Abstract
Stability and resilience against environmental perturbations are critical properties of medical and environmental biofilms and pose important targets for their control. Biofilm stability is determined by two mutually exclusive processes: attachment of cells to and detachment from the biofilm matrix. Using Shewanella oneidensis MR-1, an environmentally versatile, Fe(III) and Mn(IV) mineral-reducing microorganism, we identified mxdABCD as a new set of genes essential for formation of a three-dimensional biofilm. Molecular analysis revealed that mxdA encodes a cyclic bis(3′,5′)guanylic acid (cyclic di-GMP)-forming enzyme with an unusual GGDEF motif, i.e., NVDEF, which is essential for its function. mxdB encodes a putative membrane-associated glycosyl transferase. Both genes are essential for matrix attachment. The attachment-deficient phenotype of a ΔmxdA mutant was rescued by ectopic expression of VCA0956, encoding another diguanylate cyclase. Interestingly, a rapid cellular detachment from the biofilm occurred upon induction of yhjH, a gene encoding an enzyme that has been shown to have phosphodiesterase activity. In this way, it was possible to bypass the previously identified sudden depletion of molecular oxygen as an environmental trigger to induce biofilm dissolution. We propose a model for c-di-GMP as a key intracellular regulator for controlling biofilm stability by shifting the state of a biofilm cell between attachment and detachment in a concentration-dependent manner.
Most microbes in nature are assumed to exist as surface-associated communities in biofilms (10, 11, 50). Biofilms greatly affect their environment, whether that environment is a human host, a wastewater treatment plant, or pristine soils and sediments, and significant research has focused on understanding and controlling the resilience and stability of such biofilms (12, 35). From a microbe's point of view, the decision either to remain associated with or to sever ties to and exit a biofilm confers profound consequences to its lifestyle. Stability and resilience of a three-dimensional biofilm are controlled by two diametrically opposed states: attachment and detachment. These mutually exclusive states have in common a change in how cells associate with the biofilm matrix. The biofilm matrix consists of exopolymeric substances, such as polysaccharides, DNA, and proteins, but also of biofilm cells (for a review, see reference 45). Despite extensive research over the past decade on biofilms, on adhesion of microbial cells to a substratum surface during initial contact, and on the biofilm matrix, the molecular mechanisms of how biofilm cells stick to a biofilm and how such cells detach are largely unknown. For the purpose of this discussion, we define “adhesion” as the binding of a cell to a substratum, while the term “attachment” is used to indicate the binding of a cell to a biofilm matrix.
Although biofilm disintegration is observed frequently and considered part of a developmental biofilm program, only recently have systematic investigations provided some insights into the molecular events involved in detachment (16, 41, 48). Environmental cues, such as changes in oxygen or carbon substrate concentration, pH, or other chemical parameters have been reported to induce detachment of mature biofilms (15, 41, 48). Consequently, detachment could be an active process where an environmentally controlled, direct activation of a “detachase” initiates severing of bonds between cells and the biofilm matrix. Indeed, exopolysaccharide lyases and DNases have been implicated in cell dispersal from biofilms (2, 7, 13, 23, 24, 34, 54). Alternatively, detachment could be a passive process. In this case, the opposite of detachment, i.e., attachment, could require constant (enzymatic) activity, and a sudden cessation of such attachment activity could result in instant detachment. It is also conceivable that a combination of “attachment activity” and of detachase might be involved in physiologically controlled detachment. Recently, quantitative, high-resolution confocal laser scanning microscopy (CLSM) in conjunction with a mutant analysis has provided some insights into the detachment process (16, 41, 47, 48).
Previously, we reported a genetic analysis of surface adhesion and biofilm formation, as well as a physiological and molecular characterization of detachment in Shewanella oneidensis MR-1, which is a facultative Fe(III) and Mn(IV) mineral-reducing soil bacterium and plays a critical role in mineral dissolution, heavy-metal (im)mobilization, and pollution degradation (31, 33, 47, 48, 52). In a genetic screen for biofilm defective mutants, we identified type IV pili as critical in mediating the initial adhesion of cells to the substratum during surface colonization (47). Furthermore, and similar to other biofilm microbes, flagellum motility was found to be important in controlling the architecture of biofilms (47). We also discovered that rapid detachment of individual S. oneidensis cells could be initiated by a sudden decrease in molecular oxygen but not in electron donor concentration in a hydrodynamic flow chamber system (48). Single-cell resolution CLSM revealed that detachment occurs as the separation of individual cells and of cells in small groups throughout all layers in a biofilm. Such oxygen-dependent detachment can be induced simply by stopping the flow of an aerobic, oxygen-limited growth medium in the hydrodynamic biofilm system (48). Notably, a stop of flow of only 5 min releases up to 50% of the detachable cell mass. Here, we report the identification of a new, putative exopolysaccharide synthesis gene cluster that links the attachment and detachment of a single cell to the biofilm matrix.
A molecular system, which was first discovered and studied for the control of extracellular cellulose biosynthesis in Gluconacetobacter xylinus, formerly Acetobacterium xylinum (38, 39), has been implicated in autoaggregation of planktonic cells and in biofilm formation in several microorganisms (27, 43, 49). In G. xylinus, the membrane-bound cellulose synthase is allosterically activated by the secondary messenger molecule, cyclic di-GMP cyclic-bis(3′,5′)guanylic acid (cyclic di-GMP, abbreviated here c-di-GMP) (38). c-di-GMP is synthesized by diguanylate cyclase and degraded by phosphodiesterase activities, respectively (9, 38, 46). The recent discoveries of (i) cellulose production in many different biofilm-forming bacteria (27, 43), (ii) numerous GGDEF domain or EAL domain-containing proteins with diguanylate cyclase or phosphodiesterase activities (1, 36, 43, 49), and (iii) aggregation and biofilm defects in mutants defective in these proteins suggest that exopolysaccharide production relevant to biofilm formation may be allosterically controlled by c-di-GMP. Cellular c-di-GMP signaling in processes other than polysaccharide production has been reported as well. For example, virC in Vibrio anguillarum is involved in fish infection (30), and pleD is involved in Caulobacter crescentus in swarmer-to-stalk cell differentiation (1, 36). Cell cycle signals in this microorganism were suspected to control the activity of PleD, a GGDEF-containing, polarly localized response regulator, which is required for polysaccharide production of the holdfast (1). Notably, Simm et al. reported a decreased or dramatically increased flagellum-dependent swarming motility in adrA-expressing (GGDEF domain) and yhjH-expressing (EAL domain) Salmonella enterica serovar Typhimurium, respectively, which led the authors to suggest that c-di-GMP regulates the transition between sessility and motility (43). However, detachment of S. oneidensis cells from the biofilm matrix is independent of flagellum motility and proceeds within a few minutes after application of the environmental cue. In this work, we present data that single-cell attachment to and detachment from the biofilm matrix are linked by c-di-GMP.
