Abstract
The recently identified CprK branch of the CRP (cyclic AMP receptor protein)-FNR (fumarate and nitrate reduction regulator) family of transcriptional regulators includes proteins that activate the transcription of genes encoding proteins involved in reductive dehalogenation of chlorinated aromatic compounds. Here we report the characterization of the CprK1 protein from Desulfitobacterium hafniense, an anaerobic low-G+C gram-positive bacterium that is capable of reductive dechlorination of 3-chloro-4-hydroxyphenylacetic acid (Cl-OHPA). The gene encoding CprK1 was cloned and functionally overexpressed in Escherichia coli, and the protein was subsequently purified to homogeneity. To investigate the interaction of CprK1 with three of its predicted binding sequences (dehaloboxes), we performed in vitro DNA-binding assays (electrophoretic mobility shift assays) as well as in vivo promoter probe assays. Our results show that CprK1 binds its target dehaloboxes with high affinity (dissociation constant, 90 nM) in the presence of Cl-OHPA and that transcriptional initiation by CprK1 is influenced by deviations in the dehaloboxes from the consensus TTAAT----ATTAA sequence. A mutant CprK1 protein was created by a Val→Glu substitution at a conserved position in the recognition α-helix that gained FNR-type DNA-binding specificity, recognizing the TTGAT----ATCAA sequence (FNR box) instead of the dehaloboxes. CprK1 was subject to oxidative inactivation in vitro, most likely caused by the formation of an intermolecular disulfide bridge between Cys11 and Cys200. The possibility of redox regulation of CprK1 by a thiol-disulfide exchange reaction was investigated by using two Cys→Ser mutants. Our results indicate that a Cys11-Cys200 disulfide bridge does not appear to play a physiological role in the regulation of CprK1.
Transcriptional regulators of the CRP (cyclic AMP [cAMP] receptor protein)-FNR (fumarate and nitrate reduction regulator) family play an important role in modulating the expression of numerous metabolic genes in many facultatively or strictly anaerobic bacteria. These structurally related regulators evolved to enable an efficient response to a wide range of environmental signals in organisms that are often phylogenetically distinct. In Escherichia coli, CRP is a positive regulator of catabolite repression, directly sensing elevated cAMP levels caused by glucose starvation and activating the expression of genes involved in the utilization of alternative carbon sources (26). E. coli FNR senses the changes in redox state (e.g., the decreasing availability of oxygen) through a polynuclear iron-sulfur center whose presence enhances DNA binding, resulting in transcriptional activation of genes involved in anaerobic respiration (38). Apart from these archetypes of the CRP-FNR superfamily, there are several members that use other effector molecules that can stimulate or repress the expression of metabolic genes by causing structural changes in the DNA-binding domain of the corresponding regulator (18, 27). Binding of carbon monoxide to CooA from Rhodospirillum rubrum is necessary for the ability to regulate the expression of genes for CO-oxidative growth (19). NtcA, a regulator of nitrogen balance in Anabaena spp., is activated by 2-oxoglutarate (39), while the FNR-like protein (FLP) in Lactococcus lactis is activated by the formation of an intramolecular disulfide bridge between Cys5 and Cys102 caused by oxidative stress (14).
A branch of the CRP-FNR family that has recently emerged, CprK, includes transcriptional regulators that mediate the response to halogenated aromatic compounds. Halogenated hydrocarbons with a high degree of substitution are chemically very stable under anaerobic conditions; they are persistent and can cause toxic effects for a long time (43). Several recently discovered strictly anaerobic bacteria are capable of halorespiration, a novel respiratory pathway in which halogenated hydrocarbon compounds serve as terminal electron acceptors (13). During halorespiration, one or more halogen atoms are reductively removed from the halogenated hydrocarbon while energy is conserved via electron transport-coupled phosphorylation. An important group among halorespiring bacteria consists of members of the genus Desulfitobacterium, which belong to the low-G+C gram-positive bacteria and show high metabolic versatility in their capability to degrade halogenated phenol derivatives. Desulfitobacterium dehalogenans can reductively dechlorinate halogenated phenolic compounds at the ortho position (41), while Desulfitobacterium hafniense DCB-2 can dechlorinate 3,5-dichlorophenol at the meta position in addition to ortho-substituted compounds (10). The reductive dehalogenase CprA has been isolated from D. dehalogenans; it catalyzes the dechlorination of 3-chloro-4-hydroxyphenylacetic acid (Cl-OHPA) at the ortho position (42). The corresponding cprA gene was isolated and found to be located in a chlorophenol reductive dehalogenase gene cluster containing cprTKZEBACD open reading frames (37). Transcription of the cpr gene cluster was initiated from three promoters under halorespiring conditions, resulting in cprT, cprZE, and cprBA or cprBACD transcripts, while cprK was constitutively transcribed at a basic level. The deduced amino acid sequence of the protein encoded by cprK showed significant homology to members of the CRP-FNR family, suggesting a possible role in transcriptional regulation of the cpr gene cluster. In addition, putative regulator binding sites were identified in the three promoter regions that resembled the FNR consensus TTGAT----ATCAA (37). Recently, biochemical studies have proved that CprK from D. dehalogenans is a transcriptional activator that binds Cl-OHPA with high affinity, resulting in an active DNA-binding conformation that enables CprK to bind to a specific DNA sequence, TTAATacgcACTAA, located in the promoter region of cprBA (31). CprK was subject to inactivation by diamide, which converts free thiols to disulfides, suggesting a putative role of redox regulation in activation of CprK mediated by one or more of the five cysteines present in the protein.
The recently elucidated genome sequence of D. hafniense DCB-2 (www.jgi.doe.gov) revealed the presence of several cprK homologues (36). The cprK1 gene is situated in a cpr gene cluster that contains cprTK1ZEBA1C open reading frames, among which the product of the cprA1 gene, a Cl-OHPA reductive dehalogenase, has been isolated and characterized (11). This reductive dehalogenase was isolated from D. hafniense cells grown on pyruvate as an electron donor and Cl-OHPA as an electron acceptor. It was reported that, when the cells were grown in a medium containing fumarate instead of Cl-OHPA as an electron acceptor, no dehalogenation activity could be detected, suggesting strong induction of the corresponding cprA1 gene by Cl-OHPA. In our study, we have functionally overexpressed the gene encoding CprK1 and purified the protein in order to study its interaction in vivo and in vitro with three putative target sequences (dehaloboxes) in the promoter regions of the cprT, cprZ, and cprB genes. Next, we have investigated the DNA-protein contact in detail, using a CprK1 mutant with altered DNA-binding specificity. Finally, the question of possible redox regulation of this CprK homologue was also addressed with the help of CprK1 Cys→Ser mutants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Desulfitobacterium hafniense DCB-2 was grown in phosphate-bicarbonate buffered medium with a low chloride concentration (21) under N2/CO2 (80:20) headspace at 37°C. Electron donors and acceptors were added to the medium from aqueous, anaerobic, sterile stock solutions. D. hafniense was grown fermentatively with 40 mM pyruvate, or under halorespiring conditions in the presence of 20 mM pyruvate and 20 mM Cl-OHPA. Escherichia coli DH5α was used as a host for cloning vectors. Two overproduction strains were used in this study: E. coli BL21(DE3), to obtain recombinant protein for purification, and E. coli JM109(DE3), for in vivo promoter activity measurements. All E. coli strains were grown in Luria-Bertani (LB) medium at 37°C (unless stated otherwise), and kanamycin (30 μg/ml) or erythromycin (200 μg/ml) was added when appropriate.
