Abstract
Bordetella pertussis and Bordetella bronchiseptica, gram-negative respiratory pathogens of mammals, possess a heme iron utilization system encoded by the bhuRSTUV genes. Preliminary evidence suggested that expression of the BhuR heme receptor was stimulated by the presence of heme under iron-limiting conditions. The hurIR (heme uptake regulator) genes were previously identified upstream of the bhuRSTUV gene cluster and are predicted to encode homologs of members of the iron starvation subfamily of extracytoplasmic function (ECF) regulators. In this study, B. pertussis and B. bronchiseptica ΔhurI mutants, predicted to lack an ECF σ factor, were constructed and found to be deficient in the utilization of hemin and hemoglobin. Genetic complementation of ΔhurI strains with plasmid-borne hurI restored wild-type levels of heme utilization. B. bronchiseptica ΔhurI mutant BRM23 was defective in heme-responsive production of the BhuR heme receptor; hurI in trans restored heme-inducible BhuR expression to the mutant and resulted in BhuR overproduction. Transcriptional analyses with bhuR-lacZ fusion plasmids confirmed that bhuR transcription was activated in iron-starved cells in response to heme compounds. Heme-responsive bhuR transcription was not observed in mutant BRM23, indicating that hurI is required for positive regulation of bhu gene expression. Furthermore, bhuR was required for heme-inducible bhu gene activation, supporting the hypothesis that positive regulation of bhuRSTUV occurs by a surface signaling mechanism involving the heme-iron receptor BhuR.
To overcome iron deficiency in the host environment, bacterial pathogens have evolved high-affinity iron uptake mechanisms (58). Numerous bacterial species utilize siderophores that scavenge extracellular iron and also remove transferrin- and lactoferrin-bound iron (39, 52). Specific cell surface receptors that allow direct removal of iron from host proteins, including transferrin, lactoferrin and hemoproteins, are also produced by some bacteria (27, 46, 62, 71).
The Fur (ferric uptake regulator) protein, with ferrous iron as corepressor, represses transcription of iron uptake genes when intracellular iron concentrations are sufficient (5). In some cases, Fur derepression under iron-limiting conditions is the only requirement for expression of iron-regulated genes; however, certain iron acquisition systems also require positive regulation for full expression of iron transport functions. Three classes of positive regulators controlling expression of iron uptake genes in gram-negative bacteria have been described and include AraC-like regulators (8, 25, 30, 57), two-component signal transduction systems (19, 60), and extracytoplasmic function (ECF) σ factors (2, 10, 37). These regulators activate transcription of iron transport genes in response to iron starvation and the presence of the cognate iron compound.
The Escherichia coli ferric citrate uptake (Fec) system is a well-characterized example of an iron transport system that is positively regulated by an ECF σ factor. Transcriptional activation of fec genes involves signaling through the FecA outer membrane receptor to the FecR cytoplasmic membrane protein (23, 35). The activity of the ECF σ factor FecI is modulated by an undefined mechanism through direct physical interaction with FecR (23, 45), so that FecI-dependent transcription of fec genes occurs only under iron starvation conditions in the presence of the cognate inducer ferric citrate (10).
Host hemoproteins represent an abundant pool of iron (56) that is utilized by some bacterial pathogens. Heme iron utilization systems have been described for a number of gram-negative bacteria, including Serratia marcescens (40), Neisseria spp. (42, 67), Pseudomonas aeruginosa (41, 55), Yersinia spp. (66, 69), and Shigella dysenteriae (48). The most commonly described system consists of a heme-specific, TonB-dependent outer membrane receptor, a periplasmic heme-binding protein, and an ATP-binding cassette permease which delivers heme to the cytoplasm (27, 71). This type of heme uptake system is negatively regulated at the transcriptional level by iron through the Fur repressor (40, 48, 55, 65, 69).
Bordetella pertussis and Bordetella bronchiseptica are closely related gram-negative respiratory pathogens of mammals capable of acquiring iron through production and utilization of the native siderophore alcaligin (13, 50), by utilization of nonnative siderophores (xenosiderophores) such as enterobactin (7) and ferrichrome (6), and by utilization of host heme compounds (1, 53). We have shown that the bhuRSTUV gene cluster encodes functions required for heme iron utilization in B. pertussis and B. bronchiseptica (70) (Fig. 1). The bhuR gene encodes the outer membrane heme receptor, and the other deduced components of the heme system consist of a putative heme utilization factor (BhuS), the periplasmic heme-binding protein (BhuT), a cytoplasmic membrane permease protein (BhuU), and an ATP-binding protein (BhuV). Open reading frames predicted to encode an ECF σ factor, HurI, and cytoplasmic membrane regulator, HurR, were identified immediately upstream of the B. pertussis and B. bronchiseptica bhuRSTUV genes (70). The more distantly related avian respiratory pathogen Bordetella avium possesses an apparently orthologous heme utilization system encoded by the rhuIR and bhuRSTUV genes (51), and expression of bhuR was shown to be activated by RhuI, a HurI homolog (36).
FIG. 1.
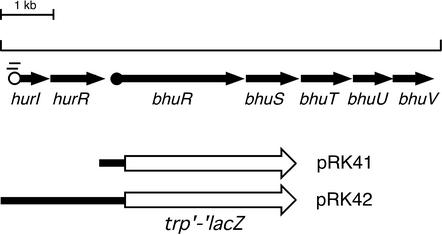
Features of Bordetella hur-bhu genetic system and bhuR-lacZ transcriptional fusions. The bhu genes encode a heme utilization system in B. pertussis and B. bronchiseptica (70). The arrows represent the spatial limits and direction of transcription of genes. The open circle represents the predicted σ70-dependent hurI promoter, and vertical bars indicate predicted overlapping Fur-binding sites. The solid circle upstream of bhuR indicates the location of sequences similar to ECF σ factor −10 and −35 elements predicted to comprise a HurI-dependent promoter. Transcriptional bhuR-lacZ and hurIR bhuR-lacZ fusions were constructed and maintained on low-copy plasmids pRK41 and pRK42, respectively. Solid bars represent portions of the hur-bhu genetic region, and the open arrow represents the promoterless trp′-′lacZ gene.