MATERIALS AND METHODS
Growth conditions and media.
Escherichia coli strains (Table 1) were grown in Luria-Bertani (LB) medium at 37°C, and Shewanella oneidensis MR-1 strains were grown at 30°C in LB, lactate medium (LM) (47), or mineral medium (MM) with the following final composition: 485 μM CaCl2 · 2H2O, 5 μM CoCl2, 0.2 μM CuSO4 · 5H2O, 57 μM H3BO3, 1.27 mM K2HPO4, 0.73 mM KH2PO4, 1.0 mM MgSO4 · 7H2O, 1.3 μM MnSO4, 67.2 μM Na2EDTA, 3.9 μM Na2MoO4 · 2H2O, 1.5 μM Na2SeO4, 150 mM NaCl, 2 mM NaHCO3, 5 μM NiCl2 · 5H2O, 1 μM ZnSO4, 9 mM (NH4)2SO4, 15 mM lactate, and 5 mM HEPES, pH 7.4 (modified after reference 31). If required, the medium was solidified with 1.5% (wt/vol) agar and supplemented with 30 μg/ml chloramphenicol, 10 μg/ml gentamicin, 25 μg/ml kanamycin, and/or 20 μg/ml tetracycline. Gene induction from the pARA and pLacTac vectors was achieved by addition of 0.2% (wt/vol) l-arabinose or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), if not indicated otherwise.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and phenotype | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α-λpir | φ80dlacZΔM15 Δ(lacZYA-argF) | 29 |
| U196 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1/λpir | ||
| S17-λpir | thi pro recA hsdR [RP4-2Tc::Mu-Km::Tn7] λpir Tpr Smr | 44 |
| HB101 | Smr; recA thi pro leu hsdRM+ | 26 |
| S. oneidensis | ||
| MR-1 | Shewanella oneidensis MR-1 wild type | 51 |
| AS93 | S. oneidensis MR-1 tagged with enhanced GFP in a mini-Tn7 construct; Gmr Cmr | 47 |
| AS140 | AS93 ΔmxdA; Gmr Cmr | This work |
| AS141 | AS93 ΔmxdB; Gmr Cmr | This work |
| AS142 | AS93 ΔmxdC; Gmr Cmr | This work |
| AS143 | AS93 ΔmxdD; Gmr Cmr | This work |
| AS145 | AS93 pARA-yhjH; Gmr Cmr Kmr | This work |
| AS146 | AS93 pARA-VCA0956; Gmr Cmr Kmr | This work |
| AS147 | AS93 pME6041; Gmr Cmr Kmr | This work |
| AS151 | AS141 pLacTac-VCA0956; Gmr Cmr Tcr | This work |
| AS152 | AS140 pLacTac-SO4180(mxdA); Cmr Tcr Gmr | This work |
| AS154 | AS140 pLacTac-VCA0956; Gmr Cmr Tcr | This work |
| AS160 | AS140 pME6031; Gmr Cmr Tcr | This work |
| Plasmids | ||
| pGP704-Sac28-Km | mobRP4+ori-R6K sacB; suicide plasmid for in-frame deletions; Kmr | Chengyen Wu, unpublished data |
| pGP704-Sac28-Km-mxdA | mxdA in-frame deletion fragment in pGP704-Sac28-Km | This study |
| pGP704-Sac28-Km-mxdB | mxdB in-frame deletion fragment in pGP704-Sac28-Km | This study |
| pGP704-Sac28-Km-mxdC | mxdC in-frame deletion fragment in pGP704-Sac28-Km | This study |
| pGP704-Sac28-Km-mxdD | mxdD in-frame deletion fragment in pGP704-Sac28-Km | This study |
| pSM2360 | mob+, delivery plasmid for mini-Tn7-lacIq-Plac-gfp; Gmr Cmr Apr | Søren Molin, unpublished data |
| pUX-BF13 | mob+ori-R6K; helper plasmid providing the Tn7 transposition functions in trans; Apr | 4 |
| RK600 | ori-ColE1 RK2-mob+RK2-tra+; Cmr helper plasmid in matings | 26 |
| pMal-c2 | ColE1 ori lacIqmalE lacZα; Apr | New England Biolabs, Massachusetts |
| pBAD42 | araC; PBAD promoter; Cmr | J. Beckwith, unpublished data |
| pME6031 | repA oriVpVSIoriVp15AoriT; Tcr | 18 |
| pME6041 | repA oriVpVSIoriVp15AoriT; Kmr | 18 |
| pARA-VCA0956 | VCA0956 under control of araC-PBAD in pME6041 | This work |
| pARA-yhjH | yhjH under control of araC-PBAD in pME6041 | This work |
| pLacTac | lacIq1-Ptac in pME6031; Tcr | This work |
| pLacTac-VCA0956 | VCA0956 in pLacTac; Tcr | This work |
| pLacTac-SO4180(mxdA) | Cmr Tcr | This work |
Strain constructions in S. oneidensis MR-1.
All genetic work was carried out according to standard protocols or following the manufacturer's instructions. Kits for the isolation and/or purification of plasmid and chromosomal DNA or PCR fragments were obtained from QIAGEN (Valencia, CA), and enzymes were purchased from New England Biolabs (Beverly, MA), if not indicated otherwise.
In-frame deletion mutants were constructed in S. oneidensis MR-1 AS93 essentially as previously reported (48). Briefly, DNA fragments of the N- and C-terminal regions of the selected genes were amplified by PCR, introducing suitable restriction sites. The fragments were digested with BamHI and SalI, respectively, and subsequently ligated. An aliquot of the ligation mixture was used as a template in a second PCR amplification reaction with the forward primer of the upstream fragment and the reverse primer of the downstream fragment of the corresponding genes to generate the mutant allele. The products were purified, digested with NcoI and SacI, and ligated into the suicide vector pGP704-Sac28-Km. The resulting plasmids (pGP704-Sac28-Km-mxdA to -mxdD) were introduced into S. oneidensis MR-1 AS93 by mating with E. coli S-17I λpir, and mutants generated by single-crossover events were selected on LB plates containing kanamycin and gentamicin. To select for double recombinants, single colonies were grown overnight in the absence of antibiotics and plated on LB containing 8% (wt/vol) sucrose. Kanamycin-sensitive mutants were then screened for the gene deletion by colony PCR using primers up- and downstream of the deletion's location. The resulting mutants with mutations in mxdABCD were lacking amino acids (aa) 25 to 447, 7 to 274, 111 to 379, and 35 to 107, respectively.
