Abstract
The LysR-type transcriptional regulator CbbR controls the expression of the cbb and gap-pgk operons in Xanthobacter flavus, which encode the majority of the enzymes of the Calvin cycle required for autotrophic CO2 fixation. The cbb operon promoter of this chemoautotrophic bacterium contains three potential CbbR binding sites, two of which partially overlap. Site-directed mutagenesis and subsequent analysis of DNA binding by CbbR and cbb promoter activity were used to show that the potential CbbR binding sequences are functional. Inverted repeat IR1 is a high-affinity CbbR binding site. The main function of this repeat is to recruit CbbR to the cbb operon promoter. In addition, it is required for negative autoregulation of cbbR expression. IR3 represents the main low-affinity binding site of CbbR. Binding to IR3 occurs in a cooperative manner, since mutations preventing the binding of CbbR to IR1 also prevent binding to the low-affinity site. Although mutations in IR3 have a negative effect on the binding of CbbR to this site, they result in an increased promoter activity. This is most likely due to steric hindrance of RNA polymerase by CbbR since IR3 partially overlaps with the −35 region of the cbb operon promoter. Mutations in IR2 do not affect the DNA binding of CbbR in vitro but have a severe negative effect on the activity of the cbb operon promoter. This IR2 binding site is therefore critical for transcriptional activation by CbbR.
Xanthobacter flavus is a chemoautotrophic bacterium which uses the Calvin cycle to assimilate carbon dioxide (9, 12). The energy to drive carbon dioxide fixation is provided by the oxidation of compounds such as methanol, formate, and H2. The majority of the genes encoding the Calvin cycle enzymes constitute three transcriptional units: the cbb and gap-pgk operons and the tpi gene. The cbb operon encodes the key enzymes of the Calvin cycle, ribulose bisphosphate carboxylase/oxygenase and phosphoribulokinase, and in addition a number of enzymes required for the regeneration of ribulose bisphosphate. Glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase are encoded by the gap-pgk operon and play a role in both the Calvin cycle and glycolysis. The tpi gene encodes triosephosphate isomerase (10, 11, 13, 14, 24). During heterotrophic growth on, for instance, succinate, the cbb operon is not expressed and the gap-pgk operon is transcribed at a low constitutive level. A transition from heterotrophic to autotrophic growth is accompanied by a rapid induction of the cbb operon and a superinduction of the gap-pgk operon (11, 14). The first two genes of the cbb operon encode the CO2-fixing enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO).
The induction and superinduction of, respectively, the cbb and gap-pgk operons are completely dependent on the presence of the transcriptional regulator CbbR, which is encoded upstream and whose gene is transcribed divergently from the cbb operon (14, 25). This transcriptional regulator is encountered in many photoautotrophic and chemoautotrophic bacteria, where it controls transcription of the cbb operon (7). CbbR of X. flavus is a dimer in solution and binds to two sites in the cbb promoter, most likely as a dimer of identical subunits (27). DNase I protection studies showed that CbbR binds with high and low affinity to two DNA regions located, respectively, between nucleotides −75 and −50 and between nucleotides −44 and −29 relative to the transcriptional start site of the promoter of the cbb operon. The addition of NADPH, but not NADP, NADH, or NAD, to the DNA binding assay buffer resulted in a threefold increase in the affinity of CbbR for the cbb promoter (27), which was also observed for CbbR of Hydrogenophilus thermoluteolus (22). DNA binding studies using circular permutated DNA fragments showed that the binding of CbbR to the cbb promoter induced a bend in the DNA of 64o. The addition of NADPH to the assay buffer resulted in a partial relaxation of the DNA-bending angle by 9o (27). It is therefore likely that the in vivo transcription of the cbb operon is controlled by the intracellular concentration of NADPH. This hypothesis is supported by the observation that, following a transition from heterotrophic to autotrophic growth conditions, intracellular NADPH concentrations rapidly increase to a level which saturates CbbR in vitro, followed by induction of the cbb operon (26).