The similarity of the B. pertussis and B. bronchiseptica HurI and HurR proteins to the ECF regulators FecI and FecR of E. coli, as well as the observation that production of the BhuR outer membrane heme receptor was heme responsive, led to the hypothesis that the bhu genes are positively regulated in response to heme compounds (70). In the present study, we show that the B. pertussis hurI gene is required for optimal heme-iron utilization as well as heme-responsive bhuR transcription and BhuR protein production. Furthermore, we found that bhuR is required for heme-inducible bhuR transcription. These results, along with similarities between the Bordetella heme utilization systems and the E. coli Fec system, support a model of iron-repressible, heme-inducible bhu gene transcription.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bordetella strains and recombinant plasmids are listed in Table 1. Plasmid vector pGEM3Z (Promega, Madison, Wis.) was used in routine cloning procedures, and suicide plasmid vector pSS1129 (64) was used to construct mutant Bordetella strains. Broad-host-range plasmids pRK415 (≈5 to 8 copies/cell) (34) and pBBR1MCS (≈30 copies/cell) (3, 38) were used in the construction of plasmids for complementation of Bordetella mutant strains or to carry reporter gene fusions.
TABLE 1.
Bordetella strains and recombinant plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| UT25Sml | B. pertussis; spontaneous streptomycin-resistant derivative of wild-type strain UT25 | 12 |
| PM8 | B. pertussis UT25Sml ΔhurI | This study |
| B013N | B. bronchiseptica; spontaneous nalidixic acid-resistant derivative of wild-type B013 | 4 |
| BRM21 | B013N bhuRΩp3Z77; BhuR−; heme utilization deficient; Kanr Ampr | 70 |
| BRM23 | B. bronchiseptica B013N ΔhurI | This study |
| pRK37 | pRK415 carrying 1.3-kb B. pertussis UT25 hurI gene and predicted promoter region; Tetr | This study |
| pRK40 | pRK415 with 3.3-kb EcoRI-HindIII trp′-′lacZ insert fragment; Tetr | This study |
| pRK41 | pRK40 with 0.5-kb B. pertussis UT25 ′hurR bhuR′ insert fragment; bhuR-lacZ transcriptional fusion; Tetr | This study |
| pRK42 | pRK40 with 2.1-kb B. pertussis UT25 hurIR bhuR′ insert fragment; hurIR bhuR-lacZ transcriptional fusion; Tetr | This study |
| pBB29 | pBBR1MCS with 8-kb XhoI-SalI fragment containing B. pertussis UT25 ′hurIR bhuRSTUV; Chlr | This study |
| pBB33 | pBBR1MCS carrying 1.3-kb BamHI-HindIII B. pertussis UT25 hurI fragment subcloned from pRK37; Chlr | This study |
| pBB34 | pBBR1MCS with 4.3-kb HindIII-KpnI fragment containing B. pertussis UT25 ′hurIR bhuRS′; Chlr | This study |
Bacterial culture conditions.
E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates. B. pertussis and B. bronchiseptica strains were cultured at 37°C on Bordet Gengou (BG) agar (9) and LB agar, respectively. Stainer-Scholte (SS) broth (63), modified as described previously (61), was used for growth of Bordetella strains in defined liquid medium. Iron-depleted SS medium was prepared by treatment with Chelex100 (Bio-Rad, Richmond, Calif.) as described previously (4) and contained no iron supplements; iron-replete SS broth contained 36 μM FeSO4. Stock solutions of bovine hemin chloride (Sigma, St. Louis, Mo.) and human hemoglobin (Sigma) were prepared as described previously (70) and added to liquid cultures at final concentrations of 5 μM and 1.25 μM, respectively, unless otherwise indicated. The media used to culture B. bronchiseptica and B. pertussis for growth stimulation bioassays were LB agar and modified LB agar (Pertussis LB), respectively (70). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml; and tetracycline, 15 μg/ml (for B. bronchiseptica and E. coli) or 10 μg/ml (for B. pertussis).
Genetic methods.
General cloning procedures were performed with host strain E. coli DH5α (Invitrogen, Carlsbad, Calif.). Triparental matings for the transfer of plasmids from DH5α donors to Bordetella recipients have been described previously (12); mobilization functions were provided by DH5α harboring plasmid pRK2013 (26).
The hurI nucleotide sequence of B. pertussis UT25 was determined on both DNA strands. Nucleotide sequencing was performed by the Advanced Genetic Analysis Center at the University of Minnesota. Oligonucleotide primers were synthesized by Invitrogen or Integrated DNA Technologies (Coralville, Iowa). Nucleotide and protein sequence data were analyzed as described previously (70). Other nucleotide sequence data were from the B. pertussis Tohama I or B. bronchiseptica RB50 genome sequences produced by the Bordetella Sequencing Groups (http://www.sanger.ac.uk/Projects/B_pertussis and http://www.sanger.ac.uk/Projects/B_bronchiseptica) at the Sanger Centre. Servers from the Sanger Centre and the National Center for Biotechnology Information (NCBI) at the National Library of Medicine were used for database searches.
Southern hybridization analysis of B. pertussis genomic DNA from wild-type and candidate hurI mutant strains was performed as described previously (59). The hurI-specific DNA hybridization probe was radiolabeled with [α-32P]dCTP (ICN Radiochemicals, Irvine, Calif.) by the random priming method (24) with the Random Primers DNA labeling system (Invitrogen).
PCR was used to amplify B. pertussis DNA regions, including the hurI gene, the presumptive bhuR promoter region (GenBank accession number AY032627) identified by similarity to other ECF σ factor promoters (22, 49), and the hurIR bhuR′ DNA fragment. PCR primers included appropriate adapters containing restriction enzyme sites to facilitate cloning. PCRs were carried out with Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.) essentially as described previously (11). The DNA template for all PCR amplifications was 300 ng of cosmid pCPbhu1 DNA (70), containing the entire B. pertussis hurIR bhuRSTUV genetic region.
