Abstract
Bacterial nitrous oxide (N2O) reductase is the terminal oxidoreductase of a respiratory process that generates dinitrogen from N2O. To attain its functional state, the enzyme is subjected to a maturation process which involves the protein-driven synthesis of a unique copper-sulfur cluster and metallation of the binuclear CuA site in the periplasm. There are seven putative maturation factors, encoded by nosA, nosD, nosF, nosY, nosL, nosX, and sco. We wanted to determine the indispensable proteins by expressing nos genes from Pseudomonas stutzeri in the nondenitrifying organism Pseudomonas putida. An in silico study of denitrifying bacteria revealed that nosL, nosX (or a homologous gene, apbE), and sco, but not nosA, coexist consistently with the N2O reductase structural gene and other maturation genes. Nevertheless, we found that expression of only three maturation factors (periplasmic protein NosD, cytoplasmic NosF ATPase, and the six-helix integral membrane protein NosY) together with nosRZ in trans was sufficient to produce catalytically active holo-N2O reductase in the nondenitrifying background. We suggest that these obligatory factors are required for Cu-S center assembly. Using a mutational approach with P. stutzeri, we also studied NosA, the Cu-containing outer membrane protein previously thought to have Cu insertase function, and ScoP, a putative membrane-anchored chaperone for CuA metallation. Both of these were found to be dispensable elements for N2O reductase biosynthesis. Our experimental and in silico data were integrated in a model of N2O reductase maturation.
Nitrous oxide (N2O) reductase, the nosZ gene product, transforms nitrous oxide to N2 as part of a respiratory mode of bacterial energy conservation. The biosynthesis of the two Cu centers of this enzyme depends on accessory proteins whose exact functions and positions in the NosZ maturation pathway are not known. This is at least in part attributable to the fact that the structure of the catalytic site, CuZ, remained unknown until very recently. This site turned out to be the first example of a biologically active Cu-S cluster in which four solely histidine-liganded Cu atoms are bridged by a sulfide ion (4, 44). How this conjugated metal center is assembled has become one of the most intriguing questions associated with N2O reductase. CuA, the other metal center, serves as the site for electron transfer to CuZ and is a dicysteinyl-bridged binuclear Cu site (5, 28). N2O reductase was among the early examples for which metal processing was recognized as part of overall enzyme biosynthesis. We conceptualized that insertion of Cu into N2O reductase requires the bacterium to handle the toxic metal along a discrete route to prevent the occurrence of free Cu inside the cell (58). This principle is now condensed in the term metallochaperone and has been studied best in eukaryotic Cu delivery systems (for a review see reference 39).
By using random Tn5 mutagenesis, several nos loci for Cu center biosynthesis were detected in Pseudomonas stutzeri (49, 57). Analysis of these loci led to the finding that there is a three-component assembly complex, NosDFY, whose putative ATP/GTPase, NosF, and general arrangement at and on both sides of the cytoplasmic membrane are similar to those of bacterial ABC transporters (12, 58, 59). Inactivation of any protein of this complex leads to an N2O reductase with a low Cu content and no Cu-S site. In particular, NosZ from the nosD promoter mutant MK402 has been studied in detail and has been shown to exhibit only the properties of a CuA protein (44, 45, 56, 58).
An additional maturation component was identified in NosA, a Cu-containing outer membrane protein of P. stutzeri JM300 (30, 31, 37). NosA was thought to be necessary to insert Cu into N2O reductase. The insertase or metallochaperone function was also attributed to the Cu-containing protein NosL, which has the features of a lipoprotein of the outer membrane (18, 34).
In certain denitrifying bacteria the nos gene cluster harbors the nosX gene. The precise role of this gene in NosZ biosynthesis has not been clarified, although it is necessary for N2O utilization by Sinorhizobium meliloti (6) and Paracoccus denitrificans (46). The latter bacterium carries in addition to nosX a homologous gene, nirX, and mutagenesis of both genes is required to generate a Nos− phenotype. Electron paramagnetic resonance hyperfine-coupling characteristics of cell extract from a nosX nirX double mutant indicated that the CuA site is altered or lacking, but the exact molecular defect is unknown. Finally, a putative assembly factor is represented by ScoP, which is a homologue of Sco1 from yeast (14, 51). The yeast factor is necessary for CuA assembly of cytochrome c oxidase (21, 38, 48). Evidence is accumulating that bacterial Sco1 homologues also function as Cu-processing factors (8, 32, 33).
Thus, we currently have a total of seven factors or candidate proteins for NosZ maturation. Given this situation, we wanted to define the minimal set of essential components. To do this, we pursued a strategy to express nosZ of P. stutzeri and its regulatory gene, nosR, together with genes encoding assembly factors in the nondenitrifying organism Pseudomonas putida. By using a heterologous background we intended to differentiate specific nos functions from potential organismal adaptations and to identify those functions for which a rescue pathway may exist. Our experimental data, together with the results of an in silico study, allowed us to formulate an integrated model for NosZ maturation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used in this study are shown in Table 1. Escherichia coli DH10B was grown in Luria-Bertani (LB) medium at 37°C and 240 rpm on a gyratory shaker. Pseudomonads were grown in LB medium at 30°C and 240 rpm for use in recombinant DNA work. For other purposes an asparagine- and citrate-containing medium, AC medium, was used, and this medium was supplemented with 5 μM Cu (11). P. putida was grown in a modified M9 medium containing 20 mM sodium citrate as the carbon source (42). This medium was supplemented with 7.5 μM Fe, 0.1 μM Mo, and 5 μM Cu. When necessary for strain maintenance, antibiotics were added at the following final concentrations: streptomycin, 200 μg/ml of medium; kanamycin, 50 μg/ml; ampicillin, 100 μg/ml (E. coli) or 50 μg/ml (P. putida); and gentamicin, 10 μg/ml (E. coli) or 15 μg/ml (P. putida).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | Cloning host | Gibco BRL |
| P. putida | Former type strain | DSM50906 |
| P. aeruginosa PAO | 25 | |
| P. stutzeri | ATCC 14405 | |
| MK21 | Spontaneous Smr mutant of P. stutzeri | 57 |
| MK404 | MK21 nosD::Tn5 | 59 |
| MK413 | MK21 nosR::Tn5 | 13 |
| MK417 | MK21 nosY::Tn5 | 59 |
| MK498P | MK21 ΔscoP::Kmr | This study |
| MK499A | MK21 nosA::Kmr | This study |
| MK4211(pSZ) | MK21 ΔnosZ::Kmr complemented with nosZ expression vector pSZ | 7 |
| Plasmids | ||
| cDEN1 | Cosmid clone of a Sau3A genomic library in pJA1 carrying P. stutzeri nos genes | 3 |
| c14 | Cosmid clone of a Sau3A genomic library in pJA1 carrying scoP of P. stutzeri | 14 |
| c61 | Cosmid clone carrying nosA of P. stutzeri | This study |
| pBSL15 | Source of Kmr cassette | 1 |
| pNS200 | pBR325 derivative with a 4.2-kb HindIII fragment carrying nosDFY | 59 |
| pUC18 | Cloning vector; Apr | 54 |
| pUC18nosAH | 6-kb HindIII fragment from cosmid c61 carrying nosA cloned into pUCP18 | This study |
| pUCP22 | Cloning vector; Apr Gmr | 53 |
| pUCP22RE | 8.8-kb Eco47III-XbaI fragment with nosRZDFYL and tatE cloned into pUCP22 | 24 |
| pUCP22RL | 8.6-kb Eco47III-XbaI fragment with nosRZDFYL cloned into pUCP22 | 24 |
| pUCP22RY | 8.1-kb Eco47III-XbaI fragment with nosRZDFY cloned into pUCP22 | This study |
| pUCP22RZ | 5.3-kb Eco47III-SmaI fragment with nosRZ cloned into pUCP22 | This study |
| pWM20 | Template for PCR amplification of the nosX probe from S. meliloti | 26 |
DNA techniques.