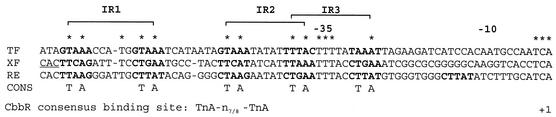
LysR-type proteins generally bind to inverted repeats containing the LysR motif T-N11-A (5). Inspection of the intergenic region between cbbR and cbbL, encoding the large subunit of RuBisCO, revealed the presence of three repeats containing the LysR motif. An alignment of the X. flavus cbb promoter with those from Thiobacillus ferrooxidans and Ralstonia eutropha showed that the sequences and relative locations of these repeats are conserved. This led to the recognition of the CbbR binding motif TNA-N7-TNA (Fig. 1) (19). The high-affinity CbbR binding site (R site) contains one CbbR binding motif (IR1), whereas the low-affinity binding site (A site) contains two, partially overlapping CbbR binding motifs (IR2 and IR3). The fact that all three of these motifs are located on the same side of the DNA helix suggests that they may be important for DNA binding and/or transcriptional activation of the cbb promoter. However, this does not imply that these are sufficient for DNA binding.
FIG. 1.
Alignment of DNA sequences of the cbbR-cbbL intergenic regions of X. flavus (XF), T. ferrooxidans (TF), and R. eutropha (RE). The CbbR binding motif is given below the alignment. ∗, identical nucleotides. Brackets, positions of LysR motifs. Inverted and direct repeats are in boldface. Positions of the −35 and −10 regions of the cbb promoter are indicated. +1, cbbL transcription start site. The start codon of the cbbR gene of X. flavus is underlined.
This paper focuses on the role that these CbbR binding motifs play in DNA binding by CbbR and transcriptional activation of the cbb promoter. The data presented in this paper show that all three CbbR binding motifs are important for transcriptional regulation by CbbR and that different functions can be assigned to each of the three.
MATERIALS AND METHODS
Media and growth conditions.
Escherichia coli strains DH5α (Bethesda Research Laboratories) and S17-1 (20) were grown on Luria-Bertani medium at 37°C. X. flavus strains H4-14 (9) and R22 (25) were grown in minimal media supplemented with gluconate (10 mM), succinate (10 mM), or methanol (0.5% [vol/vol]) at 30°C as described previously (12). X. flavus was grown on a mixture of gluconate (5 mM) and formate (20 mM) in a 3-liter batch fermentor with automatic titration with formic acid (25% [vol/vol]) to maintain a constant pH. When appropriate, the following supplements were added: ampicillin, 100 μg ml−1; X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galoactopyranoside), 20 μg ml−1; isopropyl-β-d-thiogalactopyranoside (IPTG), 0.1 mM; tetracycline, 12.5 (E. coli) or 7 μg ml−1 (X. flavus); kanamycin, 5 μg ml−1. Agar was added for solid media (1.5% [wt/vol]).
Mobilization of plasmids.
Mobilization of plasmids to X. flavus by using E. coli S17-1 containing the appropriate plasmids was performed as described by Simon et al. (20).
DNA manipulations.
Plasmid DNA was isolated via the alkaline lysis method of Birnboim and Doly (1). DNA-modifying enzymes were obtained from Boehringer Mannheim and were used according to the manufacturer's instructions. DNA fragments were isolated from agarose gels by using the Geneclean DNA purification kit from Bio 101. Other DNA manipulations were done in accordance with standard protocols. Oligonucleotides were obtained from Eurogentec. Amplification by PCR was carried out with Pwo DNA polymerase as recommended by the manufacturer (Boehringer).
Nucleotide sequencing.
Nucleotide sequencing was done with dye primers by the cycle sequencing method with Thermosequenase kit RPN 2538 from Amersham Pharmacia Biotech AB. The samples were run on the A.L.F.-Express sequencing robot.
Construction of promoter fusion vectors.
