Abstract
Twelve genes encoding key components of Clostridium cellulolyticum cellulosomes are clustered. Among them, the first, second, and fifth genes encode the assembly factor CipC and the two major cellulases Cel48F and Cel9E, respectively. Cellulolytic clones were selected from the noncellulolytic cipC insertional mutant trans-complemented with a cipC expression vector, in which one homologous recombination event between the 3′ end of the chromosomal cipC gene and the plasmidic cipC gene has restored the cluster continuity. The absence of the enzymes encoded by the cluster in the cipC mutant was thus only due to a strong polar effect, indicating that all genes were transcriptionally linked. Two large transcripts were detected in cellulose-grown cells by Northern hybridization: a 14-kb messenger which carries the cipC-cel48F-cel8C-cel9G-cel9E coding sequences and, in a smaller amount, a 12-kb messenger which carries the genes located in the 3′ part of the cluster. Four smaller transcripts were found in large amounts: a cipC-cel48F bicistronic one and three monocistronic ones, cipC, cel48F, and cel9E. The cipC-cel48F and cel48F messengers were shown to be stable. Analysis by reverse transcription-PCR suggested transcriptional linkage of all of the open reading frames. The production of a primary very large transcript covering the entire cluster was hypothesized. Primer extension analysis has identified two putative transcriptional start sites located 638/637 and 194 nucleotides upstream of the cipC translational start. The processing of the primary transcript would lead to the production of several secondary messengers displaying different stabilities, contributing to fine tuning of expression of individual genes of the operon.
Clostridium cellulolyticum, an anaerobic mesophilic bacterium, produces high-molecular-weight cellulolytic complexes named cellulosomes (6). These extracellular complexes act very efficiently on crystalline cellulose, liberating soluble oligosaccharides which are used by the bacteria as carbon and energy sources (4). Each cellulosome is composed of up to nine proteins: a scaffolding protein (CipC) and eight enzymes. These proteins can be assembled via high-affinity binding between specific modules: the cohesin modules carried by the CipC protein and the dockerin modules borne by the cellulosomal enzymes (19, 20). Twenty-two different dockerin-bearing proteins have been identified either by gene cloning or by far-Western blot analysis of the cellulolytic system with a biotinylated cohesin-bearing protein as a probe (16). The composition of the cellulosomes is thus heterogeneous and appears to be regulated by the different amounts of the available enzymes (22, 23).
Twelve genes of this enzymatic system are located in a large cluster spanning 26 kb (2). The gene encoding CipC is the first gene of this cluster. It is immediately followed by 11 genes (cel48F-cel8C-cel9G-cel9E-orfX-cel9H-cel9J-man5K-cel9M-rgl11Y-cel5N) coding for eight cellulases (Cel proteins), one mannanase (Man5K), and one pectinase (Rgl11Y) (21, 22). Cel48F and Cel9E are the major enzymatic components of the complexes (7, 27). Several genes encoding cellulosomal cellulases are located outside of the cip-cel cluster (cel5A, cel5D, and cel44O) (14, 29). Nevertheless, the cellulases encoded by the cluster were found to be essential for the building of cellulosomes efficient in crystalline cellulose degradation (16). Indeed, the insertional mutant strain cipCMut1 and the trans-complemented strain cipCMut1(pSOScipC) were both severely affected in their cellulolytic abilities compared to the wild-type strain (16). The latter strain produces complexes assembled on the trans-produced CipC protein. These complexes, although containing at least a dozen of dockerin-bearing proteins, do not contain any of the enzymes encoded by the cip-cel gene cluster.
The transcriptional organization of the cip-cel cluster and the regulation of the expression of the genes coding for the various components of the cellulolytic system of C. cellulolyticum are poorly known. Available data are mainly from the study of the first insertional mutant of this system, cipCMut1 (16). Indeed, the apparent polar effect of the cipC mutation suggests that numerous genes, including those extremely distant from cipC, such as rgl11Y, may all be transcriptionally linked in the cluster. Northern blotting of wild-type RNAs probed with a digoxigenin-labeled cipC antisense RNA showed that the first two genes (cipC and cel48F) were cotranscribed. One of the major messengers found was long enough (7.5 kb) to carry both the cipC open reading frame (ORF) (4,640 bases) and the following one, the cel48F ORF (2,168 bases), suggesting that cipC and cel48F might be partly translated from a polycistronic common mRNA. Moreover, the transcriptional linkage of two other genes of the cluster, cel8C and cel9G, was demonstrated in a previous study (1).
The present study aims to complete the data on the transcriptional organization of this key cip-cel gene cluster, by Northern hybridization with highly radiolabeled probes, reverse transcription (RT)-PCR analysis and primer extension analysis.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α was used as the recipient strain for the recombinant plasmids (derivatives of pSPT18 and pGEMT-Easy) (Roche Applied Science, Promega). It was grown at 37°C in Luria-Bertani medium, supplemented with ampicillin (100 μg ml−1) when required. Clostridium cellulolyticum (ATCC 35319) and the mutant strains cipCMut1, cipCMut1(pSOSzero), and cipCMut1(pSOScipC) were cultured anaerobically at 32°C on basal medium (BM) (8) supplemented with MN300 cellulose (5 g liter−1) (Serva). The medium used for the mutant strain cultures was supplemented with cellobiose (0.4 g liter−1) (Sigma-Aldrich). Erythromycin (10 μg ml−1) was added in transformant cultures. Recombinant clones were isolated from the cipCMut1(pSOScipC) strain on solid BM supplemented with agar (15 g liter−1), cellobiose (2 g liter−1), and erythromycin (10 μg ml−1). Cells were embedded into 4 ml of 8 g liter−1 agar and 20 g liter−1 MN300 cellulose-containing BM and overlaid onto solid medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F−endA1 hsdR17(rK− mK+) supE44 thi-1 λ−gyrA96 relA1 Δ(lacZYA argF)U169 (φ80 lacZ ΔM15) recA1 | Roche Diagnostics |
| Clostridium cellulolyticum | ||
| ATCC 35319 | Wild type | 24 |
| cipCMut1 | cipC::ISCce1::ISCce2 | 16 |
| cipCMut1 (pSOSzero) | cipCMut1 pSOSzero transformant | This study |
| cipCMut1 (pSOScipC) | cipCMut1 pSOScipC transformant | 16 |
| Plasmids | ||
| pSOSzero | pSOS95 derivative with entire expression cassette deleted | 23 |
| pSOScipC | pSOS952 derivative expression vector bearing cipC gene | 16 |
| pZ41 | pUC18 derivative carrying 6.3-kb PvuII fragment bearing cel9G gene | 1 |
| pLM26 | pUC13 derivative carrying 5-kb HindIII/Sau3AI fragment bearing cel8C | 1 |
| pETFc | E. coli expression vector bearing cel48F gene | 26 |
| pSPT18 | Cloning vector; Apr | Roche Diagnostics |
| pSPTcipC | pSPT18 derivative carrying 1.1-kb HindIII/BamHI fragment of cipC | 16 |
| pSPTcel48F | pSPT18 derivative carrying 0.9-kb BamHI/Asp718 fragment of cel48F | This study |
| pSPTcel8C | pSPT18 derivative carrying 0.9-kb PvuII fragment of cel8C | This study |
| pSPTcel9G | pSPT18 derivative carrying 0.75-kb HindIII/BglII fragment of cel9G | This study |
| pSPTcel9E | pSPT18 derivative carrying 0.9-kb EcoRI-digested PCRa fragment of cel9E | This study |
| pSPTcel9H | pSPT18 derivative carrying HincII/EcoRV-digested PCRa fragment of cel9H | This study |
| pSPTcel9J | pSPT18 derivative carrying PstI/PvuII-digested PCRa fragment of cel9J | This study |
| pSPTrgl11Y | pSPT18 derivative carrying SspI/HincII-digested PCRa fragment of rgl11Y | This study |
| pGEM-T Easy | Cloning vector; Apr | Promega |
| pGEMorfX | pGEM-T Easy derivative carrying PCRa fragment of orfX | This study |
| pGEMman5K | pGEM-T Easy derivative carrying BspEI/NcoI-digested PCRa fragment of man5K | This study |
| pGEMcel9M | pGEM-T Easy derivative carrying PCRa fragment of cel9M | This study |
| pGEMcel5N | pGEM-T Easy derivative carrying PCRa fragment of cel5N | This study |
| pGEM16S | pGEM-T Easy derivative carrying PCRa fragment of 16S gene | This study |
The primers used to synthesize the PCR fragments are listed in Table 2.
