| ATPγS | 7 | 97 | 42 | 43 |
| GTP | 11 | 98 | 27 | 27 |
| dATP | 5 | 98 | 24 | 24 |
| ADP | 10 | 97 | 21 | 21 |
| ATP (no Mg2+)c | NDd | 92 | 19 | 21 |
H-MinD (2.0 μM) was added to mixtures containing phospholipid vesicles (0.5 mg/ml) and nucleotide (500 μM) in RB. MgCl2 was added to 1 mM, and mixtures were incubated for 15 min at 30°C and then subjected to centrifugation for 1 min at 16,000 × g. The percentages of H-MinD and phospholipid recovered in the pellet fractions were measured as described in Materials and Methods. The last column shows the percentages of H-MinD in the pellet fractions after adjustment for the incomplete sedimentation of phospholipids.
Measured values.
MgCl2 was omitted from the reaction.
ND, not detected.
ATP hydrolysis.
ATPase activity was assayed by measuring the conversion of [α-32P]ATP to [α-32P]ADP, by using thin-layer chromatography essentially as described previously (7). Mixtures (30 μl) in RB (25 mM Tris-Cl, 50 mM KCl; pH 7.5) contained [α-32P]ATP, phospholipid vesicles, and purified proteins as indicated. Mixtures were incubated at 30°C for 5 min, and reactions were started by addition of MgCl2 to 5 mM. At the times indicated, 1 μl of the mixture was spotted onto a polyethyleneimine cellulose plate. Plates were developed with 0.5 M LiCl in 1 M formic acid, and radioactivity in the ADP and ATP spots was measured by using a Packard Instant Imager.
RESULTS
Functionality of proteins used in the present study.
For the in vitro studies described below, we made use of His6-tagged versions of MinC, MinD, and MinE. Gfp-MinC-H is a MinC derivative in which residues 1 to 4 have been replaced with Gfpmut2 (6) and the linker peptide ASMTGGQQMGRIP. In addition, the peptide LEH6 is fused to the C-terminal residue. H-MinD contains the tag MRGSH6 fused to the N-terminal starting methionine of native MinD, and MinE-H contains the tag EH6 in place of the C-terminal lysine residue of native MinE.
To ensure that these derivatives are functional in vivo, we constructed plasmids pLL19 [cI(ts) PλR::gfp-minC-h], pLL61 (Plac::h-minD minE), and pLL67 (Plac::h-minD minE-h). Plasmid pLL19 expresses gfp-minC-h from the λR promoter in a temperature-dependent fashion (23). Plasmid pLL61 encodes h-minD and native minE under control of the lac promoter, and pLL67 is identical to pLL61 except that minE is replaced with minE-h. Plasmids pLL19 and either pLL61 or pLL67 were introduced into strain LL1 (ΔminCDE), and transformants were grown at 37°C in the absence or presence of IPTG. In the absence of IPTG, both LL1/pLL19/pLL61 and LL1/pLL19/pLL67 cells showed the classical minicell phenotype, and Gfp-MinC-H was dispersed throughout the cytoplasm. In contrast, both strains reverted to a wild-type division pattern when grown in the presence of IPTG, and the Gfp-MinC-H fusion showed typical dynamic behavior, with average pole-to-pole oscillation cycles of 51 s (LL1/pLL19/pLL61) and 63 s (LL1/pLL19/pLL67) (Fig. 1 and Table 1). The somewhat slower oscillation observed in LL1/pLL19/pLL67 cells correlated with a somewhat less effective suppression of minicell formation, suggesting that addition of the tag to MinE reduced its cellular level and/or activity to some degree (13, 36). Nevertheless, all three tagged proteins were clearly functional in vivo.
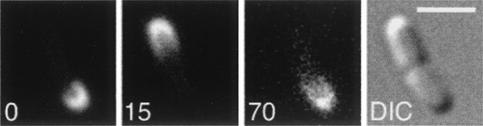
FIG. 1.
Pole-to-pole oscillation of Gfp-MinC-H in the presence of H-MinD and MinE-H. Fluorescence and differential interference contrast (DIC) images showing the dynamics of Gfp-MinC-H in a cell of strain LL1/pLL19/pLL67 [ΔminCDE/cI(ts) PλR::gfp-minC-h/Plac::h-minD minE-h] are depicted. Cells were grown in the presence of IPTG (25 μM) as described in the legend to Table 1. Times are indicated in seconds. Bar, 2 μm.
TABLE 1.
In vivo functionality of His-tagged Min proteinsa
| Strain | Cells grown with: |
||||
|---|---|---|---|---|---|
| Glucose |
IPTG |
||||
| Phenotype (%PS) | Loca- tion | Phenotype (%PS) | Loca- tion | Avg cycle time (range) | |
| LL1/pLL19/pLL61 | Min− (ND) | C | WT (2) | O | 51 (40-65) |
| LL1/pLL19/pLL67 | Min− (43) | C | WT (11) | O | 63 (44-90) |
Cells of strain LL1/pLL19 [ΔminCDE/cI(ts) PλR::gfp-minC-h] carrying either pLL61 (Plac::h-minD minE) or pLL67 (Plac::h-minD minE-h) were grown at 37°C for 4.6 mass doublings to an OD600 of 0.4 to 0.5 in the presence of either 0.1% glucose or IPTG at 10 μM (pLL61) or 25 μM (pLL67). The location of Gfp-MinC-H was determined by fluorescence microscopy of live cells. The oscillation cycle time represents the average of 20 events and is given in seconds. Division phenotypes were determined by DIC microscopy of chemically fixed cells. The percentages of polar septa (%PS) were based on the determination of the positions of at least 100 septa. Locations: C, cytoplasmic; O, oscillating. WT, wild-type division pattern. Most cells were rods of normal size, and <12% of septa were misplaced at a pole. Min− refers to a minicell division pattern, i.e., a mixture of rods, short filaments, and minicells; >40% of septa were misplaced at a pole. ND, not determined.
The tagged Min proteins were overexpressed in ΔminCDE strains and purified. In addition, we purified a mutant derivative of MinE-H (*MinE-H), which lacks residues 2 to 33 corresponding to a domain required for interaction with MinD (22, 33, 47).
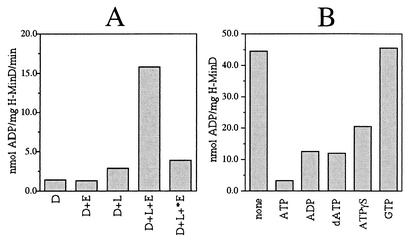
Native MinD was previously shown to be a weak ATPase (7), whose ATPase activity is stimulated by the simultaneous presence of phospholipid vesicles and MinE (19). To ensure our preparations of H-MinD and MinE-H contained active protein, we measured ATPase activity of H-MinD in the absence or presence of MinE-H and in the absence or presence of the large phospholipid vesicles described further below (Fig. 2A). Addition of either lipid vesicles or MinE-H alone stimulated H-MinD-dependent ATPase activity only marginally (twofold or less). As was observed with the native forms of MinD and MinE (19), however, the presence of both vesicles and MinE-H stimulated the ATPase activity of H-MinD almost 12-fold. Specific activities, furthermore, were similar to those reported for the native protein (7, 19). As expected, *MinE-H failed to stimulate the ATPase activity of H-MinD, even in the presence of lipid vesicles (Fig. 2A).
FIG. 2.
MinD ATPase assays. (A) Stimulation of H-MinD ATPase activity by phospholipid and MinE-H. Proteins (each at 12 μM) were added to RB containing 1 mM [α-32P]ATP (0.33 μCi/nmol) and 0.5 mg of phospholipid vesicles/ml as indicated. Mixtures were placed at 30°C, and reactions were started by the addition of MgCl2 to 5 mM. The amounts of ADP produced after 40 min were used to calculate specific activities. D, H-MinD; E, MinE-H; *E, *MinE-H; L, phospholipid vesicles. (B) Nucleotide competition. H-MinD (5 μM) was added to RB containing 100 μM [α-32P]ATP (3.3 μCi/nmol) and a 1 mM concentration of the indicated unlabeled nucleotide. Mixtures were placed at 30°C, MgCl2 was added to 5 mM, and the amounts of ADP produced after 60 min were determined.
Based on these results, we conclude that the tagged Min proteins retained both the physiological and biochemical activities of the native proteins.
MinD associates with phospholipid membranes in an ATP-dependent fashion.