The intergenic region between cbbR and cbbL containing the cbb promoter on plasmid pTZ00 (27) was mutated and amplified by site-directed mutagenesis PCR (17) using mutant oligonucleotides and the oligonucleotides CR2 (5′-CATAGGATCCGGAGGCCGCGGCGAGC-3′) and Preind (5′-CGCGAATTCGTGTCCTTGGGCTGGTAG-3′), containing, respectively, BamHI and EcoRI restriction sites. The resulting DNA fragments were digested with BamHI and EcoRI and ligated into pBluescript KSII (Stratagene) or pTZ19U (Bio-Rad) digested with the same enzymes. The nucleotide sequences of the resulting plasmids were determined to verify that unwanted mutations had not been introduced in the PCR. The plasmids were digested with BamHI and EcoRI and ligated into the promoter-probe vector pBC3 (11), which was digested with the same enzymes. The resulting plasmids, with mutant cbb promoter cbbL-lacZ fusion plasmids, were mobilized to X. flavus strains H4-14 and R22 by using E. coli S17-1.
An approach similar to that described above was followed to create a lacZ fusion with the 5′ end of cbbR by using the primers CBBRFUSIEBA (5′-AAAGGATCCGCGCGAGGATATCGGTGTCC-3′) and CBBRFUSIEEC (5′-ATCGAATTCATCTGCGCGGTCACGGCGGGCGG-3′). The resulting plasmid, pBCfCbbR, containing the cbbR-lacZ fusion was mobilized to X. flavus strains H4-14 and R22 by using E. coli S17-1.
Enzyme assays.
Cell extracts were prepared by using a French pressure cell as described previously (11). β-Galactosidase activity was determined as described by Miller except that cell extracts were used instead of cell suspensions (15). RuBisCO activity was determined by measuring the incorporation of 14CO2 into acid-stable compounds (4). Protein was determined as described by Bradford with bovine serum albumin as the standard (2).
Preparation and labeling of DNA fragments used in binding studies.
32P-labeled DNA fragments containing either wild-type or mutant cbbR-cbbL intergenic-region DNA were obtained as described earlier (27).
Gel retardation assay.
Gel retardation assays were performed as described previously with 32P-labeled DNA fragments (10,000 cpm), purified CbbR, and in some experiments NADPH (final concentration, 200 μM) in an assay volume of 20 μl (27). The samples were subjected to nondenaturing gel electrophoresis using 6% acrylamide gels in Tris-borate buffer and run at 4°C and 10 V/cm. Following drying, the gel was analyzed by autoradiography. The radioactivity in the gel was quantified with a Cyclone phosphorimager by using the program Optiquant, version 03.00 (Canberra Packard Instrument Co.).
RESULTS
CbbR binding to the R site of the cbb promoter.
We previously (19) reported that the promoters of the cbb operons of R. eutropha, T. ferrooxidans, and X. flavus have sequence similarities in the region protected by CbbR from DNase I (Fig. 1). Based on these sequence comparisons we proposed a CbbR binding motif: TNA-N7-TNA (19). The R site of the promoter of the cbb operon contains one CbbR binding motif sequence (IR1), while the A site contains two partially overlapping binding motifs (IR2 and IR3) (Fig. 1).