Standard protocols described previously (24) were used for genomic DNA extraction, plasmid DNA preparation and purification, agarose gel electrophoresis, dephosphorylation, and ligation of DNA. Restriction enzymes were used as recommended by the manufacturers. Transformation was done by electroporation. For Southern blot analysis the alkaline capillary transfer method was used (9). DNA hybridization was done overnight with the gene probes listed in Table 2. Detection was done by using chemiluminescence with the CDP-Star reagent (Roche Biochemicals) combined with the EasyHyb system from the same manufacturer or as described previously (20). Gene probes were prepared in a two-step PCR. The first amplification was done with the templates listed in Table 2. Purified PCR fragments were labeled in a second PCR by using a digoxigenin-labeled nucleotide mixture (Roche Biochemicals).
TABLE 2.
Primers used for PCR amplification and Southern or Northern hybridization
| Probe or gene | Primer | Sequence | Template | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| apbE | Forward | 5′-CCGACCATGGGCAGCAGCTN-3′ | P. putida | 40 | This study |
| Reverse | 5′-ATCGAAGGTGTGCGAATAGN-3′ | ||||
| NA | Forward | 5′-GGCGCCGATGGTCGTTA-3′ | c61 | 56 | This study |
| Reverse | 5′-CCGGCCTTGAGTTCGACA-3′ | ||||
| ND | Forward | 5′-GTATCAAGGCCAGTTCACCA-3′ | pNS200 | 50 | This study |
| Reverse | 5′-TCATCAGGATGCCGTAGTTC-3′ | ||||
| NL | Forward | 5′-ACTGGCCGTGTTGCTCGCTT-3′ | cDEN1 | 45 | This study |
| Reverse | 5′-GCAGCAGCGCCTGATCGATT-3′ | ||||
| NR | Forward | 5′-TTCGAGATGGCGATCTTCACTGC-3′ | cDEN1 | 56 | This study |
| Reverse | 5′-TCAGGGTTCCACCACTTG-3′ | ||||
| NX | Forward | 5′-CGATCGCGTCGTATTCG-3′ | pWM20 | 56 | 26 |
| Reverse | 5′-ATGCGGACAGCCGAACT-3′ | ||||
| NZ | Forward | 5′-GTTGCTGCCACGGCTCTC-3′ | cDEN1 | 55 | This study |
| Reverse | 5′-GTCGGCGTCGGTGTTGTC-3′ | ||||
| OC | Forward | 5′-CGCGCCGACCTCCTACATCT-3′ | P. aeruginosa | 60 | This study |
| Reverse | 5′-GAACACCGCGTCCTTGCTCC-3′ | ||||
| scoP | Forward | 5′-GGACATCTGCCCGACCA-3′ | P. putida | 50 | This study |
| Reverse | 5′-CAGGTTACCGCTGTGATCCA-3′ | ||||
| TatC1 | Forward | 5′-GGTCTGGGGCTTCATCGC-3′ | P. aeruginosa | 57 | 24 |
| Reverse | 5′-CCATGCCGACCACGAAAC-3′ |
RNA analysis.
Cells were grown under denitrifying conditions with 5 μM Cu or without added Cu (the concentration of adventitious Cu in the medium was ≤0.7 μM). RNA was isolated from fresh cells with a total RNA extraction kit (Roche Diagnostics) or was extracted by the hot phenol method from cells frozen in liquid nitrogen (52). Northern blotting was performed as described previously (52). Hybridization and detection of digoxigenin-labeled probes were carried out according to the instructions of the EasyHyb system (Roche Diagnostics).
Cloning and sequencing of nosA and scoP.
A P. stutzeri cosmid library (3) was screened for an oprC homologue by Southern hybridization with probe OC at 55°C. An approximately 6-kb HindIII fragment was cloned into pUC18 with E. coli DH10B as the host, generating pUC18nosAH. A stretch of ca. 2.9 kb was sequenced on both strands by primer walking by using a dye terminator kit (Amersham Pharmacia Biotech) with an ALFexpress sequencer according to instructions of the manufacturer. The previously described partial sequence of scoP was completed. The gene was designated orf193; it is located on cosmid c14 downstream of fnrA (14).
Mutagenesis of nosA and scoP.
nosA of MK21 was inactivated by insertion of a kanamycin resistance cassette, Kmr, into the single XhoI restriction site in the orientation opposite that of nosA, generating strain MK499A. The mutation was verified by Southern hybridization with the nosA probe NA, as well as by transcriptional analysis. scoP was inactivated by replacing an internal 160-bp XcmI-BsaAI fragment with a Kmr cassette in the same orientation, generating strain MK498P. The mutation was verified by Southern blot analysis.
Construction of nosZ expression vectors.
Vector pUCP22RZ was constructed by cloning the nosRZD′-carrying Eco47III-SmaI fragment into Ecl136II- and SmaI-digested pUCP22. The pUCP replicon is functional in pseudomonads. pUCP22RY was assembled as described previously for pUCP22RL and pUCP22RE (24). The forward primer, 5′-ACGTGCGCAGATCAGCAATAACC-3′, was located 34 bp upstream of the SmaI site in nosD that was used for construction of pUCP22RZ. The reverse primer, 5′-GTACTATCTAGACCGCACACGTGACACTCG-3′ (nucleotides 129 to 146 of nosL), was designed to add an XbaI site (underlined) 176 bp downstream of the nosY stop codon. The SmaI- and XbaI-digested PCR product was ligated into pUCP22RZ that was digested with the same restriction enzymes, resulting in plasmid pUCP22RY(nosRZDFY). Due to primer design this vector also encoded the 49 N-terminal amino acids of the 191 amino acids of NosL. This fragment did not include the cysteine and histidine residues for Cu binding (34). The P. putida expression strains harboring plasmids pUCP22RZ, pUCP22RY, pUCP22RL, and pUCP22RE were designated RZ, RY, RL, and RE, respectively.
Cell extract, cell fractionation, gel electrophoresis, and enzyme detection.
The size of the inoculant in aerobic LB medium was increased from 3 to 20 ml. Cells were grown by using a two-phase growth mode in 100 ml of minimal medium in a 300-ml flask. The initial optical density at 660 nm was 0.03. The aerobic growth phase (shaking speed, 240 rpm) was changed to O2-limited conditions when the optical density reached approximately 0.3. The shaking speed was reduced to 120 rpm, and sodium nitrate was added to a final concentration of 0.1%. After incubation overnight cells were harvested under cold conditions at 5,000 × g. The cell pellet was washed once with 25 mM Tris-HCl (pH 7.5) and suspended in the same buffer. The cells were disrupted by two rounds of pulsed sonication (2 min each). Insoluble material was separated by centrifugation at 15,000 × g for 20 min. The supernatant was used for gel electrophoresis and Western blot analysis of NosZ.
Fractionation of cells into periplasm and cytoplasm fractions was done by adapting the partial lysozyme digestion method (40). Freshly harvested cells were washed twice with 200 mM Tris-HCl (pH 7.5) and suspended on ice at a 1:1.5 (wt/vol) ratio in 200 mM Tris-1 M sucrose (pH 7.5). For each 1 g of biomass 150 μl of 0.1 mM EDTA (pH 7.6) and 3 ml of a lysozyme solution (2 mg/ml) were added. Spheroblast formation was monitored with a microscope and was stopped by adding 150 μl of 1 M MgCl2 per g of cell mass. The periplasm was separated from spheroblasts by centrifugation at 4,000 × g, and outer membrane components were removed by ultracentrifugation at 80,000 × g for 1 h. The spheroblasts were washed once in 200 mM Tris-0.5 M sucrose-50 mM MgCl2 and lysed by addition of distilled water and sonication as described above for whole cells. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a 5% polyacrylamide stacking gel and a 10% polyacrylamide separating gel was used for protein separation. Immunochemical detection of NosZ was done with polyclonal antiserum and protein A-horseradish peroxidase conjugate (17) either colorimetrically with 4-chloro-1-naphthol or by chemiluminescence with Luminol reagent (Pierce).