To assess the association of purified H-MinD with phospholipids in vitro, we devised a simple and rapid phospholipid vesicle sedimentation assay. A mixture of total E. coli phospholipids containing rhodamine-labeled phosphatidylethanolamine as a tracer (0.5% of total phospholipid) was used to prepare large phospholipid vesicles. The method of preparation yielded a mixture of uni- and multilamellar vesicles of various sizes, as judged by light (see below) and electron microscopy (not shown). The majority of these vesicles were sufficiently large to be readily sedimented during a short centrifugation step.
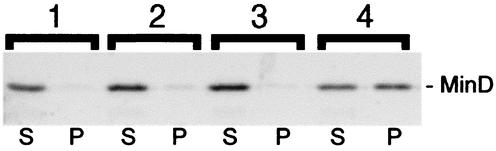
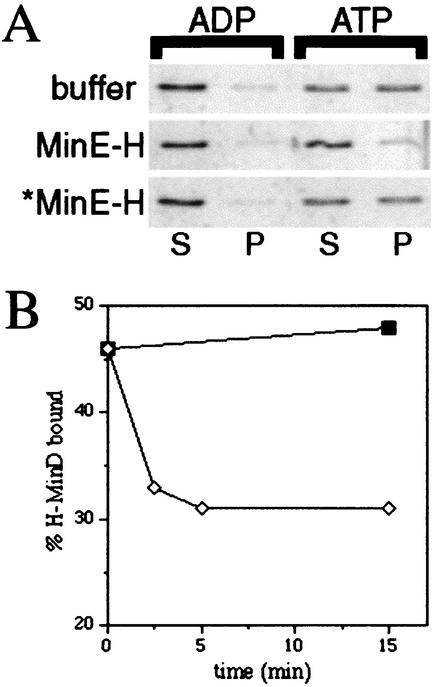
In a typical experiment, purified H-MinD (to 2 μM) was added to RB (25 mM Tris-Cl, 50 mM KCl; pH 7.5) containing 500 μM ADP or ATP and either no lipid or 0.5 mg of phospholipid vesicles/ml. MgCl2 was added to 1 mM, and the mixtures were incubated at 30°C. After 15 min, the mixtures were subjected to centrifugation for 1 min at 16,000 × g, and aliquots of the supernatant and pellet fractions were used to determine the amounts of phospholipid and/or H-MinD in each. The results of one such experiment are shown in Fig. 3. Of the reactions containing ADP and ATP, respectively, 97 and 99% of phospholipid was recovered in the pellet fractions. Interestingly, 52% of H-MinD cosedimented with the vesicles in the presence of ATP. In contrast, only 4 to 7% of the protein was found in the pellet fractions of the reactions containing vesicles and ADP or containing either nucleotide but no lipid (Fig. 3). This result indicated that the ATP-bound form of MinD (MinD.ATP) had a significantly higher affinity for phospholipid membrane than the ADP-bound form (MinD.ADP).
FIG. 3.
ATP-dependent association of MinD with phospholipid vesicles. SYPRO Ruby-stained gel showing sedimentation of H-MinD in the presence of lipid vesicles and ATP. Purified H-MinD (to 2 μM) was added to RB containing a 500 μM concentration of either ADP (1, 2) or ATP (3, 4) and either no lipid (1, 3) or 0.5 mg/ml of phospholipid vesicles (2, 4). MgCl2 was added to 1 mM, and mixtures were incubated at 30°C for 15 min. Mixtures were subjected to centrifugation (16,000 × g for 1 min at room temperature), and equivalent aliquots of the supernatant (S) and pellet (P) fractions were subjected to SDS-PAGE.
During the course of the present study, essentially the same experiment as that shown in Fig. 3 was performed repeatedly as part of the experiments described further below. In eight separate experiments using comparable conditions (i.e., 2 μM H-MinD, 500 μM ATP or ADP, 0.5 to 1.0 mg of lipid/ml, 1 mM MgCl2), the fractions of H-MinD cosedimenting with vesicles in the presence of ADP ranged from 7 to 25%, with a mean (x) of 15.0% and a standard deviation (σ) of 5.7%, whereas in the presence of ATP the fractions ranged from 50 to 62% (x = 56.2%, σ = 3.5%). In each case, the amount of H-MinD cosedimenting with vesicles in the presence of ATP was severalfold higher (x = 4.3, σ = 1.6, range = 2.3 to 7.4) than in the presence of ADP. The variability in the amounts of H-MinD cosedimenting with vesicles in the presence of ADP suggested some aspecific binding and/or trapping of H-MinD by some batches of phospholipid vesicles, which were prepared fresh daily. To avoid this and other potential sources of nonspecific variability, care was taken throughout the present study to perform critically related sets of sedimentation assays as part of self-contained experiments, with the exact same batches of reagents.
ADP and dATP compete to a similar extent with ATP for binding to MinD (7). To assess whether the poorly hydrolyzable ATP analogue ATPγS is also capable of interacting with H-MinD, we tested its ability to compete with [α-32P]ATP in ATPase assays. As indicated in Fig. 2B, ATPγS did compete with [α-32P]ATP for interaction with H-MinD, albeit to a lesser extent than unlabeled ADP, dATP, and ATP itself. In contrast, GTP did not compete noticeably.
We then compared the association of H-MinD with membrane in the presence of ATP, ADP, dATP, ATPγS, or GTP. After incubation in the absence of lipid, a small amount of H-MinD (5 to 13%) was recovered in the pellet fraction regardless of the nucleotide used in the reaction (Table 2). In the presence of lipids, this fraction increased further in all cases and also in the absence of nucleotide (not shown). Nevertheless, the fraction of H-MinD that cosedimented with lipid vesicles was again clearly dependent on the specific nucleotide present in the reaction. Thus, whereas ∼25% of H-MinD was present in the pellet after incubation with ADP, dATP, or GTP, 51% of H-MinD was recovered in the pellet after incubation with ATP. After adjustment for the incomplete sedimentation of phospholipid (93%), this value increased to 55% (Table 2).
We previously determined that the binding of MinD to nucleotide requires metal. Consistent with this requirement, ATP failed to stimulate association of H-MinD with the vesicles when Mg2+ was omitted from the reaction (Table 2).
Interestingly, the association of H-MinD with vesicles was also stimulated by ATPγS, indicating that the binding of MinD.ATP to membrane does not require hydrolysis of the nucleotide. Although ATPγS clearly stimulated H-MinD binding to vesicles, ATPγS was consistently somewhat less effective than ATP (Table 2; see also below). This may be partly or wholly due to a relatively low affinity of MinD for ATPγS, as is indicated by the nucleotide competition experiments (Fig. 2B).
In a recent independent study, Hu et al. (17) similarly showed that native MinD binds phospholipid vesicles in the presence of ATP or ATPγS. Both that study and the present results support a simple model in which MinD that is bound to ATP and Mg2+ (MinD.ATP) assembles on the membrane by direct interactions with phospholipids, while the ADP-bound form (MinD.ADP) is cytoplasmic.
Cooperative binding of MinD to phospholipid vesicles.
To characterize the interaction of H-MinD with vesicles in more detail, we varied the concentrations of H-MinD, ATP, and vesicles in the mixture, as well as the time of their incubation. These experiments showed that neither the phospholipid nor the ATP concentration used in the reactions above were limiting the amount of H-MinD cosedimenting with the vesicles (results not shown). Interestingly, however, the binding of H-MinD to vesicles was markedly stimulated by increasing the concentration of the protein in the reaction.
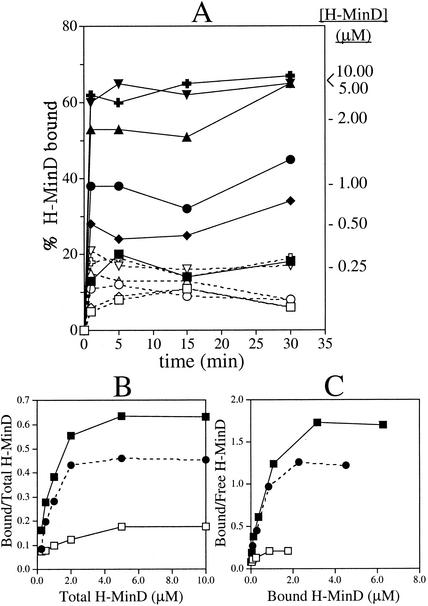
Figure 4A shows time course experiments in which H-MinD at various concentrations was incubated for various times with vesicles in the presence of either ATP or ADP. Within the concentration range tested (0.25 to 10.00 μM), binding of H-MinD to vesicles reached equilibrium within 1 min (excluding the 1-min centrifugation period), indicating that binding was relatively rapid.