To construct a system for controlled gene induction in S. oneidensis MR-1, genes of interest (gfp [for green fluorescent protein], VCA0956, and yhjH) were amplified by PCR using DNA from the corresponding chromosomal DNA (Vibrio cholerae ElTor N16961) (19), for VCA0956 and E. coli K12-MG1655 (5) for yhjH. Introduced NheI and PstI restriction sites were used to clone the products into pBAD42 (J. Beckwith, unpublished data). A fragment containing the repressor encoding gene araC, the corresponding inducible promoter region, and gfp, VCA0956, or yhjH was then released by NsiI and PstI restriction. The fragment was gel purified and ligated into vector pME6041 digested with PstI, resulting in plasmids pARA-gfp, pARA-VCA0956, and pARA-yhjH. Orientation of the fragment relative to the transcriptional terminator flanking the multiple cloning site of pME6041 was ensured by restriction analysis. A second inducible system was constructed based on the lacIq1-Ptac promoter system. A fragment containing the lacIq gene and the tac promoter was amplified from the vector pMAL-c2 (New England Biolabs), introducing BamHI and PstI restriction sites at the 5′ and 3′ ends, respectively. Additionally, the primer design yielded a mutation in the −35 region of the lacIq promoter, to obtain a higher expression of the repressor and thus a tighter repression of the system (17). The fragment was digested with BamHI and PstI and ligated into the broad host range vector pME6031 that was treated with the same enzymes, resulting in vector pLacTac. Genes to be cloned into this vector were amplified by PCR using chromosomal DNA of the corresponding microorganism, introducing PstI and EcoRI restriction sites at the 5′ and 3′ ends, respectively. Plasmids were introduced by electroporation (32). Functionality of both inducible systems was tested in the S. oneidensis MR-1 wild-type strain by induction of gfp in planktonic cultures (data not shown).
RNA extraction and reverse transcription-PCR (RT-PCR).
S. oneidensis MR-1 cells were grown in 50 ml MM for 7, 24, and 40 h, respectively, before 25 ml of culture was centrifuged for 5 min at 4°C and 4,000 × g. The cell pellet was resuspended in 1 ml ice-cold AE buffer (20 mM sodium acetate, 1 mM EDTA, pH 5.2) and centrifuged again for 5 min at 4°C and 15,000 × g. After resuspension of the pellet in 500 μl ice-cold AE buffer, 900 μl of phenol-chloroform-isoamylalcohol (25:24:1) preheated to 60°C and 10 μl of a 25% (wt/vol) sodium dodecyl sulfate solution were added and incubated for 10 min at 60°C. Following centrifugation at 4°C and 15,000 × g, the aqueous phase was transferred into a fresh tube, 62.5 μl of a 2 M sodium acetate solution (pH 5.2) and 500 μl phenol-chloroform-isoamylalcohol were added, followed by another centrifugation. This step was repeated until no interphase was visible. RNA was precipitated by the addition of 2.5 volumes of ethanol (96% vol/vol), incubation for 30 min at −80°C, and centrifugation at 4°C and 15,000 × g. Following two washing steps with 75% (vol/vol) ethanol, the RNA was dried at room temperature. Prior to the reverse transcriptase (RT) reaction, the RNA was subjected to a DNA digest. The RNA sediment was resuspended in 400 μl water, and 50 μl DNase buffer (40 mM Tris-HCl [pH 8], 10 mM NaCl, 6 mM MgCl2, and 10 mM CaCl2), and 5 μl DNaseI (10 U/μl) was added. The reaction mixture was incubated for 1 h at 37°C. Subsequently, the RNA was precipitated as described above; this step was repeated three times.
The RT reaction was carried out using the SuperScriptIII kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions at 25°C for 5 min, 1 h at 50°C, and 15 min at 70°C using 1 μg RNA in a 20-μl final volume. The product was used in a PCR using suitable primers, and RNA without RT treatment was used as a negative control.
Biofilm cultivation and image acquisition.
Biofilms were cultivated at 30°C in LM medium in three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm. Microscope coverslips (Fisher Scientific, Pittsburgh, PA) were used as a colonization surface, glued with silicone (GE Sealants and Adhesives, Hunterville, NC) onto the channels, and left to dry for 24 h at room temperature prior to use. Assembly, sterilization, and inoculation of the flow system were carried out essentially as previously described (47). Experiments were carried out in triplicate in at least two independent experiments.
For switching medium, the flow was arrested briefly, and the medium was exchanged in the bubble trap and the upstream tubing. This process took no longer than 1 min, and control channels, where the medium flow was stopped in parallel without changing the medium, ensured that the observed effects were not due to that short arrest. CLSM was performed at defined spots close to the inflow before and after the treatment.
Stop-of-flow-induced detachment was carried out as described previously (48). Briefly, the medium flow was arrested for 15 min and subsequently resumed for the same amount of time. CLSM images were taken immediately before the stop of flow and after 15 min of flow.
Microscopic visualization and image acquisition of biofilms were conducted at the Stanford Biofilm Research Center using an upright Zeiss LSM510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany) equipped with the following objectives: 10×/0.3 Plan-Neofluar, 20×/0.5 W Achroplan, and 40×/1.2 W C-Apochromat. For displaying biofilm images, CLSM images were processed using the IMARIS software package (Bitplane AG, Zürich, Switzerland) and Adobe Photoshop. Biofilm parameters such as biomass and average biofilm thickness were quantified with the COMSTAT program (20).
For complementation studies, biofilm experiments were carried out with 96-well microtiter plate assays using crystal violet, essentially as previously described (47). Strains were allowed to grow for 16 h prior to processing and spectrophotometrical quantification at 570 nm with a VERSAmax tunable microplate reader (Molecular Devices, California).
Extraction and quantification of c-di-GMP from Shewanella oneidensis.
Cells were grown at 30°C in 30 ml LM supplemented with 40 mM lactate and 0.2% l-arabinose to an optical density at 600 nm of 0.45, centrifuged, and washed with phosphate-buffered saline. The pellets were then resuspended in phosphate-buffered saline. c-di-GMP was extracted by heat and ethanol in triplicate (3, 43). Cells were heated at 100°C for 5 min, and then ethanol was added to a final concentration of 65%. Samples were centrifuged, the supernatant was retained, and the extraction was repeated. Combined supernatants were dried using a Speed-Vac and then stored for subsequent liquid chromatography-mass spectrometry (LC-MS) analysis of the c-di-GMP.
For LC-MS analysis, dried samples were resuspended in 50 μl of 10 mM ammonium acetate buffer, vortexed, ultrasonicated, and centrifuged, and the supernatant was retained. This was repeated, and the supernatants were combined.