The promoterless lacZ gene used to construct bhuR-lacZ transcriptional fusions was derived from mini-Tn5 lacZ1 (20). The trp′-′lacZ fragment was cloned as a 3.3-kb EcoRI-HindIII DNA fragment to the broad-host-range plasmid pRK415, resulting in plasmid pRK40 (Table 1). A 0.5-kb fragment containing the predicted bhuR promoter region, spanning the 3′ end of hurR, the hurR-bhuR intergenic region, and the 5′ bhuR region, was PCR amplified with primers containing EcoRI adapter ends. This 0.5-kb bhuR promoter region was cloned to the EcoRI site upstream of the trp′-′lacZ cassette in pRK40 to create the bhuR-lacZ transcriptional fusion plasmid pRK41. A 2.1-kb fragment containing hurIR and the 5′ bhuR region was PCR amplified with primers containing EcoRI adapters and similarly cloned to plasmid pRK40 to construct hurIR bhuR-lacZ fusion plasmid pRK42. The same antisense bhuR primer was used in PCRs to amplify the 0.5-kb bhuR promoter and the 2.1-kb hurIR bhuR′ fragment used for construction of reporter plasmids pRK41 and pRK42, respectively, so that the bhuR-lacZ fusion junctions were identical.
Construction of Bordetella ΔhurI mutant strains.
An in-frame deletion of 297 bp of hurI coding sequence was constructed by PCR overlap extension as described previously (11, 32). Sequences flanking the desired hurI deletion were PCR amplified with primer pair Hur1 (5′-GGGGGATCCGTTCGCGCTCACAATGTC-3′) and Hur2 (5′-GAACGCCTGGCGCACTTTTTGCAGCCAGCCGTGATG-3′) and primer pair Hur3 (5′-CATCACGGCTGGCTGCAAAAAGTGCGCCAGGCGTTC-3′) and Hur4 (5′-GGGGAATTCGGTCGGCTTCGAGGTAGA-3′). The Hur2 and Hur3 primers were designed so that the Hur1 and Hur2 and the Hur3 and Hur4 PCR products had complementary 21-nucleotide overlaps at the termini (spanning the region to be deleted). These PCR products were combined at a 1:1 molar ratio, denatured, and annealed, and the ΔhurI fragment was amplified with the outside primers Hur1 and Hur4 which carried restriction enzyme site adapters for cloning of the PCR product to allelic exchange vector pSS1129. The resulting plasmid, pSS9, was mated to B. pertussis strain UT25Sm1, and the ΔhurI mutation was transferred to the B. pertussis chromosome by homologous recombination as described by Stibitz (64) to produce ΔhurI mutant PM8. Allelic exchange was verified by PCR and by Southern hybridization analysis with a hurI-specific DNA probe (data not shown).
To construct a B. bronchiseptica ΔhurI mutant by allelic exchange, plasmid pSS9 was delivered to B. bronchiseptica B013N by electroporation, and plasmid integrants were selected based on ampicillin and gentamicin resistance. These integrants were passaged without antibiotic selection for several generations to allow the second crossover needed for allelic exchange. Passaged cells were plated for isolated colonies and then replicated onto LB agar and LB agar containing gentamicin to score for loss of pSS9-derived gentamicin resistance. Gentamicin-sensitive strain BRM23 was determined by PCR analysis to contain the correct ΔhurI mutation (data not shown).
Hemin and hemoglobin growth stimulation bioassays.
Growth stimulation bioassays of hemin and hemoglobin utilization by B. pertussis and B. bronchiseptica strains were performed as described previously (13, 70). Images of bioassay plates were obtained with an AGFA Arcus II flatbed scanner.
Heme-responsive BhuR protein production.
B. bronchiseptica strains B013N (wild type), BRM21 (bhuRΩp3Z77) (70), BRM23 (ΔhurI), and BRM23 (pRK37) (ΔhurI/hurI+) were grown on LB agar for 24 h and inoculated to iron-replete SS broth (with tetracycline at a final concentration of 15 μg/ml for BRM23[pRK37]). Cells were grown for 24 h with shaking and then subcultured 1:200 to one iron-replete and two iron-depleted SS cultures. After 18 h of growth, hemin was added to one iron-depleted culture at a final concentration of 1.25 μM, and all cultures were grown for 4 additional h. Cells were harvested, and the total membrane fractions were prepared and analyzed as described previously (33). Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels and visualized by Coomassie blue staining.
β-Galactosidase assays.
B. bronchiseptica strains carrying low-copy-number bhuR-lacZ reporter plasmid pRK41 or pRK42 or control plasmid vector pRK40 were grown in iron-replete SS medium and then subcultured as described for BhuR protein production. Eighteen hours after subculture, hemin was added to one iron-depleted culture at a final concentration of 5 μM. The iron-replete and both iron-depleted cultures were grown for 8 additional h and assayed for β-galactosidase activity by the method of Miller (47), as modified by Brickman and coworkers (15). β-Galactosidase activities represent the means of triplicate assays (n = 3 ± 1 standard deviation). The results reported are representative of at least two separate experimental trials.
Nucleotide sequence accession number.
The GenBank accession number for the B. pertussis UT25 hurI gene is AF508979.
RESULTS
Determination of B. pertussis UT25 hurI nucleotide sequence.
To analyze hurI and its role in bhu gene regulation and to examine potential differences between the sequenced B. pertussis Tohama I strain and our clinical isolate UT25, the nucleotide sequence of the UT25 hurI gene was determined. The UT25 hurI open reading frame encodes a 169-amino-acid protein with a molecular mass of 19.3 kDa. A TBLASTN search at the NCBI database confirmed that the HurI protein is similar to members of the ECF family of σ factors. The UT25 HurI protein displays 41% and 46% similarity to the FecI and PupI σ factors of E. coli and Pseudomonas putida, respectively (GenBank accession numbers: FecI, M63115; PupI, X77918). The B. pertussis UT25 hurI coding sequence is 100% identical to that of B. pertussis Tohama I hurI, 99% identical to the B. bronchiseptica RB50 hurI gene, and 72% identical to the B. avium rhuI gene (GenBank accession number AY095952). B. pertussis and B. bronchiseptica HurI amino acid sequences are 100% identical, while the B. avium RhuI protein shows 69% similarity to HurI.
Upstream of the UT25 hurI gene, putative −10 (5′-TAAAAT-3′) and −35 (5′-TTGCAT-3′) regions similar to the E. coli σ70 promoter consensus sequence (29) were identified. Overlapping inverted repeat sequences with 67% and 73% identity to the consensus Fur-binding sequence (16, 21) were found in the predicted −10 region. This observation is consistent with previous experiments that demonstrated strong functional Fur binding in this region (70), suggesting that hurI expression is iron regulated.