DNA isolation, cloning, and molecular techniques.
Chromosomal DNA was obtained from C. cellulolyticum, using the genomic DNA purification kit (Promega). Large- and small-scale plasmid purification from E. coli was performed, using kits from QIAGEN and Promega. Restriction enzymes and DNA-modifying enzymes were purchased from Promega and Roche Applied Science and used as recommended by the manufacturer. DNA sequences located upstream from the ORF cipC and downstream of the ORF cel5N in the genome of C. cellulolyticum were amplified by inverse PCR (18). Total chromosomal DNA of the strain was digested by a restriction enzyme cutting the gene near the unknown sequence. To determine the sequence upstream from cipC and downstream from cel5N, DNA was digested, by HinfI, and DraI, respectively. The resulting fragments were ligated and used as templates for PCR amplification with divergent primer pairs (Table 2): Cip-IPCRdir/Cip-IPCRrev and N-IPCRdir/N-IPCRrev, respectively (18). The inverse PCR products were then purified using the Nucleospin extract purification kit (Macherey Nagel) and cloned into the pGEMT-Easy vector prior to sequence analysis. DNA sequencing was performed by Genome Express (Grenoble, France). Recombinant chromosome analysis was performed by PCR using primers Cip-PCRdir, F-PCRrev, ISCce1-PCRdir, ISCce1-PCRrev, and Sos-PCRdir (Table 2) with genomic DNA from the strain cipCMut1 or cipCMut1(pSOScipC) clones as the template.
TABLE 2.
Primers used in this study
| Primer namea | 5′-to-3′ sequence | Positionb |
|---|---|---|
| Cip-RTdir | TGTTGTTAAATCAGGTAATGTAGG | 4291 |
| F-RTrev | TCTTCCGTGCATTGCTTCCAACC | 294 |
| F-RTdir | GCGAAATCAAGTTCAATGCTGACGG | 1426 |
| C-RTrev | ATCAGCTGCACTTGAAGCGATG | 102 |
| C-RTdir | AATGATGGTGCTATAGATGC | 1226 |
| G-RTrev | TGCAGATGTATATGACATAGGTAG | 318 |
| G-RTdir | CACTTCCAGCATAGATGTTAAGG | 2058 |
| E-RTrev | GAGCAGTTATCTTGTACTTAGG | 220 |
| E-RTdir | GTCAGTGACTCCTCCAACTA | 2463 |
| X-RTrev | AAGCAGATGCAGTTGGTGATG | 154 |
| X-RTdir | GATACAACAGGTCTGGTCAG | 472 |
| H-RTrev | TCTGTTAATTGAGCATCTCC | 227 |
| H-RTdir | TATTGATGCATTGGACGTTG | 2067 |
| J-RTrev | GCCTGTGACATATTAGTTCC | 280 |
| J-RTdir | AAAGTGTATGGAAATGAACCG | 2049 |
| K-RTrev | GAGTATCATTGTCAACATCACC | 115 |
| K-RTdir | CAGAGCATCCTGACGTAGTC | 1251 |
| M-RTrev | ATTCGTACAATGACCAACCT | 336 |
| M-RTdir | GTTCTTTGGAGGTACGATTG | 1374 |
| Y-RTrev | ACCAGAGGTAATTGGAGTGG | 246 |
| Y-RTdir | GGTCAATCAACAGAGTTAGATAC | 1933 |
| N-RTrev | GTTTGGTTTACCAGTCATCC | 333 |
| Cip-PE1 | CCTAAAGACTTTTTACGCATTG | 20 |
| Cip-PE2 | TTTAGGCATACTGATATAACTGAA | −121 |
| Cip-PE3 | TTAAGCCGTTTACCTCAGGTGC | −512 |
| F-PE1 | ATTCTTACTCATTTTACACCTTCC | 12 |
| F-PE2 | AGAGTAAAATCTGCTCATATGTTT | −35 |
| C-PE1 | AACCTTTGATCATGTTCTTACCT | 13 |
| G-PE1 | TCTTAAGCAACAAGCCATCACTCC | 11 |
| E-PE1 | AACCTTTTTTTCATATTTACCTCC | 14 |
| E-RCdir | TGGTATTGCAACCTGGACCGTAA | 1158 |
| E-RCrev | GAAGATCTAACAGTGTGATTTTTC | 2658 |
| X-RCdir | GGTAAACCATTTTACATCTGATT | 63 |
| X-RCrev | TTCAGTATCTCCAAATGCACC | 638 |
| H-RCdir | GCTTTTAACTATGGGGAGGCTT | 103 |
| H-RCrev | GCAAAATCGAGAGCATCAATA | 2183 |
| J-RCdir | CCTCCTTTCAACTACGCAGAAGCATTT | 106 |
| J-RCrev | TTTACCTTCGTCATACATAACCGCAAC | 1809 |
| K-RCdir | GATTCCATATGGCTGCGACTACAT ACAAA | 72 |
| K-RCrev | GGGCTCGAGTCTGACTACGTCAGGATG | 1272 |
| M-RCdir | GGGGTGCACCCGAGGAGCAAA | 476 |
| M-RCrev | AACAATCGTACCTCCAAAGAAC | 1395 |
| Y-RCdir | AATTCCATATGACGGCAGCATCGG CACGC | 76 |
| Y-RCrev | GGGCTCGAGCGAAAGCAAACTTGCT TTTAG | 2030 |
| N-RCdir | GGCAGACAGAGGAATAGGGTT | 255 |
| N-RCrev | TATCAATCCGCCTGTATCACCTG | 1257 |
| 16S-RCdir | CCGAATTCGTCGACAACAGAGTTT GATCCTGGCTCAG | 8 (E. coli) |
| 16S-RCrev | TACGGTTACCTTGTTACGAC | 1494 (E. coli) |
| 16S-RS | CCGTCAATTCCTTTGAGTTT | 907 (E. coli) |
| F-RS | ACTAGTATCCAACGGAG | 471 |
| Cip-IPCRdir | GTAAGCACCTGAGGTAAACGG | −536 |
| Cip-IPCRrev | CCATTTATGTAATTAAATGACAAG | −649 |
| N-IPCRdir | CCAAAATGTCACGTTTAGCAAAGGA AGTAT | 862 |
| N-IPCRrev | GATACTTATCCAAAGCTGCCTC | 879 |
| N-S1 | CTTTGGAAGCATACATGAGG | 323/orf4 |
| N-S2 | CTCATGGAATTCAGGAACTAT | 528/orf5 |
| Cip-S1 | TACTAAAATCACTTATTGTCAGTAC | 650/orf2 |
| Cip-S2 | GCATAGCACATATGACCTTC | 819/orf2 |
| Cip-S3 | TGGCATTGGTAGCTCAGGAT | 433/orf1 |
The primer's name includes the letter or name of the gene followed by the analysis for which it was designed: RT, RT-PCR; PE, primer extension; RC, riboprobe cloning; RS, riboprobe synthesis; IPCR, inverse PCR; and S, sequencing. Dir and rev indicate direct and reverse primers, respectively.