Purification of NosZ and activity measurements.
For purification of NosZ from P. putida, we started with 100 to 150 g of cell mass obtained as follows. Three 2-liter flasks containing 1 liter of M9 medium each were inoculated from agar slants. The flasks were incubated aerobically overnight and used to inoculate a 50-liter batch culture. The carboy was sparged with air at a flow rate of 0.5 liter min−1 through grade D2 sinter glass disks. Sodium nitrate (5.3 g) was added once the optical density reached approximately 0.3. Cells were harvested after 24 h by continuous-flow centrifugation. A typical yield was about 60 to 80 g per batch. P. putida RZ did not grow well in a 50-liter culture and was therefore cultured in 2-liter flasks. Cells were frozen in liquid nitrogen and stored at −20°C until they were used. NosZ was isolated under a protective argon atmosphere by using the protocol established for P. stutzeri (11). The enzyme was monitored in the chromatographic fractions immunochemically. The yield of NosZ from the initial cell mass ranged from 4 to 20 mg; a high yield was not associated with a particular expression strain. The NosZ activity of whole cells was measured with 50 mM citrate as the electron donor by gas chromatography (7); the purified enzyme was measured spectrophotometrically with photoreduced benzyl viologen (11). One unit of enzyme activity was defined as 1 μmol of N2O reduced per min. The Cu contents of NosZ, culture media, and chromatographic fractions were determined by flame atomic absorption spectroscopy. UV-visible spectra were recorded with an HP-8453 diode array photometer (Hewlett-Packard).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported here have been deposited in the EMBL nucleotide sequence databank under accession numbers AJ507426 (nosA) and Z26044 (scoP).
RESULTS
P. putida as the nondenitrifying host for nos genes from P. stutzeri.
Our first objective was to find a suitable organism for heterologous NosZ biosynthesis. We focused on the genus Pseudomonas to avoid problems with codon usage and to work in a Nos− background with an otherwise high level of similarity to P. stutzeri. P. putida DSM50906, the former type strain, was selected as the host from 10 pseudomonads tested for their inventory of nos genes by Southern hybridization. The probes used for hybridization and their sources are listed in Table 2; their locations in the corresponding genes of source organisms are shown in Fig. 1. P. putida tested negative for all P. stutzeri nos genes except nosA, which showed that this nondenitrifying organism did not provide specific functions for N2O respiration. On the other hand, P. putida gave a strong hybridization signal for tatC (data not shown). Since NosZ is translocated to the periplasm via the Tat translocon (24), a tat strain would not be suitable for nosZ expression. Furthermore, we also obtained experimental evidence for the presence in this strain of sco and apbE genes by PCR amplification and/or Southern hybridization. Our work was initiated before data for the genome of P. putida KT2440 became available. The sequence of KT2440 confirmed our experimental findings for strain DSM50906 and showed that valid information for the former type strain may be deduced from information for the apparently very similar organism KT2440.
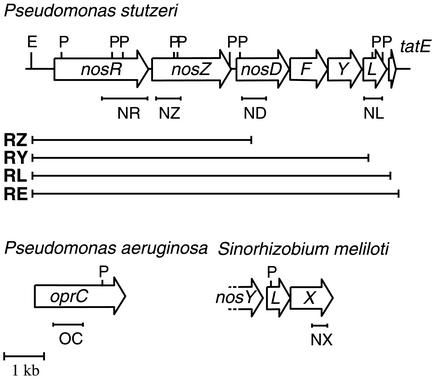
FIG. 1.
nos gene cluster of P. stutzeri with maturation genes and the gene combinations used for expression in P. putida. RZ, RY, RL, and RE are the fragments cloned into pUCP22 that were used for heterologous expression. The locations of the DNA probes (NR, NZ, ND, NL, OC, and NX) and the corresponding source genes and host bacteria are indicated. Restriction sites: P, PstI; E, Eco47III.
Cloning and inactivation of nosA.
It has been suggested that the outer membrane protein NosA is required for NosZ biosynthesis and has a Cu insertase function for NosZ of P. stutzeri JM300 (31, 37). A homologue of NosA, OprC, was isolated from Pseudomonas aeruginosa (55), but its supposed role in NosZ biosynthesis was not studied further. Because of the presence of the potential maturation gene nosA in P. putida, we set out to isolate this gene first from P. stutzeri ZoBell and to study whether it is necessary for NosZ maturation in an organism in which NosZ is well characterized. We screened a genomic cosmid library of P. stutzeri (3) with the P. aeruginosa oprC probe and identified a homologous locus on cosmid c61. A 6-kb HindIII fragment was cloned into pUC18, resulting in pUC18nosAH. The subclone was sequenced and analyzed for coding regions. A 2,157-bp open reading frame (ORF) had the ability to encode a 719-amino-acid protein with an Mr of 79,552, the expected mass of NosA. High levels of sequence similarity with OprC from P. aeruginosa and NosA from P. stutzeri JM300 showed that we had isolated the homologous gene. We also found that an ORF on the complementary strand, immediately downstream of nosA, encodes a putative transport protein with similarity to homologues in Brucella melitensis (SwissProt accession no. Q8YH60), Xylella fastidiosa (Q9PG65), Vibrio cholerae (Q9KV91), and Yersinia pestis (AAM85288).
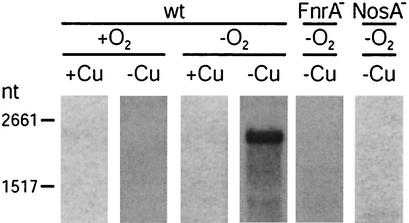
We studied nosA transcription in P. stutzeri by Northern blot analysis. The principal regulatory signals for nosA expression were absence of oxygen and absence of Cu (Fig. 2). Hardly any transcriptional activity was found in aerobic cells in medium that was supplemented with Cu (5 μM) or in aerobic unsupplemented medium composed of pro analysis ingredients (Cu concentration, ≤0.7 μM). Under O2-limiting conditions nosA was derepressed only in the absence of Cu. Transcription of nosA was not increased further by shifting cells to denitrification conditions. O2-dependent regulation involved the Crp-Fnr-type regulator FnrA, since the fnrA mutant MKR2 showed no nosA transcript (Fig. 2).
FIG. 2.
Oxygen and copper repress transcription of nosA from P. stutzeri. Strains were grown in AC medium under aerobic (+O2) or O2-limiting (−O2) conditions with 5 μM Cu (+Cu) or without Cu supplementation (−Cu) (residual Cu concentration, ≤0.7 μM Cu). wt, MK21 having wild-type features; FnrA−, mutant MKR2; NosA−, mutant MK499A. Total RNA was isolated and subjected to Northern blot analysis with the nosA probe NA (Table 2) as described in Materials and Methods. For calibration, digoxigenin-labeled RNA molecular weight marker I from Roche Diagnostics was used. nt, nucleotides.
To investigate whether NosA is necessary for NosZ biosynthesis, we constructed a nosA knockout mutant (see Materials and Methods). No transcript was found in mutant MK499A under conditions of maximal nosA transcription, which verified the mutational event (Fig. 2). MK499A exhibited no phenotype with respect to N2O reduction. The N2O consumption by whole cells of the mutant was comparable to that by parent strain MK21 under both Cu-deficient and Cu-supplemented conditions (data not shown). When nosA was repressed by Cu, no effect on NosZ synthesis was observed in the wild type. NosA was therefore excluded from the minimal set of specific factors required for NosZ biosynthesis.
Role for ScoP?