Samples were analyzed at the Stanford Mass Spectrometry Facility. An Agilent 1100 high-performance liquid chromatography system equipped with an autosampler and degasser was used for solvent delivery and sample introduction. Samples (10 μl or 20 μl) were injected into a reverse-phase C18 Targa column (2.1 by 40 mm; Dp, 3 μm). The column was eluted at 30°C at a flow rate of 0.5 ml/min with the following gradient: 0 to 0.3 min, 0% B; 1.5 min, 90% B; 1.6 to 2.0 min, 0% B; 3.0 min, 90% B; 4.0 to 8.0 min, 0% B (A, 20 mM ammonium acetate; B, acetonitrile).
The Quattro Premier (MicroMassWaters) triple-quadrupole mass spectrometer equipped with electrospray ion source was used for peak detection. The collision energy was 30 eV, and the cone voltage was 35 V. c-di-GMP was detected in the multiple-reaction monitoring mode with the following transitions: 691.1→151.9, 691.1→248.0, and 691.1→539.8.
c-di-GMP was synthesized as previously described (21) and used as an aqueous solution in 10 mM ammonium acetate as an external authentic standard. A calibration curve was plotted for concentrations of 0, 31.25, 62.5, 125.0, 250.0, and 500.0 fmol/μl in triplicate; the graph was linear in this range, with a correlation coefficient, R2, of >0.99. The limit of detection of this method was 10 to 15 fmol.
RESULTS
Identification of an EPS biosynthesis operon in S. oneidensis MR-1.
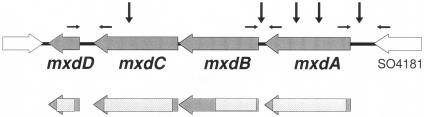
To elucidate the connection between attachment and detachment, we focused on components controlling attachment of S. oneidensis cells to biofilms. Based on the S. oneidensis genome sequence, no genes encoding enzymes for exopolysaccharide (EPS) biosynthesis with significant similarity to those in Pseudomonas aeruginosa, E. coli, or Vibrio cholerae were obvious. With a genetic analysis using Tn5 mutagenesis coupled to a 96-well-based screen, we previously identified several mutants defective in biofilm formation (47). Among the mutants isolated were five with transposon insertions mapping to five independent positions of the gene cluster designated SO4180-4177, which we subsequently named mxdABCD (for “biofilm matrix deficient”) because of the mutants' biofilm phenotype (see below). Two insertions were found in mxdA, one in the promoter region of mxdA, one in the intergenic region between mxdA and mxdB, and one in mxdC (Fig. 1). Analysis of the amino acid sequences of these open reading frames revealed that MxdA is predicted to be a 462-aa protein containing a C-terminal region (aa 321 to 461) with weak homology to a GGDEF domain. MxdB is predicted to be a membrane-associated 403-aa protein with homology to glycosyl transferases of the family GT 2 type, and MxdC is predicted to be a 351-aa membrane-associated protein with homology to efflux pump proteins. MxdD is oriented in the same direction as the previous genes and is predicted to be a 118-aa membrane-associated protein without homology to any known protein. The MxdB amino acid sequence was 25% identical and 42% similar to that of AcsAB, the cellulose synthase of G. xylinus, over a range of 192 amino acids. The orientation and sequences of the mxd genes were highly similar and homologous, respectively, only to the Vibrio parahaemolyticus RIMD genes VPA0392 to -94.
FIG. 1.
Organization of the mxdABCD genes in S. oneidensis MR-1. Horizontal arrows indicate the positions of primers used in transcription analyses, and vertical arrows mark the positions of transposon insertions that led to the identification of the gene cluster. Hatched areas indicate the in-frame deletions generated (see Materials and Methods). The mxdABCD genes correspond to the SO gene annotations designated SO4180-4177 (The Institute for Genomic Research).
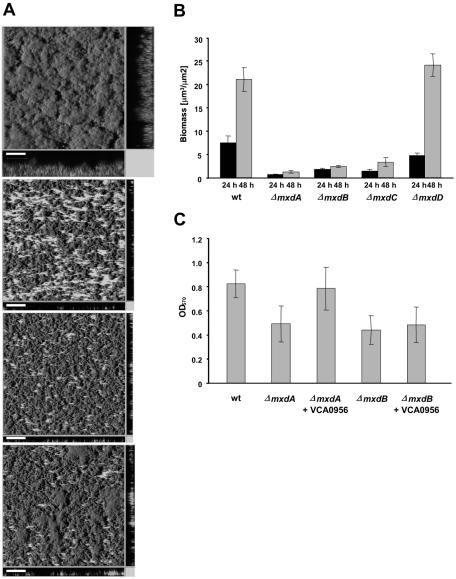
To examine the function of these genes in biofilm formation, we constructed in-frame deletion mutants of all genes and analyzed their phenotype in biofilms grown under hydrodynamic conditions (47). As Fig. 2A shows, deletion mutants of mxdA, mxdB, and mxdC exhibited strong defects in biofilm formation and were severely impacted in developing a three-dimensional architecture. The initial adhesion of these mutants appeared to be similar to the wild type (data not shown). The mutant biofilms were arrested at the stage of a cell monolayer or few cell layers and did not seem to progress from this stage even after 48 h of incubation (Fig. 2A). The most severe phenotype was visible for the ΔmxdA mutant (Fig. 2A). Quantification of biofilm biomass revealed that biofilms of ΔmxdA, ΔmxdB, and ΔmxdC had between 84 and 94% less biomass than wild-type biofilms (Fig. 2B) after 48 h. Deletion mutant ΔmxdD showed a delayed phenotype but progressed to a wild-type-like architecture after 48 h. The in-frame deletion of mxdA but not of mxdB could be complemented by VCA0956, a known c-di-GMP-forming diguanylate cyclase (Fig. 2C). The wild-type biofilm phenotype of both mutants could be restored by wild-type gene expression in trans (see Fig. 4) or by knock-in gene reconstructions with the wild-type alleles (data not shown). These data, together with the sequence analysis, suggested that the mxd genes might encode a gene cluster essential for biofilm matrix formation in S. oneidensis.
FIG.2.
Involvement of the mxdABCD genes in cell attachment and three-dimensional biofilm architecture. (A) Biofilm phenotypes of (from top to bottom) AS93 (wild-type control), ΔmxdA, ΔmxdB, and ΔmxdC in hydrodynamic flow chambers. Images display shadow projections of biofilms formed after 48 h. x-z and y-z cross-sectional images at selected positions in the biofilm are shown at the bottom and right side, respectively. Scale bar, 70 μm. (B) Quantification of biomass of the ΔmxdABCD mutants. CLSM images of the experiments shown in panel A were quantified by COMSTAT (20). (C) Complementation of ΔmxdA and ΔmxdB by GGDEF-encoding VCA0956. Biofilm formation in 96-well microtiter plates was measured by crystal violet staining of cells attached to the walls, as described in Materials and Methods.