The position indicated refers to the position of the primer 5′ nucleotide on the corresponding gene, with the nucleotide numbering beginning from the first codon of the gene. The sequences of the corresponding genes are described in GenBank under accession no. U40345, U30321, M87018, AF316823, and DQ156106.
Protein analysis.
Proteins (5 μg) eluted from residual cellulose (Fc fraction) as described by Maamar et al. (16) were separated by 7.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis using a Phast system apparatus (Amersham Biosciences). The proteins were stained with Coomassie blue or electrotransferred onto nitrocellulose BA83 membrane (Schleicher & Schuell). The Western blot was incubated with a polyclonal rabbit antiserum raised against Cel9E and then revealed using the enhanced chemiluminescence detection system (Amersham Biosciences).
RNA isolation.
Total RNAs were isolated from cells grown on cellulose-containing BM as described previously (16). The culture (600 ml) was stopped at the end of the exponential phase of growth (6 days). The cells were collected by pipetting, taking care to not disturb the sedimented cellulose. After centrifugation, cells were resuspended in 2.5 ml of lysis buffer (30 mM Tris-HCl, pH 8, 100 mM NaCl, 5 mM EDTA, 1% SDS) and RNAs were purified as previously described (16). Total RNAs were quantified by spectrophotometric analysis at 260 nm.
Northern blot analysis.
RNAs were denatured in RNA sample buffer (50% formamide, 40 mM MOPS [morpholinopropanesulfonic acid; pH 7], 10 mM sodium acetate, 1 mM EDTA, 2.2 M formaldehyde, 8.33% glycerol) at 65°C for 5 min and separated by electrophoresis through 0.8% agarose gel containing formaldehyde (0.22 M) in running buffer (40 mM MOPS [pH 7], 10 mM sodium acetate, 1 mM EDTA, 0.22 M formaldehyde). RNAs were transferred overnight to a positively charged nylon membrane (Roche Applied Science) by capillary transfer using 20× SSC buffer (1.5 M NaCl, 0.15 M sodium citrate [pH 7]; Promega), and hybridized with an excess of α-32P-labeled antisense RNA or Cy5-labeled cDNA probes in the Ultra-Hyb hybridization solution (Ambion) at 68°C or 42°C overnight, respectively. For the radioactive probes, Hyperfilm (Amersham Biosciences) were used for autoradiography. For Cy5-labeled cDNA probes, the Fluorescent Science Imaging System FLA-5000 (Fujifilm) with the red laser diode (635 nm) and the light pollution reduction filter was used for detection.
The different antisense RNA probes were synthesized from the linearized appropriate constructs (pSPTcipC, pSPTcel48F, pSPTcel8C, pSPTcel9G, pSPTcel9E, pSPTcel9H, pSPTcel9J, pSPTrgl11Y, pGEMorfX, pGEMman5K, pGEMcel9M, pGEMcel5N, and pGEM16S) by in vitro runoff transcription using the SP6 or the T7 RNA polymerase (Roche Applied Science). The reaction mix (20 μl) consisted of the linearized vector (1 μg), 1× transcription buffer, rNTP (Promega) (rATP, rCTP, and rGTP at 0.19 mM each and rUTP at 0.019 mM), RNase inhibitor (Rnasin Promega, 40 U), [α-32P]UTP (Amersham Biosciences; 300 μCi), and RNA polymerase (20 U). The reaction mixture was incubated 2 h at 37°C, and then subjected to a DNase I, RNase-free treatment (20 U; Roche Applied Science) during 15 min at 37°C to eliminate the vector. The reaction was stopped with 4 μM EDTA (pH 7.5) and incubated 10 min at 65°C to inactivate the enzymes. The RNA probes were purified on chromatography columns (Micro Bio-Spin Chromatography Column P-6; Bio-Rad).
The Cy5-labeled cDNA probes were obtained from in vitro-synthesized sense RNA (see information on in vitro runoff transcription above) by using SuperScript III reverse transcriptase (Invitrogen) and the primers F-RS and 16S-RS for cel48F and 16S rRNA, respectively. Before each reverse transcription, the RNAs were denatured for 5 min at 65°C. The reaction mixture (40 μl) consisted of 2.5 μM the specific primer; 166.5 μM each dATP, dTTP, and dGTP; 25 μM dCTP; 37.5 μM Cy5-dCTP; 10 μg RNA; 1× RT buffer, 0.1 M dithiothreitol, 40 U of RNase inhibitor, and 200 U of reverse transcriptase. The mix was incubated for 2 h at 50°C with addition of 200 U of enzyme after 1 h of incubation. The reaction was stopped by heating 15 min at 70°C, and then the reaction mixture was treated with DNase-free RNase (Roche Applied Science) and Rnase H (Promega). The Cy5-labeled cDNAs were purified on affinity column (Genomics; Millipore) and concentrated on exclusion column (Microcon YM-30; Millipore). The Cy5-labeled cDNA probes were quantified by spectrophotometric analysis at 260 nm. The frequency of incorporation of labeled nucleotides was calculated from the optical density at 650 nm. To validate the probe's functionality, a range of several dilutions of each Cy5-labeled probe were done. After UV cross-linkage on a nylon membrane, the probes were quantified (see “RNA quantification”).
RNA quantification.
The software used was Multi Gauge, version 2.0 (Science Lab 2002). After probing with the Cy5-labeled cDNA probes, the intensities of the regions corresponding to the different RNA species were measured in linear arbitrary units (LAU). For each RNA, a background region (BG) was measured in LAU. The RNA quantities were expressed in (LAU − BG)/mm2. cipC-cel48F and cel48F mRNA quantities were normalized with the 16S rRNA.
RT-PCR.
RT and the following PCRs were performed from 100 ng of total RNA with the Titan One Tube RT-PCR kit (Roche Applied Science). The reaction mix (50 μl) consisted of 0.2 mM deoxynucleotide triphosphate (dNTP), 5 mM DTT, 5 U of RNase inhibitor, 0.4 μM each direct and reverse primer, 100 ng of RNA, 1× RT-PCR buffer, and 1 μl of enzyme mixture containing the avian myeloblastosis virus reverse transcriptase and the Taq and Tgo DNA polymerases. Reaction mixtures were incubated at 50°C for 30 min; the cDNA products were then amplified by PCR. The PCR negative and positive controls were done with the Taq-Tgo DNA polymerase mix, by using RNA and genomic DNA as templates, respectively.
Primer extension.