Construction and phenotypic analysis of Bordetella hurI mutants.
To determine the role of hurI in heme utilization, ΔhurI mutant strains PM8 (B. pertussis) and BRM23 (B. bronchiseptica) were constructed. An in-frame deletion derivative of the B. pertussis hurI gene which lacked 297 bp internal to the hurI coding sequence was constructed by PCR overlap extension. The resulting altered HurI protein would lack a 99-amino-acid sequence that includes σ factor regions predicted to be involved in core RNA polymerase binding and −10 promoter sequence recognition (44). Bordetella ΔhurI mutants PM8 and BRM23 were tested for the ability to use hemin and hemoglobin in growth stimulation bioassays.
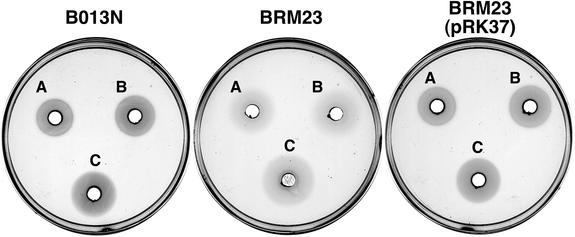
The wild-type B. bronchiseptica strain, B013N, was capable of utilizing both hemin and hemoglobin as iron sources, exhibiting well-defined, dense zones of growth surrounding wells containing either compound (Fig. 2). In contrast, isogenic B. bronchiseptica ΔhurI mutant BRM23 was significantly less proficient in utilization of hemin and hemoglobin, as demonstrated by weak, diffuse zones of growth. Genetic complementation of BRM23 with the hurI+ plasmid pRK37 restored a wild-type heme utilization phenotype (Fig. 2), indicating that the BRM23 heme utilization defect was due to the mutation in hurI. BRM23 harboring plasmid vector control pRK415 remained defective in heme utilization (data not shown). A similar defect in heme utilization was observed in B. pertussis ΔhurI mutant strain PM8, and the mutant phenotype was restored to wild type by hurI+ plasmid pRK37 (data not shown). These bioassay results indicate that hurI encodes a product that is required for optimal utilization of heme iron compounds by B. bronchiseptica and B. pertussis. The phenotypes of the ΔhurI mutant strains are consistent with the hypothesis that hurI encodes a positive regulator which functions to increase expression of heme transport genes.
FIG. 2.
Heme utilization by B. bronchiseptica strains. Growth stimulation bioassays with solutions of bovine hemin chloride or human hemoglobin added to wells: A, 50 μM hemin; B, 100 μM hemin; C, 25 μM hemoglobin. Strains: B013N, wild-type B. bronchiseptica parent strain; BRM23, ΔhurI; BRM23(pRK37), BRM23 carrying plasmid-borne hurI.
Analysis of hurI-dependent BhuR production.
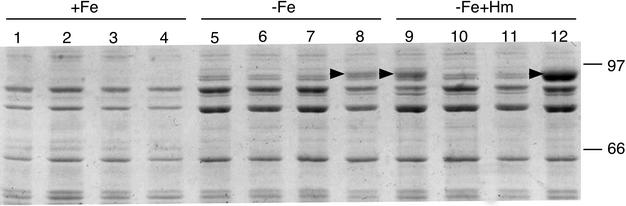
Heme-responsive BhuR protein production was previously observed in wild-type B. bronchiseptica strain B013N (70). To determine if this increased BhuR production in response to hemin is mediated by hurI, BhuR levels in total membrane fractions from wild-type and mutant strains were examined by SDS-PAGE after growth in hemin-containing media. B. bronchiseptica strains B013N (wild type), BRM21 (bhuR), BRM23 (ΔhurI), and BRM23(pRK37) (ΔhurI/hurI+) were grown in iron-replete SS medium or iron-depleted SS medium with or without hemin supplementation. None of the cells grown in iron-replete medium appeared to produce proteins of the predicted 90-kDa molecular mass of BhuR (Fig. 3). Cells grown in iron-depleted medium without hemin supplementation produced multiple iron-regulated membrane proteins in the 80- to 97-kDa molecular mass range, making resolution of the BhuR protein difficult. However, BhuR production was observed in BRM23(pRK37) cells carrying the hurI gene on a low-copy-number plasmid (Fig. 3, lane 8).
FIG. 3.
hurI-dependent, heme-responsive BhuR protein production. Total envelope proteins from B. bronchiseptica cell fractions were separated by SDS-PAGE. Arrows indicate the migration position of the BhuR protein. Samples are grouped according to the culture conditions used: +Fe, iron-replete; −Fe, iron-depleted; −Fe+Hm, iron-depleted with hemin added to a 1.25 μM final concentration after 18 h of growth. Strains: B013N, wild-type parent strain (lanes 1, 5, and 9); BRM21, bhuR (lanes 2, 6, and 10); BRM23, ΔhurI (lanes 3, 7, and 11); BRM23(pRK37), BRM23 carrying plasmid-borne hurI (lanes 4, 8, and 12).
Wild-type B. bronchiseptica B013N showed a pattern of BhuR production consistent with previous results (70): BhuR was present in low abundance in the membrane fractions of iron-starved wild-type cells (Fig. 3, lane 5), while iron-starved cells exposed to hemin produced significant levels of BhuR (Fig. 3, lane 9). As expected, the bhuR mutant BRM21 failed to produce BhuR protein under any growth condition tested (Fig. 3, lanes 2, 6, and 10). Similarly, BhuR was not detected in membrane fractions of the ΔhurI mutant strain BRM23 (Fig. 3, lanes 3, 7, and 11). In contrast, BRM23 cells carrying the hurI+ plasmid pRK37 produced some BhuR under low-iron growth conditions and significantly elevated levels when starved for iron and exposed to hemin (Fig. 3, compare lanes 8 and 12).
These data indicate that production of BhuR is iron repressed, hurI dependent, and heme inducible. Furthermore, the BhuR overproduction phenotype in BRM23(pRK37) cells carrying five to eight copies of hurI is consistent with the proposed function of HurI as a regulator that directs transcription from the bhuR promoter.
bhuR transcription is heme responsive.