FIG. 4.
Self-enhanced association of MinD with phospholipid vesicles. (A) Time course membrane-binding assays at various H-MinD concentrations. MgCl2 (1 mM) was added to RB containing 1.0 mg of phospholipid vesicles/ml, a 500 μM concentration of either ADP (open symbols) or ATP (filled symbols), and H-MinD at 0.25 μM (squares), 0.50 μM (diamonds), 1.00 μM (circles), 2.00 μM (triangles), 5.00 μM (crosses), or 10.00 μM (inverted triangles). After 1, 5, 15, and 30 min, aliquots were immediately fractionated by centrifugation for 1 min, and the percentages of H-MinD cosedimenting with vesicles were determined. The percentages of bound H-MinD at the four time points were averaged for each reaction to generate panels B and C. The average values for the ATP-containing reactions are indicated at the right of the panel. (B and C) Analyses of the equilibrium binding data from panel A. Panel B shows the fraction of bound H-MinD as a function of the H-MinD concentration in the reaction. Panel C shows Scatchard plots of the same data. Symbols: ▪, reactions in the presence of ATP; □, reactions in the presence of ADP; •, the amount of bound H-MinD in the presence of ADP was subtracted from that in the presence of ATP before plotting of the data.
Interestingly, analyses of the binding isotherms showed that the binding of H-MinD to vesicles did not conform to a typical Langmuir adsorption reaction. In Fig. 4B the fraction of bound H-MinD at equilibrium is plotted as a function of the total H-MinD concentration in the reaction. If H-MinD were to bind the phospholipid surface in a simple adsorption reaction, the amount of bound H-MinD should increase with increasing concentrations of total H-MinD, whereas the fraction of bound H-MinD should decrease (28). In reality, however, both the amount and the fraction of H-MinD.ATP found associated with vesicles at equilibrium sharply increased with the total H-MinD concentration in the reaction. For example, whereas only ∼16% of H-MinD.ATP associated with the vesicles when the protein was added to 0.25 μM, this fraction increased to ∼65% when H-MinD was added to 5.0 μM (Fig. 4A, B). Thus, increased concentrations of H-MinD stimulated its binding to the vesicles, implying cooperativity in the association of the protein with the phospholipid surface.
Cooperative binding is similarly obvious from Scatchard analyses of the equilibrium binding data. As shown in Fig. 4C, a plot of the bound to free H-MinD ratio versus bound H-MinD shows a pronounced convex curvature, a finding indicative of cooperative binding (28).
At concentrations of total H-MinD of >5 μM, the fraction of bound H-MinD.ATP did not increase further but remained at ∼65% (Fig. 4 and data not shown). Based on experiments in which we measured the ability of H-MinD in supernatant fractions to bind a fresh supply of vesicles, we estimate that 10 to 15% of our purified H-MinD preparation consisted of inactive protein (not shown). Accordingly, the fraction of bound H-MinD could be increased somewhat further by enrichment for active protein in a prior round of cosedimentation with lipid vesicles. For the experiments in Table 3, H-MinD (5 μM) was incubated for 10 min at 30°C with vesicles (1 mg/ml) and a 100 μM concentration of either ATP or ATPγS. After sedimentation for 1 min, 66 and 51% of H-MinD, respectively, were recovered in the pellet fractions of the mixtures containing ATP and ATPγS. The pellets were resuspended in RB containing 500 μM ATP, ATPγS, or ADP. Mixtures were next incubated for 10 min at 30°C and then sedimented again (25 min at 16,000 × g) to determine the fractions of bound H-MinD. Whether vesicles had been prefractionated with H-MinD.ATP or H-MinD.ATPγS in the first round, the results were very similar. Thus, ca. 77, 60, or 22% of H-MinD cosedimented with either variety of prefractionated vesicles upon incubation with ATP, ATPγS, or ADP, respectively, in the second round (Table 3). The finding that incubation with ADP led to release of H-MinD from both types of predecorated vesicles further demonstrated that binding of both H-MinD.ATP and H-MinD.ATPγS to membrane is a reversible interaction.
TABLE 3.
Effect of prior copurification with vesicles on the fraction of membrane-associating H-MinDa
| Reaction | Nucleotide |
% H-MinD in pellet |
||
|---|---|---|---|---|
| Step 1 | Step 2 | Step 1 | Step 2 | |
| 1 | ATP | ATP | 66 | 76 |
| 2 | ATP | ATPγS | 66 | 58 |
| 3 | ATP | ADP | 66 | 21 |
| 4 | ATPγS | ATP | 51 | 77 |
| 5 | ATPγS | ATPγS | 51 | 62 |
| 6 | ATPγS | ADP | 51 | 23 |
In step 1, H-MinD (5 μM) was incubated in RB with phospholipid vesicles (1.0 mg/ml), MgCl2 (1 mM), and either ATP or ATPγS (100 μM) for 10 min at 30°C. Mixtures were sedimented for 1 min (16,000 × g). The pellet fractions were resuspended in the original volume of RB containing 1 mM MgCl2 and incubated for 5 min at 30°C. In step 2, the suspensions were split into aliquots to which ATP, ATPγS, or ADP was added to 500 μM. Mixtures were incubated for another 10 min and subjected to centrifugation for 25 min at 16,000 × g. As indicated, 66% (ATP) or 51% (ATPγS) of H-MinD fractionated with the vesicles in the first step. Consequently, the total concentration of H-MinD in the reactions during the second step was 3.3 μM (reactions 1 to 3) or 2.6 μM (reactions 4 to 6).
MinE stimulates dissociation of MinD from phospholipid membrane.
It has been proposed that MinE stimulates dissociation of MinD from the membrane (13, 16, 19, 25, 29). To determine the effect of the MinE protein on the MinD-phospholipid association in our in vitro system, purified H-MinD (2 μM) was incubated with nucleotide (500 μM) and phospholipid vesicles (0.5 mg/ml) as described above. After 10 min, purified MinE-H (2 μM), *MinE-H (2 μM), or MinE storage buffer was added and, after an additional 5 min, lipid vesicles and associated protein were pelleted by centrifugation for 1 min.
As in the experiments described above, only a small fraction of H-MinD (∼10%) cosedimented with the vesicles when ADP was used as the nucleotide, regardless of whether or not MinE-H had been added to the reaction, and a significant fraction of H-MinD (59%) was found associated with phospholipid after incubation with ATP and when MinE-H was omitted from the reaction (Fig. 5A). Interestingly, however, addition of MinE-H led to a sharp reduction in the amount of H-MinD (to 19%) associated with the vesicles (Fig. 5A). Time course experiments showed that the fraction of bound H-MinD reached a new, and lower, plateau within 2.5 to 5 min upon addition of MinE-H to the mixtures (Fig. 5B).
FIG. 5.
MinE-stimulated dissociation of MinD from vesicles. (A) SYPRO Ruby-stained gels showing the effect of MinE-H on the binding of H-MinD to vesicles. MgCl2 (1 mM) was added to RB containing 2 μM H-MinD, 0.5 mg of phospholipid vesicles/ml, and a 500 μM concentration of either ADP or ATP. After 10 min, MinE storage buffer (50 mM HEPES-KOH, 0.1 mM EDTA, 10% glycerol; pH 7.2), 2 μM MinE-H, or 2 μM *MinE-H was added as indicated. After an additional 5 min, mixtures were fractionated by centrifugation for 1 min, and equivalent aliquots of the supernatant (S) and pellet (P) fractions were subjected to SDS-PAGE. (B) Time course of MinE-H-stimulated dissociation of H-MinD from vesicles. MgCl2 (1 mM) was added to RB containing 2 μM H-MinD, 0.5 mg of phospholipid vesicles/ml, and 500 μM ATP. After 10 min, either buffer (▪) or 2 μM MinE-H (⋄) was added. At the times indicated, aliquots were fractionated by centrifugation for 1 min, and the percentages of membrane-bound H-MinD were determined.
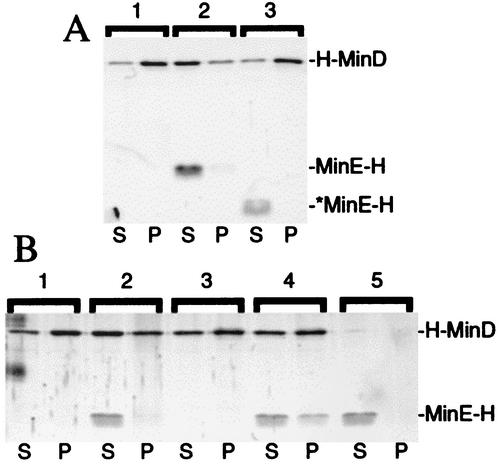
Consistent with the finding that the N-terminal domain of MinE is required for its interaction with MinD (22, 33, 47), the addition of *MinE-H failed to stimulate dissociation of H-MinD from the vesicles (Fig. 5A, 52% of H-MinD in pellet).