Total RNA was reverse transcribed by using the Superscript III reverse transcriptase (Invitrogen) and a radioactive 5′-end-labeled oligonucleotide primer. Primers were end labeled by incubating 5 pmol of the primer with 10 U of T4 polynucleotide kinase (Biolabs), 60 μCi of [γ-32P]ATP (6,000 Ci/mmol; Amersham Biosciences), 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM dithiothreitol, and 0.1 M spermidine at 37°C for 30 min. One microliter of radiolabeled primer which had previously been purified on Micro Bio Spin chromatography column (Bio-Rad) was annealed with 15 μg of RNA and incubated at 50°C for 50 min in a 20-μl reaction mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 1 mM of each deoxynucleotide triphosphate, and 200 U of reverse transcriptase. Extension products were analyzed on an 8% polyacrylamide sequencing gel. Their migrations were compared to the migration of the 25-bp DNA Step Ladder (Promega), also labeled as previously described. To map the exact transcriptional start site, sequencing reaction mixtures were used as ladders. The sequencing reactions were performed on recombinant plasmids containing the C. cellulolyticum DNA region analyzed by primer extension. The Thermo Sequenase Cycle Sequencing kit (Amersham Biosciences) was used for all sequencing reactions according to the supplier's protocol.
RESULTS
cipCMut1(pSOScipC) recombinant clone studies.
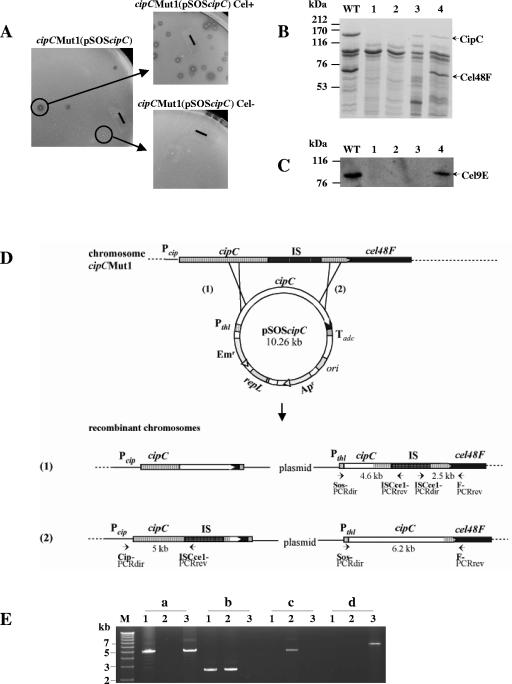
Cellulolysis is severely affected in C. cellulolyticum strain cipCMut1 (16), which contains a disrupted cipC gene (Fig. 1D). Indeed, neither the mutant strain nor the cipC trans-complemented strain cipCMut1(pSOScipC) were found to produce any of the enzymatic proteins encoded by the genes following cipC in the cip-cel cluster of genes (16). This result strongly suggests that all genes of the cluster are transcriptionally linked. In order to confirm (i) this hypothesis and (ii) that the noncellulolytic phenotype (Cel−) observed was only due to a strong polar effect, recombinant cellulolytic clones (Cel+) were selected from the cipCMut1(pSOScipC) trans-complemented strain (Fig. 1A). The analysis of the cipC-cel48F chromosomal region of Cel+ clones by PCR (Fig. 1E) revealed that they carried the entire plasmid. This structure resulted from one homologous recombination event between the chromosomal cipC gene and the plasmidic cipC gene, in the 3′-end region of the genes. The integration of the plasmid at this place restored the cluster continuity (Fig. 1D, case 2). Some Cel− recombinant clones were also found to result from the insertion of the plasmid in the 5′ end of the chromosomal cipC gene (Fig. 1D, case 1), leaving the interrupted cipC gene copy in front of cel48F. The cellulolytic system produced by the cipCMut1(pSOScipC) Cel+ strains contains, in addition to the CipC protein, the two major cellulosomal cellulases Cel48F (Fig. 1B, lane 4) and Cel9E (Fig. 1C, lane 4), whereas the cipCMut1(pSOScipC) Cel− clones produce only CipC (Fig. 1B and C, lane 3). Therefore, in the Cel+ strains,the expression of cipC and of the following genes is under the control of the plasmidic thiolase promoter located in front of the cip-cel wild-type cluster. The restoration of the Cel+ phenotype in certain plasmid-chromosome recombinants asserts that the absence of the cluster-encoded enzymes in the cipCMut1 strain was only due to the insertional inactivation of cipC, confirming the polar effect and showing that the genes of the cluster are transcriptionally linked.
FIG. 1.
Recombinant cipCMut1(pSOScipC) clone analysis. (A) Phenotype of cipCMut1(pSOScipC) stains. A cellulolytic strain (Cel+) was selected from the cipC trans-complemented cipC mutant strain cipCMut1(pSOScipC) on cellulose-containing solid medium. The solid bars correspond to 6 mm. (B) SDS-polyacrylamide gel electrophoresis analysis and (C) Western blot analysis with antiserum raised against Cel9E of the Fc fraction from wild-type (lane WT), cipCMut1 (lane 1), cipCMut1(pSOSzero) (lane 2), cipCMut1(pSOScipC) Cel− (lane 3), and cipCMut1(pSOScipC) Cel+ (lane 4) strains. (D) Schematic representation of possible homologous recombination events between the mutated chromosomal cipC gene and the wild-type plasmidic cipC gene. Pcip, promoter region of the cipC gene; IS, insertion element ISCce1::ISCce2 (15); Pthl, promoter region of the constitutive thiolase gene and Tadc, transcriptional terminator region of the acetoacetate decarboxylase gene. Both Pthl and Tadc are from Clostridium acetobutylicum ATCC 824 (30). Small solid arrows show primers used for PCR analysis. (E) PCR analysis of cipCMut1(pSOScipC) recombinant clones followed by 0.7% agarose gel electrophoresis. PCR was carried out with primers Cip-PCRdir and ISCce1-PCRrev (a), ISCce1-PCRdir and F-PCRrev (b), Sos-PCRdir and ISCce1-PCRrev (c), and Sos-PCRdir and F-PCRrev (d) with 20 ng of genomic DNA of cipCMut1 (lanes 1), cipCMut1(pSOScipC) Cel− (lanes 2), and cipCMut1(pSOScipC) Cel+ (lanes 3). The fragments of 15.3, 12.5, and 8.9 kb, expected in lanes a2, b3, and d2, respectively, are too large to be synthesized by PCR with the Expand long template PCR system (Roche Applied Science). M, DNA molecular weight marker X (Roche Applied Science).
Transcript size.
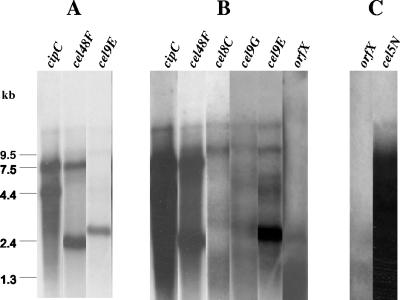
In order to study the transcriptional organization of the genes belonging to the cluster, Northern blot hybridization experiments were performed with the different gene-specific radioactive (32P) riboprobes. Total RNA was isolated from C. cellulolyticum wild-type cells grown in MN300 cellulose-containing basal medium during 6 days at 32°C. The results of hybridization on 10-μg RNA blots showed several transcripts carrying the genes cipC, cel48F, cel8C, cel9G, and cel9E (Fig. 2A and B). In the first part of the cluster, four transcripts were found to be very abundant: the monocistronic transcripts of cipC (4.9 kb), cel48F (2.3 kb), and cel9E (2.8 kb) and the polycistronic transcript cipC-cel48F (7.6 kb) (Fig. 2A). Besides these major transcripts, the five probes corresponding to the five first genes of the cluster annealed to some other rare messengers. The largest messenger detected by each of the probes is approximately 14 kb long (Fig. 2B). Although this size could not be accurately determined, it is in good agreement with the expected size of the cipC-cel9E part of the cluster (13.5 kb). A second large transcript of around 10 kb was detected by the probes cel48F, cel8C, cel9G, and cel9E. Moreover, signals ranging from 4 to 6 kb were observed with the cel9G and cel9E probes. They might result from hybridization with degraded messengers. In contrast to cipC, cel48F, and cel9E, no monocistronic transcript was shown with the probes cel8C and cel9G.