DNA isolation, cloning, and site-directed mutagenesis.
D. hafniense chromosomal DNA was isolated by first mechanically disrupting the cells by bead beating and then purifying by using the Bio 101 DNA isolation kit according to standard protocols provided by the manufacturer (Geneclean). Desired DNA fragments were PCR amplified from D. hafniense genomic DNA as a template, digested with restriction endonucleases, purified from an agarose gel, and finally ligated into a linearized pET24d expression vector (the cprK1 gene and its mutant derivatives) or into pAK80 (promoter fragments). The plasmids and primers used in this study are listed in Tables 1 and 2, respectively. Different protocols were used to introduce single-nucleotide changes into the DNA sequence: the ExSite (Stratagene) method was used to create CprK1(V192E) by replacing a GTC codon with GAG; QuikChange (Stratagene) was used to replace TGT codons with TCT, resulting in the CprK1(C11S) and CprK1(C200S) mutants; and finally, the overlap extension protocol (20) was used to change both half-sites of the inverted repeat in DB3 from 5′-TTAAT-3′ into 5′-TTGAT-3′.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pET24d | 5.3-kb expression vector, pMB1 ori, Kanr, T7 promoter | Novagen |
| pAK80 | 11.0-kb shuttle vector, p15A/L. lactis ori, Eryr, promoterless lacLM genes | 22 |
| pWUR89 | cprK1 gene cloned in frame with six-His translational fusion in pET24d using primers BG1379 and BG1380 | This work |
| pWUR166 | 266-bp cprE-cprB intergenic region cloned into pAK80 using BG1704 and BG1743, resulting in DB3::lacLM promoter fusion | This work |
| pWUR168 | 322-bp cprK1-cprZ intergenic region cloned into pAK80 using BG1702 and BG1742, resulting in DB2::lacLM promoter fusion | This work |
| pWUR171 | 164-bp cprK1-cprT intergenic region cloned into pAK80 using BG1699 and BG1782, resulting in DB1::lacLM promoter fusion | This work |
| pWUR172 | CprK1(V192E)-encoding plasmid, pWUR89 derivative | This work |
| pWUR175 | pWUR166 with TTAAT(N4)ATTAA→TTGAT(N4)ATCAA substitution resulting in DB3FNR::lacLM promoter fusion | This work |
| pWUR176 | cprK1 gene cloned into pET24d using primers BG1379 and BG1814 | This work |
| pWUR210 | CprK1(C11S)-encoding plasmid, pWUR176 derivative | This work |
| pWUR211 | CprK1(C200S)-encoding plasmid, pWUR176 derivative | This work |
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequencea | Orientationb |
|---|---|---|
| BG1379 | GCGCGCCATGGCTGTTGAAGGTTTGGGCAAGG | S |
| BG1380 | CGCGCGGATCCGAGTAATACGATGTTTGTTCAG | A |
| BG1699 | GCGGATCCACCCATGAGAAATTTGTGCATGG | S |
| BG1702 | GCGGATCCAGCTGCTCATTGTTCAAAGC | A |
| BG1704 | GCGGATCCGCTATAAAAACTAAAAATGTACC | A |
| BG1742 | GCGCAAGCTTGATAAGAAAAAGAATAAAA TCATCG | S |
| BG1743 | GCGCAAGCTTGGCCAAGGTCGCCATGACG | S |
| BG1748 | GGTTGAGAAATTCAGGTAAAG | S |
| BG1749 | GGATCACATACGCAAGTATTAATG | A |
| BG1782 | GCGCAAGCTTCGTTTCACTTTGTGATATTGAC | A |
| BG1814 | CGCGCGGATCCTAGTAATACGATGTTTGTTCAG | A |
| BG1942 | GCAGTCTTTATGCTCCGAAATG | S |
| BG1943 | GTGATATTGACTATACCG | A |
| BG1944 | CCTGCTTCAAAAAATATCTCC | S |
| BG1945 | CTAATACATAAAAAGAAGCTG | A |
Introduced endonuclease restriction sites are underlined.
S, sense primer; A, antisense primer.
Overproduction and purification of wild-type and mutant CprK1.
E. coli BL21(DE3) carrying one of the pET24d derivatives (Table 1) was grown in LB medium at 37°C until the A600 reached 0.6. At this point, 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the medium and incubation was continued at 20°C overnight (O/N) to facilitate overproduction of CprK1 in the soluble protein fraction. Cells were collected by centrifugation at 4°C, then resuspended in 1/20 volume buffer A (50 mM sodium phosphate buffer [pH 7.0], 100 mM NaCl) and disrupted by a French press at 100 MPa. Cell debris was removed by centrifugation at 4°C and 16,000 × g for 30 min. The supernatant was applied to a HiPrep 16/10 heparin FF column (Amersham Biosciences) that was preequilibrated with buffer A. Unbound proteins were washed with 8 column volumes of buffer A, and CprK1 was subsequently step-eluted using buffer B (50 mM sodium phosphate buffer [pH 7.0], 300 mM NaCl). Fractions that contained the recombinant protein were pooled and concentrated using Vivaspin 10,000-molecular-weight-cutoff devices (Vivascience). Concentrated samples were applied to a Tricorn Superdex 200 10/300 GL gel filtration column (Amersham Biosciences) preequilibrated with buffer C (50 mM Tris-HCl [pH 7.0], 100 mM NaCl). Denaturing gel electrophoresis of the protein samples was done in a 10% polyacrylamide gel. Protein concentrations were determined using the method developed by Bradford (6).
EMSA.
Three D. hafniense promoter regions were PCR amplified using Pfu DNA polymerase from D. hafniense genomic DNA as a template. The 60-bp cprK1-cprT intergenic region with dehalobox DB1 was amplified by primers BG1942 and BG1943. The 63-bp cprK1-cprZ intergenic region with dehalobox DB2 was amplified using primers BG1944 and BG1945. Dehalobox DB3, centered in a 52-bp cprE-cprB intergenic region, was amplified using primers BG1748 and BG1749. PCR-amplified fragments were loaded onto a nondenaturing 10% polyacrylamide gel, excised, and extracted by a modified “crush-and-soak” method. NEW buffer (10 mM Tris-HCl [pH 8.0], 750 mM NaCl, 10 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 40 μg/ml glycogen) was used to incubate the crushed gel piece O/N at 50°C with continuous shaking (1,400 rpm). After incubation, the gel pieces were separated from the aqueous phase by using a Bio-Spin chromatography column (Bio-Rad); then the supernatant was treated with phenol-chloroform, followed by ethanol precipitation of DNA. The promoter fragments were radioactively labeled at the 5′ end using [γ-32P]ATP and T4 polynucleotide kinase. Unincorporated radioactive nucleotides were removed with the help of Bio-Spin P30 columns (Bio-Rad). Purified CprK1 protein for electrophoretic mobility shift assay (EMSA) experiments was obtained from fractions eluted from the heparin chromatography column and buffer exchanged into 50 mM Tris-HCl buffer (pH 7.5). EMSA reaction mixtures (20 μl) contained 1× POP buffer (20% glycerol, 50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 2.5 mM EDTA, 250 mM NaCl), 1 mM dithiothreitol, 1 μg poly(dG-dC)-poly(dG-dC), 25 μM Cl-OHPA or 250 μM 4-OHPA, purified CprK1, and approximately 1 nM 32P-labeled DNA. The reaction mixtures were first incubated at 24°C for 30 min to allow complex formation, then loaded onto a 6% polyacrylamide gel buffered with 89 mM Tris and 89 mM boric acid (ca. pH 8.1), and electrophoresed at a constant current of 10 mA at 4°C, followed by drying and autoradiography.