The same effect of MinE on MinD-membrane association was evident when MinE-H was added to vesicles that had been predecorated with H-MinD (Fig. 6 and Table 4). After incubation of predecorated vesicles with either *MinE-H or a buffer control for 5 min in the presence of ATP, ∼82% of H-MinD remained associated with vesicles after sedimentation for 1 min. Upon incubation with MinE-H, however, only 50% of H-MinD still cosedimented with vesicles. This percentage decreased still further when the centrifugation step was extended from 1 to 25 min. Also, in this case more than 80% of H-MinD remained bound to membrane when incubated with buffer or *MinE-H, but only ∼32% did so upon incubation with MinE-H. Both the MinE-H-stimulated dissociation of H-MinD from vesicles and the enhancing effect of an extended centrifugation period were quite reproducible (Table 4), although in some experiments (e.g., Fig. 5A) the effects of MinE-H were more pronounced than in others. Why a longer centrifugation period resulted in a somewhat more efficient MinE-dependent dissociation of H-MinD is not clear. Time course experiments indicated that the effect is not merely due to a longer total incubation period of the reagents (Fig. 5B and results not shown). The segregation of vesicles to the bottom of the reaction vessel may simply reduce the efficiency by which H-MinD molecules that have been released can reassociate with the vesicles, but other possibilities exist as well.
FIG. 6.
Stabilization of a MinE-MinD-membrane complex in the presence of ATPγS. SYPRO Ruby-stained gels showing the effect of MinE-H on, and the interaction of MinE-H with, prefractionated H-MinD-membrane complexes are shown. (A) Vesicles were decorated with H-MinD.ATP and treated with buffer (lane 1), MinE-H (lane 2), or *MinE-H (lane 3) as described in the legend to Table 4. (B) Vesicles were incubated with H-MinD (lanes 1 to 4) or without (lanes 5) and in the presence of either ATP (lanes 1 and 2) or ATPγS (lanes 3 to 5). Vesicles were harvested by centrifugation and resuspended in RB with the same nucleotide, and either 1 μM MinE-H (lanes 2, 4, and 5) or the same volume of buffer (lanes 1 and 3) was then added. After incubation, mixtures were refractionated, and equivalent aliquots of the supernatant (S) and pellet (P) fractions were subjected to SDS-PAGE. For additional details, see the text and the legend to Fig. 7.
TABLE 4.
MinE-H stimulated dissociation of H-MinD from predecorated vesiclesa
| Reaction | Condition | Mean % protein (SD) in pellet at: |
|||||
|---|---|---|---|---|---|---|---|
| 1 min |
25 min |
||||||
| H-MinD | MinE-H | *MinE-H | H-MinD | MinE-H | *MinE-H | ||
| 1 | Spin 1 | 66.0 (1.0) | 67.5 (1.5) | ||||
| 2 | P1 + buffer | 82.0 (1.0) | 87.0 (2.0) | ||||
| 3 | P1 + MinE-H | 50.0 (2.1) | 16.3 (1.5) | 31.8 (4.3) | 4.5 (1.1) | ||
| 4 | P1 + *MinE-H | 81.0 (0.0) | 0.0 (0.0) | 82.0 (2.0) | 0.5 (0.5) | ||
In reaction 1, H-MinD (5 μM) was incubated with vesicles (1 mg/ml) in RB plus 500 μM ATP and 1 mM MgCl2 for 10 min at 30°C. Vesicles were sedimented (16,000 × g for 1 min; spin 1). The pellet (P1) was resuspended in the original volume of RB plus 500 μM ATP and 1 mM MgCl2, and the percentages of H-MinD and phospholipid were determined. For reactions 2 to 4, resuspended P1 was supplemented with MinE storage buffer (reaction 2), MinE-H (reaction 3), or *MinE-H (reaction 4), and mixtures were incubated for 5 min at 30°C. Vesicles were sedimented (16,000 × g) for either 1 or 25 min, and the percentages of phospholipid, H-MinD, MinE-H, and *MinE-H in the pellet fractions were determined. MinE-H and *MinE-H were used at 5 μM, and the H-MinD/MinE-H and H-MinD/*MinE-H ratios in reactions 3 and 4 were ∼0.67. The mean percent values and standard deviations of two (reactions 1, 2, and 4) or four (reaction 3) independent experiments obtained with two different preparations of H-MinD and MinE-H are given. Values were adjusted for incomplete sedimentation of vesicles. More than 93% of the phospholipid was recovered in the pellet fraction in each case.
Only small amounts of the MinE-H protein (and virtually none of the *MinE-H protein) were recovered in the pellet fractions in these experiments (Fig. 6A and Table 4). Nevertheless, the amounts of MinE-H cosedimenting with the vesicles reproducibly increased with the amount of H-MinD in the pellet fractions. Thus, whereas ∼4.5% of MinE-H (4.5 pmol) was recovered in the pellet, together with ∼32% of H-MinD (21.4 pmol), in the 25-min centrifugation experiments, ∼16% of MinE-H (16.0 pmol) cosedimented with 50% of H-MinD (33.5 pmol) in the 1-min centrifugation experiments (Table 4). These results are consistent with previous work showing that, in vivo, MinE is recruited to the membrane in a MinD-dependent fashion (35). The fact that only small amounts of MinE-H were found to cosediment with the vesicles in these experiments was also not unexpected. If MinE both requires membrane-bound MinD to associate with the vesicles itself and stimulates dissociation of MinD from the membrane, the association of MinE with MinD-decorated vesicles is expected to be short-lived because its action at the membrane will result in its own dissociation from the membrane.
Role of nucleotide hydrolysis in MinD/MinE dissociation from the membrane.
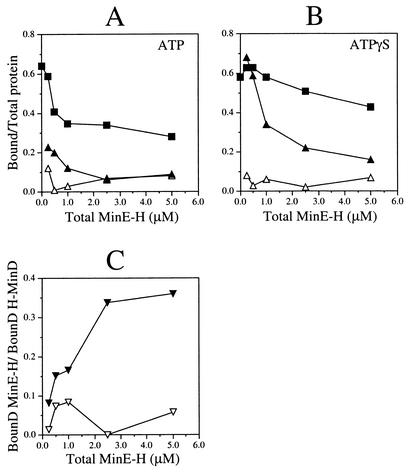
To assess the role of nucleotide hydrolysis in the MinE-stimulated dissociation of MinD.ATP from phospholipids, we compared the ability of MinE-H to dislodge H-MinD from vesicles that were decorated with either H-MinD.ATP or H-MinD.ATPγS. Decorated vesicles (at ∼3.0 μM H-MinD) were incubated with the appropriate nucleotide and with various amounts of MinE-H for 5 min, and the fractions of proteins cosedimenting with vesicles were plotted as a function of MinE-H concentration. Interestingly, membrane-bound H-MinD.ATPγS proved significantly more resistant to the action of MinE-H than membrane-bound H-MinD.ATP (Fig. 6B and 7). For example, incubation of the H-MinD.ATP-decorated vesicles with 1.0 μM MinE-H reduced the fraction of bound H-MinD from a maximum of 64% (no MinE-H added) to 35%, a value close to the minimum of 28% reached after incubation with 5.0 μM MinE-H (Fig. 6B and 7A). In contrast, membrane-bound H-MinD.ATPγS appeared to be impervious to MinE-H at up to 1.0 μM, and the fraction of bound H-MinD dropped only moderately from 58% (no MinE-H added) to 43% upon incubation with 5.0 μM MinE-H (Fig. 6B and 7B).
FIG. 7.