FIG. 2.
Northern blot analysis of transcripts carrying the genes of the cip-cel cluster. Ten micrograms (A and B) or 40 μg (C) of total RNA purified from cells grown on cellulose-containing medium was subjected to electrophoresis through 0.8% agarose-0.22% formaldehyde-containing gels, transferred to nylon membranes, and then hybridized with various gene-specific radioactive riboprobes. One hour (A) or 4 h (B and C) of exposure revealed major and minor messengers, respectively. Each lane shows the results of the hybridization with the indicated gene-specific probe. The order of the lanes in the figure follows the order of the genes in the cluster.
The probes orfX, cel9H, cel9J, man5K, cel9M, rgl11Y, and cel5N did not reveal any defined messenger. In an attempt to detect the rare messenger(s), Northern blotting analysis was performed again using 40 μg of RNA instead of 10 μg. Unfortunately, the abundance of RNAs on the membranes engendered a high background noise, and it was thus not possible to distinguish any specific signal in the area of the blots containing RNAs 1 to 10 kb long. However, a big transcript larger than 12 kb long was detected with probes of the 3′ moiety of the cluster excluding orfX (e.g., Fig. 2C). This RNA could carry the information of all the genes of the second part of the cluster. The DNA strand carrying the genes cel9H to cel5N is 11.8 kb long, and we know that cel5N is the last gene of the cip-cel cluster. Indeed, looking at the region following cel5N, a gene (orf4) encoding an LysR family protein was found (GenBank accession no. DQ156106), but in the opposite direction from the cel5N ORF.
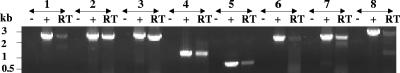
The alternative method of RT-PCR analysis was used to investigate transcriptional linkage in the cip-cel cluster. One reverse primer designed to hybridize to the beginning of each ORF was used to synthesize cDNA molecules from RNAs (Table 2). PCR amplifications were performed by adding a direct primer which binds to the cDNA within the preceding ORF. RT-PCR products of the predicted length were generated for all primer pairs spanning intergenic regions from cipC to cel5N (e.g., for the cel9H-cel9J intergenic region in Fig. 3, lane 5, RT). To explore the putative transcriptional association of three consecutive genes, the same cDNA template was subjected to PCR amplification with the same reverse primer and a direct primer that binds two ORFs upstream. An RT-dependent product was obtained with the pair of primers cipC-RTdir/C-RTrev, indicating that the cipC, cel48F, and cel8C genes are transcriptionally coupled (Fig. 3, lane 1, RT). Similarly, it was shown that cel48F, cel8C, and cel9G (Fig. 3, lane 2, RT), and that cel8C, cel9G, and cel9E (Fig. 3, lane 3, RT) are linked, respectively. A specific RT-PCR product was also obtained in mixtures containing the E-RTdir/H-RTrev pair of primers (Fig. 3, lane 4, RT), indicating that messengers carrying cel9E, orfX, and at least the beginning of cel9H also exist. It was nevertheless not possible to obtain any RT-PCR product, neither with the pairs G-RTdir/X-RTrev and X-RTdir/J-RTrev nor with the pair H-RTdir/K-RTrev (data not shown). No apparent internal RNA secondary structures which could explain these results were found within the cel9E, cel9H, and cel9J ORFs (Fig. 4). Conversely, cel9J, man5K, and cel9M were shown to be carried by the same messenger, as man5K, cel9M, and rgl11Y and as cel9M, rgl11Y, and cel5N (Fig. 3, RT in lanes 6, 7, and 8, respectively).
FIG. 3.
RT-PCR analysis of the genes of the cip-cel cluster. The cDNA reverse transcripts obtained from total RNAs were subjected to PCR amplification with pairs of primers (Table 2) in order to reveal transcriptional links between two or three successive genes. The reaction products were analyzed by electrophoresis in 0.7% agarose gel (lanes RT). The pairs of primers which were used are as follows: cipC-RTdir/C-RTrev (lanes 1), F-RTdir/G-RTrev (lanes 2), C-RTdir/E-RTrev (lanes 3), E-RTdir/H-RTrev (lanes 4), H-RTdir/J-RTrev (lanes 5), J-RTdir/M-RTrev (lanes 6), K-RTdir/Y-RTrev (lanes 7), and M-RTdir/N-RTrev (lanes 8). −, PCRs performed on RNAs in the absence of RT; +, PCRs performed on genomic DNA templates.
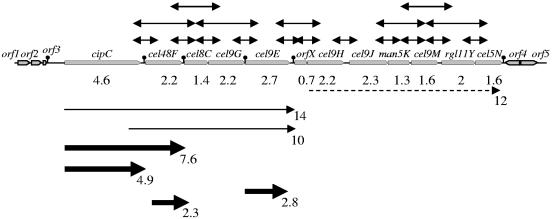
FIG. 4.
Schematic representation of the gene arrangement in the 26-kb cip-cel cluster. The 12 genes of the cluster are indicated by gray arrows. orf1 and orf2 code for putative β-lactamase and alkaline phosphatase, respectively. orf3 presents 44% similarity to a hypothetical protein-encoding sequence from Clostridium perfringens (GenBank accession no. BA000016.3 gi 47118322). orf4 and orf5 encode a putative transcriptional regulator of the LysR family and an acyl coenzyme A thioesterase module, respectively. Arrows over the map indicate the transcriptional links revealed by RT-PCR analyses. Major transcripts revealed by Northern blotting are indicated below the map by thick black arrows. Minor transcripts are shown by thin and dotted lines. Sizes in kb are indicated. Putative mRNA secondary structures are indicated downstream of orf3, cipC, cel48F, cel9G, cel9E, and cel5N. Their ΔG, calculated by the Zuker algorithm (32), are −20.4, −24.3, −34.5, −13.9, −27.5, and −21.9 kcal/mol, respectively.
Very large RNA species were clearly detected by Northern blotting (Fig. 4). Moreover, several RT-PCR products covering three successive genes were obtained, even in the 3′ moiety of the cluster. These results strongly support the hypothesis that all the genes of the cluster are cotranscribed in a very large primary messenger which might be subject to processing.
Identification of potential transcription initiation sites.