The HurI protein is predicted to be an ECF σ factor which directs transcription from a promoter upstream of bhuR when iron-starved cells are exposed to heme compounds. Since the hur-bhu systems of B. pertussis and B. bronchiseptica are nearly identical at the nucleotide sequence level and BhuR production was heme responsive and hurI dependent in B. bronchiseptica (70) (Fig. 3), analyses of B. pertussis bhuR promoter activity were facilitated by using bhuR-lacZ transcriptional fusions (Fig. 1) in the more readily cultivated species B. bronchiseptica.
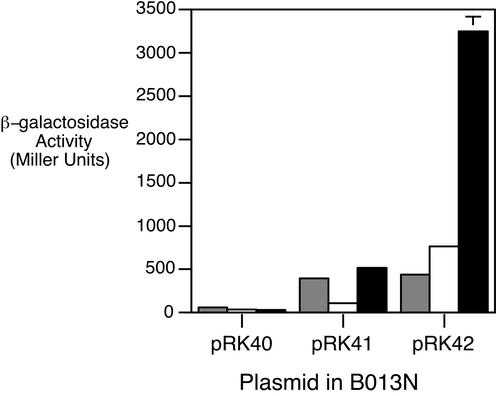
Strains were grown in iron-replete SS medium and iron-depleted SS medium with or without hemin. Wild-type B. bronchiseptica B013N cells carrying the control plasmid vector pRK40 exhibited low levels of β-galactosidase activity under all growth conditions tested (Fig. 4). Iron-replete B013N cells carrying the bhuR-lacZ fusion (pRK41) produced approximately 400 Miller units of β-galactosidase activity, while the bhuR transcriptional activity of the iron-depleted strain was nearly fourfold lower. This pattern of expression is similar to that of the positively regulated Bordetella alcaligin siderophore system: in the absence of alcaligin inducer, iron-starved cultures exhibit lower alcABCDER transcriptional activity than parallel iron-replete cultures (11, 14). However, supplying iron-starved B013N(pRK41) with 5 μM hemin resulted in a fivefold increase in β-galactosidase activity over that of iron-starved cells that were not exposed to hemin. Iron starvation was a prerequisite for heme-inducible bhuR expression, since cells grown in iron-replete SS broth did not exhibit elevated bhuR transcriptional activity in response to hemin (data not shown).
FIG. 4.
Heme-regulated bhuR transcription in wild-type B. bronchiseptica. Bacteria were grown in iron-replete or iron-depleted SS broth with or without hemin and assayed for β-galactosidase activity as described in Materials and Methods. Bars represent β-galactosidase activities of B013N carrying the plasmid vector control (pRK40), bhuR-lacZ (pRK41), or hurIR bhuR-lacZ (pRK42) reporter fusion plasmid. Gray bars, iron-replete; open bars, iron-depleted; solid bars, iron-depleted with hemin supplementation.
The hur and bhu genes are transcribed in the same direction, and some bhu-specific transcripts may actually initiate at the predicted upstream iron-regulated hurI promoter. Although cells carrying the pRK41 bhuR-lacZ fusion plasmid exhibited heme-inducible transcription under iron-limiting conditions, it was reasoned that removal of the bhuR promoter region from the natural genetic context of the hurIR bhuRSTUV gene cluster may alter bhuR transcription. Therefore, another reporter fusion plasmid which carries hurIR bhuR-lacZ (pRK42) (Fig. 1) was constructed and analyzed in wild-type strain B013N (Fig. 4). B013N(pRK42) cells demonstrated iron-repressible and heme-inducible expression of bhuR, which correlated with the results from the BhuR protein analyses (Fig. 3). Iron starvation resulted in a twofold increase in bhuR transcriptional activity compared with that of iron-replete cells. Transcription of bhuR-lacZ was further increased fourfold upon addition of heme to iron-depleted cells. Iron-replete B013N(pRK42) cells did not exhibit enhanced bhuR-lacZ expression in the presence of hemin (data not shown), indicating that both iron starvation and the presence of heme are required for optimal bhuR expression.
Heme-responsive bhuR transcription is hurI dependent.
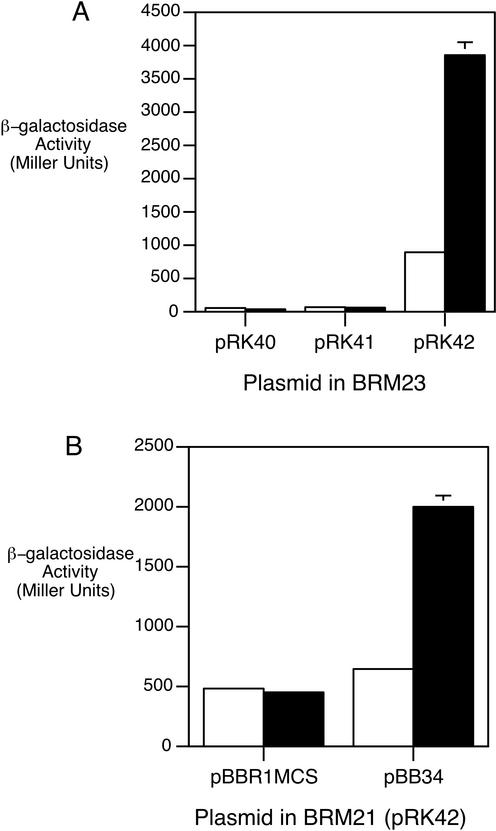
To determine whether heme-responsive bhuR transcription is mediated by the putative ECF σ-factor HurI, bhuR promoter activity was monitored in B. bronchiseptica ΔhurI mutant BRM23. BRM23 cells carrying the plasmid-borne bhuR-lacZ fusion (pRK41) were grown in medium with or without added hemin. Iron-replete cultures of ΔhurI strain BRM23 exhibited levels of bhuR-lacZ transcriptional activity similar to those of wild-type B013N (data not shown). Iron-depleted BRM23(pRK41) cells yielded negligible levels of β-galactosidase activity, and bhuR transcription was not enhanced in response to hemin, indicating that hurI is required for heme-responsive bhuR transcription (Fig. 5A). Complementation of the BRM23 ΔhurI mutant with plasmid pRK42 carrying hurIR in cis to the bhuR-lacZ reporter fusion restored heme-responsive bhuR transcriptional activation: iron-starved BRM23(pRK42) cells exposed to hemin exhibited a fourfold increase in β-galactosidase activity over the level in iron-starved cells grown in the absence of hemin. These results are consistent with the BhuR protein studies (Fig. 3), in which BRM23 complemented with hurI in trans showed increased levels of BhuR production compared with ΔhurI mutant BRM23. These data demonstrate that heme-inducible bhuR transcriptional activation is absolutely dependent on hurI.