Recruitment of MinE to MinD-decorated vesicles. MgCl2 (1 mM) was added to phospholipid vesicles (1 mg/ml) in RB containing a 500 μM concentration of either ATP (A) or ATPγS (B), and either 5 μM H-MinD or no H-MinD. After incubation for 10 min, vesicles were sedimented for 1 min and then resuspended in the original volume of RB containing 1 mM MgCl2 and 500 μM ATP (A) or ATPγS (B). Aliquots were next supplemented with various concentrations of MinE-H (0, 0.25, 0.50, 1.00, 2.50, or 5.0 μM), and the mixtures were incubated for 5 min. Vesicles were sedimented for 25 min, and the percentages of phospholipid, H-MinD, and MinE-H in the pellet fractions were determined. (A and B) The fractions of bound H-MinD (▪), bound MinE-H in the presence of H-MinD (▴), and bound MinE-H in the absence of MinD (▵) were plotted as a function of MinE-H concentration. (C) Ratios of bound MinE-H to bound H-MinD in the presence of either ATP (▿) or ATPγS (▾) as a function of MinE-H concentration. Before calculation of each ratio, the amount of bound MinE-H in the reaction lacking H-MinD was subtracted from the amount of bound MinE-H in the corresponding reaction containing H-MinD.
As described above (Table 4), the fractions of cosedimenting MinE-H and H-MinD appeared to be correlated. Upon incubation of H-MinD.ATP-decorated vesicles with a low concentration of MinE-H (0.25 μM), 23% of MinE-H and 59% of H-MinD cosedimented with vesicles. However, bound fractions of both proteins dropped rapidly at higher concentrations of MinE-H. For example, when MinE-H was added to 2.5 μM, only 6% of MinE-H was recovered in the pellet, together with 34% of H-MinD (Fig. 7A). Interestingly, significantly higher fractions of MinE-H were retained by the vesicles decorated with H-MinD.ATPγS (Fig. 6B and 7B). When it was added to 0.25 μM, 68% of MinE-H cosedimented with 63% of H-MinD, and when it was added to 2.5 μM, 21% of MinE-H cosedimented with 54% of H-MinD. Less than 10% of MinE-H sedimented when H-MinD was omitted from any of these reactions, demonstrating that the association of MinE-H with the vesicles was indeed dependent on H-MinD (Fig. 6B and 7A and B).
In Fig. 7C the effect of ATPγS on the binding of H-MinD and MinE-H to vesicles is displayed by plotting the ratio of bound MinE-H to bound H-MinD against the total MinE-H concentration. In the presence of ATP, this ratio already reached a small maximal value of 0.084 at 1.0 μM MinE-H. In the presence of ATPγS, however, the ratio only began to plateau at 2.5 μM MinE-H and reached a value of up to 0.36 at 5.0 μM MinE-H. From the latter value we deduce that as few as three molecules of H-MinD are sufficient to tether one molecule of MinE-H to the phospholipid surface under these conditions.
These results show that MinE-H can be directly recruited to MinD-decorated phospholipid vesicles in vitro. Furthermore, they indicate that hydrolysis of the nucleotide is required for efficient MinE-stimulated release of MinD and, hence, MinE itself, from the membrane. Similar conclusions were reached in the recent study by Hu et al. (17).
MinD-dependent recruitment of the division inhibitor MinC to membrane.
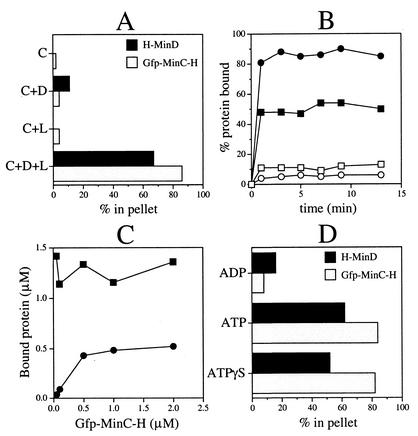
MinD is thought to recruit MinC to the membrane in vivo through a direct interaction with the C-terminal D-domain of MinC (DMinC) (18, 20, 23, 34). To determine whether this recruitment can be mimicked in vitro, purified H-MinD (2 μM) was allowed to assemble on phospholipid vesicles (0.5 mg/ml) in the presence of ATP (500 μM) for 5 min, and purified Gfp-MinC-H was added to 0.5 μM. After an additional 10 min, mixtures were separated into soluble and insoluble fractions by centrifugation for 1 min. The amounts of H-MinD and phospholipid in each fraction were determined as described above, and the amounts of Gfp-MinC-H were determined by measuring Gfp fluorescence (Fig. 8A).
FIG. 8.
Recruitment of MinC to MinD-decorated vesicles as shown in vesicle cosedimentation assays. The amounts of phospholipids and proteins in the pellet and supernatant fractions were determined. The values represent cosedimenting protein in percentages of total added to the reaction (A, B, and D) or in amounts (C). (A) Cosedimentation of Gfp-MinC-H with phospholipid vesicles in the presence of H-MinD. MgCl2 (1 mM) was added to RB containing 500 μM ATP, 2 μM H-MinD, and/or 0.5 mg of phospholipid vesicles/ml, as indicated. After incubation for 5 min, Gfp-MinC-H (to 0.5 μM) was added. After an additional 10 min, vesicles were sedimented for 1 min. Abbreviations: D, H-MinD; C, Gfp-MinC-H; L, phospholipid vesicles. (B) Time course of complex formation between membrane, MinD-H, and Gfp-MinC-H. H-MinD (to 2 μM) was added to RB containing 0.5 μM Gfp-MinC-H, 0.5 mg of phospholipid vesicles/ml, 1 mM MgCl2, and a 500 μM concentration of either ADP (open symbols) or ATP (filled symbols). From each reaction, aliquots were removed at the indicated time points and immediately fractionated by centrifugation for 1 min. Symbols: squares, H-MinD; circles, Gfp-MinC-H. (C) Binding of Gfp-MinC-H to H-MinD-decorated vesicles is saturable. MgCl2 (1 mM) was added to RB containing 500 μM ATP, 2 μM H-MinD, and 0.5 mg of phospholipid vesicles/ml, and mixtures were incubated for 5 min. Gfp-MinC-H was added to 0.05, 0.10, 0.50, 1.00, or 2.00 μM. After an additional 10 min, vesicles were sedimented for 1 min. Symbols: ▪, H-MinD; •, Gfp-MinC-H. (D) Recruitment of Gfp-MinC-H to H-MinD.ATPγS-decorated vesicles. MgCl2 (1 mM) was added to RB containing 2 μM H-MinD, 0.5 mg of phospholipid vesicles/ml, and 500 μM ADP, ATP, or ATPγS. After incubation for 5 min, Gfp-MinC-H was added to 0.1 μM. After an additional 5 min, vesicles were sedimented for 1 min.
Very little of the Gfp-MinC-H protein (<4%) was recovered in the pellet fractions when H-MinD and/or lipid vesicles were omitted from the reactions, showing that Gfp-MinC-H remained soluble in this assay system when either of these components was missing. In contrast, 86% of Gfp-MinC-H was recovered in the pellet fraction when both H-MinD and phospholipid were present in the reaction, indicating that Gfp-MinC-H bound to H-MinD-decorated vesicles (Fig. 8A). Time course experiments showed that the binding reaction of Gfp-MinC-H reached equilibrium within 1 min of addition of H-MinD to the mixtures (Fig. 8B). Thus, both the assembly of H-MinD on the vesicles and the consequent association of Gfp-MinC-H are relatively rapid processes (Fig. 8B).
Furthermore, titration experiments showed that H-MinD.ATP-decorated vesicles were close to being saturated with Gfp-MinC-H at the concentrations of the division inhibitor (0.5 μM) and H-MinD (2.0 μM) used in these experiments. Thus, when Gfp-MinC-H was used at 0.5 μM or above, 13 to 16 pmol of the protein cosedimented together with 35 to 41 pmol (58 to 68%) of H-MinD (Fig. 8C). Similar to what we deduced for the recruitment of MinE-H above, this result indicates that no more than three membrane-bound MinD monomers are sufficient to recruit one monomer of MinC.
Figure 8D shows that Gfp-MinC-H was also efficiently recruited to vesicles decorated with H-MinD.ATPγS, indicating that hydrolysis of the nucleotide by MinD is not required for recruiting MinC to the membrane.
Recruitment of Gfp-MinC-H to H-MinD-decorated phospholipid vesicles could also be readily visualized by fluorescence microscopy. Figure 9A and B show vesicles that were incubated with 2 μM H-MinD, 0.1 μM Gfp-MinC-H, and a 500 μM concentration of either ADP or ATP. In reactions containing ATP, Gfp-MinC-H was clearly seen to decorate lipid vesicles (Fig. 9B), whereas reactions containing ADP resulted in a homogeneous fluorescent haze in the surrounding buffer (Fig. 9A). As expected, similar homogenous fluorescence was observed in reactions containing ATP but lacking H-MinD (not shown).