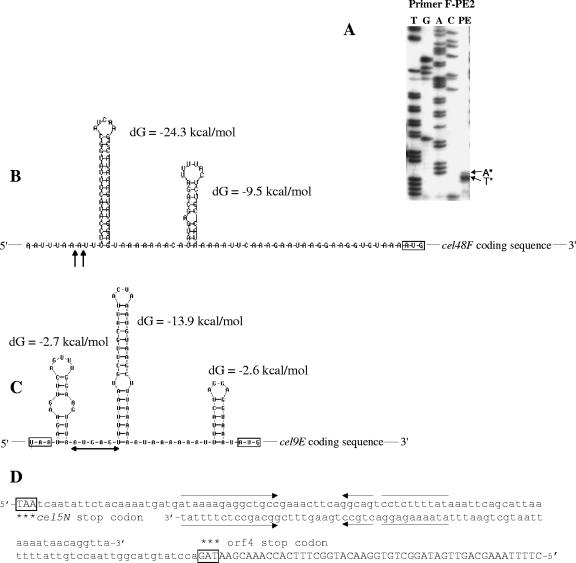
To identify promoters upstream of cipC, primer extension analysis was performed on total RNAs extracted from cells that had been grown in cellulose-containing medium. Primer extension reactions were carried out with primers cip-PE1, cip-PE2, and cip-PE3 (Table 2), which are complementary to the −2/+20, −121/−144, and −512/−538 regions, respectively (Fig. 5A). No short elongation product was obtained with cip-PE1 (Fig. 6). The cip-PE3 and cip-PE2 primers were used to precisely map the distal and proximal putative transcription start areas S1 (Fig. 5B) and S2 (Fig. 5C), respectively. The distal start nucleotides were mapped to the point 637 and 638 nucleotides (nt) upstream of the ATG codon (Fig. 5A). The deduced −35 and −10 sequences matched well the consensus for Bacillus σA and clostridial promoter sequence (−35, TTGACA; −10, TATAAT) (10, 31). One direct repeat region was found downstream of this transcriptional start site (Fig. 5A). We do not know yet if this region is involved in any regulation process. S2 was located 194 nt upstream of the cipC translational initiation codon. The −35 and −10 regions (P2) of this proximal putative start site showed low homology to the consensus for σ factors of gram-positive bacteria. This 5′ end might result from an effective transcription start site or might correspond to a degradation product of the primary mRNA. We know that the −392 to −25 promoter region carrying P2 does not promote high level of transcription in C. cellulolyticum when used into an expression vector (22). On the other hand, the long leader sequence and the presence of repeats in the promoter region suggest that some type of control may occur at the posttranscriptional level.
FIG. 5.
Promoter region sequence (A) and primer extension analysis (B and C) to map the transcription start sites of cipC. End-labeled primers cip-PE3 (B) and cip-PE2 (C) were used to identify the putative P1 and P2 promoter start sites S1 and S2. The same primers were used to generate the sequencing ladder by the dideoxy method (lanes marked T, G, A, and C). Primer extension products (PE) are shown to the right of the corresponding sequencing ladder on an 8% (wt/vol) sequencing gel. In panel A, the −35 and −10 regions of the putative P1 and P2 promoters are indicated in bold italics, the cip-PE1 (Fig. 6), cip-PE2, and cip-PE3 annealing regions are highlighted, the possible start sites are marked by S1 and S2 over angled arrows, and inverted or direct repeats are indicated by arrows. SD, Shine-Dalgarno sequence.
FIG. 6.
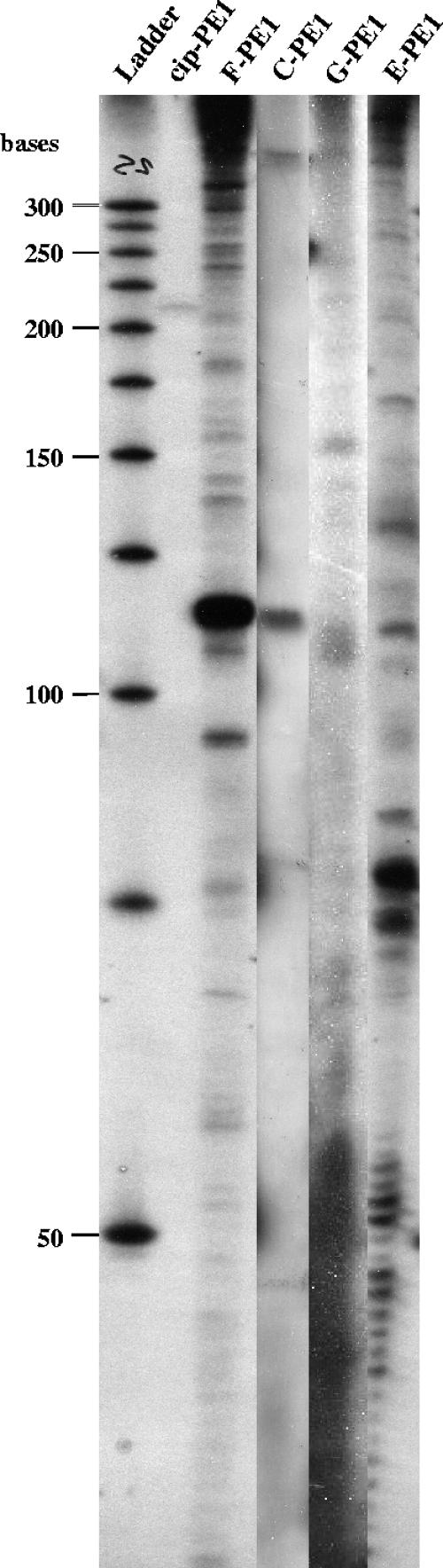
Primer extension from cipC, cel48F, cel8C, cel9G, and cel9E mRNAs. The experiments were performed with 15 μg of total RNAs and primers covering the 5′ end of the various ORFs (cip-PE1 for cipC, F-PE1 for cel48F, C-PE1 for cel8C, G-PE1 for cel9G, and E-PE1 for cel9E) The sizes of the reverse transcripts were determined by using a labeled 25-bp DNA Step Ladder (Promega).
To determine whether a promoter was present upstream of the cel48F gene, primer extension analysis was carried out on RNA using the radiolabeled primer F-PE1, which was complementary to a region 12 nt downstream of the initiation codon of cel48F (Table 2). A ladder pattern, including high-molecular-weight bands, was obtained when the cDNA products were analyzed (Fig. 6), which suggests that the cipC and cel48F genes form an operon. Nevertheless, one band of the ladder pattern of around 115 nt long was found to be especially intense (Fig. 6). This primer extension endpoint, located 103 nt upstream of the ATG codon (Fig. 7A), might result from mRNA processing or from the presence of secondary mRNA structures that lead to RT stalling. Primer extension analyses were also undertaken to identify putative 5′ ends of transcripts upstream of cel8C, cel9G, and cel9E. The patterns obtained showed several bands of various intensities, including high-molecular-weight products (Fig. 6). One band each of the cel8C and cel9E patterns appeared as a major product and might indicate mRNA processing sites. All of these results suggest that some primer extension products, in particular the minor products, might result from reverse transcriptase pausing or hairpin loop formation and do not represent transcriptional start points. High-molecular-weight products indicate that the corresponding genes are located inside large polycistronic mRNAs. Intergenic regions of the 5′ moiety of the cluster contain four putative stable RNA secondary structures which are different from Rho-independent terminator structures (no U stretch at the 3′ end) (Fig. 4). Two of them are located near the putative processing sites upstream of cel48F and cel9E (Fig. 7B and C). One can point out that these putative processing sites are located at the 5′ extremity of a secondary structure which might protect the secondary messenger from degradation and explain their high stability (see below and Fig. 8B). One putative stable RNA secondary structure was also found downstream of cel48F and cel9E (Fig. 4). They might also contribute to stabilize the 3′ ends of the monocistronic cel48F and cel9E messengers.
FIG. 7.
Study of the putative processing sites upstream of cel48F and cel9E. (A) Reverse transcriptase mapping of the processing site upstream of cel48F (primer F-PE2). Lanes T, G, A, and C show the dideoxy sequencing ladder obtained with the same primer as used for the reverse transcriptase analysis (lane PE). The mapped 5′ ends are marked by asterisks. Putative mRNA secondary structures in the cipC-cel48F (B), cel9G-cel9E (C), and cel5N-orf4 (D) intergenic regions are shown as calculated by the Zuker algorithm (32). dG, ΔG. AUG and stop codons are boxed. Vertical arrows indicate the putative processing sites deduced from panel A. In panel B, the horizontal arrow indicates the preferred putative processing region deduced from Fig. 6. In panel C, putative Rho-independent terminator sequences are present in both strands. Inverted repeats are indicated by facing arrows. Both stop codons are boxed. The 3′ end of the orf4 coding sequence is capitalized.