FIG. 5.
Heme-responsive bhuR transcription is hurI and bhuR dependent. B. bronchiseptica strains were cultured as described in Materials and Methods. Bars indicate β-galactosidase activities of cultures grown under different conditions: open bars, iron-depleted; solid bars, iron-depleted with hemin supplementation. (A) Strain BRM23 (ΔhurI) carrying plasmid vector control pRK40, bhuR-lacZ reporter plasmid pRK41, or complementing hurIR bhuR-lacZ reporter plasmid pRK42. (B) Strain BRM21 (bhuR) carrying pRK42 and vector control pBBR1MCS or bhuR+ plasmid pBB34.
BhuR is required for heme-responsive bhuR transcriptional activation.
The signaling process leading to transcriptional activation of the fecABCDE genes in E. coli has been shown to require the ferric citrate outer membrane receptor protein FecA (28). Recognition of ferric citrate by FecA results in signaling through its N-terminal domain to the FecR cytoplasmic membrane protein and subsequently to release or activation of the FecI σ factor for transcription of the fec genes. The BhuR N terminus is highly similar to that of FecA (70), suggesting that BhuR may be involved in a similar signaling cascade resulting in transcriptional activation of bhu genes in response to heme inducer.
To examine the effect of a bhuR mutation on the transcriptional responsiveness of the bhuR promoter, bhuR-lacZ transcriptional activity was examined in B. bronchiseptica bhuR mutant strain BRM21 (Fig. 5B). Iron-starved BRM21 cells carrying the hurIR bhuR-lacZ fusion plasmid pRK42 along with a compatible plasmid vector control, pBBR1MCS, failed to show heme-inducible bhuR expression. However, plasmid pBB34 (bhuR+) restored heme-inducible bhuR transcription to BRM21(pRK42), indicating that BhuR is required for heme-responsive transcriptional activation. The chromosomal bhuR mutation of BRM21 results in a defect in heme iron utilization and is also polar on the downstream bhuSTUV genes (70). Heme iron utilization was not fully restored to BRM21 by bhuR+ plasmid pBB34 but was restored to wild-type levels by plasmid pBB29, carrying bhuRSTUV (data not shown). These results indicate that BhuR is involved in the heme-signaling cascade resulting in bhuR transcriptional activation but that transport of heme or bhuSTUV-encoded functions may not be required for this process.
DISCUSSION
To initiate and sustain a productive infection, Bordetella species must acquire iron in the iron-restricted host environment. Production and utilization of the native siderophore alcaligin (13, 50) and utilization of xenosiderophores that may be present on host mucosa may allow B. pertussis and B. bronchiseptica to scavenge iron. Host molecules, including transferrin, lactoferrin, and heme compounds, also represent potential sources of iron that could be accessed during colonization and persistence within a host. The bhu genetic system of B. pertussis and B. bronchiseptica was identified and demonstrated to be required for heme utilization (70).
Heme iron utilization systems similar to the Bordetella bhu system have been described for a number of gram-negative bacterial pathogens, including Vibrio cholerae (54), Yersinia pestis (69), Yersinia enterocolitica (65), and P. aeruginosa (55). The genes encoding heme transport systems in these organisms are negatively regulated by iron through the Fur repressor; however, no apparent Fur-binding activity was detected in the B. pertussis bhuR promoter region (70), which suggested that the mechanism of Bordetella bhu gene regulation may differ from that of other characterized systems. BhuR protein was not produced in substantial amounts under iron-limiting conditions, but it was produced in much greater abundance when iron-starved cultures were exposed to hemin. The identification of the hurIR genes immediately upstream of the bhu gene cluster led to the hypothesis that the bhu system is positively regulated under iron-limiting conditions in the presence of the cognate substrate, heme.
Based upon similarities in gene organization and predicted amino acid sequences between the Bordetella hur-bhu system and the well-characterized E. coli fec system, along with results from the present study, we propose a model for regulation of the Bordetella bhu genes. Under iron starvation conditions, Fur derepression of the promoter upstream of hurI would allow σ70-directed transcription of the hurIR genes and perhaps low-level expression of bhuRSTUV by readthrough transcription. Higher levels of heme-responsive bhuRSTUV transcription are predicted to originate from a HurI-dependent promoter upstream of bhuR. As proposed for FecI and FecR, HurI would remain inactive through its interaction with HurR until the BhuR outer membrane receptor binds heme, transferring the receptor occupancy signal through HurR to the HurI σ factor, allowing it to activate transcription at the HurI-dependent promoter upstream of the bhu genes. In the present study, using two different bhuR-lacZ reporter plasmids, we showed that bhuR expression is enhanced four- to fivefold when iron-starved cells are exposed to heme. Furthermore, the BhuR heme receptor is required for hurI-dependent bhuR transcription, indicating its role in the signaling cascade.
B. avium possesses a gene cluster, rhuIR bhuRSTUV (GenBank accession number AY095952), that is required for heme utilization and is similar to the B. pertussis and B. bronchiseptica hurIR bhuRSTUV gene clusters (51). The rhuI gene is predicted to encode an ECF σ factor which is 69% identical to HurI. B. avium carrying rhuI in multicopy showed increased BhuR production and increased expression of a chromosomal bhuR::phoA fusion under both iron-replete and iron-depleted culture conditions, suggesting that RhuI activates expression of the B. avium bhu genes. Similar to our findings in B. bronchiseptica, expression of a plasmid-borne bhuR-lacZYA fusion in B. avium was enhanced by heme compounds, and this responsiveness was BhuR dependent (36). A requirement for RhuI in heme utilization and heme-dependent bhu transcription in B. avium has not been reported.