FIG. 9.

Microscopy of phospholipid vesicles showing MinD-mediated recruitment and MinE-stimulated release of MinC. MgCl2 (1 mM) was added to RB containing 2 μM H-MinD, 0.5 mg of phospholipid vesicles/ml, and 500 μM ADP (A and C) or ATP (B and D to G). After incubation for 5 min, 0.1 μM Gfp-MinC-H was added. Incubation was continued for an additional 10 min with either no further additions (A to D) or the addition of MinE storage buffer (E), 2 μM MinE-H (F), or 2 μM *MinE-H (G) 5 min after the addition of Gfp-MinC-H. For panels A and B, aliquots of the reactions were immediately placed on microscope slides and imaged by DIC and fluorescence microscopy. For panels C to G, vesicles were collected by centrifugation for 25 min and imaged immediately after resuspension in RB containing 1 mM MgCl2 and 500 μM ADP (C) or ATP (D-G). Bar: 2.7 μm (A and B) or 2.0 μm (C to G).
Thus, the MinD-dependent membrane recruitment of MinC that is observed in vivo (20, 23, 34) can be mimicked in vitro with a minimal set of purified components.
MinE-mediated release of both MinC and MinD from phospholipid vesicles.
The experiments above showed that H-MinD.ATP recruited both Gfp-MinC-H and MinE-H to phospholipid vesicles and that MinE-H stimulated the release of H-MinD and itself from the vesicles. In a reaction involving all three Min proteins, therefore, the MinE-stimulated release of MinD from vesicles would be expected to cause MinC to be dislodged as well. To test this, H-MinD (2 μM) was allowed to decorate vesicles for 5 min in the presence of 500 μM ATP, and Gfp-MinC-H was added to 0.1 μM. After incubation for another 5 min, either buffer or MinE-H (to 2 μM) was added, and incubation was continued for a subsequent period of 5 min. Mixtures were then fractionated by sedimentation for 1 min, and the fractions were analyzed.
As predicted, MinE-H stimulated the release of both Gfp-MinC-H and H-MinD proteins from the vesicles (Fig. 10A). Thus, the addition of MinE-H caused a drop in bound Gfp-MinC-H from 86 to 57% and a drop in bound H-MinD from 60 to 39%. Moreover, the MinE-H-stimulated release of both proteins was more efficient when the centrifugation step was extended from 1 to 25 min. In this case, the addition of MinE-H caused drops in bound Gfp-MinC-H and H-MinD from 85 to 13% and from 53 to 21%, respectively (Fig. 10B). As expected, addition of *MinE-H instead of MinE-H had no effect on the amounts of cosedimenting proteins (not shown).
FIG. 10.
MinE-stimulated release of both MinC and MinD from phospholipid vesicles. MgCl2 (1 mM) was added to RB containing 2 μM H-MinD, 0.5 mg of phospholipid vesicles/ml, and 500 μM ADP or ATP. After incubation for 5 min, Gfp-MinC-H was added to 0.1 μM. After an additional 5 min, mixtures were supplemented with either 2 μM MinE-H or an equal volume of MinE storage buffer. After incubation for another 5 min, vesicles were sedimented for 1 min (A) or 25 min (B). Values represent the fractions (in percentages) of H-MinD (solid bars) and Gfp-MinC-H (shaded bars) cosedimenting with the lipid vesicles.
The effect of MinE-H on the association of Gfp-MinC-H with vesicles could also be monitored qualitatively by inspection of the sedimented vesicles by fluorescence microscopy. Vesicles decorated with H-MinD.ATP and Gfp-MinC-H showed a clear fluorescent signal around the vesicles, and the addition of buffer or *MinE-H prior to sedimentation had no obvious effect on this signal (Fig. 9D, E, and G). In contrast, the addition of MinE-H markedly reduced the vesicle-associated signal to close to background (Fig. 9F).
MinE-mediated specific release of MinC from MinC-MinD.ATPγS-membrane complexes.
Both genetic studies in E. coli and yeast two-hybrid assays indicated that MinE is capable of interfering with the interaction between MinC and MinD (8, 10, 22). Direct evidence for such an activity of MinE came from experiments in which we compared the ability of MinE-H to stimulate the release of Gfp-MinC-H from vesicles that had been predecorated with Gfp-MinC-H and H-MinD in the presence of either ATP or ATPγS (Fig. 11). Decorated vesicles were incubated with the appropriate nucleotide and various amounts of MinE-H for 5 min, and the fractions of proteins cosedimenting with vesicles were plotted as a function of MinE-H concentration (Fig. 11A and B).
FIG. 11.
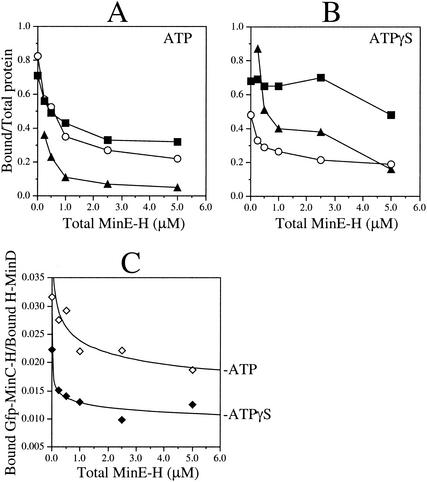
MinE stimulates the specific release of Gfp-MinC-H from H-MinD.ATPγS-decorated lipid vesicles. (A and B) MgCl2 (1 mM) was added to RB containing 5 μM H-MinD, 1 mg of phospholipid vesicles/ml, and 500 μM ATP (A) or ATPγS (B). After incubation for 5 min, Gfp-MinC-H was added to 0.1 μM. After an additional 5 min, vesicles were sedimented for 1 min and resuspended in the original volume of RB containing 1 mM MgCl2 and 500 μM ATP (A) or ATPγS (B). Decorated vesicles were supplemented with MinE-H to various concentrations (0, 0.25, 0.50, 1.00, 2.50, or 5.00 μM) and incubated for 5 min. Vesicles were sedimented for 25 min, and the fractions of cosedimenting H-MinD (▪), MinE-H (▴), and Gfp-MinC-H (○) were determined and plotted as a function of the MinE-H concentration in the reaction. (C) The data from panels A (ATP, ⋄) and B (ATPγS, ⧫) displayed as the ratio of bound Gfp-MinC-H to bound H-MinD versus the total MinE-H concentration.
In the presence of ATP, MinE-H caused the release of both H-MinD and Gfp-MinC-H as described above. Release was already significant upon addition of MinE-H to 0.25 μM and was close to maximal for both H-MinD (71 to 33%) and Gfp-MinC-H (83 to 27%) when MinE-H was used at 2.5 μM (Fig. 11A). As in reactions lacking Gfp-MinC-H (Fig. 7), H-MinD resisted release in the presence of ATPγS, and the fractions of MinE-H cosedimenting with vesicles in the presence of ATPγS were significantly greater than those in the presence of ATP at every MinE-H concentration tested (Fig. 11B).
In the same reactions containing ATPγS, in contrast, Gfp-MinC-H was readily released from the vesicles in a MinE-H-dependent manner. Similar to the reactions containing ATP (Fig. 11A), moreover, release of Gfp-MinC-H in the presence of ATPγS was close to maximal (48 to 22%) upon addition of MinE-H to 2.5 μM (Fig. 11B). In Fig. 11C the relevant data are presented as plots of bound Gfp-MinC-H to bound H-MinD ratios versus the MinE-H concentration. The shape of the plots obtained with ATP and ATPγS are very similar, implying that MinE-H dislodged Gfp-MinC-H from the H-MinD.ATPγS-membrane complexes quite efficiently.
We conclude that MinE can dislodge MinC from the MinC-MinD-membrane complex, even when dissociation of MinD from the membrane is inhibited by blocking nucleotide hydrolysis.
DISCUSSION
In this study we obtained in vitro evidence for the following. (i) The ATP form of MinD readily associates with phospholipid vesicles in the presence of Mg2+, whereas the ADP form does not. (ii) MinD.ATP binds membrane in a cooperative fashion. (iii) Both MinC and MinE can be recruited by MinD.ATP-decorated vesicles. (iv) MinE stimulates dissociation of MinD.ATP from the membrane, and this requires hydrolysis of the nucleotide. (v) MinE stimulates dissociation of MinC from MinD.ATP-decorated vesicles, even when ATP hydrolysis is blocked.