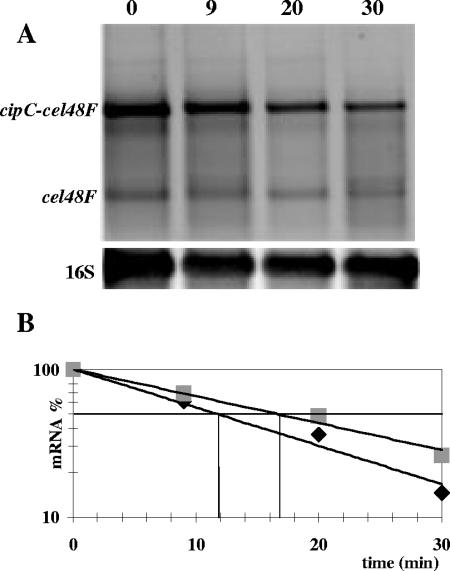
FIG. 8.
Detection of cipC-cel48F and cel48F mRNA stabilities by Northern blot analyses. (A) Total RNAs were purified from cells grown at 32°C before addition of rifampin. RNAs were extracted at the indicated times (0, 9, 20, and 30 min) after addition of rifampin. The blotted RNAs were hybridized with cel48F- and 16S-specific Cy5-labeled cDNA probes. (B) Half-life determination of the cipC-cel48F (▪) and cel48F (▴) transcripts. Each RNA was quantified from the intensity of the corresponding area minus background contribution using the Multi Gauge, version 2.0, software. In order to compare the quantities of the two specific-mRNAs in the four samples, these quantities were normalized relatively to the extremely stable 16S RNA. This experiment has been performed twice with samples collected at different times (data not shown). Half-lives determined from each mRNA species from the two different experiments were equal.
In the 3′-end region of the cip-cel cluster, cel5N is followed by orf4 transcribed on the opposite strand. Two putative Rho-independent terminator structures encoded by the two complementary strands were found in the intergenic region (Fig. 7D). These structures might be responsible for the transcription termination of the cluster and of orf4.
Stability of the cipC-cel48F and cel48F messengers.
The cipC-cel48F and cel48F messengers were found to be abundant (Fig. 2A and B). This abundance might reflect high stabilities. If these mRNAs are from the result of processing events, the monocistronic cel48F mRNA might be derived from the larger cipC-cel48F mRNA. In order to investigate the stability of these messengers and to test this last hypothesis, 500 μg of rifampin were added to a 6-day-old culture of C. cellulolyticum on MN300 cellulose, in order to stop transcription. Total RNAs, purified from aliquots of the culture after 9, 20, and 30 min of incubation at 32°C, were analyzed after Northern blotting using Cy5-labeled cel48F and 16S cDNA probes (Fig. 8A). 16S RNA, cipC-cel48F mRNA, and cel48F mRNA were quantified by measuring the fluorescence of the corresponding hybridized probes. As shown in Fig. 8A, both transcripts decreased together after the addition of rifampin but at different rates; the quantity of the cel48F mRNA was found to decrease more slowly (half-life of 17 min) compared to the large cipC-cel48F mRNA (half-life of 12 min) (Fig. 8B). These results suggest that the cel48F transcript is not a processed product of the longer cip-cel48F messenger. Both transcripts might be the products of the 14-kb messenger.
DISCUSSION
To better understand which mechanisms govern the production of the key cellulosomal proteins from Clostridium cellulolyticum, we attempted to characterize the major mRNAs that are synthesized from the genes belonging to the cip-cel cluster and identify the putative promoter regions.
Long messengers bearing the information of several successive genes were detected by Northern blotting. The first five genes were found to be cotranscribed in a 14-kb-long mRNA. Many messengers carrying the information of one, two, or three of these five genes were also detected. The abundance of some of these transcripts indicates that the genes cipC, cel48F, and cel9E appeared to be the most expressed (Fig. 4). Conversely, the genes located in the second moiety were found to be poorly expressed. Indeed, we only detected a small amount of a messenger which covers a large 3′ moiety of the cluster and might be the main translated species that produces the Cel9H-to-Cel5N proteins. We were not able to detect by Northern blotting any RNA large enough to carry all of the genes of the cluster (26 kb). Such RNA is presumably too unstable to be detected. Indeed, a quickly in vivo-degraded messenger would be in too small a quantity to be detected after purification. Nevertheless, all of the intergenic regions were found to be carried by RNA species (RT-PCR analyses). Recently, Han et al. (12) reported a large (12 kb long) messenger covering four genes of the “cel” cluster of another mesophilic cellulolytic strain of Clostridium cellulovorans. Four transcription units for the first eight genes were described by the authors. Concerning C. cellulolyticum, the polar effect observed in the cipCMut1 strain, on the one hand, and the various different messengers identified in this study, on the other hand, strongly suggest that the cip-cel cluster constitutes a very large operon.
Two different hypotheses might be proposed. In the first one, only one promoter region located upstream of cipC controls the expression of all the genes of the cluster as a single unit, the various messengers detected being produced either by transcriptional attenuation and/or by processing of the primary transcript. In the second hypothesis, internal promoters exist: for example, in front of cel48F and/or cel9E. In such a case, then the expression from this promoter(s) would be dependent on the expression of the first genes of the cluster in a cellulose-containing medium. Indeed, none of the enzymes encoded by the genes of the cluster could be detected in the cipCMut1 strain (16). Primer extension experiments did not reveal any clear start point either in front of cel48F or in front of cel8C, cel9G, or cel9E. Conversely, the presence of large cDNAs in the primer extension reactions suggest (i) that these genes are included in polycistronic messengers and (ii) that the major band(s) which corresponds to messengers carrying the gene on its 5′-end part might result from processing events. Nevertheless, we cannot exclude that some internal promoters might be activated, depending on the culture conditions. Indeed, promoter/operator regions located in front of the mannanase gene man5K, the pectinase gene rgl11Y, or orfX might be the target for specific regulators. Such a hypothesis will be explored in future experiments by fusion with a reporter gene.
The stability of the cipC-cel48F and cel48F messengers was found to be high (half-lives of 12 and 17 min, respectively). In Bacillus subtilis, in early stationary phase, 80% of the messengers have a half-life of less than 7 min and only 3% display a half-life of more than 15 min (11). In E. coli and Lactococcus lactis, in exponential phase, the mean half-lives are around 7 and 6 min, respectively (25, 28). After the addition of rifampin, which blocked transcriptional events, both cipC-cel48F and cel48F transcript amounts decreased; we did not observe any increase of the cel48F monocistronic messenger correlated with a decrease of the bicistronic messenger which could clearly indicate that the former was obtained from the latter. Nevertheless, we could observe that different mRNA preparations exhibit very different ratios of these two mRNA species (Fig. 2A, lane cel48F, and Fig. 8A). These two observations suggest that both messengers might be issued from processing events from long primary transcripts and accumulate because of their higher stabilities. The presence of a stem-loop structure at the 5′ end of the cel48F processed monocistronic mRNA (Fig. 7B) might protect it from exonucleolytic degradation and thus participate in its remarkable stability, as has been shown for the B. subtilis aprE and gapA transcripts (3, 17). This phenomenon constitutes a posttranscriptional regulation process and might greatly contribute to the final equilibrium between the different proteins encoded by the cluster. Nevertheless, we cannot exclude that the monocistronic cel48F messenger might also be a derivative of the bicistronic mRNA by a low rate of processing. Indeed, in the experiment with rifampin, at time zero, the proportions of the large and small mRNAs were found to be of 90% and 10%, respectively, whereas 30 min later the proportions were estimated to be 70% and 30%, respectively. Such a hypothesis was proposed in the case of the dnaK operon of B. subtilis (13). In E. coli, there are two major endoribonucleases involved in mRNA processing, RNase E and RNase III (9). In B. subtilis, two paralogous endoribonucleases (J1 and J2) which share functional homologies with RNase E from E. coli, have been characterized (5). Orthologues of these two RNases were found in many clostridia. Similar RNA maturation processes are thus expected to occur in these bacteria. Nevertheless, no sequence similarity could be found between processing sites of the cip-cel cluster and the processing sites found in the B. subtilis dnaK operon (13) and gapA operon (17) (data not shown). On the other hand, sequence alignment of the different putative cluster processing sites did not reveal any consensus sequence which might be recognized by one specific C. cellulolyticum RNase. The processing mechanism should be characterized in the future.