B. pertussis and B. bronchiseptica ΔhurI mutants were defective in utilizing heme iron sources in growth stimulation bioassays; the zones of growth were much less turbid and their perimeters were less defined than those of the wild-type parental strains. Perhaps the loss of hurI affects the kinetics of heme uptake or efficiency of heme iron utilization due to the low abundance of BhuR receptor on the cell surface. In the model proposed for bhu gene regulation, low-level HurI-independent bhuR expression in the absence of inducer would be required in order to have some BhuR present at the cell surface to sense the availability of heme in the environment. If this were the case, basal levels of BhuR produced in the absence of bhu gene activation (e.g., in a hurI mutant) may be sufficient to supply modest growth-promoting levels of heme to the cells. HurI-dependent bhu gene activation may be more critical in environments where heme concentrations are much lower and efficient scavenging of heme iron is essential for cell growth.
BhuR expression analysis and bhuR-lacZ reporter fusions demonstrated that heme-responsive bhuR expression is hurI dependent. Cells carrying hurI on low-copy-number plasmids overproduced the BhuR protein and exhibited higher levels of bhuR-lacZ transcription than wild-type cells carrying a single copy of hurI. Differences in levels of bhuR-lacZ expression in iron-replete compared with iron-depleted cultures were correlated with the presence or absence of the hurIR genes carried in cis to the bhuR-lacZ fusion. Iron-starved wild-type cells carrying the hurIR bhuR-lacZ fusion plasmid pRK42 exhibited approximately twofold more β-galactosidase activity than iron-replete cultures. In contrast, iron-starved wild-type cells carrying bhuR-lacZ plasmid pRK41 showed a fourfold reduction in activity compared with iron-replete cultures. It is possible that transcription from the iron-regulated hurI promoter on pRK42 reads through bhuR-lacZ, resulting in higher levels of expression under iron-limiting conditions. Furthermore, alterations in the stoichiometry between the HurI and HurR regulators and bhuR promoter targets could have a major influence on bhuR-lacZ expression.
The loss of heme-responsive bhuR transcription in B. bronchiseptica ΔhurI mutant BRM23 demonstrated that HurI is required for bhuR transcriptional activation. Interestingly, although complementation of BRM23 (bhuR-lacZ) cells with compatible, medium-copy-number (≈30 copies/cell) hurI+ plasmid pBB33 restored iron-regulated bhuR-lacZ expression, transcriptional activation did not occur in response to hemin (data not shown). If HurR acts as an anti-σ factor that sequesters or inactivates HurI in the absence of heme, excessive levels of HurI production in cells carrying pBB33 might overwhelm the negative influence of HurR and thus result in the observed inducer-independent transcription initiation at the bhuR promoter. The pup system of P. putida WCS358 is regulated by the PupI σ factor and PupR regulator in a manner analogous to the E. coli Fec system (37), in that transcription of pseudobactin siderophore uptake genes is pupI dependent and pseudobactin responsive. Similar to the heme-independent expression of bhuR-lacZ in BRM23 carrying ≈30 copies of hurI, P. putida pupI mutants genetically complemented by the pupI gene in multicopy show a pattern of inducer-independent pup gene transcription (37).
Experiments analyzing bhuR-lacZ expression in B. bronchiseptica bhuR mutant BRM21 demonstrated that the BhuR outer membrane receptor itself is required for signaling, resulting in heme-responsive bhu gene transcriptional activation, but that heme uptake may not be required for this process. The transport and signaling processes are also separable in the E. coli Fec system, as evidenced by fecA mutants which retain the ability to transport ferric citrate but no longer respond to its presence (28). The extended N-terminal regions of FecA and the P. putida PupB receptors are critical for inducer-responsive signaling resulting in fec (35) and pup (37) transcriptional activation, respectively. This N-terminal region is unique to outer membrane receptors whose expression is regulated by ECF σ factors (10). The BhuR N terminus is highly similar to the corresponding regions of FecA and PupB (70), suggesting similar functional signaling properties among these receptors.
Innate iron restriction in the mammalian host would be expected to lead to derepression of Fur-regulated promoters, resulting in uninduced basal levels of expression of all Bordetella iron acquisition systems. Siderophore-mediated iron transport may be most critical during early stages of infection, when organisms must colonize an intact mucosal surface which may not provide a ready source of heme iron. However, if colonization is successful, the elaborated Bordetella toxins, including pertussis toxin (for B. pertussis) (68), dermonecrotic toxin (43), tracheal cytotoxin (18), and adenylate cyclase/hemolysin (17, 31), would presumably damage the epithelium and other host cells, providing a source of heme iron for Bordetella cells. Priority regulation of Bordetella iron acquisition systems according to iron source availability may allow faster adaptation to a changing host environment and thus a greater degree of pathogenic success.
Acknowledgments
We thank Timothy Brickman for helpful discussions and critical reading of the manuscript and Mark Anderson for assistance with replica plating of potential Bordetella mutants. We acknowledge Michael Walker for technical assistance.