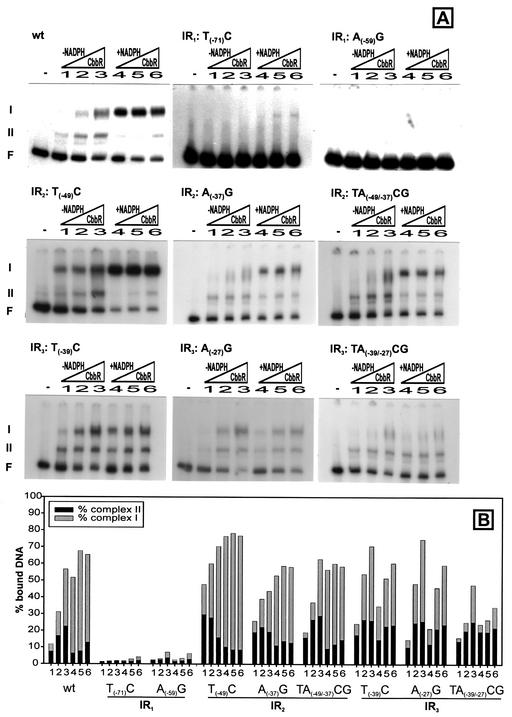
The ability of CbbR to bind to DNA templates was analyzed by gel retardation followed by quantitation of the percentage of DNA bound to CbbR. Two DNA-protein complexes of low and high mobilities were observed when a wild-type template was used as a binding substrate (Fig. 2A, wild type). According to our previous interpretation the high-mobility complex (complex II) is due to the binding of one CbbR dimer to the high-affinity binding site of the cbb operon promoter; the low-mobility complex (complex I) results from the binding of an additional CbbR dimer to the low-affinity binding site (25, 27). As was observed previously, the affinity of CbbR for its cognate binding sites is increased in the presence of NADPH (Fig. 2A, wild type). The presence of high- and low-affinity binding sites was further examined by using DNA fragments harboring only IR1 or IR2 and IR3. Only a single high-mobility DNA-protein complex is observed when a 20-bp DNA fragment containing IR1 (SRΔ16) (Fig. 3) is used as template, indicating the binding of single CbbR dimer. However, CbbR binding to a DNA fragment (SR8) (Fig. 3) harboring IR2 and IR3 was not observed (data not shown), even though this DNA fragment is protected in a DNase I footprint (27). This strongly suggests that the perfect inverted repeat IR1 represents a high-affinity binding site, whereas IR2 and IR3 represent a low-affinity site.
FIG. 2.
(A) Gel retardation assays of wild-type (wt) and mutant IR1, IR2, and IR3 cbb promoters performed with identical increasing amounts (as indicated by the triangles) of purified CbbR (34, 68, or 136 ng of CbbR per assay) with or without the CbbR inducer NADPH (final concentration, 200 μM). F, free (unbound) DNA; I, complex I (DNA bound by two CbbR dimers); II, complex II (DNA bound by one CbbR dimer). (B) Percentages of bound DNA in complex I and complex II for wild-type and mutated cbb promoters.
FIG. 3.
Site-directed mutagenesis of the cbb promoter of X. flavus. Mutation positions are given relative to the transcriptional start site of the cbb promoter (+1). The half-sites of the three inverted repeats are in boldface. The start codon of cbbR is underlined. The DNA fragments used to determine the binding of CbbR to IR1 (SRΔ16) or IR2 and IR3 (SR8) are indicated by solid bars above the nucleotide sequence. Every tenth nucleotide upstream from the transcriptional start site is indicated by a dot above the nucleotide sequence.
To determine whether the CbbR binding motifs in the R site are important for CbbR binding and promoter activity, single and double point mutations were introduced in the conserved CbbR nucleotides by site-directed mutagenesis, resulting in mutant cbb promoters with mutations T(−71)A, TA(−71/−69)CG, and A(−59)G (Fig. 3). Point mutations in the CbbR binding motif of the R site virtually abolished DNA binding by CbbR (Fig. 2). Limited DNA binding activity was visible only at high CbbR concentrations and in the presence of NADPH (4 to 6% bound DNA). Interestingly, these mutations not only affected binding to the R site harboring IR1 but also prevented the binding of CbbR to the A site of the cbb operon promoter containing IR2 and IR3.