In vivo promoter probe assay.
E. coli JM109(DE3) cells were cotransformed with a pET24d expression vector carrying the wild-type cprK1 gene or one of its mutated derivatives and a pAK80 plasmid carrying either the DB1::lacLM, DB2::lacLM, DB3::lacLM, or DB3FNR::lacLM promoter fusion (Table 1). Each experiment was carried out in triplicate. A 50-ml volume of LB medium was supplemented with kanamycin (30 μg/ml) and erythromycin (200 μg/ml), inoculated with 1% O/N-grown bacterial culture, and then placed on a rotary shaker at 37°C until an A600 of 0.3 was reached. At this point, 0.1 mM IPTG was added to the culture, as well as 20 mM Cl-OHPA when appropriate, and a time zero sample was taken. The Erlenmeyer flasks were transferred to a 20°C incubator and were continuously shaken O/N. β-Galactosidase activity was measured as described by Sambrook et al. (33), except that prior to measurement of adsorption of o-nitrophenol at 420 nm, reaction mixtures were centrifuged for 2 min at 14,000 × g to remove cell particles. Hence, correction with the A550 values was not necessary. Throughout this study, 1 Miller unit was defined as follows: 1,000 × A420/(t · v · OD600) where t is reaction time, v is sample volume, and OD600 is the optical density of the culture at 600 nm at the time the sample was taken.
RESULTS AND DISCUSSION
Desulfitobacterium hafniense cpr gene cluster.
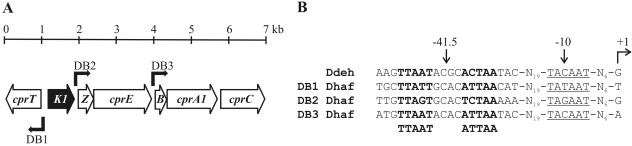
The gene coding for CprK1 protein accession number ZP_00558871.1) is located in the 7-kb chlorophenol reductive dehalogenase gene cluster of the Desulfitobacterium hafniense genome (Fig. 1A). In analogy to the situation in D. dehalogenans (37), members of the cpr gene cluster code for proteins with known or putative functions in halorespiration: cprT encodes a putative trigger factor (proline cis/trans isomerase), the product of cprE has homology to GroEL-type chaperones, CprB is a probable membrane anchor for the product of the downstream cprA1, a Cl-OHPA reductive dehalogenase (11), and cprC is expected to code for a NosR/NirI-type transcriptional regulator. Apart from the orthologous gene in the D. dehalogenans cpr gene cluster, no homologue of cprZ could be identified in sequence databases. The deduced amino acid sequence of the protein encoded by cprK1 is 232 amino acids long and has a molecular size of 26.5 kDa with an isoelectric point of 8.1. CprK1 shows 94% sequence similarity (89% identity) to D. dehalogenans CprK, 41% similarity (20% identity) to E. coli FNR, and 44% similarity (21% identity) to E. coli CRP. Three putative CprK1 binding sites (dehaloboxes) were identified as 14-bp inverted repeats in the intergenic regions of cprT-cprK1 (DB1), cprK1-cprZ (DB2), and cprE-cprB (DB3) (Fig. 1B). They all contain a 5-bp palindromic sequence that is separated by 4 spacing nucleotides. The deduced consensus sequence for dehaloboxes is TTAAT----ATTAA, which closely resembles the FNR consensus TTGAT----ATCAA (17) and more distantly resembles the target sequence of CRP, TGTGA------TCACA (26). The three dehaloboxes are centered at −41.5 relative to putative Pribnow boxes (−10), suggesting that these promoters share similar characteristics with class II promoters (7). Sequence alignment of D. hafniense dehaloboxes (DB1, DB2, and DB3) with the corresponding cprE-cprB intergenic region from D. dehalogenans, for which the transcription start site has been mapped (37) (Fig. 1B), further confirms that these promoters belong to class II.
FIG. 1.
(A) Desulfitobacterium hafniense chlorophenol reductive dehalogenase gene cluster. (B) Alignment of three D. hafniense intergenic regions containing 14-bp inverted repeats termed “dehaloboxes” (DB1 to DB3) with the corresponding cprE-cprB intergenic region from D. dehalogenans. Positions of dehaloboxes (boldfaced), putative Pribnow boxes (underlined), and the transcription start site previously mapped in D. dehalogenans are indicated with arrows above the DNA sequence alignment. The proposed consensus sequence for dehaloboxes is given below the sequence alignment.
Overproduction and purification of CprK1.
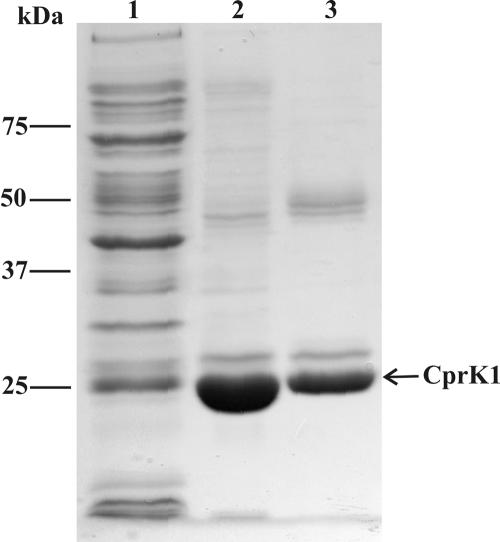
A 699-bp DNA fragment coding for CprK1 was PCR amplified from D. hafniense genomic DNA using primers BG1379 and BG1814 and cloned into a pET24d overexpression vector, resulting in plasmid pWUR176. E. coli BL21(DE3) cells carrying pWUR176 were grown aerobically until early-exponential-growth phase; then overproduction of CprK1 was induced upon addition of 0.1 mM IPTG, followed by O/N cultivation at 20°C. The recombinant protein contributed to approximately 15% of the total cell protein content, and O/N cultivation of the cells at a low temperature resulted in approximately 50% CprK1 being produced in the soluble cytoplasmic fraction. CprK1 was purified in two steps (Fig. 2). First, cell extracts were applied to a HiPrep heparin column, a general affinity chromatography matrix for DNA-binding proteins. Weakly bound CprK1 was eluted by increasing the concentration of NaCl from 100 mM to 300 mM. Pooled fractions that contained CprK1 were concentrated and applied to a Tricorn Superdex gel filtration column. The total yield of purified CprK1 was 3 mg protein per liter of bacterial culture. Purified recombinant CprK1 had the expected molecular size of 26.5 kDa (estimated from an SDS-polyacrylamide gel electrophoresis gel). Gel filtration experiments indicated that under these conditions CprK1 exists as a dimer (data not shown).