Bacterial homologues of yeast Sco1 are involved in Cu processing for heme-copper oxidases. Since homologues of sco1 are also part of P. putida genomes (see above), we used the same approach that was used with nosA. In P. stutzeri a sco1 homologue, designated scoP (formerly orf193), is located downstream of fnrA and close to a ccoNOQP gene cluster encoding a cbb3-type oxidase (14, 51). The previously described partial sequence of orf193 was completed. Sco1 proteins have a conserved CxxxCP motif and a histidine residue, which are important for Cu binding. Sco1 homologues of P. stutzeri and P. putida both harbor these critical sequences.
scoP of P. stutzeri was mutagenized by replacing an internal fragment with a kanamycin resistance cassette. The deletion removed the region coding for the functionally important cysteine motif (data not shown). The resulting mutant, MK498P, was not affected in terms of growth on N2O (in AC medium sparged with N2O and not supplemented with Cu). When assayed by gas chromatography, it showed the same rate of N2O reduction as the wild type. Thus, ScoP is not an obligatory factor for NosZ maturation. Overall, the studies with P. stutzeri allowed us to consider the presence of NosA and Sco1 homologues in P. putida as a supportive but not an indispensable background.
Expression of nos genes in P. putida.
We constructed a set of expression vectors to examine which factors are essential for NosZ biosynthesis (Fig. 1). The pUCP22RZ vector carried nosR and nosZ and allowed us to test whether Cu incorporation occurred in the P. putida host in the absence of maturation factors. Two other expression vectors, pUCP22RY and pUCP22RL, carried the nos gene cluster of P. stutzeri either with or without nosL and allowed us to probe whether NosL has a specific role in NosZ maturation. The nos genes were all under the control of their native promoters. Plasmid pUCP22RE carried the transport gene tatE in addition to the nos genes (Fig. 1).
The DNA fragments were amplified by PCR prior to cloning, which may have caused accidental mutations. Therefore, we performed complementation studies with nos mutants to ensure the integrity of these genes. All of the expression vectors were able to restore N2O reduction in vivo in MK413 (nosR::Tn5) and MK4211 (ΔnosZ::Kmr), and all of the expression vectors except pUCP22RZ complemented MK404 (nosD::Tn5) and MK417 (nosY::Tn5) (data not shown). We could not test the functionality of NosL by complementation because the nosL mutant MK424 exhibits a Nos+ phenotype (18). However, as no requirement was established for NosL (see below), this did not affect our conclusions.
The growth conditions included low oxygen tension in the presence of nitrate to enable inorganic nitrate metabolism of the host strain. We used these conditions as a precautionary measure without specifically addressing the regulatory requirements in the host. Nitrate utilization has been reported previously for P. putida (19), and the genome of strain KT2440 contains the structural gene for assimilatory nitrate reductase. P. putida strain DSMZ 1088-260 was described as a heterotrophic nitrifier that reduces nitrate to nitrite with evolution of some NO under anaerobic conditions (15). A nanomolar concentration of NO was shown to be effective for inducing nosZ transcription (52).
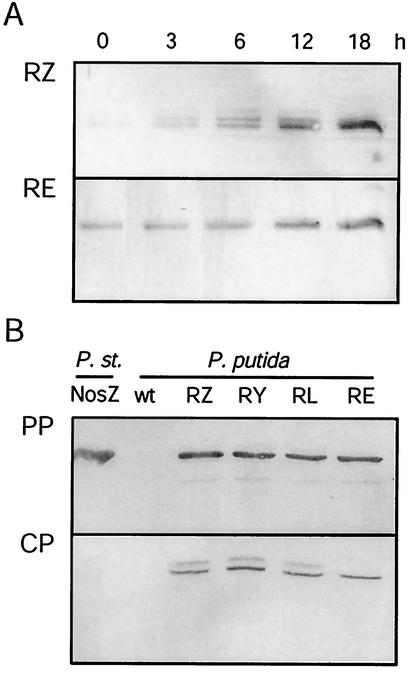
Traces of NosZ were present in P. putida RZ grown under aerobic conditions (Fig. 3A). Under denitrifying conditions enzyme synthesis was enhanced and led to a continuous increase in the NosZ concentration in the growing culture over 18 h. There was no difference in the overall strength of nosZ expression between strains RZ and RE of P. putida. However, only one band was detected in Western blot analysis with the pUCP22RE construct, whereas the RZ derivative (Fig. 3A), as well as RY and RL, produced two protein bands accompanied by weak satellite signals.
FIG. 3.
NosZ is expressed in P. putida and translocated to the periplasm. (A) Cell extracts from growing cultures of P. putida strains RZ and RE were analyzed by Western blotting over an 18-h period. Coexpression of tatE in strain RE resulted in complete translocation of NosZ. (B) Western blot analysis of periplasm (PP) and cytoplasm (CP). RZ, RY, RL, and RE are the different nos gene combinations expressed in P. putida DSM50906 (see Fig. 1). wt, nontransformed wild type; P. st., processed NosZ protein from P. stutzeri MK21. For the conditions used for cell growth, cell fractionation, preparation of cell extract, and immunoblotting see Materials and Methods.
NosZ is translocated to the periplasm of P. putida.
Formation of two NosZ species with different masses is related in P. stutzeri to defects in transport and processing of the signal peptide. To probe for the location of NosZ in P. putida, the cytoplasmic and periplasmic cell compartments were isolated and analyzed immunochemically. With all constructs we found the largest amount of NosZ in its processed form in the periplasm (Fig. 3B). Some NosZ contamination or nonspecific cleavage of pre-NosZ was apparent in the cytoplasmic fraction. In strains RZ, RY, and RL we found unprocessed NosZ in the cytoplasm; however, we did not find unprocessed NosZ in the cytoplasm of strain RE, which contains tatE in addition to the nos genes. Thus, coexpression of tatE was not essential, but it ensured complete export of the enzyme to the periplasm.
P. putida synthesized active NosZ with three coexpressed maturation factors.
Since NosZ was located in the expression strains in its innate functional compartment, we wanted to determine whether these strains were capable of N2O utilization. Activity measurements were obtained by gas chromatography by using whole cells and citrate as the electron donor. Data were collected over a 2-h period. No N2O consumption was detected with the P. putida wild type or any of the expression strains. Although NosZ reached its functional site, whole cells were not able to respire N2O. We isolated the periplasm of P. putida RL and fractionated it by gel permeation chromatography on Sephacryl S-300 to analyze NosZ for the presence of Cu as described previously (7). A Cu peak was clearly associated with NosZ (data not shown). Thus, the absence of cellular N2O reduction was not due to a lack of prosthetic Cu. A likely explanation for the lack of in vivo activity is absence of an appropriate electron donor; i.e., endogenous electron donors of P. putida are not able to couple to NosZ. We attempted to complement the defect by transforming approximately 20-kb fragments of genomic P. stutzeri DNA cloned in vector pUCP22RY into P. putida, but we could not convert the heterologous host into an N2O-respiring bacterium.
As the next step we wanted to determine whether enzyme purified from the expression strains exhibited in vitro activity. All recombinant enzymes except the enzyme obtained from P. putida RZ reduced N2O (Table 3). Bleaching of benzyl viologen was proportional to the amount of NosZ in the assay mixture. The specific activity ranged from 0.7 to 2 U · mg of protein−1 and was in the range of values found with enzyme preparations from P. stutzeri (11, 44). Holo-NosZ contains a total of 12 Cu atoms in a dimeric molecule. The numbers of Cu atoms in the isolated NosZ proteins of P. putida strains were lower (8.9 Cu atoms for RL and 9.6 Cu atoms for RE) but were reasonably close to the theoretical value.
TABLE 3.