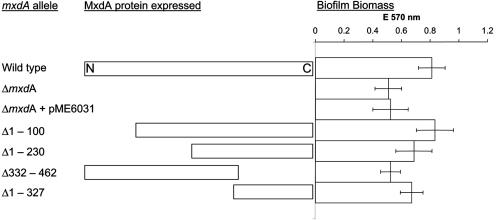
FIG. 4.
Truncation analysis of MxdA. 5′ and 3′ deletions were introduced in mxdA, and strains expressing these alleles in a ΔmxdA genetic background were tested for biofilm formation by rescuing the ΔmxdA phenotype. Rectangles on the left indicate the size and region of the expressed protein relative to the wild type; bars on the right indicate biofilm biomass formed in a 96-well plate assay. The MxdA NVDEF region spans amino acids at positions 358 to 362.
The transcriptional organization of the mxdABCD genes was examined by RT-PCR. When RNA prepared from cells grown to late exponential phase in MM containing 40 mM lactate (see Materials and Methods) was used as a template, RT-PCR products were obtained for primer pairs probing for a contiguous mRNA between mxdA and mxdB and between mxdB and mxdC (Fig. 1 and results not shown). The reading frames of mxdB and mxdC overlapped by one base and were therefore not probed. No RT-PCR product was observed from the primer pair combination SO4181 (upstream of mxdA) and mxdA. Preliminary transcriptional analysis revealed that mxdABCD mRNA was present in cells in late exponential and stationary phases but not in early or mid-exponential-growth phases (data not shown).
The role of c-di-GMP in biofilm formation and detachment.
The finding of a potential GGDEF protein (MxdA) in the vicinity of a putative glycosyl transferase (MxdB) was intriguing; we speculated that in S. oneidensis the activity of an EPS synthase, such as MxdB, might also be regulated by c-di-GMP, which is synthesized by GGDEF domain proteins, conceivably, MxdA. To examine this hypothesis, we tested whether the biofilm phenotype of ΔmxdA could be rescued by expressing VCA0956 in trans from an IPTG-inducible promoter. VCA0956 was previously shown to have c-di-GMP-forming diguanylate cyclase activity (see below) (49). As evident from Fig. 2C, ectopic expression of this GGDEF protein complemented the ΔmxdA mutation to wild-type levels. However, VCA0956 did not complement a ΔmxdB mutation, suggesting that the diguanylate cyclase activity or any other activity associated with VCA0956 is not sufficient for rescue of the biofilm phenotype.
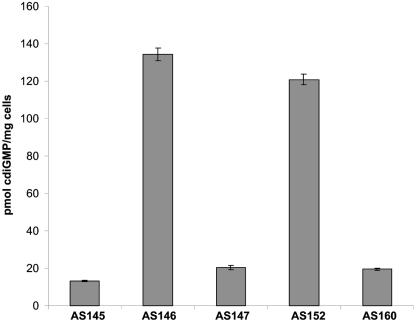
We then tested whether MxdA might encode a diguanylate cyclase by examination of the intracellular level of c-di-GMP. LM-grown cells of strain AS152, which were induced with arabinose, were harvested; c-di-GMP was extracted and quantified by LC-MS analysis as described in Materials and Methods. Authentic c-di-GMP was used as a standard. As Fig. 3 shows, S. oneidensis strains carrying overexpressed VCA0956 (AS146) or mxdA (AS152) contained intracellular c-di-GMP levels that were about fivefold higher than those in empty vector carrying wild-type control strain (AS160) or the yhjH-overexpressing strain (AS145) (see below). A truncation analysis of mxdA was conducted and revealed that the region with the most critical function, based on rescue of the mxdA biofilm phenotype, was a C-terminal region containing a NVDEF sequence at positions 358 to 362, which appears to resemble a modified GGDEF domain. Truncation of the first 230 amino acids resulted in intermediate complementation of the mxdA biofilm phenotype in a 96-well titer plate (Fig. 4). However, a mxdA allele where only the C-terminal NVDEF domain was deleted (Δ332-462) was unable to rescue the mutant phenotype. On the other hand, expression of the NVDEF domain alone was sufficient to partially confer rescue (Fig. 4). These data strongly suggest that MxdA can function as a diguanylate cyclase with an essential, modified GGDEF-like NVDEF domain. Moreover, these data provide the first indication that domains with a consensus motif significantly different from GGDEF, e.g., NVDEF can act as diguanylate cyclase in vivo.
FIG. 3.
Intracellular content of c-di-GMP. c-di-GMP content in planktonically grown cells were determined as described in Materials and Methods and is expressed per milligram (wet weight) of cells. The strains assayed included AS145 (AS93 plus pARA:yhjH), AS146 (AS93 plus pARA:VCA0956), AS147 (AS93 plus empty vector), AS152 (AS140 plus pLacTac:mxdA), and AS160 (AS140 plus empty vector). Error bars represent one standard deviation.
To test directly whether manipulation of the intracellular c-di-GMP concentration affects attachment and detachment of S. oneidensis, we constructed two strains with altered intracellular c-di-GMP concentration, strain AS146, which carried VCA0956, and AS145, which carried yhjH, an S. enterica serovar Typhimurium gene encoding an EAL domain-containing enzyme with c-di-GMP-hydrolyzing phosphodiesterase activity (43), under the control of an inducible PBAD promoter. We examined the phenotypes of these strains, including the empty vector-containing (pME6041) wild-type strain AS147, in the hydrodynamic biofilm system. Inoculum cells were grown in LB medium containing 25 μg/ml kanamycin, diluted in LM to an optical density at 600 nm of 0.01, and injected into the flow chambers to seed the glass surface. After a 40-min incubation, flow was initiated with LM containing 25 μg/ml kanamycin and 0.2% arabinose as an inducer. Figure 5 displays representative images of 24-h-old biofilms. AS146 biofilms displayed a dramatically increased thickness compared to AS147. Strain AS146 was determined to have a growth rate nearly identical to that of AS147 in planktonic culture. It is therefore likely that the increased thickness observed in the AS146 biofilms was due to increased retention of cells in the biofilm. S. oneidensis wild-type cells constantly shed from biofilms to a small extent during normal growth (data not shown). Cessation or reduction of such shedding could retain cells in the biofilm, as observed here. In contrast, analysis of biofilms formed by cells of yhjH-expressing strain AS145 resulted in a dramatically different phenotype. After the 40-min incubation period following the injection of AS145 cells into the flow chamber, the cell density on the glass surface was indistinguishable from that of the control (AS147) or AS146 strain (data not shown). However, 30 min after the flow of the arabinose-containing medium was started, the adhering cells began to separate from the surface. As a result, practically no biofilm of AS145 had formed after 24 h under inducing conditions (Fig. 5). This observation cannot be explained by the slightly lower growth rate of induced AS145 cells (ca. 70% of that of AS147 in planktonic culture). Rather, this unexpected finding indicates that induction of yhjH and, by inference, of lower c-di-GMP levels dramatically interferes with the maintenance of cell adhesion to the glass substratum. On the other hand, an increased level of cellular c-di-GMP leads to growth of a thick biofilm.