FIG. 2.
Purification of CprK1. The SDS-PAGE gel shows consecutive steps of CprK1 purification: Lane 1, cell extract (35 μg); lane 2, heparin affinity chromatography (35 μg); lane 3, gel filtration chromatography (12 μg).
In vitro DNA-binding properties of CprK1.
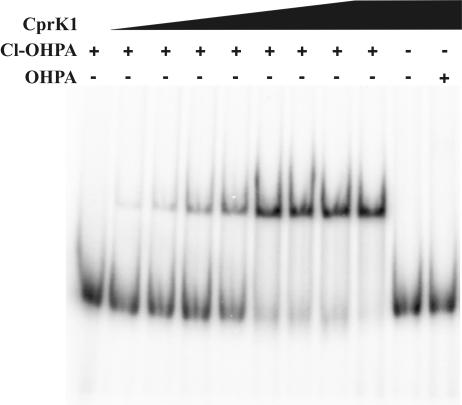
The interaction between the putative transcriptional regulator CprK1 and DNA was first investigated in detail by EMSA using a 52-bp cprE-cprB intergenic region that contained dehalobox DB3, a 14-bp perfect inverted repeat (TTAATacacATTAA), as a putative binding target. Binding reactions with increasing amounts of purified CprK1 protein were carried out in the presence of 25 μM Cl-OHPA, a compound that is known to be a substrate for the Cl-OHPA reductive dehalogenase enzyme encoded by the downstream cprA1 gene. Formation of protein-DNA complexes was observed at CprK1 concentrations as low as 10 nM, indicating a high affinity of CprK1 for its target DNA (Fig. 3). All target DNA was in the complexed form when 200 nM CprK1 was added to the EMSA binding mixture. By calculating the ratio between free DNA and CprK1-bound DNA, a dissociation constant (KD) of approximately 90 nM was determined for the CprK1-DB3 complex. Binding of CprK1 to DNA was observed only in the presence of Cl-OHPA. When Cl-OHPA was omitted from the binding reaction mixture or when it was replaced by its dechlorinated derivative, 4-OHPA, we could not observe any protein-DNA complex formation, which indicates a very specific allosteric effect of Cl-OHPA on CprK1. Titrating a constant amount of CprK1 (150 nM) with increasing amounts of Cl-OHPA in the binding mixtures (from 0.25 μM to 50 μM) resulted in a binding constant of Cl-OHPA to CprK1 of approximately 0.4 μM. These results correlate well with that obtained for D. dehalogenans CprK (31). CprK shows high affinity (KD, 190 nM) to a 193-bp cprB promoter fragment from D. dehalogenans that contains a TTAATacgcACTAA inverted repeat. DNA binding of CprK was observed only in the presence of Cl-OHPA, not with 4-OHPA, 2-Cl-PA, or cAMP. The affinity of CprK to Cl-OHPA was determined under two conditions: in the presence of target DNA (KD, 0.4 μM) and by isothermal titration calorimetry measurement in the absence of DNA (KD, 3.4 μM) (31).
FIG. 3.
Interaction of purified CprK1 with the cprE-cprB intergenic region containing dehalobox DB3. An electrophoretic mobility shift assay was performed in the presence (+) or absence (−) of 25 μM Cl-OHPA or 250 μM 4-OHPA using approximately 1 nM 32P-labeled DNA and increasing amounts of CprK1 protein. Concentrations of CprK1 in lanes are as follows, from left to right: 0, 10, 20, 50, 100, 200, 300, 500, 1,500, 1,500, and 1,500 nM.
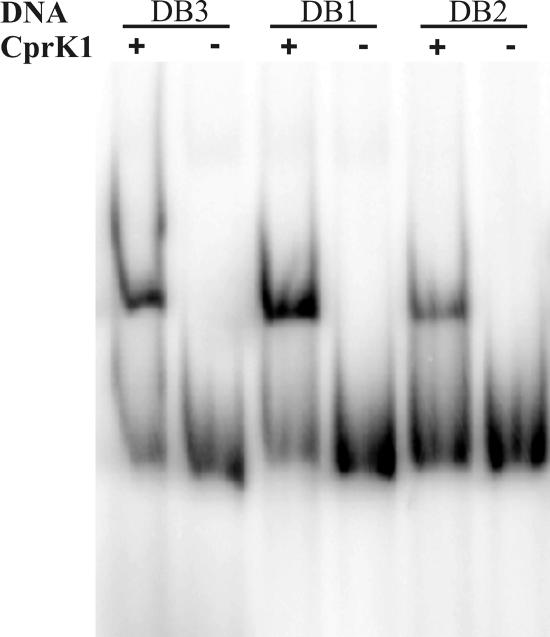
Besides the detailed study of the interaction between dehalobox DB3 and CprK1, we also carried out EMSA experiments with two other predicted target sites for CprK1. These target sites are: dehalobox DB1, in the center of the 60-bp intergenic region of cprT-cprK1, and dehalobox DB2, in the center of the 63-bp intergenic region of cprK1-cprZ. The binding reactions were carried out in the presence of 25 μM Cl-OHPA, 50 to 90 cps 32P-labeled DNA, and 100 nM CprK1. The results clearly show that all intergenic regions contain a binding site for CprK1, but under the conditions used, CprK1 shows different affinities toward them (Fig. 4). CprK1 binds dehalobox DB1 with similar affinity as it does the well-studied DB3, while clearly less protein-DNA complex is formed with DB2 as a target DNA sequence at the same CprK1 concentration. The optimal binding site (DB3) for CprK1 contains a perfect inverted repeat, TTAAT----ATTAA, that we propose to be the consensus sequence for dehaloboxes. Deviation from this consensus is likely to influence the DNA-binding affinity of CprK1. DB1 has one mismatch at position 4 compared to the consensus sequence (T replaces the preferred A), while DB2, the least optimal binding site, has two substitutions (A is replaced by G and T is replaced by C at the twofold-symmetry-related positions 4 and 11, respectively) (Fig. 1). Decreased binding affinity is probably caused by the elimination of a positive interaction (H bond) between the side chain amino group of Arg196 and the C-2 atom of thymine at positions 4 and 11 when G · C/C · G base pairs replace the A · T/T · A base pairs. The identity of the four bases between the inverted repeats can also influence the binding affinity, although they do not directly interact with the regulator protein (35). However, findings that FNR-regulated class I or class II promoters have highest activity when the middle two base pairs in the spacing nucleotides are A or T are not applicable in the case of the dehaloboxes studied here.
FIG. 4.
Interaction of CprK1 with dehaloboxes DB1, DB2, and DB3. An electrophoretic mobility shift assay was carried out in the presence (+) or absence (−) of 100 nM CprK1 protein, 25 μM Cl-OHPA, and one of the three 32P-labeled DNA-containing dehaloboxes DB1, DB2, and DB3 as indicated above the gel.
In vivo interaction of CprK1 with E. coli RNA polymerase.