These results partially overlap with those of Hu et al. (17), who also found that MinD binds phospholipid vesicles in the presence of ATP or ATPγS, that MinE is recruited to MinD-decorated vesicles, and that MinE-stimulated nucleotide hydrolysis causes dissociation of both proteins from the vesicles. The results have implications for the mechanism of MinD/MinE-driven protein oscillation, as well as for the mechanism(s) by which MinE prevents MinC/MinD-mediated division inhibition.
Initial observations on Min protein dynamics suggested that oscillation of MinD involved the release of the protein from the membrane at one end of the cell, followed by cooperative reassembly onto the membrane at the other end (36). Moreover, they suggested a role for MinE in disassembly of MinD from the membrane, as indicated by the findings that MinD fails to oscillate in the absence of MinE, that the oscillation frequency is inversely related to the MinD to MinE ratio in the cell, and that the movements of the two proteins appear to be physically coordinated (11, 13, 36, 39). Important support for these proposals came from the finding that MinE stimulated MinD's ATPase activity in a phospholipid-dependent fashion, suggesting that MinD might bind membrane directly and that MinE might modulate this interaction by stimulating the conversion of MinD.ATP to MinD.ADP (19).
In addition, a different type of support, as well as novel insights into possible mechanisms of protein oscillation, have come from theoretical models aimed at rationalizing the observed dynamic behaviors of MinD and MinE. Three such models have recently been proposed independently (16, 25, 29). In all three the suggested mechanism of MinD and MinE dynamics is self-organizing and independent of any topological determinants other than the membrane. The value of these models is in demonstrating that the oscillating behavior of the Min proteins can, in principle, result from a limited set of properties of the proteins themselves. In addition, they provide explanations for the dynamic patterns of the Min proteins that are observed under specific circumstances, such as in filamentous or spherical cells (4, 11, 13, 34-36), which are otherwise difficult to understand. Although the models differ distinctly in several aspects, all three assume that MinD and MinE undergo coupled reaction-diffusion type interactions which require (i) binding of MinD to the membrane, (ii) MinD-mediated recruitment of MinE to the membrane, and (iii) MinE-dependent release of MinD from the membrane. The present results, as well as the recent results from Hu et al. (17), provide direct evidence for these properties of MinD and MinE and also show that the MinE-stimulated dissociation of MinD from the membrane is dependent on conversion of the ATP-bound form of MinD to the ADP-bound form.
In addition, our equilibrium binding experiments provide direct evidence for cooperativity in the manner by which MinD assembles on the phospholipid surface. This finding is consistent with yeast two-hybrid studies showing MinD self-interaction (23, 42), as well as with the observation that the turnover rate of ATP by MinD in the presence of both MinE and phospholipid increases with increasing MinD concentration (19). Cooperativity is also indicated by the recent observation that, at saturating concentrations of MinD, the protein is able to form a packed helical array on the surface of phospholipid vesicles, which is accompanied by a conversion of the vesicle shape from spherical to cylindrical (17). How this phenomenon is related to the cooperativity in the present binding experiments is unclear. We did not observe any MinD-dependent increase in tube-shaped membrane vesicles under the conditions of our binding studies (not shown). Possibly, the formation of such tubes occurs only when the ratio of MinD to phospholipid surface is very high and/or when vesicles are sufficiently small (17).
In two of the dynamic models for Min protein oscillation, the association of MinD with the membrane is explicitly assumed to be a self-enhancing process (25, 29). Our binding studies and the other accumulated evidence (17, 19) support this assumption. We note, however, that the present evidence does not yet distinguish between the possibilities that MinD self-interaction takes place in solution, on the membrane, or both. Association of MinD with membrane is likely to involve multiple types of self-interaction (e.g., monomer to dimer or dimer to polymer), which may preferentially take place either in the cytoplasm or on the membrane surface. Additional work is required to clarify this point.
To date, the sole known purpose of the MinD and MinE proteins is to precisely target the activity of the division inhibitor MinC within the cell. The MinD/MinE-driven oscillation cycle of MinC likely ensures that the time-averaged activity of its N-terminal ZMinC domain is maximal at the cell ends and minimal at the cell center, forcing Z ring assembly to the desired site at midcell (13, 16, 25, 29). However, targeting of MinC by MinD is not merely important for keeping ZMinC away from the membrane at midcell, but also to significantly enhance ZMinC activity at noncentral sites. In the absence of MinD, MinC remains in the cytoplasm and fails to inhibit Z ring assembly unless the concentration of MinC is artificially elevated over 25-fold (10, 20, 34). Therefore, MinD-mediated targeting is also needed for elevating the local concentration of ZMinC on the membrane to a level sufficiently high to effectively counteract FtsZ assembly. We recently showed that in vivo complexes of MinD with the C-terminal binding domain of MinC (DMinC) possess a specific affinity for one or more septal ring components, whereas neither MinC or MinD by themselves do. This finding indicates that, beyond recruiting MinC to the membrane, MinD in wild-type cells additionally serves in targeting the division inhibitor more specifically to sites on the membrane that contain undesired nascent FtsZ assemblies that lie off-center (23). Here, we have shown that recruitment of MinC to MinD-decorated vesicles can be readily mimicked in vitro in the presence of both ATP and ATPγS. These results confirm yeast two-hybrid studies indicating that MinC binds MinD directly (15, 18, 22, 23, 27, 42) and also show that hydrolysis of the nucleotide in the MinD.ATP-membrane complex is not required for this interaction.
Consistent with the findings that MinC failed to bind vesicles in the absence of MinD and that MinE stimulated the dissociation of MinD from vesicles, we also showed that addition of MinE to MinC-MinD-membrane complexes that were formed in the presence of ATP resulted in the release of both MinC and MinD from the vesicles. Interestingly, however, addition of MinE to such complexes prepared in the presence of ATPγS resulted in the specific release of MinC. Thus, in contrast to the MinE-stimulated membrane release of MinD, the MinE-stimulated release of MinC does not require ATP hydrolysis by MinD. These results reinforce the notion that MinE acts to suppress MinC/MinD-mediated division inhibition in more than one way (36) and indicate that suppression of MinC activity involves two biochemical steps that may occur in consecutive order. In the first step, binding of MinE to the MinC-MinD.ATP-membrane complex causes the release of MinC. Our titration experiments indicate that MinC and MinE each can bind membrane-associated MinD with about the same stoichiometry. One attractive possibility is that MinE efficiently competes with DMinC for the same, or overlapping, binding site(s) on a MinD dimer or oligomer. Alternatively, MinE may induce a conformational change in MinD prior to ATP hydrolysis, which significantly lowers its affinity for DMinC without causing membrane dissociation of MinD itself. In the second step, MinE stimulates nucleotide hydrolysis by MinD, resulting in the release of MinD.ADP from the membrane. This energy-requiring step is proposed to be critical for MinD/MinE-driven oscillation of MinC and, thus, for ensuring that MinC activity is specifically kept away from the cell center (13, 16, 17, 19, 25, 29). In addition, release of MinD from the membrane in this step would simultaneously cause the release of any MinC that might still be associated, for whatever reason, after the first step.
In aggregate, the present and previous work suggest an order of events during half an oscillation cycle of the Min proteins, as depicted in Fig. 12. In vivo, a portion of MinE colocalizes with the MinD “tube” (also called “polar zone”) while a large proportion concentrates in the E ring at the rim of the MinD tube (11, 13, 35, 39). Although MinE is known to form dimers (24), the accumulation of protein in the E ring suggests that it also forms higher-order complexes. Cooperativity in the association of MinE with MinD-membrane complexes is explicitly assumed in one reaction-diffusion model to explain the formation of the E ring, as well as to produce MinD/MinE oscillation patterns in silico that closely mimic those observed in vivo (29). In Fig. 12, we similarly assume that cytoplasmic MinE is not only recruited to the membrane through MinD-MinE interactions but also recruited through MinE-MinE interactions. Further work is needed to test the validity of this specific assumption.
FIG. 12.