Characteristics of recombinant NosZ proteins from P. putida
| Strain | Absorbance (nm)a | NosZ species | Sp act (U/mg of protein)b |
|---|---|---|---|
| RZ | 480, 535, 785 | Type V (CuA) | 0.026 ± 0.028 (3) |
| RY | 485sh, 539, 630sh, 782 | Type I (CuA and CuZ) | 1.224 ± 0.180 (4) |
| RL | 480sh, 538, 625sh, 780 | Type I | 0.662 ± 0.093 (4) |
| RE | 485sh, 535, 637, 777 | Types I and II (CuZ∗) | 2.007 ± 0.340 (4) |
sh, shoulder.
Mean ± standard deviation (number of measurements).
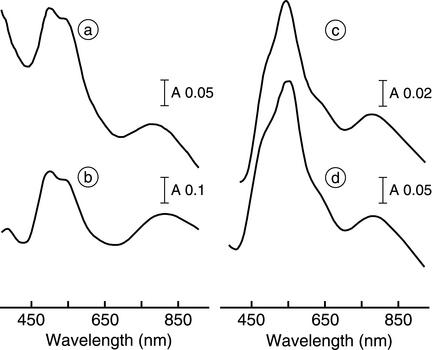
UV-visible absorption spectra of the purified enzyme were recorded. NosZ from P. putida RZ had the spectral properties of a CuA-only protein (Fig. 4, spectrum a, and Table 3). The same features were found with NosZ from the nosD mutant MK402 (45) or a reconstituted recombinant enzyme isolated from E. coli (50). The properties were also nearly identical to the properties of an engineered soluble CuA-binding domain of cytochrome c oxidase from P. denitrificans after reconstitution with CuCl2 (Fig. 4, spectrum b). In this case absorption maxima were present at 363, 480, 530, and 808 nm (29). In accordance with the spectral properties, we found a metal content of 3.8 Cu atoms, as expected for a CuA-only NosZ.
FIG. 4.
Electronic absorption spectra of recombinant NosZ proteins. Spectrum a is the spectrum for NosZ isolated from P. putida strain RZ expressing nosR and nosZ. The isolated protein exhibits a CuA-type spectrum. Spectrum b is the spectrum for a CuA-type protein, represented by the soluble domain from P. denitrificans cytochrome c oxidase subunit II and reconstituted in vitro with Cu(II) (29). Spectrum c is the spectrum for NosZ isolated from P. putida strain RY expressing nosRZDFY. The isolated protein exhibits the spectrum of the purple species (type I) of NosZ, which represents the high-activity enzyme form. Spectrum d is the spectrum for the purple species of NosZ isolated from denitrifying P. stutzeri MK4211(pSZ) (7). All spectra were recorded in 50 mM Tris-HCl (pH 7.5).
Absorption maxima at 540 and 780 nm with a slight shoulder at 480 nm are characteristic of the purple form (type I) of NosZ isolated under anaerobic conditions (11, 45). NosZ obtained from P. putida RY displayed a spectrum that was nearly superimposable with the spectrum of the purple enzyme from P. stutzeri (Fig. 4, spectra c and d). NosZ from P. putida RL and NosZ from P. putida RE (slightly more pronounced) also exhibited a small peak around 630 nm. This peak is indicative of a spectral contribution from the pink form (type II) of NosZ. The pink form is generated during aerobic purification. It seems to be a mixture of species that have lost part of CuZ and have reacted with oxygen to form a CuZ* species having a yet-to-be-defined structural modification (43). P. putida had to be aerated during the entire growth cycle, which may have affected the enzyme and resulted in partial oxygen damage. Overall, NosZ from P. putida showed the same variability in Cu content, enzyme activity, and spectral variations as P. stutzeri. We attribute enzyme lability and the bulk of catalytic site heterogeneity to the Cu-S cluster.
DISCUSSION
NosZ maturation, a matter of topology.
In initiating this study it was important to resolve the transport of NosZ by finding evidence for the Tat translocon in host strain DSM50906 by Southern hybridization. In all P. putida strains carrying a nosZ expression plasmid the enzyme was found in the periplasm; i.e., the Tat system of P. putida processed NosZ without requiring any specific recognition factor for the reductase (Fig. 3B). In P. putida RZ part of NosZ was found in its preform, indicating that there was saturation of the translocation system or some difficulty in the processing of the heterologous protein. This was relieved by coexpression of tatE from P. stutzeri. The findings for P. putida parallel those reported previously for P. stutzeri (24), in which tatE seems to have a dedicated but not obligatory role in NosZ translocation within the Tat translocon.
Formulating the sequence of assembly events requires knowledge concerning in which cellular compartment the process takes place. By subjecting cells to Cu deficiency in a metal-extracted medium it is possible to separate NosZ export and Cu insertion and show that the maturation process is periplasmic and not cytoplasmic (36). We confirmed the previous findings by fractionating P. stutzeri cell compartments, performing Western blotting, and measuring the activity of whole cells by gas chromatography. Cu-deficient cells of P. stutzeri export NosZ to the periplasm but exhibit no or little NosZ activity. The activity is restored by adding Cu to cells arrested for protein synthesis (data not shown). This means that periplasmic NosZ undergoes Cu-dependent maturation posttranslocationally. On the other hand, NosZ can be retained in the cytoplasm by mutating the Tat-specific signal peptide (17) or by inactivating the principal transport gene, tatC (24). In either case cytoplasmic NosZ does not incorporate Cu.
NosZ synthesis by P. putida and an integrated model for enzyme maturation.
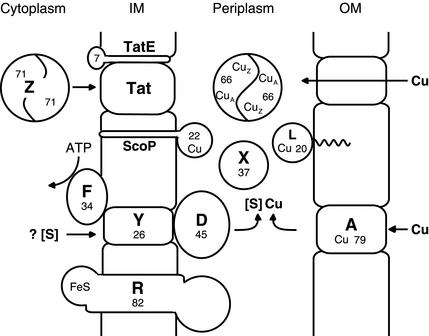
We propose a topological model for NosZ maturation which integrates the current experimental data and in silico evidence (Fig. 5). Apo-NosZ is exported prior to and independent of cofactor insertion. A CuA-only NosZ protein is formed by P. putida RZ in the absence of P. stutzeri-specific nos maturation functions. In contrast, CuZ assembly depends on the coexpression of nosDFY, and only under such conditions was a catalytically active NosZ protein obtained with the spectral features of both the CuA and CuZ species (NosZ type I). This attributes to NosDFY a role in the assembly of the CuZ center, and its function is more likely to be in the provision of sulfur than that of Cu. CuA can be reconstituted in vitro into the apoprotein from exogenous Cu, whereas attempts to do this for CuZ have failed. This fact can be explained by the lack of an appropriate sulfur source. We propose that sulfur is provided from a cytoplasmic source through the action of the NosDFY ABC transporter system. Cu from the medium may pass through NosA or another cation-permeable pore and is delivered to the site of Cu cluster biosynthesis by a Cu chaperone, possibly NosL. Pathways for Cu and sulfur donation thus converge in the periplasm for Cu-S cluster formation. Beyond the minimal requirement for NosDFY in the heterologous host, proteins of P. putida may provide rescue functions, and considering these proteins helped us address further requirements for NosZ maturation. The nonobligatory components of our model and the arguments for why they should be considered are discussed below.
FIG. 5.
Components of the maturation process for N2O reductase and topology. Single uppercase letters indicate the products of nos genes. The numbers indicate the approximate protein masses (in kilodaltons). Cu-containing proteins are indicated. NosZ is shown as a dimer, but otherwise no inferences about stoichiometries of protein complexes are drawn, nor is the composition of the Tat translocon indicated other than to show the supportive role of TatE. NosF has ATPase activity (Honisch and Zumft, unpublished data). NosD belongs to a protein family with carbohydrate and sugar hydrolase signatures (10). [S] is a sulfur donor whose chemical nature is not known. NosL is shown with a lipid anchor. Cu may enter the periplasm via NosA or another porin. The membrane-bound NosR carries putative FeS groups in its cytoplasmic domain and may have other functions in addition to acting as a transcriptional regulator for nosZ. IM and OM, cytoplasmic and outer membranes, respectively. For further discussion see the text.