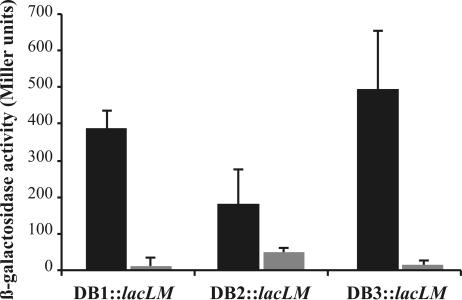
We have shown that CprK1 can bind to its target dehaloboxes (DB1, DB2, and DB3) in vitro in the presence of Cl-OHPA. In order to investigate whether this protein is capable of initiating transcription in concert with the RNA polymerase holoenzyme, we have constructed a fusion between three D. hafniense promoters containing DB1, DB2, or DB3 and the β-galactosidase-encoding lacLM genes on plasmid pAK80 (22). The resulting promoter-fusion products were DB1::lacLM on pWUR171 (containing a 164-bp cprK1-cprT intergenic region), DB2::lacLM on pWUR168 (containing a 322-bp cprK1-cprZ intergenic region), and DB3::lacLM on pWUR166 (containing a 266-bp cprE-cprB intergenic region). E. coli JM109(DE3) cells were cotransformed with one of the pAK80 constructs and with pWUR176 containing the cprK1 gene under the control of an IPTG-inducible T7 promoter. During the promoter probe assay, CprK1 production in aerobically grown E. coli JM109(DE3) cells was induced by the addition of 0.1 mM IPTG followed by O/N growth at 20°C under two different conditions: in the presence of 20 mM Cl-OHPA or without additional effector. The pattern of β-galactosidase expression mediated by CprK1 was similar for all three promoters (Fig. 5). There was little detectable β-galactosidase activity in cells grown in the absence of effector molecules. Maximal activity was observed in cells cultured in a medium supplemented with 20 mM Cl-OHPA. Control experiments were done with IPTG-induced cells carrying an empty pET24d expression vector instead of pWUR176. Under these growth conditions, E. coli strain JM109(DE3) exhibits low-level (4 to 6 Miller units) native β-galactosidase background activity (data not shown), possibly originating from phospho-β-galactosidase encoded by the bgi operon. The measured β-galactosidase activities correlate well with the affinity of CprK1 toward these dehaloboxes, deduced by the ratio of free versus complexed DNA in electrophoretic mobility shift assays, with DB3::lacLM having the highest promoter activity and DB2::lacLM showing the lowest.
FIG. 5.
In vivo DNA-binding properties of CprK1. In E. coli cells CprK1 was overproduced in the presence of one of the three D. hafniense promoters containing dehalobox DB1, DB2, or DB3 fused with the lacLM genes. Aerobic E. coli cultures were grown under two different conditions: in the presence of 20 mM Cl-OHPA (solid bars) or in the absence of the effector (shaded bars). Promoter activity was expressed by measuring β-galactosidase activity (Miller units).
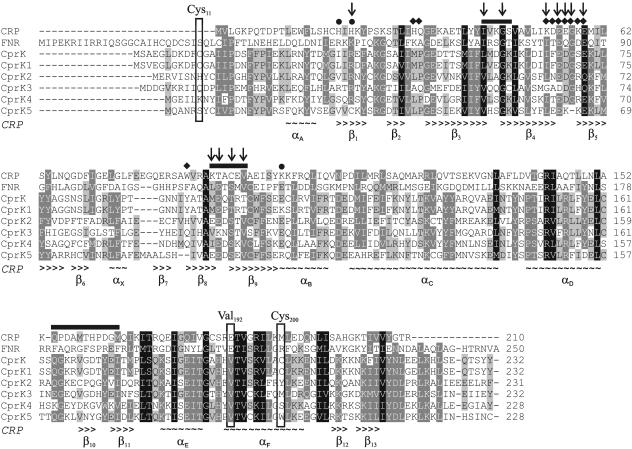
In vivo promoter probe experiments showed that CprK1 is capable of Cl-OHPA-dependent initiation of transcription by enabling the recruitment of the E. coli RNA polymerase. The −41.5 position of the target sites (dehaloboxes) of CprK1 relative to the putative Pribnow boxes (−10) places these halorespiration-inducible promoters within class II promoters (Fig. 1B). The class II-type contact between E. coli CRP or FNR and the RNA polymerase complex (RNAP) has been thoroughly investigated, and three contact sites (activating regions [AR]) have been identified in the regulators (15). AR1 makes contact with a conserved region (“287 determinant”) of the C-terminal domain of the RNAP α subunit (αCTD) (5), while AR2 interacts with four negatively charged amino acids of the N-terminal domain of the α subunit (αNTD) (8). The third possible interaction site (AR3) in CRP or FNR targets a conserved region in the σ70 factor of the RNA polymerase holoenzyme (28, 32). This conserved region of σ70 (“region 4”) contains five positively charged amino acids. Sequence alignments of the corresponding subunits of the E. coli and D. hafniense RNAP show that amino acids for AR1 and AR3 contact are conserved in D. hafniense RNAP, while the characteristic negatively charged amino acids for AR2 contact are missing. Using sequence alignment of D. dehalogenans CprK and five D. hafniense CprK homologues with E. coli CRP and FNR, and using the secondary structure of CRP (34), we have identified conserved amino acids that are likely to be involved in the regulatory protein-RNAP interaction (Fig. 6). These amino acids in CprK1 are residues 56 to 60 (β3-β4 turn in CRP) and residues 99 to 104 (β8-β9 turn in CRP), which aligned well with AR1 structures of FNR, the positively charged surface-exposed residue Arg35 (AR2), and the conserved residues Ile66, Glu68, Asp69, Gly70, and Glu72, three of which are negatively charged (AR3).
FIG. 6.
Sequence alignment of E. coli CRP and FNR proteins with D. dehalogenans CprK and five D. hafniense CprK homologues (CprK1 to CprK5). Essential FNR/CRP residues (AR) previously identified to be involved in RNA polymerase contact are indicated above the alignment as follows: rectangles, AR1; circles, AR2; diamonds, AR3. The secondary structure of E. coli CRP is given below the sequence alignment (∼, α-helix; >, β-sheet). Vertical arrows indicate amino acids that are conserved in most CprK homologues and correspond to AR1, AR2, or AR3. In boxes, amino acids are highlighted that correspond to the position of the nonconserved Cys11 and Cys200 residues in D. hafniense CprK1, capable of intermolecular disulfide bridge formation, and to the position of Val192 in the recognition α-helix of CprK1, which is conserved in all CprK homologues and corresponds to the Glu181 and Glu209 residues, essential for specific DNA binding in CRP and FNR, respectively. Protein accession numbers (based on the D. hafniense genome sequence, version of 24 June 2005) are as follows: ZP_00558871.1 (CprK1), ZP_00558892.1 (CprK2), ZP_00558887.1 (CprK3), ZP_00560926.1 (CprK4), and ZP_00559167.1 (CprK5).
Transcriptional activation by an FNR-type mutant protein, CprK1(V192E).