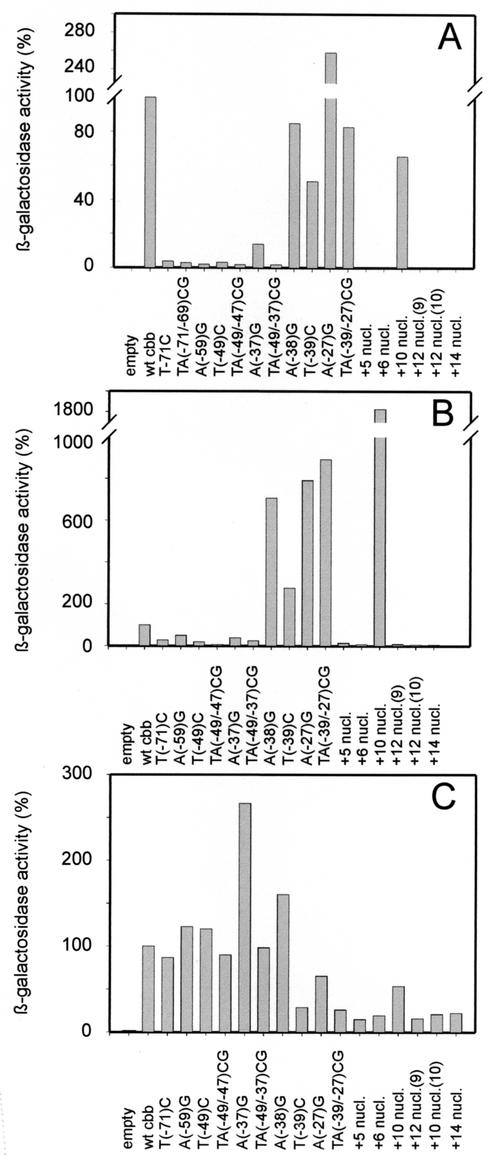
To assess the effect of these mutations on the activity of the cbb operon promoter, fusions with the reporter gene lacZ were constructed. The activity of β-galactosidase in X. flavus harboring a lacZ fusion with either the wild-type cbb promoter or mutated promoters was determined following autotrophic growth on methanol-containing medium (Fig. 4A). High activities were observed when the wild-type cbb promoter drove the expression of lacZ. However, the point mutations introduced in IR1 reduced the activity of the cbb promoter to 2 to 4% of that of the wild type. These results clearly show that the CbbR binding motif in the R site is essential for binding and subsequent transcriptional activation of the promoter of the cbb operon by CbbR.
FIG. 4.
In vivo activation of wild-type (wt) and cbb promoter mutants. (A and B) Normalized levels of β-galactosidase activities expressed by cbbL-lacZ fusions in X. flavus H4-14 grown autotrophically (A) or heterotrophically (B). β-Galactosidase activities driven by the wild-type cbb promoter were, respectively, 7,285 and 250 nmol per min per mg of protein and were set at 100%. Empty, promoter probe vector pBC3 without an insert. (C) Normalized levels of β-galactosidase activities expressed by cbbL-lacZ fusions in X. flavus R22 grown heterotrophically. The β-galactosidase activity driven by the wild-type cbb promoter was 59 nmol per min per mg of protein and was set at 100%.
CbbR binding to the A site: IR2.
The A site of the cbb promoter contains two partially overlapping CbbR binding motifs, IR2 and IR3. Interestingly, the right site of IR2 is also the left site of IR3 (Fig. 1). To determine whether IR2 is important in DNA binding by CbbR, the CbbR binding motif of IR2 was changed by single and double point mutations [T(−49)C, A(−37)G, and TA(−49/−37)CG; Fig. 3]. Analysis of the mutant cbb promoters with gel retardation assays showed that the mutations did not inhibit binding by CbbR (Fig. 2A). CbbR displayed an increased affinity for the DNA template carrying the T(−49)C mutation, whereas the binding of CbbR to DNA fragments carrying the A(−37)G and TA(−49/−37)CG mutations was comparable to that of the wild type (Fig. 2B). Although the mutations did not negatively affect in vitro DNA binding by CbbR, they had a dramatic effect on the activity of the cbb operon promoter (Fig. 4A). The β-galactosidase activity in cell extracts of X. flavus harboring a fusion between lacZ and the cbb promoter carrying the T(−49)A or TA(−49/−37)CG mutation was only 2% of that of the wild type following autotrophic growth. The A(−37)G mutation caused an 86% reduction in cbb promoter activity. These results indicate that, although DNA binding by CbbR is not affected in the mutant IR2 cbb promoters, IR2 is important for activation of the cbb promoter in vivo.