NosA, the putative component of a metal uptake system.
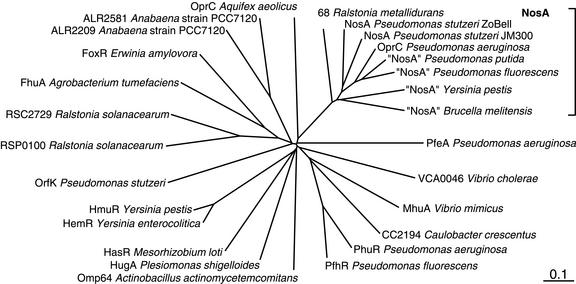
A phylogenetic tree shows that the NosA proteins occur in a tight subcluster of a larger family of siderophore-binding or heme receptor proteins, such as HasR, FhuA, and other proteins (Fig. 6). A relationship between NosA and TonB-dependent proteins was noted previously (30, 55). A TonB-box C motif exists near the C terminus, but a corresponding TonB box near the N terminus has not been found. Other than the association with the genes encoding NosX and NosL (see below), NosA is not consistently associated with nos genes. Conspicuously, NosA homologues occur in the nondenitrifying bacteria Yersinia pestis CO92, (41), P. putida KT2440, and Pseudomonas fluorescens PFO-1. The high degrees of amino acid sequence identity of these proteins (up to 54%) with NosA from denitrifiers suggest that there is functional uniformity.
FIG. 6.
Phylogenetic tree of NosA proteins and homologues. The SwissProt data bank and genome project databases were searched with P. aeruginosa OprC by using FASTA3. A cutoff value of 10−4 was used. The tree was constructed with CLUSTAL X and TreeView 1.6.6. Several bacteria contain more than one member of this family.
The observed regulatory responses allow the conclusion that the outer membrane protein, NosA, functions in anaerobic metabolism, and since it is repressed by Cu, its role seems to be limited to conditions in which the Cu supply is low. NosA might be involved in Cu uptake and represent the outer membrane pore for Cu ion or Cu chelator passage (30) rather than provide a Cu insertase activity for NosZ. The promoter region has a CTTCCCGAAA sequence similar to that of the binding region for the Cu-dependent regulator CopR of Pseudomonas syringae (35). Furthermore, the nosA promoter has a degenerate FNR box, TTGAC-N1-GTCAA, and because of the spacing of the palindromes, only one half-site may act as a recognition site for the FnrA regulator for the upregulation observed under anaerobic conditions (Fig. 2).
NosA was purified as a Cu-containing protein. Its spectral properties and mode of Cu binding are not known (30). Alignment of the NosA proteins reveals a set of conserved Cys, His, and Met residues which could bind Cu as blue or type 1 Cu. Neisseria gonorrhoeae has in AniA an outer membrane protein that functions as a nitrite reductase and has a type 1 Cu electron transfer site (27); however, NosA has no sequence similarity to AniA. While chemically induced nosA mutants of P. stutzeri JM300 synthesize a virtually Cu-free N2O reductase (37), the nosA knockout mutant MK499A of P. stutzeri ATCC 14405 displayed no phenotype with respect to NosZ activity and Cu content (this study). It is possible that the inability of JM300 mutants to reduce N2O was due to the lack of protein E (30), which might be a component of the NosDFY assembly apparatus.
ScoP, a candidate protein for CuA assembly.
P. putida RZ synthesizes a NosZ protein whose spectral features clearly show that only CuA is metallated (Fig. 4, spectrum a). It is feasible that the biosynthesis of CuA was dependent on a host function for synthesis of the same center of cytochrome aa3. P. putida KT2440 has an aa3-type oxidase. Studies of cytochrome aa3 biogenesis in yeast support the hypothesis that incorporation of CuA is catalyzed by Sco1 and the Cu chaperone Cox17. Cu(I) is thought to be transferred from Cox17 to Sco1, which inserts Cu into CuA of subunit II (23, 38). In Bacillus subtilis the Sco1 homologue YmpQ affects cytochrome c oxidase but not menaquinol oxidase, thus favoring a role in CuA synthesis (32). The soluble domain of the Sco1 homologue PrrC from Rhodobacter sphaeroides has thiol disulfide oxidoreductase activity which can be used for Cu mobilization (33). Thus, with respect to a Sco requirement, we found a homologue in each of the available genomes of nosZ-harboring strains. It is interesting in this context that Rhodobacter capsulatus contains the homologue SenC (accession no. Q52720), and even though this bacterium has no oxidase with a CuA center, it disposes over NosZ. However, in spite of these multiple lines of indirect evidence, the ScoP protein was dispensable for NosZ biosynthesis in P. stutzeri. We presume that loss of ScoP did not result in a recognizable phenotype because metallation of CuA also proceeds spontaneously or a substitute protein involved in Cu processing takes over.
nosL is a constant partner of nos gene clusters.
Sixteen individually analyzed denitrifiers and entire genomes show that in each case a nosDFYL gene cluster is present and conserved. nosL is cotranscribed with nosDFY in P. stutzeri (U. Honisch and W. G. Zumft, unpublished data), which indicates that NosL has a function related to NosZ maturation. NosL was purified from Achromobacter cycloclastes as a Cu-containing protein. While the Cu(I) site of NosL is remarkably stable in the presence of oxygen, the Cu(II) form has little affinity for Cu and releases the metal (34). These properties support the hypothesis that NosL has a metallochaperone role and that the putative function is to guide Cu from the site of periplasmic entry to N2O reductase. Nevertheless, the plausible role of NosL in NosZ maturation is not obligatory. Coexpression of nosL was not a requirement for a functional NosZ in P. putida.
Random Tn5 mutagenesis and selection for loss of growth on N2O resulted in mutants with insertions in each gene of the nosRZDFYL cluster except nosL (6, 26, 49). Since NosL is a nonselectable marker, this suggests that there is a functional substitute for NosL or NosL is dispensable. Also, a nosL mutant lacks a recognizable phenotype (18). No nosL homologue was detected in DSM50906 by Southern hybridization, and none was evident in the genome of KT2440, which suggests that P. putida has several ways to process Cu for its Cu proteins, which may provide a rescue function for NosZ biosynthesis.
ApbE as a functional NosX homologue in NosZ maturation.
In individual studies of nos gene clusters, a nosX gene was found in S. meliloti and several other bacteria but not in the well-studied denitrifiers P. stutzeri and P. aeruginosa. Since the absence of nosX in the pseudomonads would bring into question the role of this gene in encoding an essential maturation component, we addressed the distribution of nosX in genomes of denitrifiers in silico and also searched for a potential rescue function.
NosX proteins exhibit high sequence similarity among themselves but not with other proteins (46), although sequence similarity to RnfF has been noted (6). RnfF is a membrane-bound periplasmic protein belonging to an R. capsulatus complex presumably involved in electron transport for nitrogen fixation (47). A data bank search revealed that P. putida KT2440 carries along with apbE a potential nosX homologue; the same gene was amplified by PCR from strain DSM50906. ApbE is a 36-kDa monotopic inner membrane protein, and most of its soluble domain is located in the periplasm (2). A periplasmic location but not membrane association is essential for the ApbE function directed at ThiH. The latter protein is a putative FeS protein involved in the last step in synthesis of the 4-methyl-5-β-hydroxyethylthiazole monophosphate moiety of thiamine monophosphate. ApbE is thought to be involved in the redox-dependent synthesis or repair of ThiH as part of a membrane-associated complex (22). ApbE shows sequence similarity to RnfF, and most importantly, rnfF is able to complement a Salmonella enterica serovar Typhimurium apbE mutation (2). We argue that NosX and ApbE are functionally interchangeable members of the same protein family in order to account for the dispensability of nosX coexpression in the P. putida background. The mass of NosX is comparable to the mass of ApbE, and NosX is predicted to be periplasmic because of a signal sequence with features for Tat targeting (46). Location of NosX in the outer cell compartment is consistent with the site of Cu cluster biosynthesis. The release of Cu and/or sulfide from the corresponding donor molecules requires redox steps, and NosX may fulfill such a role.