The dehalobox consensus sequence (TTAAT----ATTAA) is highly similar to the consensus binding sequence of FNR (TTGAT----ATCAA) except that at the twofold-related positions 3 and 12, base pairs A · T/T · A are conserved in all dehaloboxes (Fig. 1B), while base pairs G · C/C · G are present without exception in FNR boxes. Recognition by wild-type FNR is abolished if the TTGAT----ATCAA motif is changed into TTAAT----ATTAA by a base pair substitution at positions 3 and 12 (4). It is known that in the recognition α-helix of FNR, Glu209 is involved in making essential hydrogen bonds with cytosine at positions 3 and 12 of the FNR box sequence (15). This glutamate (E209-RS) is highly conserved in other FNR homologues. However, at the same position in CprK1 and all other CprK homologues, a valine is present (V192-RS) (Fig. 6). In order to see if a single amino acid change in the recognition α-helix of CprK1 would result in altered DNA target sequence specificity, we have replaced valine at position 192 by a glutamate. CprK1 was mutated by replacing a GTC codon with GAG in pWUR89, a plasmid carrying the wild-type cprK1 gene cloned in frame with a six-His translational fusion. The resulting pWUR172 plasmid codes for CprK1(V192E), a protein that has FNR-type conserved residues in its recognition α-helix (E192-RS).
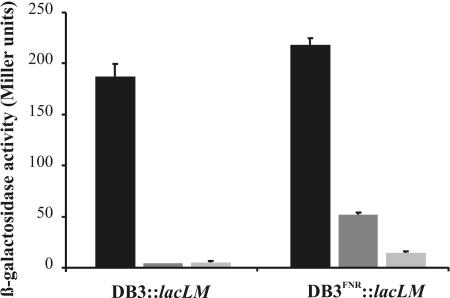
E. coli JM109(DE3) cells were cotransformed with pWUR166 containing the DB3::lacLM promoter fusion and either the pWUR89 or the pWUR172 plasmid. Cells were grown aerobically until mid-exponential-growth phase, when production of wild-type CprK1 or CprK1(V192E) was induced by addition of 0.1 mM IPTG, and cells were allowed to grow O/N at 20°C in the presence of 20 mM Cl-OHPA. Activation of transcription by CprK1(V192E) from the TTAATacacATTAA motif-containing promoter was completely abolished (Fig. 7). The β-galactosidase activity measured in cultures overproducing CprK1(V192E) was not higher than the control background level.
FIG. 7.
Transcriptional activation by the FNR-type mutant CprK1(V192E). In vivo promoter probe assays were carried out with aerobically grown E. coli JM109(DE3) cells overproducing wild-type CprK1 (solid bars) or CprK1(V192E) (dark shaded bars) or carrying an empty pET24d expression vector (light shaded bars). All experiments were done in the presence of 20 mM Cl-OHPA with cells carrying either a DB3::lacLM promoter fusion or a DB3FNR::lacLM promoter fusion.
In order to see if the Val192Glu substitution in CprK1 has enabled this protein to recognize an FNR box, we mutated dehalobox DB3 (TTAATacacATTAA) at both half-sites of the inverted repeat into a consensus FNR-binding site, resulting in DB3FNR (TTGATacacATCAA). A 266-bp cprE-cprB intergenic region containing DB3FNR was fused to the promoterless lacLM reporter genes. The resulting construct, designated pWUR175, is identical to pWUR166 except for the single-base-pair substitutions in each half-side of the dehalobox. The loss of activity of CprK1(V192E) was restored more than 13-fold (to approximately 25% of the activity of wild-type CprK1) on a promoter containing the TTGAT----ATCAA motif in DB3FNR::lacLM (Fig. 7). Interestingly, wild-type CprK1 showed similar activity on promoters containing either DB3 or DB3FNR, suggesting a lack of preference of Val192 toward A · T or G · C base pairs at position 3 and toward T · A or C · G base pairs at position 12 in its target sequence. Likewise, an FNR mutant in which Glu209 was replaced by a valine also gained the ability to interact with an altered FNR box containing A · T at position 3 and T · A at position 12 (3).
Similar observations were made with a mutant CRP. In the recognition α-helix of CRP, a glutamate at position 181 (RE181---R) is essential for the recognition of the G · C base pair at position 4 and C · G at position 13 in its target sequence, TGTGA------TCACA (15) (Fig. 6). The twofold-related positions 4 and 13 in CRP boxes correspond to positions 3 and 12 in FNR boxes and dehaloboxes. It was shown that replacement of Glu181 in CRP by valine or leucine results in the complete elimination of specificity between G · C, A · T, C · G, and T · A bases at the corresponding positions 4 and 13 (12).
It is very likely that CprK1 could recognize a motif with any of the four nucleotides at positions 3 and 12. However, despite the lack of a favorable interaction of Val192 in CprK1 with any of the nucleotides, strong conservation of the A · T base pair at position 3 and the T · A base pair at position 12 occurs in dehaloboxes (Fig. 1). Another transcriptional regulator, NtcA from Anabaena sp. strain PCC 7120, shares similar features. In NtcA a valine is conserved at the position that corresponds to Val192 in CprK1, and the deduced NtcA-binding DNA motif (TGTA--------TACA) also has conserved A · T/T · A base pairs in the corresponding positions 4 and 13 (23).
A homology search showed that the genome sequence of D. hafniense contains at least two FNR homologues protein accession numbers ZP_00560130 and ZP_00558676) with an E-RS motif in their recognition α-helices that can probably recognize an FNR box but not a dehalobox. We hypothesize that the conservation of A · T/T · A base pairs at the twofold-related positions 3 and 12, respectively, was fixed in dehaloboxes to eliminate cross talk between structurally related but functionally different CRP/FNR-type regulators, i.e., to avoid false induction of halorespiration genes by these FNR homologues. On the other hand, since wild-type CprK1 is theoretically able to bind both to dehaloboxes and to FNR boxes in the presence of Cl-OHPA (Fig. 7), the same issue of cross talk mediated by CprK1 remains open. Since the two FLPs in D. hafniense do not contain the conserved cysteine residues of E. coli FNR (except for a cysteine in ZP_00560130 that corresponds to Cys122 of E. coli FNR) (16), they are not likely to sense redox changes through the incorporation of an Fe-S cluster. Thus, the identity of possible effector molecules for these FNR homologues, as well as their target genes, remains a question. It is likely that the target promoters of the FNR homologues contain more than one regulator binding site (complex bacterial promoters, reviewed by Barnard et al. [2]); therefore, either cross talk of CprK1 with other FNR homologues can be prevented by the existence of a repressor that does not allow binding of, and transcriptional initiation by, CprK1 on FNR-regulated promoters or, alternatively, transcriptional initiation requires an interaction of two regulators that is possible only between the FNR homologue and the unknown regulator and not with CprK1.
Is CprK1 subject to redox regulation?
The crystal structure of D. hafniense CprK1 shows the presence of two intermolecular disulfide bridges that connect Cys11 of monomer A with Cys200 in monomer B and vice versa (M. G. Joyce et al., unpublished data). Electrophoretic mobility shift assays have already indicated that oxidized CprK1 is not able to bind its target DNA (data not shown), an observation similar to those made with CprK from D. dehalogenans by Pop et al. (31). Since these experiments were performed in vitro, the question remained whether the Cys11-Cys200 disulfide bridge in CprK1 is physiologically relevant. In other words, is CprK1 subject to redox regulation in the cytoplasm by a disulfide-thiol exchange reaction? If this were the case, then replacement of the two cysteine residues with serine would result in a constantly active CprK1 protein that would be insensitive to oxidation and would activate transcription in vivo more efficiently than wild-type protein.