Support for this study was provided by Public Health Service grants R01 AI-31088 (S.K.A.) and T32 AI-07421 (C.K.V.) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Agiato, L. A., and D. W. Dyer. 1992. Siderophore production and membrane alterations by Bordetella pertussis in response to iron starvation. Infect. Immun. 60:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angerer, A., S. Enz, M. Ochs, and V. Braun. 1995. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. Fecl belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol. Microbiol. 18:163-174. [DOI] [PubMed] [Google Scholar]
- 3.Antoine, R., and C. Locht. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6:1785-1799. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, S. K., and M. O. Clements. 1993. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J. Bacteriol. 175:1144-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 6.Beall, B. 1998. Two iron-regulated putative ferric siderophore receptor genes in Bordetella bronchiseptica and Bordetella pertussis. Res. Microbiol. 149:189-201. [DOI] [PubMed] [Google Scholar]
- 7.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 8.Beaumont, F. C., H. Y. Kang, T. J. Brickman, and S. K. Armstrong. 1998. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J. Bacteriol. 180:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bordet, J., and O. Gengou. 1906. Le microbe de la coqueluche. Ann. Inst. Pasteur (Paris) 20:731-741. [Google Scholar]
- 10.Braun, V. 1997. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch. Microbiol. 167:325-331. [DOI] [PubMed] [Google Scholar]
- 11.Brickman, T. J., and S. K. Armstrong. 2002. Bordetella interspecies allelic variation in AlcR inducer requirements: identification of a critical determinant of AlcR inducer responsiveness and construction of an alcR(Con) mutant allele. J. Bacteriol. 184:1530-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brickman, T. J., and S. K. Armstrong. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J. Bacteriol. 178:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brickman, T. J., J. G. Hansel, M. J. Miller, and S. K. Armstrong. 1996. Purification, spectroscopic analysis and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. Biometals 9:191-203. [DOI] [PubMed] [Google Scholar]
- 14.Brickman, T. J., H. Y. Kang, and S. K. Armstrong. 2001. Transcriptional activation of Bordetella alcaligin siderophore genes requires the AlcR regulator with alcaligin as inducer. J. Bacteriol. 183:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brickman, T. J., B. A. Ozenberger, and M. A. McIntosh. 1990. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212:669-682. [DOI] [PubMed] [Google Scholar]
- 16.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Confer, D. L., and J. W. Eaton. 1982. Phagocyte impotence caused by an invasive bacterial adenylate cyclase. Science 217:948-950. [DOI] [PubMed] [Google Scholar]
- 18.Cookson, B. T., A. N. Tyler, and W. E. Goldman. 1989. Primary structure of the peptidoglycan-derived tracheal cytotoxin of Bordetella pertussis. Biochemistry 28:1744-1749. [DOI] [PubMed] [Google Scholar]
- 19.Dean, C. R., S. Neshat, and K. Poole. 1996. PfeR, an enterobactin-responsive activator of ferric enterobactin receptor gene expression in Pseudomonas aeruginosa. J. Bacteriol. 178:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (Fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enz, S., V. Braun, and J. H. Crosa. 1995. Transcription of the region encoding the ferric dicitrate-transport system in Escherichia coli: similarity between promoters for fecA and for extracytoplasmic function sigma factors. Gene 163:13-18. [DOI] [PubMed] [Google Scholar]
- 23.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 25.Fetherston, J. D., S. W. Bearden, and R. D. Perry. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol. Microbiol. 22:315-325. [DOI] [PubMed] [Google Scholar]
- 26.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genco, C. A., and D. White Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 28.Harle, C., I. Kim, A. Angerer, and V. Braun. 1995. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 14:1430-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinrichs, D. E., and K. Poole. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hewlett, E. L., and V. M. Gordon. 1988. Adenylate cyclase toxin of Bordetella pertussis, p. 193-210. In A. C. Wardlaw and R. Parton (ed.), Pathogenesis and immunity in pertussis. Wiley, New York, N.Y.
- 32.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 33.Kang, H. Y., T. J. Brickman, F. C. Beaumont, and S. K. Armstrong. 1996. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J. Bacteriol. 178:4877-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 35.Kim, I., A. Stiefel, S. Plantor, A. Angerer, and V. Braun. 1997. Transcription induction of the ferric citrate transport genes via the N terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol. Microbiol. 23:333-344. [DOI] [PubMed] [Google Scholar]
- 36.Kirby, A. E., D. J. Metzger, E. R. Murphy, and T. D. Connell. 2001. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect. Immun. 69:6951-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koster, M., W. van Klompenburg, W. Bitter, J. Leong, and P. Weisbeek. 1994. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 13:2805-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop 2nd, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800-802. [PubMed] [Google Scholar]
- 39.Lankford, C. E. 1973. Bacterial assimilation of iron. Crit. Rev. Microbiol. 2:273-331. [Google Scholar]
- 40.Letoffe, S., J. M. Ghigo, and C. Wandersman. 1994. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc. Natl. Acad. Sci. USA 91:9876-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letoffe, S., V. Redeker, and C. Wandersman. 1998. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol. Microbiol. 28:1223-1234. [DOI] [PubMed] [Google Scholar]
- 42.Lewis, L. A., E. Gray, Y. P. Wang, B. A. Roe, and D. W. Dyer. 1997. Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23:737-749. [DOI] [PubMed] [Google Scholar]
- 43.Livey, I., and A. C. Wardlaw. 1984. Production and properties of Bordetella pertussis heat-labile toxin. J. Med. Microbiol. 17:91-103. [DOI] [PubMed] [Google Scholar]
- 44.Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahren, S., S. Enz, and V. Braun. 2002. Functional interaction of region 4 of the extracytoplasmic function sigma factor FecI with the cytoplasmic portion of the FecR transmembrane protein of the Escherichia coli ferric citrate transport system. J. Bacteriol. 184:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mietzner, T. A., and S. A. Morse. 1994. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu. Rev. Nutr. 14:471-493. [DOI] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Mills, M., and S. M. Payne. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 177:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 50.Moore, C. H., L. A. Foster, D. G. Gerbig, D. W. Dyer, and B. W. Gibson. 1995. Identification of alcaligin as the siderophore produced by Bordetella pertussis and B. bronchiseptica. J. Bacteriol. 177:1116-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy, E. R., R. E. Sacco, A. Dickenson, D. J. Metzger, Y. Hu, P. E. Orndorff, and T. D. Connell. 2002. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect. Immun. 70:5390-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 53.Nicholson, M. L., and B. Beall. 1999. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology 145:2453-2461. [DOI] [PubMed] [Google Scholar]
- 54.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 55.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 56.Panter, S. S. 1994. Release of iron from hemoglobin. Methods Enzymol. 231:502-514. [DOI] [PubMed] [Google Scholar]
- 57.Pradel, E., N. Guiso, and C. Locht. 1998. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J. Bacteriol. 180:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J. E., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 60.Schmitt, M. P. 1999. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J. Bacteriol. 181:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider, D. R., and C. D. Parker. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 63.Stainer, D. W., and M. J. Scholte. 1970. A simple chemically defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63:211-220. [DOI] [PubMed] [Google Scholar]
- 64.Stibitz, S. 1994. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 235:458-465. [DOI] [PubMed] [Google Scholar]
- 65.Stojiljkovic, I., and K. Hantke. 1992. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11:4359-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 67.Stojiljkovic, I., V. Hwa, L. de Saint Martin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 68.Tamura, M., K. Nogimori, S. Murai, M. Yajima, K. Ito, T. Katada, M. Ui, and S. Ishii. 1982. Subunit structure of islet-activating protein, pertussis toxin, in conformity with the A-B model. Biochemistry 21:5516-5522. [DOI] [PubMed] [Google Scholar]
- 69.Thompson, J. M., H. A. Jones, and R. D. Perry. 1999. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun. 67:3879-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]