FIG. 5.
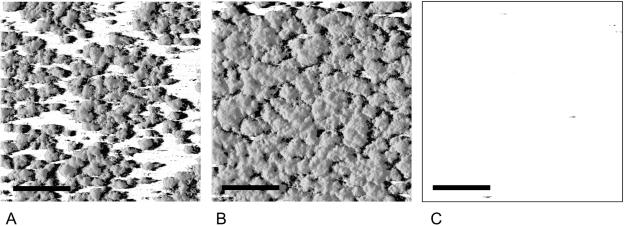
Effects of VCA0956 and yhjH expression on S. oneidensis biofilm architecture. Images display shadow projections of biofilms formed after 24 h of AS147 (A), AS146 constitutively overexpressing VCA0956 (B), and AS145 constitutively overexpressing yhjH (C). The inocula were grown in LB containing 25 μg/ml kanamycin, diluted to an optical density at 600 nm of 0.01, and injected into the flow chamber. After 40 min, the flow of LM containing 25 μg/ml kanamycin and 0.2% arabinose as inducer was initiated. Scale bar, 200 μm.
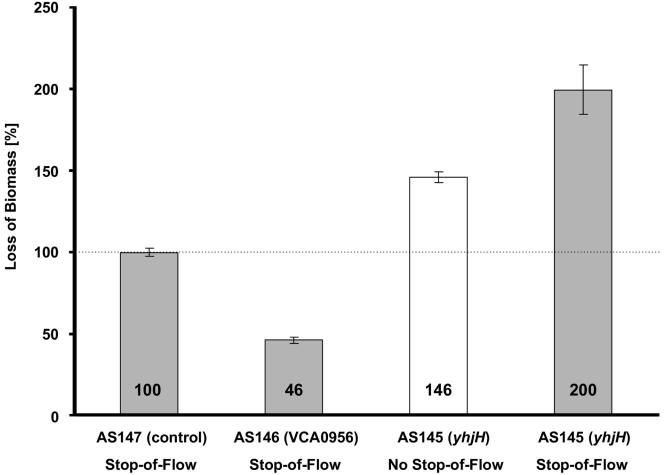
So far, our results collectively suggest a strong correlation between c-di-GMP levels and cellular attachment to the biofilm matrix. This control might be mediated through the putative glycosyl transferase MxdB. This hypothesis predicts that lowering of cellular c-di-GMP level might be sufficient to induce detachment of biofilms in the absence of an external environmental cue. We had previously shown that a decrease in molecular oxygen concentration functions in S. oneidensis biofilms as an external signal for detachment (48). To test this prediction, we grew biofilms of AS145 and AS146 in the hydrodynamic flow chamber in the absence of the inducer arabinose. After 20 h of growth, the flowthrough medium was amended with 0.2% arabinose, and biofilms were monitored by CLSM (Fig. 6). In contrast to AS147 and AS146, cells of AS145 began to detach from the biofilms in the absence of a stop of flow at 60 to 70 min after induction. After 90 min, 18% of the biofilm biomass was lost; and after 120 min, 38% of the biomass was lost. In contrast, wild-type biofilms harboring the empty plasmid (AS147) and biofilms of AS146 continued to increase in thickness after induction. A subsequent stop-of-flow treatment of AS146 biofilms induced detachment of only 50% of the wild type, while the same treatment released twice as much biomass from AS145 biofilms (Fig. 6). These observations suggest that not only attachment but also detachment are controlled by c-di-GMP. Furthermore, induction of a gene encoding an EAL-containing protein with c-di-GMP-hydrolyzing phosphodiesterase activity can bypass the requirement for an external environmental cue as the physiological signal and induce detachment.
FIG. 6.
Effects of VCA0956 and yhjH expression on developed S. oneidensis wild-type biofilms. Displayed are the amounts of biomass detached relative to the detached biomass of wild-type AS147 (set to 100%). Biofilms of strains AS147, AS145, and AS146 were grown in flow chambers for 20 h prior to induction with 0.2% (wt/vol) l-arabinose. After 90 min of induction, detachment was induced by a stop of flow (gray bars). The white bar indicates biomass detached from AS145 after 120 min without a stop of flow.
DISCUSSION
In this work, we examined the connection between attachment of biofilm cells to and detachment from the biofilm matrix in S. oneidensis and provided data suggesting that both processes are linked by the mxd genes and by c-di-GMP. Figure 7 summarizes the key components and their mode of interaction in controlling the transitioning of biofilm cells between attachment and detachment.
FIG. 7.
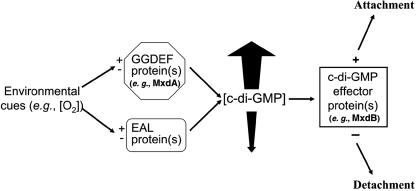
Model for control of attachment and detachment by c-di-GMP in S. oneidensis. An environmental cue is sensed by a sensor protein(s), which modulates the enzymatic activity of a c-di-GMP-forming diguanylate cyclase(s), such as GGDEF domain-containing proteins and MxdA, and/or of c-di-GMP-hydrolyzing phosphodiesterase(s), such as EAL domain-containing proteins. An altered general or localized c-di-GMP pool allosterically affects the activity of proteins or enzymes involved in attachment and/or detachment, such as MxdB. MxdA is postulated to be a key c-di-GMP-forming enzyme and to function in (structural) context with the putative glycosyl transferase MxdB. The c-di-GMP level (of a general intracellular or a localized pool) can be controlled in several ways: via activation and/or inhibition of a diguanylate cyclase(s) or by inhibition and/or activation of phosphodiesterases. Upon activation, MxdA catalyzes the formation of c-di-GMP, which stimulates the polysaccharide synthesis activity of MxdB, resulting in increased attachment. Downregulation of MxdA or activation of a phosphodiesterase(s) and subsequent decreased glycosyl transferase activity could result in decreased attachment (i.e., detachment). At this point, it is unclear whether detachment is due to the activation of a detachase or the inhibition of an attachment activity such as MxdB by low c-di-GMP levels.