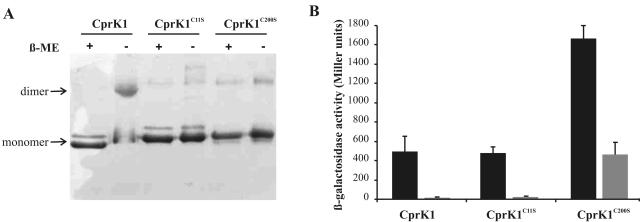
We have created two single-amino-acid substituted mutants of CprK1 by replacing a TGT codon with TCT in pWUR176, resulting in plasmid pWUR210, encoding the CprK1(C11S) variant, and plasmid pWUR211, encoding the CprK1(C200S) protein. Both mutant proteins were overproduced and purified. SDS-PAGE of protein samples that were heat denatured in the presence or absence of 5% β-mercaptoethanol proved that CprK1(C11S) and CprK1(C200S) are not capable of intermolecular disulfide bridge formation (Fig. 8A). However, gel filtration experiments showed that CprK1(C11S) and CprK1(C200S) still existed as homodimers in solution, which indicated that the disulfide bridge is not essential for dimer formation. To test their in vivo activity, we cotransformed E. coli JM109(DE3) cells with pWUR166 containing the DB3::lacLM fusion and either pWUR210 or pWUR211. E. coli cells were grown aerobically until mid-exponential phase; then CprK1 production was induced by addition of 0.1 mM IPTG and was continued in the presence or absence of 20 mM Cl-OHPA at 20°C. β-Galactosidase activity was measured after O/N growth. There was no significant difference between the transcriptional activation mediated by wild-type CprK1 and that mediated by CprK1(C11S) (Fig. 8B). The mutant CprK1(C200S) had 340% of wild-type activity in the presence of Cl-OHPA, but its effector-free activity was also significantly higher (28 times) than the wild-type background, indicating most likely that the CprK1(C200S) mutant was overproduced to a greater extent in the soluble cytoplasmic fraction than the wild type or the CprK1(C11S) mutant. This hypothesis is supported by in vitro observations in EMSA experiments where CprK1(C200S) did not show increased DNA-binding affinity compared to equal amounts of wild-type CprK1 under reducing conditions (data not shown). In conclusion, in vivo transcriptional activation experiments by CprK1 and its single-amino-acid substituted derivatives CprK1(C11S) and CprK1(C200S) suggest that recombinant CprK1 does not contain a disulfide bridge in the cytoplasm of aerobically cultivated E. coli cells.
FIG. 8.
Effect of redox state of CprK1 on its activity. (A) SDS-PAGE gel of purified wild-type CprK1, CprK1(C11S), and CprK1(C200S) proteins (5.8 μg) that were heat denatured in the presence (+) or absence (−) of 5% β-mercaptoethanol (β-ME). CprK1(C11S) and CprK1(C200S) proteins each contain a C-terminal six-His tag, causing a slight difference in their mobilities compared to that of wild-type CprK1 on SDS-PAGE gels but leaving other properties of the proteins unaffected. (B) In vivo promoter probe assay using E. coli JM109(DE3) cells harboring plasmid pWUR166 with a DB3::lacLM promoter fusion and a pET24d plasmid derivative that overproduces either wild-type CprK1, CprK1(C11S), or CprK1(C200S) protein. Cells were grown either in the presence of 20 mM Cl-OHPA (solid bars) or in the absence of the effector (shaded bars).
There is further evidence that supports this hypothesis. First of all, the midpoint redox potential of E. coli cytoplasm is ≤−250 mV (40), which strongly disfavors the formation of stable disulfide bonds in proteins. Thus, it is generally accepted that in the absence of enzymatic systems (e.g., thiol-disulfide oxidoreductases), the formation of cysteine-cysteine linkages in the cytoplasm is extremely slow (24). However, an increasing number of transcriptional regulators that undergo a temporary thiol-disulfide exchange reaction or thiol modification during oxidative stress has been identified. Active FLP from Lactobacillus casei contains an intramolecular disulfide bridge (14). In CrtJ homologues, two cysteine residues are conserved (Cys249 and Cys420) that form a disulfide bridge in the active CrtJ protein characterized from Rhodobacter capsulatus (30). OxyR from E. coli is a well-studied regulator of genes involved in antioxidative response (1). The crystal structure of the oxidized active form of OxyR shows the presence of a disulfide bridge between Cys199 and Cys208, two residues that are 17 Å apart in the reduced protein (9). Kim et al. questioned the physiological relevance of this disulfide bridge in activation of the protein and regarded it as merely an artifact of the lengthy crystallization process (25). Instead, they proposed a multiple-activation-state model where Cys199 of OxyR can be modified into a sulfenic acid, S-nitrosothiol, or a mixed glutathione disulfide intermediate according to different environmental signals. A similar event could have occurred during the crystallization of CprK1 under aerobic conditions, resulting in the formation of the possibly nonphysiological intermolecular disulfide bridges between Cys11 and Cys200. These residues are conserved only in D. hafniense CprK1 and D. dehalogenans CprK, not in other D. hafniense CprK homologues, although these proteins do show an unusually high cysteine content (Fig. 6). The only cysteine that is shared among four CprK homologues (including CprK1) and CprK is Cys105, which corresponds to Cys122 of FNR from E. coli. This cysteine is essential for intramolecular disulfide bond formation and is involved in Fe-S cluster binding in FNR (16). Pop et al. have investigated the role of Cys105 by replacing this residue with alanine in D. dehalogenans CprK (31). Transcriptional activation mediated by the C105A mutant was compared with that of wild-type CprK, which led to the conclusion that this mutation has no effect on the activity of this protein. Although we have shown that there is no evidence of disulfide bond formation in recombinant CprK1 when it is produced aerobically in E. coli, we cannot exclude the possibility that this protein gives a different response in D. hafniense when the cells are exposed to oxygen. It was reported that although no growth occurs in its presence, D. hafniense can tolerate oxygen and resume growth in anaerobic media after oxygen exposure for 24 h (29). In D. hafniense there are redox proteins (iron-sulfur proteins, flavins, flavoproteins, and quinones) that are potentially capable of carrying out the one-electron reduction of oxygen to superoxide, and possibly further to hydrogen peroxide. One of the redox proteins is the Cl-OHPA reductive dehalogenase (CprA1) itself, encoded in the same cpr gene cluster as CprK1. This enzyme contains a corrinoid cofactor and three [4Fe-4S] redox centers that are normally involved in the electron transfer during the reductive dehalogenation of Cl-OHPA to 4-OHPA (11). Accumulation of the highly reactive superoxide and hydrogen peroxide radicals would be toxic for the cells, so a likely defense mechanism can involve the down-regulation of the transcription of cpr genes by oxidative inactivation of CprK1 (31). However, D. hafniense cells have the capacity to deal directly with these radicals. We have found two superoxide dismutase- and two catalase-encoding genes in its genome sequence, as well as a gene whose product shows high homology to a peroxidase/catalase from Geobacillus stearothermophilus. Madsen and Licht detected catalase activity in D. hafniense cells (29), further confirming that D. hafniense directly inactivates reactive radicals produced upon O2 exposure rather than preventing their formation, which would require the down-regulation of several redox proteins.