We found that there was a consistent association of the core nosRZDFY cluster with the nosX or apbE genes as determined by a data bank search. A. cycloclastes (accession no. AF047429), Bradyrhizobium japonicum (AJ002531), B. melitensis (16), P. denitrificans (46), Rhodopseudomonas palustris (genome), and S. meliloti (6, 26) all have a nosRZDFYLX gene organization. nosX or apbE loci may also be distant from nos genes. In N. gonorrhoeae the nosX gene is 42 kb downstream of nosL and a putative apbE gene is 18 kb upstream of nosR. A single apbE locus, separated from the nos cluster, is present in the genome of P. aeruginosa (PA2993). Screening the genomes of Ralstonia metallidurans, Ralstonia solanacearum, Burkholderia mallei, and Burkholderia pseudomallei for an apbE homologue in each case resulted in an ORF approximately 1 kb upstream of nosZ in the opposite transcriptional orientation. These ORFs may represent functional nosX genes. In the latter group of bacteria nos clusters are organized so that nosR is located either immediately downstream of nosZ (R. metallidurans and R. solanacearum; nosX-nosZRDFYL) or downstream of nosL (B. pseudomallei and B. mallei; nosX-nosZDFYLR).
Acknowledgments
We thank H. Körner for valuable help with software applications and Y.-K. Chan for providing plasmid pWM20. Preliminary sequence data for the comparative in silico study were provided freely by the U.S. DOE Joint Genome Institute and the Institute for Genomic Research.
This work was supported by the Deutsche Forschungsgemeinschaft and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Alexeyev, M. F. 1995. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques 18:52-55. [PubMed] [Google Scholar]
- 2.Beck, B. J., and D. M. Downs. 1999. A periplasmic location is essential for the role of the ApbE lipoprotein in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 181:7285-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, C., and W. G. Zumft. 1992. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for denitrification. J. Bacteriol. 174:2394-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, K., K. Djinovic-Carugo, T. Haltia, I. Cabrito, M. Saraste, J. J. G. Moura, I. Moura, M. Tegoni, and C. Cambillau. 2000. Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase. Evidence of a bridging inorganic sulfur. J. Biol. Chem. 275:41133-41136. [DOI] [PubMed] [Google Scholar]
- 5.Brown, K., M. Tegoni, M. Prudencio, A. S. Pereira, S. Besson, J. J. Moura, I. Moura, and C. Cambillau. 2000. A novel type of catalytic copper cluster in nitrous oxide reductase. Nat. Struct. Biol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 6.Chan, Y.-K., W. A. McCormick, and R. J. Watson. 1997. A new nos gene downstream from nosDFY is essential for dissimilatory reduction of nitrous oxide by Rhizobium (Sinorhizobium) meliloti. Microbiology 143:2817-2824. [DOI] [PubMed] [Google Scholar]
- 7.Charnock, J. M., A. Dreusch, H. Körner, F. Neese, J. Nelson, A. Kannt, H. Michel, C. D. Garner, P. M. H. Kroneck, and W. G. Zumft. 2000. Structural investigations of the CuA centre of nitrous oxide reductase from Pseudomonas stutzeri by site-directed mutagenesis and X-ray absorption spectroscopy. Eur. J. Biochem. 267:1368-1381. [DOI] [PubMed] [Google Scholar]
- 8.Chinenov, Y. V. 2000. Cytochrome c oxidase assembly factors with a thioredoxin fold are conserved among prokaryotes and eukaryotes. J. Mol. Med. 78:239-242. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201:134-139. [DOI] [PubMed] [Google Scholar]
- 10.Ciccarelli, F. D., R. R. Copley, T. Doerks, R. B. Russelland, and P. Bork. 2002. CASH—a β-helix domain wide spread among carbohydrate-binding proteins. Trends Biochem. Sci. 27:59-62. [DOI] [PubMed] [Google Scholar]
- 11.Coyle, C. L., W. G. Zumft, P. M. H. Kroneck, H. Körner, and W. Jakob. 1985. Nitrous oxide reductase from denitrifying Pseudomonas perfectomarina, purification and properties of a novel multicopper enzyme. Eur. J. Biochem. 153:459-467. [DOI] [PubMed] [Google Scholar]
- 12.Cuypers, H., J. Berghöfer, and W. G. Zumft. 1995. Multiple nosZ promoters and anaerobic expression of nos genes necessary for Pseudomonas stutzeri nitrous oxide reductase and assembly of its copper centers. Biochim. Biophys. Acta 1264:183-190. [DOI] [PubMed] [Google Scholar]
- 13.Cuypers, H., A. Viebrock-Sambale, and W. G. Zumft. 1992. NosR, a membrane-bound regulatory component necessary for expression of nitrous oxide reductase in denitrifying Pseudomonas stutzeri. J. Bacteriol. 174:5332-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuypers, H., and W. G. Zumft. 1993. Anaerobic control of denitrification in Pseudomonas stutzeri escapes mutagenesis of an fnr-like gene. J. Bacteriol. 175:7236-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daum, M., W. Zimmer, H. Papen, K. Kloos, K. Nawrath, and H. Bothe. 1998. Physiological and molecular biological characterization of ammonia oxidation of the heterotrophic nitrifier Pseudomonas putida. Curr. Microbiol. 37:281-288. [DOI] [PubMed] [Google Scholar]
- 16.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J.-J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreusch, A., D. M. Bürgisser, C. W. Heizmann, and W. G. Zumft. 1997. Lack of copper insertion into unprocessed cytoplasmic nitrous oxide reductase generated by an R20D substitution in the arginine consensus motif of the signal peptide. Biochim. Biophys. Acta 1319:311-318. [DOI] [PubMed] [Google Scholar]
- 18.Dreusch, A., J. Riester, P. M. H. Kroneck, and W. G. Zumft. 1996. Mutation of the conserved Cys165 outside the CuA domain destabilizes nitrous oxide reductase but maintains its catalytic activity: evidence for disulfide bridges and a putative disulfide isomerase gene. Eur. J. Biochem. 237:447-453. [DOI] [PubMed] [Google Scholar]
- 19.Eberl, L., A. Ammendola, M. H. Rothballer, M. Givskov, C. Sternberg, M. Kilstrup, K. H. Schleifer, and S. Molin. 2000. Inactivation of gltB abolishes expression of the assimilatory nitrate reductase gene (nasB) in Pseudomonas putida KT2442. J. Bacteriol. 182:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engler-Blum, G., M. Meier, J. Frank, and G. A. Müller. 1993. Reduction of background problems in nonradioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal. Biochem. 210:235-244. [DOI] [PubMed] [Google Scholar]
- 21.Glerum, D. M., A. Shtanko, and A. Tzagoloff. 1996. Sco1 and Sco2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271:20531-20535. [DOI] [PubMed] [Google Scholar]
- 22.Gralnick, J., E. Webb, B. Beck, and D. Downs. 2000. Lesions in gshA (encoding gamma-l-glutamyl-l-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar typhimurium LT2. J. Bacteriol. 182:5180-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heaton, D. N., G. N. George, G. Garrison, and D. R. Winge. 2001. The mitochondrial copper metallochaperone Cox17 exists as an oligomeric, polycopper complex. Biochemistry 40:743-751. [DOI] [PubMed] [Google Scholar]
- 24.Heikkilä, M. P., U. Honisch, P. Wunsch, and W. G. Zumft. 2001. Role of the Tat transport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 183:1663-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holloway, B. W., U. Römling, and B. Tümmler. 1994. Genomic mapping of Pseudomonas aeruginosa PAO. Microbiology 140:2907-2929. [DOI] [PubMed] [Google Scholar]