Concluding remarks.
In summary, the results obtained suggest that C. cellulolyticum uses differential segmental mRNA stability to fine tune the expression of individual genes of the cip-cel operon. The promoter region, identified in front of cipC, would control the expression of the entire cluster by the way of a long 26-kb primary transcript that is processed into various secondary transcripts, which display different stabilities. One secondary bicistronic transcript and three monocistronic stable transcripts ensure the expression of the scaffoldin gene cipC and the genes coding for the two major cellulases Cel48F and Cel9E at a high level. Other secondary transcripts of lower stability would mediate the expression of the other genes at a weaker level. This work constitutes the first step of the cip-cel gene regulation study. Transcriptional fusions with a reporter gene would now be used to explore the regulation of the cipC promoter activity and to identify the putative internal promoter(s).
Acknowledgments
We thank Violaine Bonnefoy, Patrice Bruscella, Yann Denis, Pascale de Philip, Sandrine Pagès, Henri-Pierre Fiérobe, Anne Bélaich, and Jean Pierre Bélaich for fruitful discussions.
We acknowledge financial support received from the Centre National de la Recherche Scientifique and the Université de Provence.
REFERENCES
- 1.Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J. P. Belaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 2.Bélaich, A., C. Tardif, H.-P. Fiérobe, S. Pagès, and J.-P. Bélaich. 2004. The clostridial cellulosome, p. 343-359. In M. M. Nakano and P. Zuber (ed.), Strict and facultative anaerobes: medical and environmental aspects. Horizon Biosciences, Norfolk, United Kingdom.
- 3.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67:157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desvaux, M., E. Guedon, and H. Petitdemange. 2001. Kinetics and metabolism of cellulose degradation at high substrate concentrations in steady-state continuous cultures of Clostridium cellulolyticum on a chemically defined medium. Appl. Environ. Microbiol. 67:3837-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Even, S., O. Pellegrini, L. Zig, V. Labas, J. Vinh, D. Brechemmier-Baey, and H. Putzer. 2005. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 33:2141-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gal, L., S. Pages, C. Gaudin, A. Belaich, C. Reverbel-Leroy, C. Tardif, and J.-P. Belaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudin, C., A. Belaich, S. Champ, and J.-P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giallo, J., C. Gaudin, and J.-P. Bélaich. 1985. Metabolism and solubilization of cellulose by Clostridium cellulolyticum H10. Appl. Environ. Microbiol. 49:1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 10.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hambraeus, G., C. von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 12.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J. Bacteriol. 185:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homuth, G., A. Mogk, and W. Schumann. 1999. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol. Microbiol. 32:1183-1197. [DOI] [PubMed] [Google Scholar]
- 14.Maamar, H. 2003. Etude in vivo du système cellulolytique de Clostridium cellulolyticum: caractérisation du premier mutant d'insertion cipC. Ph.D. thesis, University of Aix-Marseille I, Marseille, France.
- 15.Maamar, H., P. de Philip, J.-P. Belaich, and C. Tardif. 2003. ISCce1 and ISCce2, two novel insertion sequences in Clostridium cellulolyticum. J. Bacteriol. 185:714-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maamar, H., O. Valette, H. P. Fierobe, A. Belaich, J. P. Belaich, and C. Tardif. 2004. Cellulolysis is severely affected in Clostridium cellulolyticum strain cipCMut1. Mol. Microbiol. 51:589-598. [DOI] [PubMed] [Google Scholar]
- 17.Meinken, C., H. M. Blencke, H. Ludwig, and J. Stulke. 2003. Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology 149:751-761. [DOI] [PubMed] [Google Scholar]
- 18.Ochman, H., M. M. Medhora, D. Garza, and D. L. Hartl. 1990. Amplification of flanking sequences by inverse PCR. Academic Press, New York, N.Y.
- 19.Pagès, S., A. Bélaïch, H.-P. Fierobe, C. Tardif, C. Gaudin, and J.-P. Bélaïch. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagès, S., A. Belaich, C. Tardif, C. Reverbel-Leroy, C. Gaudin, and J.-P. Belaich. 1996. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 178:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagès, S., O. Valette, L. Abdou, A. Bélaïch, and J.-P. Bélaïch. 2003. A rhamnogalacturonan lyase in the Clostridium cellulolyticum cellulosome. J. Bacteriol. 185:4727-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perret, S., A. Bélaich, H.-P. Fierobe, J.-P. Bélaich, and C. Tardif. 2004. Towards designer cellulosomes in Clostridia: mannanase enrichment of the cellulosomes produced by Clostridium cellulolyticum. J. Bacteriol. 186:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perret, S., H. Maamar, J. P. Belaich, and C. Tardif. 2004. Use of antisense RNA to modify the composition of cellulosomes produced by Clostridium cellulolyticum. Mol. Microbiol. 51:599-607. [DOI] [PubMed] [Google Scholar]
- 24.Petitdemange, E., F. Caillet, J. Giallo, and C. Gaudin. 1984. Clostridium cellulolyticum sp. nov., a cellulolytic, mesophilic species from decayed grass. Int. J. Syst. Bacteriol. 34:155-159. [Google Scholar]
- 25.Redon, E., P. Loubière, and M. Cocaign-Bousquet. 2005. Role of mRNA stability during genome-wide adaptation of Lactococcus lactis to carbon starvation. J. Biol. Chem. 280:36380-36385. [DOI] [PubMed] [Google Scholar]
- 26.Reverbel-Leroy, C., A. Belaich, A. Bernadac, C. Gaudin, J. P. Belaich, and C. Tardif. 1996. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology 142:1013-1023. [DOI] [PubMed] [Google Scholar]
- 27.Reverbel-Leroy, C., S. Pages, A. Belaich, J.-P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selinger, D. W., R. M. Saxena, K. J. Cheung, G. M. Church, and C. Rosenow. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tardif, C., A. Bélaich, H. P. Fierobe, S. Pagès, P. de Philip, and J.-P. Bélaich. 2006. Clostridium cellulolyticum: cellulosomes and cellulolysis. In I. Kataeva (ed.), Cellulosome, Nova Sciences Publishers, Inc., New York, N.Y.
- 30.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young, M., N. P. Minton, and W. L. Staudenbauer. 1989. Recent advances in the genetics of the clostridia. FEMS Microbiol. Rev. 5:301-325. [DOI] [PubMed] [Google Scholar]
- 32.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]