In conclusion, our results indicate that heterologously overproduced CprK1 does not contain a Cys11-Cys200 disulfide bridge in vivo. Consequently, in D. hafniense this protein is most likely to be the subject of activation only by Cl-OHPA and not by disulfide to thiol reduction.
Acknowledgments
This work was supported by a grant received from NWO-CW (Netherlands Organization of Scientific Research, Chemical Sciences Division).
The sequence data of the Desulfitobacterium hafniense DCB-2 genome were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). We thank Eric Johansen (Chr. Hansen, Denmark) for kindly providing pAK80 plasmid and for additional help.
REFERENCES
- 1.Aslund, F., M. Zheng, J. Beckwith, and G. Storz. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA 96:6161-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A., and S. Busby. 1994. Location and orientation of an activating region in the Escherichia coli transcription factor, FNR. Mol. Microbiol. 11:383-390. [DOI] [PubMed] [Google Scholar]
- 4.Bell, A. I., J. A. Cole, and S. J. Busby. 1990. Molecular genetic analysis of an FNR-dependent anaerobically inducible Escherichia coli promoter. Mol. Microbiol. 4:1753-1763. [DOI] [PubMed] [Google Scholar]
- 5.Benoff, B., H. Yang, C. L. Lawson, G. Parkinson, J. Liu, E. Blatter, Y. W. Ebright, H. M. Berman, and R. H. Ebright. 2002. Structural basis of transcription activation: the CAP-αCTD-DNA complex. Science 297:1562-1566. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Busby, S., and R. H. Ebright. 1997. Transcription activation at class II CAP-dependent promoters. Mol. Microbiol. 23:853-859. [DOI] [PubMed] [Google Scholar]
- 8.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 9.Choi, H., S. Kim, P. Mukhopadhyay, S. Cho, J. Woo, G. Storz, and S. Ryu. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105:103-113. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen, N., and B. K. Ahring. 1996. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 46:442-448. [Google Scholar]
- 11.Christiansen, N., B. K. Ahring, G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the 3-chloro-4-hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436:159-162. [DOI] [PubMed] [Google Scholar]
- 12.Ebright, R. H., A. Kolb, H. Buc, T. A. Kunkel, J. S. Krakow, and J. Beckwith. 1987. Role of glutamic acid-181 in DNA-sequence recognition by the catabolite gene activator protein (CAP) of Escherichia coli: altered DNA-sequence-recognition properties of [Val181]CAP and [Leu181]CAP. Proc. Natl. Acad. Sci. USA 84:6083-6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Fantroussi, S., H. Naveau, and S. N. Agathos. 1998. Anaerobic dechlorinating bacteria. Biotechnol. Prog. 14:167-188. [DOI] [PubMed] [Google Scholar]
- 14.Gostick, D. O., J. Green, A. S. Irvine, M. J. Gasson, and J. R. Guest. 1998. A novel regulatory switch mediated by the FNR-like protein of Lactobacillus casei. Microbiology 144:705-717. [DOI] [PubMed] [Google Scholar]
- 15.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44:1-34. [DOI] [PubMed] [Google Scholar]
- 16.Green, J., A. D. Sharrocks, B. Green, M. Geisow, and J. R. Guest. 1993. Properties of FNR proteins substituted at each of the five cysteine residues. Mol. Microbiol. 8:61-68. [DOI] [PubMed] [Google Scholar]
- 17.Guest, J. R., J. Green, A. S. Irvine, and S. Spiro. 1996. The FNR modulon and FNR-regulated gene expression, p. 317-338. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. R. G. Landes Company, Austin, Tex.
- 18.Harman, J. G. 2001. Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta 1547:1-17. [DOI] [PubMed] [Google Scholar]
- 19.He, Y., T. Gaal, R. Karls, T. J. Donohue, R. L. Gourse, and G. P. Roberts. 1999. Transcription activation by CooA, the CO-sensing factor from Rhodospirillum rubrum. The interaction between CooA and the C-terminal domain of the α subunit of RNA polymerase. J. Biol. Chem. 274:10840-10845. [DOI] [PubMed] [Google Scholar]
- 20.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 21.Holliger, C., G. Schraa, A. J. Stams, and A. J. Zehnder. 1993. A highly purified enrichment culture couples the reductive dechlorination of tetrachloroethene to growth. Appl. Environ. Microbiol. 59:2991-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, F., S. Wisen, M. Widersten, B. Bergman, and B. Mannervik. 2000. Examination of the transcription factor NtcA-binding motif by in vitro selection of DNA sequences from a random library. J. Mol. Biol. 301:783-793. [DOI] [PubMed] [Google Scholar]
- 24.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111-135. [DOI] [PubMed] [Google Scholar]
- 25.Kim, S. O., K. Merchant, R. Nudelman, W. F. Beyer, Jr., T. Keng, J. DeAngelo, A. Hausladen, and J. S. Stamler. 2002. OxyR: a molecular code for redox-related signaling. Cell 109:383-396. [DOI] [PubMed] [Google Scholar]
- 26.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749-795. [DOI] [PubMed] [Google Scholar]
- 27.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559-592. [DOI] [PubMed] [Google Scholar]
- 28.Lamberg, K. E., C. Luther, K. D. Weber, and P. J. Kiley. 2002. Characterization of activating region 3 from Escherichia coli FNR. J. Mol. Biol. 315:275-283. [DOI] [PubMed] [Google Scholar]
- 29.Madsen, T., and D. Licht. 1992. Isolation and characterization of an anaerobic chlorophenol-transforming bacterium. Appl. Environ. Microbiol. 58:2874-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda, S., C. Dong, D. Swem, A. T. Setterdahl, D. B. Knaff, and C. E. Bauer. 2002. Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc. Natl. Acad. Sci. USA 99:7078-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pop, S. M., R. J. Kolarik, and S. W. Ragsdale. 2004. Regulation of anaerobic dehalorespiration by the transcriptional activator CprK. J. Biol. Chem. 279:49910-49918. [DOI] [PubMed] [Google Scholar]
- 32.Rhodius, V. A., and S. J. Busby. 2000. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase σ70 subunit: application of suppression genetics. J. Mol. Biol. 299:311-324. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schultz, S. C., G. C. Shields, and T. A. Steitz. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science 253:1001-1007. [DOI] [PubMed] [Google Scholar]
- 35.Scott, C., J. D. Partridge, J. R. Stephenson, and J. Green. 2003. DNA target sequence and FNR-dependent gene expression. FEBS Lett. 541:97-101. [DOI] [PubMed] [Google Scholar]
- 36.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 37.Smidt, H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399-428. [DOI] [PubMed] [Google Scholar]
- 39.Tanigawa, R., M. Shirokane, S. Maeda Si, T. Omata, K. Tanaka, and H. Takahashi. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc. Natl. Acad. Sci. USA 99:4251-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unden, G., S. Becker, J. Bongaerts, J. Schirawski, and S. Six. 1994. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Leeuwenhoek 66:3-22. [DOI] [PubMed] [Google Scholar]
- 41.Utkin, I., C. Woese, and J. Wiegel. 1994. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int. J. Syst. Bacteriol. 44:612-619. [DOI] [PubMed] [Google Scholar]
- 42.van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 43.WHO. 1989. Chlorophenols other than pentachlorophenol, p. 208. Environmental Health Criteria 93. WHO, Geneva, Switzerland.