With genetic experiments, we showed that a diguanylate cyclase encoded by mxdA is essential for matrix attachment and development of a three-dimensional biofilm architecture (Fig. 2). The predicted MxdA gene product contains the essential C-terminal sequence NVDEF with weak homology to a GGDEF motif (Fig. 4); overexpression of mxdA increased the cellular c-di-GMP level (Fig. 3); the ΔmxdA biofilm phenotype could be rescued by VCA0956, a gene encoding a GGDEF domain-containing protein with c-di-GMP-forming diguanylate cyclase activity (Fig. 2C) (49). For these reasons, we concluded that MxdA contains diguanylate cyclase activity and when activated increases the in vivo cellular c-di-GMP level. This results in enhanced attachment and increased biofilm formation (Fig. 5). We postulate that this positive c-di-GMP signaling acts through MxdB, a putative membrane-associated glycosyl transferase (Fig. 2C). The mxdB gene product is essential for cell attachment to the matrix but not for cell adhesion to the substratum surface (Fig. 2). Massive detachment, which was qualitatively and quantitatively indistinguishable from oxygen starvation-induced detachment (43), could be induced by overexpression of yhjH encoding an EAL-domain containing protein (Fig. 5 and 6). This protein was previously shown to have phosphodiesterase activity towards c-di-GMP (43). Thus, by simply modulating the intracellular c-di-GMP pool via overexpression of a diguanylate cyclase or phosphodiesterase, respectively, we were able to shift the state of biofilm cells from attachment to detachment and vice versa under the same experimental conditions. In addition, this manipulation of the intracellular c-di-GMP pool allowed us to uncouple the physiological detachment response from the external environmental cue (Fig. 6) (48). We postulate that the mode of action of c-di-GMP in attachment and detachment is not primarily via control of gene regulation, as has been directly or indirectly postulated for several other systems, but by a direct (e.g., allosteric) control, for example, of a polysaccharide-synthesizing enzyme such as the MxdB (Fig. 7) (R. M. Saville et al., unpublished data). This speculation is based on the apparent functional similarity of the predicted mxdABCD gene products to cellulose synthase from Gluconacetobacter xylinus, including its activation by c-di-GMP. An alternative mode of action of intracellular c-di-GMP could be in indirect control of detachment and attachment by modulating cellular sensitivity to variation in local oxygen concentrations, where low c-di-GMP levels render cells more susceptible to fluctuations in oxygen. In light of these results, our previously identified dependence of the detachability of S. oneidensis biofilms on CRP, ArcA, and EtrA can therefore be attributed to an indirect role of these transcriptional regulators. For example, they may be required for controlling the expression of the molecular detachment/attachment machinery or signal transduction components, as we had previously suggested (48).
Cellular c-di-GMP levels have been implicated in bacterial biofilm formation, in most cases, via control of exopolysaccharide production (22, 27, 37, 43, 49). Recent work demonstrated that diguanylate cyclase and phosphodiesterase activities of GGDEF domain- and EAL domain-containing proteins control the intracellular concentrations of c-di-GMP in several microorganisms, including G. xylinus, Salmonella enterica serovar Typhimurium, Yersinia, and Staphylococci (6, 8, 25, 40, 42, 43). The Vibrio cholerae EAL domain-containing protein VieA negatively controls the expression of vpsABCDEFGHIJK and vpsLMNOPQ genes by controlling expression of the regulator VpsR (49). For VieS, which is hypothesized to control the phosphodiesterase activity of VieA by transphosphorylation, amino acids are thought to be the environmental trigger. However, no direct evidence has been reported (49). Previously, Simm et al. (43) presented experiments with Salmonella enterica serovar Typhimurium, E. coli, and P. aeruginosa that showed enhanced biofilm formation in adrA (GGDEF protein)-overexpressing strains and enhanced swarming motility in yhjH-expressing strains (expressing an EAL domain protein). These observations led the authors to conclude that c-di-GMP signaling controls the transitioning between sessility and motility and to infer that c-di-GMP might act by allosteric regulation. Notably, biofilm formation and the swarming behavior were observed under different physiological conditions (adherence to glass tubes versus swarming agar plates), and no direct transitioning between sessility and motility was studied. In addition, cell movement does occur in sessile biofilms (28), and motility is not required for dissolution of a biofilm, as corroborated by the observation that flagellum-dependent motility is not required for detachment in S. oneidensis (48). Other experiments in our laboratory demonstrated a direct, nontranscriptional control of detachment (R. M. Saville and A. M. Spormann, unpublished data) and strongly suggest that this link is an allosteric control based on c-di-GMP. The model of detachment proposed here resembles that of Gjermansen et al. (16). Mutations suppressing the carbon starvation-induced detachment of Pseudomonas putida cells mapped to GGDEF-EAL domain-containing proteins (16). These mutants carry an enhanced stickiness.
The apparent high abundance of GGDEF and EAL domain proteins, as predicted from the genome sequence of S. oneidensis (51 GGDEF, 27 EAL, and 20 GGDEF-EAL hybrid proteins) and other environmental microorganisms (14, 37), raises the question of how multiple diverse environmental cues can be integrated into physiologically diverse output responses, such as motility, exopolysaccharide biosynthesis, cell cycle events, and biofilm detachment via a common intracellular c-di-GMP pool. Formally, it is conceivable that c-di-GMP signaling reflects a global physiological state and that other additional cellular signals are necessary to elicit a particular physiological response. However, our overexpression experiments using VCA0956 and yhjH demonstrated that altering c-di-GMP levels was sufficient to induce an attachment or detachment response. A model alternative to signaling via a general c-di-GMP pool would be multiple, localized c-di-GMP signaling, as considered by Simm et al. (43). c-di-GMP synthesis and hydrolysis in the immediate protein vicinity of c-di-GMP effector proteins, such as MxdB, would assure specific signaling. In support of this hypothesis is the finding of multiple proteins containing GGDEF and/or EAL domains in S. oneidensis and other microbes, which could enable a rapid, localized turnover of c-di-GMP by environmental control of both enzyme activities. Such locally produced c-di-GMP molecules would exchange only minimally with a general cellular pool of free c-di-GMP. Our results and those of others on overexpressing GGDEF and EAL domain proteins do not rule out the possibility of such localized signaling, since the cellular overexpression of such enzyme activities would presumably vastly change the overall cellular c-di-GMP concentration, in turn affecting localized signaling even by a minimal exchange. If confirmed, it would also mean that microbial cells are more compartmentalized than previously thought. Another implication of our results is that proteins with sequences other than the canonical GGDEF sequence could act as diguanylate cyclases. As our data showed, the NVDEF region of MxdA is essential for its function.