- 26.Holloway, P., W. McCormick, R. J. Watson, and Y.-K. Chan. 1996. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J. Bacteriol. 178:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Householder, T. C., W. A. Bell, S. Lissenden, J. A. Cole, and V. L. Clark. 1999. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J. Bacteriol. 181:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroneck, P. M. H., W. A. Antholine, J. Riester, and W. G. Zumft. 1988. The cupric site in nitrous oxide reductase contains a mixed-valence [Cu(II), Cu(I)] binuclear center: a multifrequency electron paramagnetic resonance investigation. FEBS Lett. 242:70-74. [DOI] [PubMed] [Google Scholar]
- 29.Lappalainen, P., R. Aasa, B. G. Malmström, and M. Saraste. 1993. Soluble CuA-binding domain from the Paracoccus cytochrome c oxidase. J. Biol. Chem. 268:26416-26421. [PubMed] [Google Scholar]
- 30.Lee, H. S., A. H. T. Abdelal, M. A. Clark, and J. L. Ingraham. 1991. Molecular characterization of nosA, a Pseudomonas stutzeri gene encoding an outer membrane protein required to make copper-containing N2O reductase. J. Bacteriol. 173:5406-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, H. S., R. E. W. Hancock, and J. L. Ingraham. 1989. Properties of a Pseudomonas stutzeri outer membrane channel-forming protein (NosA) required for production of copper-containing N2O reductase. J. Bacteriol. 171:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattatall, N. R., J. Jazairi, and B. C. Hill. 2000. Characterization of YpmQ, an accessory protein required for the expression of cytochrome c oxidase in Bacillus subtilis. J. Biol. Chem. 275:28802-28809. [DOI] [PubMed] [Google Scholar]
- 33.McEwan, A. G., A. Lewin, S. L. Davy, R. Boetzel, A. Leech, D. Walker, T. Wood, and G. R. Moore. 2002. PrrC from Rhodobacter sphaeroides, a homologue of eukaryotic Sco proteins, is a copper-binding protein and may have a thiol-disulfide oxidoreductase activity. FEBS Lett. 518:10-16. [DOI] [PubMed] [Google Scholar]
- 34.McGuirl, M. A., J. A. Bollinger, N. Cosper, R. A. Scott, and D. M. Dooley. 2001. Expression, purification, and characterization of NosL, a novel Cu(I) protein of the nitrous oxide reductase (nos) gene cluster. J. Biol. Inorg. Chem. 6:189-195. [DOI] [PubMed] [Google Scholar]
- 35.Mills, S. D., C.-K. Lim, and D. A. Cooksey. 1994. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol. Gen. Genet. 244:341-351. [DOI] [PubMed] [Google Scholar]
- 36.Minagawa, N., and W. G. Zumft. 1988. Cadmium-copper antagonism in the activation of periplasmic nitrous oxide reductase of copper-deficient cells from Pseudomonas stutzeri. Biol. Metals 1:117-122. [Google Scholar]
- 37.Mokhele, K., Y. J. Tang, M. A. Clark, and J. L. Ingraham. 1987. A Pseudomonas stutzeri outer membrane protein inserts copper into N2O reductase. J. Bacteriol. 169:5721-5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nittis, T., G. N. George, and D. R. Winge. 2001. Yeast Sco1, a protein essential for cytochrome c oxidase function, is a Cu(I)-binding protein. J. Biol. Chem. 276:42520-42526. [DOI] [PubMed] [Google Scholar]
- 39.O'Halloran, T. V., and V. C. Culotta. 2000. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 275:25057-25060. [DOI] [PubMed] [Google Scholar]
- 40.Pages, J.-M., J. Anba, A. Bernadac, H. Shinagawa, A. Nakata, and C. Lazdunski. 1984. Normal precursors of periplasmic proteins accumulated in the cytoplasm are not exported post-translationally in Escherichia coli. Eur. J. Biochem. 143:499-505. [DOI] [PubMed] [Google Scholar]
- 41.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 42.Ramos-Gonzales, M. I., and S. Molin. 1998. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 180:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasmussen, T., B. C. Berks, J. N. Butt, and A. J. Thomson. 2002. Multiple forms of the catalytic centre, CuZ, in the enzyme nitrous oxide reductase from Paracoccus pantotrophus. Biochem. J. 364:807-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen, T., B. C. Berks, J. Sanders-Loehr, D. M. Dooley, W. G. Zumft, and A. J. Thomson. 2000. The catalytic center in nitrous oxide reductase, CuZ, is a copper sulfide cluster. Biochemistry 39:12753-12756. [DOI] [PubMed] [Google Scholar]
- 45.Riester, J., W. G. Zumft, and P. M. H. Kroneck. 1989. Nitrous oxide reductase from Pseudomonas stutzeri, redox properties and spectroscopic characterization of different forms of the multicopper enzyme. Eur. J. Biochem. 178:751-762. [DOI] [PubMed] [Google Scholar]
- 46.Saunders, N. F. W., J. J. Hornberg, W. N. M. Reijnders, H. V. Westerhoff, S. de Vries, and R. J. M. van Spanning. 2000. The NosX and NirX proteins of Paracoccus denitrificans are functional homologues: their role in maturation of nitrous oxide reductase. J. Bacteriol. 182:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmehl, M., A. Jahn, A. Meyer zu Vilsendorf, S. Hennecke, B. Masepohl, M. Schuppler, M. Marxer, J. Oelze, and W. Klipp. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241:602-615. [DOI] [PubMed] [Google Scholar]
- 48.Schulze, M., and G. Rödel. 1989. Accumulation of the cytochrome c oxidase subunits I and II in yeast requires a mitochondrial membrane-associated protein, encoded by the nuclear SCO1 gene. Mol. Gen. Genet. 216:37-43. [DOI] [PubMed] [Google Scholar]
- 49.Viebrock, A., and W. G. Zumft. 1987. Physical mapping of transposon Tn5 insertions defines a gene cluster functional in nitrous oxide respiration by Pseudomonas stutzeri. J. Bacteriol. 169:4577-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viebrock, A., and W. G. Zumft. 1988. Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J. Bacteriol. 170:4658-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vollack, K.-U., E. Härtig, H. Körner, and W. G. Zumft. 1999. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol. Microbiol. 31:1681-1694. [DOI] [PubMed] [Google Scholar]
- 52.Vollack, K.-U., and W. G. Zumft. 2001. Nitric oxide signaling and transcriptional control of denitrification genes in Pseudomonas stutzeri. J. Bacteriol. 183:2516-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.West, S. E. H., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 128:81-86. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Yoneyama, H., and T. Nakae. 1996. Protein C (OprC) of the outer membrane of Pseudomonas aeruginosa is a copper-regulated channel protein. Microbiology 142:2137-2144. [DOI] [PubMed] [Google Scholar]
- 56.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zumft, W. G., K. Döhler, and H. Körner. 1985. Isolation and characterization of transposon Tn5-induced mutants of Pseudomonas perfectomarina defective in nitrous oxide respiration. J. Bacteriol. 163:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zumft, W. G., and P. M. H. Kroneck. 1996. Metal-center assembly of the bacterial multicopper enzyme nitrous oxide reductase. Adv. Inorg. Biochem. 11:193-221. [Google Scholar]
- 59.Zumft, W. G., A. Viebrock-Sambale, and C. Braun. 1990. Nitrous oxide reductase from denitrifying Pseudomonas stutzeri: genes for copper-processing and properties of the deduced products, including a new member of the family of ATP/GTP-binding proteins. Eur. J. Biochem. 192:591-599. [DOI] [PubMed] [Google Scholar]