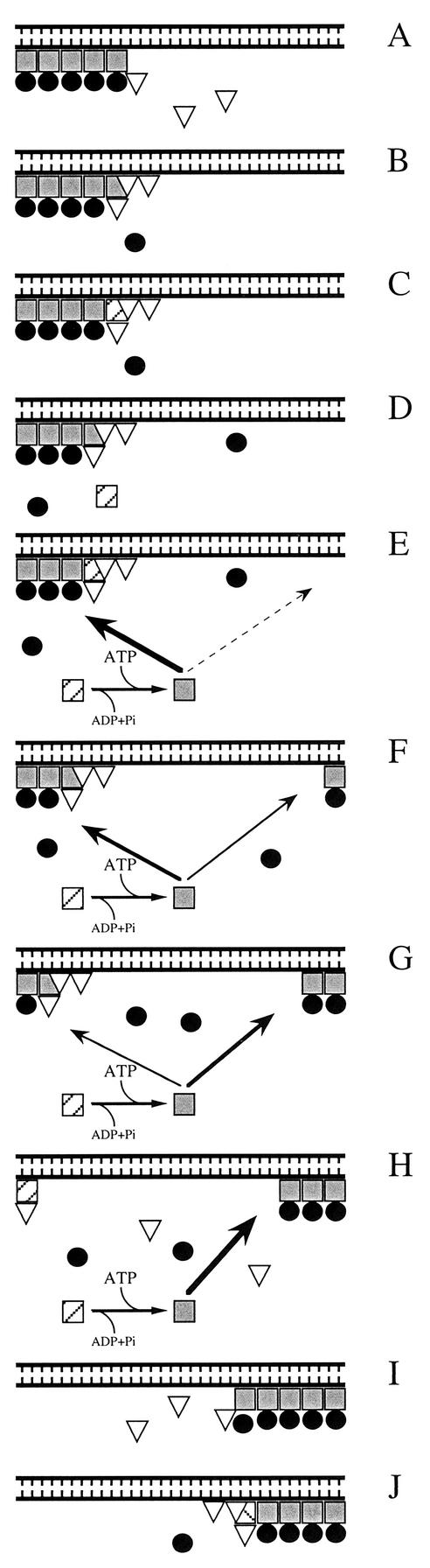
Model of ATP-driven MinCDE dynamics. Proposed order of events during one-half of an oscillation cycle of the Min proteins in E. coli. The model is based on the present and recently reported biochemical evidence (17, 19), as well as on recent cytological (11, 13, 39) and mathematical studies (16, 25, 29). Symbols: ░⃞, MinD.ATP; ▨, MinD.ADP; ▿, MinE; •, MinC. The tooth-combed lines at the top of each panel represent the phospholipid bilayer of the cytoplasmic membrane along the long axis of a cell, and the area underneath represents the cytoplasm. MinD.ATP can bind phospholipid at any site along the membrane. However, the probability it will bind any specific site increases with the amount of MinD.ATP and decreases with the amount of MinE that is already bound at or near this site. Consequently, this probability ranges from high (thick arrow) to low (dashed, thin arrow). See the text for further explanation.
In Fig. 12A, MinD.ATP has cooperatively assembled on the membrane at one cell end and has recruited the MinC division inhibitor. MinD.ATP also attracts MinE, and binding of MinE to a MinC-MinD.ATP complex results in the release of MinC (Fig. 12B). The interaction of MinE with MinD.ATP stimulates nucleotide hydrolysis (Fig. 12C), leading to dissociation of the complex. Upon dissociation, MinE binds an adjacent MinD.ATP molecule on the membrane while MinD.ADP is released into the cytoplasm (Fig. 12D). Cytoplasmic MinD.ADP exchanges the nucleotide for ATP and regains affinity for the phospholipid bilayer (Fig. 12E to H). Regenerated MinD.ATP can bind anywhere on the membrane. However, because of the cooperative nature of the interaction of MinD.ATP with membrane, the existing MinD.ATP tube effectively competes with the rest of the membrane for MinD.ATP subunits. In addition, the strength of this competition is inversely related to the distance from the exsisting tube (25, 29). Thus, the cytoplasmic MinD.ATP molecule in Fig. 12E is most likely to reassociate with the existing MinD assembly on the membrane. However, if it diffuses away sufficiently far before nucleotide exchange occurs and reassociates with the membrane at the opposite end of the cell (Fig. 12F), there is a small but significant probability that it will initiate the assembly of a second tube by capturing additional MinD.ATP, as well as MinC, from the cytoplasm (Fig. 12G). Since most of MinE is still occupied with dissassembly of the old tube, the new MinD tube grows at the expense of the old one until the latter is nearly completely disassembled (Fig. 12F to H). At this point the E ring collapses (Fig. 12H), and the released MinE reassembles on the rim of the new tube to start the disassembly process in the other half of the cell (Fig. 12I and J).
Acknowledgments
We thank Cynthia Hale for technical support, Anu Salvekar and Jay Johnson for help in strain construction, Mike Strainic and Pieter de Haseth for help with fluorometry, and Hans Meinhardt for helpful discussions.
This work was supported by NIH grant GM-57059. In addition, L.L.L. and D.M.R. were supported by NIH NRSA Institutional Training Grant T32GM08056.
REFERENCES
- 1.Adler, H. I., W. D. Fisher, A. Cohen, and A. A. Hardigree. 1967. Miniature Escherichia coli cells deficient in DNA. Proc. Natl. Acad. Sci. USA 57:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 99:6316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for colocalization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 4.Corbin, B. D., X. C. Yu, and W. Margolin. 2002. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 21:1998-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordell, S. C., R. E. Anderson, and J. Lowe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 7.de Boer, P. A. J., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1990. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl. Acad. Sci. USA 87:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 10.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, X., Y. L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hale, C. A., and P. A. J. de Boer. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale, C. A., H. Meinhardt, and P. A. J. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in E. coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale, C. A., A. C. Rhee, and P. A. J. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi, I., T. Oyama, and K. Morikawa. 2001. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard, M., A. D. Rutenberg, and S. de Vet. 2001. Dynamic compartmentalization of bacteria: accurate division in E. coli. Phys. Rev. Lett. 87:27810-27812. [DOI] [PubMed] [Google Scholar]
- 17.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99:6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli: spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 20.Hu, Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82-90. [DOI] [PubMed] [Google Scholar]
- 21.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, J., C. Cao, and J. Lutkenhaus. 1996. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, J. E., L. L. Lackner, and P. A. J. de Boer. 2002. Targeting of DMinC/MinD and DMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ-rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, G. F., Y. L. Shih, M. W. Maciejewski, N. P. Bains, B. Pan, S. L. Rowland, G. P. Mullen, and L. I. Rothfield. 2000. Structural basis for the topological specificity function of MinE. Nat. Struct. Biol. 7:1013-1017. [DOI] [PubMed] [Google Scholar]
- 25.Kruse, K. 2002. A dynamic model for determining the middle of Escherichia coli. Biophys. J. 82:618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 27.Marston, A. L., and J. Errington. 1999. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol. 33:84-96. [DOI] [PubMed] [Google Scholar]
- 28.Matthews, J. C. 1993. Fundamentals of receptor, enzyme, and transport kinetics. CRC Press, Inc., Boca Raton, Fla.
- 29.Meinhardt, H., and P. A. J. de Boer. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder, E., and C. L. Woldringh. 1989. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J. Bacteriol. 171:4303-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pichoff, S., B. Vollrath, C. Touriol, and J.-P. Bouché. 1995. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol. 18:321-329. [DOI] [PubMed] [Google Scholar]
- 34.Raskin, D. M., and P. A. J. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raskin, D. M., and P. A. J. de Boer. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91:685-694. [DOI] [PubMed] [Google Scholar]
- 36.Raskin, D. M., and P. A. J. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothfield, L., S. Justice, and J. García-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 38.Rowland, S. L., X. Fu, M. A. Sayed, Y. Zhang, W. R. Cook, and L. I. Rothfield. 2000. Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol. 182:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih, Y. L., X. Fu, G. F. King, T. Le, and L. Rothfield. 2002. Division site placement in E. coli: mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J. 21:3347-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, Q., and W. Margolin. 2001. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol. 183:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun, Q., X.-C. Yu, and W. Margolin. 1998. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol. Microbiol. 29:491-503. [DOI] [PubMed] [Google Scholar]
- 42.Szeto, J., S. Ramirez-Arcos, C. Raymond, L. D. Hicks, C. M. Kay, and J. A. Dillon. 2001. Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol. 183:6253-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szeto, T. H., S. L. Rowland, and G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183:6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teather, R. M., J. F. Collins, and W. D. Donachie. 1974. Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J. Bacteriol. 118:407-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, X.-C., and W. Margolin. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315-326. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., S. Rowland, G. King, E. Braswell, and L. Rothfield. 1998. The relationship between hetero-oligomer formation and function of the topological specificity domain of the Escherichia coli MinE protein. Mol. Microbiol. 30:265-273. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, C.-R., P. A. J. de Boer, and L. I. Rothfield. 1995. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. USA 92:4314-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]