Further implications of our model are that attachment per se might be due to environmentally controlled exopolysaccharide biosynthesis and that detachment might simply be a consequence of a controlled cessation or reduction of such activity. Thus, the molecular basis of biofilm stability and resilience could be an exopolysaccharide-based biochemistry, which mediates the reversible interaction between a biofilm cell and the biofilm matrix. Although polysaccharides have long been recognized as playing an important role in biofilms (45), we hypothesize from this work that the molecular basis for attachment could include a tight association of the exopolysaccharide-producing cell with the excreted polysaccharide, e.g., via a covalent bond to a exopolysaccharide-producing enzyme, such as the membrane-associated glycosyl transferase MxdB. The free, excreted polyalcohol end of the exported polysaccharides could then associate with the biofilm matrix via numerous hydrogen bonds to exopolysaccharides or DNA released from other cells (45, 53). Experiments testing these hypotheses are in progress. It is conceivable that such a mechanism could also exist with modifications in other microorganisms, such as Pseudomonas aeruginosa, Vibrio cholerae, and E. coli.
Acknowledgments
We thank Soeren Molin for providing pSM2360 and for many helpful discussions. We are also grateful to Lucy Shapiro and Dieter Haas and their research groups for kindly providing plasmid pBAD42 and the pME vectors, respectively.
This research was supported by grants to A.M.S. from the National Science Foundation for the Stanford EMSI and CAMPWS and to Y.H. by Grants-in-Aid for Scientific Research (no. 16011223 and 16350086) from the Ministry of Education, Science, Sports and Culture of Japan, and Mitsui Chemicals, Inc.
REFERENCES
- 1.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal.2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 2.Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert.1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 3.Amikam, D., O. Steinberger, T. Shkolnik, and Z. Ben-Ishai.1995. The novel cyclic dinucleotide 3′-5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem. J. 311:921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouillette, E., M. Hyodo, Y. Hayakawa, D. K. Karaolis, and F. Malouin. 2005. 3′,5′-Cyclic diguanylic acid reduces the virulence of biofilm-forming Staphylococcus aureus strains in a mouse model of mastitis infection. Antimicrob. Agents Chemother. 49:3109-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, A. L., J. R. Tuckerman, G. Gonzalez, R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. A. Gilles-Gonzalez. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40:3420-3426. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, J. W., Z. Lewandowski, D. Debeer, D. Caldwell, D. Korber, and G. James. 1994. Biofilms, the customized microniche. J. Bacteriol. 176:2137-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Ann. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 13.Dow, J. M., L. Crossman, K. Findlay, Y. Q. He, J. X. Feng, and J. L. Tang. 2003. iofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 100:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 15.Gjermansen, M., and T. Tolker-Nielsen. 2003. Programmed dissolution of Pseudomonas putida biofilms. Poster no. 211(A). Presented at Biofilms 2003, American Society for Microbiology.
- 16.Gjermansen, M., P. Ragas, C. Sternberg, S. Molin, and T. Tolker-Nielsen. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7:894-906. [DOI] [PubMed] [Google Scholar]
- 17.Glascock, C. B., and M. J. Weickert. 1998. Using chromosomal lacIQ1 to control expression of genes on high-copy-number plasmids in Escherichia coli. Gene 223:221-231. [DOI] [PubMed] [Google Scholar]
- 18.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 19.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 21.Hyodo, M., and Y. Hayakawa. 2005. An improved method for synthesizing cyclic bis(3′-5′)diguanylic Acid (c-di-GMP). Bull. Chem. Soc. Jpn. 77:2089-2093. [Google Scholar]
- 22.Itoh, Y., X. Wang, B. J. Hinnebusch, J. F. Preston III, and T. Romeo. 2005. Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaolis, D. K., M. H. Rashid, R. Chythanya, W. Luo, M. Hyodo, and Y. Hayakawa. 2005. c-di-GMP (3′-5′-cyclic diguanylic acid) inhibits Staphylococcus aureus cell-cell interactions and biofilm formation. Antimicrob. Agents Chemother. 49:1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 27.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:75-88. [DOI] [PubMed] [Google Scholar]
- 28.Klausen, M., A. Aaes-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1995. Sequence of a novel virulence-mediating gene, virC, from Vibrio anguillarum. Gene 164:95-100. [DOI] [PubMed] [Google Scholar]
- 31.Myers, C. R., and K. H. Nealson. 1988. Bacterial Manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 32.Myers, C. R., and J. M. Myers. 1997. Replication of plasmids with the p15A origin in Shewanella putrefaciens MR-1. Lett. Appl. Microbiol. 24:221-225. [DOI] [PubMed] [Google Scholar]
- 33.Nealson, K. H., A. Belz, and B. McKee. 2002. Breathing metals as a way of life: geobiology in action. Antonie Leeuwenhoek 81:215-222. [DOI] [PubMed] [Google Scholar]
- 34.Ott, C. M., D. F. Day, D. W. Koenig, and D. L. Pierson. 2001. The release of alginate lyase from growing Pseudomonas syringae pathovar phaseolicola. Curr. Microbiol. 42:78-81. [DOI] [PubMed] [Google Scholar]
- 35.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 36.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romling, U., M. Gomelsky, and M. Y. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 38.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. van der Marel, J. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 39.Ross, P., R. Mayer, H. Weinhouse, D. Amikam, Y. Huggirat, M. Benziman, E. de Vroom, A. Fidder, P. de Paus, L. A. Sliedregt, G. A. van der Marel, and J. H. van Boom. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933-18943. [PubMed] [Google Scholar]
- 40.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauer, K., M. C. Cullen, A. H. Rickard, L. A. Zeef, D. G. Davies, and P. Gilbert. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004.GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 44.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 45.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 46.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thormann, K. M., R. Saville, S. Shukla, and A. M. Spormann. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thormann, K. M., R. Saville, S. Shukla, and A. M. Spormann. 2005. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 187:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol, 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolker-Nielsen, T., and S. Molin. 2000. Spatial Organization of microbial biofilm communities. Microb. Ecol. 40:75-84. [DOI] [PubMed] [Google Scholar]
- 51.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 52.Ward, M. J., Q. S. Fu, K. R. Rhoads, C. H. Yeung, A. M. Spormann, and C. S. Criddle. 2003. A derivative of the menaquinone precursor 1,4-dihydroxy-2-naphthoate is involved in the reductive transformation of carbon tetrachloride by aerobically grown Shewanella oneidensis MR-1. Appl. Microbiol. Biotechnol. 63:571-577. [DOI] [PubMed] [Google Scholar]
- 53.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 54.Xun, L. Y., R. A. Mah, and D. R. Boone. 1990. Isolation and characterization of disaggregatase from Methanosarcina mazei LYC. Appl. Environ. Microbiol. 56:3693-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]