CbbR binding to the A site: IR3.
To assess the role of IR3 in CbbR binding and activation of the promoter of the cbb operon, three mutations (T(−39)C, A(−27)G, and TA(−39/-27)CG; Fig. 3) were introduced into the cbb promoter. Analysis of the results of gel retardation experiments showed that the affinity of CbbR for DNA fragments carrying the single point mutations compared to that for the wild-type fragments was not reduced (Fig. 2). However, the addition of NADPH to the reaction mixture did not increase the affinity of CbbR for the mutated binding sites. This is in sharp contrast to the increased DNA binding by CbbR in the presence of NADPH seen when wild-type cbb promoter fragments are used. The double mutation (TA(−39/−27)CG) had a strong negative effect on the formation of complex I, which is the result of CbbR binding to both the R and A sites of the cbb operon promoter. In addition, NADPH did not stimulate binding by CbbR to this mutant template.
The ability of the mutant promoters to drive expression of a cbb-lacZ fusion was tested following autotrophic growth of X. flavus on methanol. Surprisingly, mutations T(−39)C and TA(−39/−27)CG did not have any effect on the activity of the cbb promoter during autotrophic growth (Fig. 4A). Mutation A(−27)G even resulted in a 2.5-fold increase in cbb promoter activity. The activity of the mutant cbb promoters was also determined following heterotrophic growth on succinate. Although the wild-type cbb promoter is not active under these conditions, a low level of promoter activity is observed when the promoter is present in multiple copies on a plasmid (8, 10). The mutations in IR3 resulted in strongly increased cbb promoter activities compared to those for the wild type during heterotrophic growth on succinate (Fig. 4B). However, the activities observed were lower than those following growth on methanol; the mutant promoters are still induced (2.6- to 9.6-fold) by autotrophic growth conditions. IR3 partially overlaps the −35 region of the cbb promoter, which could result in a constitutive cbb promoter which is no longer dependent on CbbR activation. To rule out this possibility, the activity of the mutant promoters in X. flavus R22 was determined. This strain carries a cbbR disruption and is no longer able to activate transcription from the cbb promoter (25). While wild-type X. flavus carrying mutant IR3 binding sites showed relatively high levels of β-galactosidase activity, these activities were not observed in the CbbR mutant strain (Fig. 4C). This clearly shows that the mutations introduced in IR3 result in increased CbbR-dependent cbb promoter activity during both heterotrophic and autotrophic growth.
CbbR dimers bind to the same side of the DNA helix.
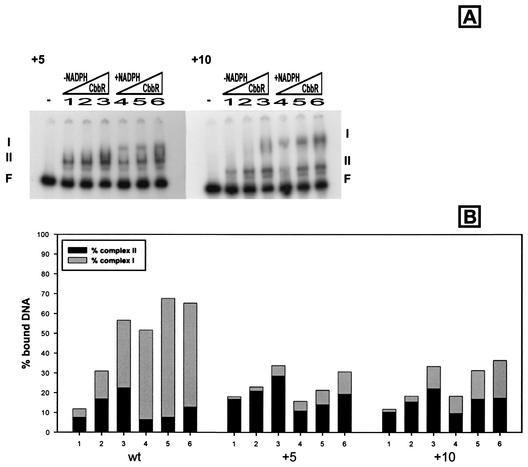
The three CbbR binding sites are located on the same side of the DNA helix, and the centers of the CbbR binding sites are separated by one, two, and three helical turns (Fig. 1). To assess the importance of helical phasing on DNA binding and cbb promoter activation by CbbR, nucleotides were inserted between IR1 and IR2. Analysis of DNA-protein interaction by gel retardation experiments showed that CbbR had a reduced affinity for DNA fragments with 5 and 10 nucleotides inserted between IR1 and IR2 (Fig. 5). However, increasing the helical phasing by 5, 6, 12, and 14 nucleotides had a stronger inhibitory effect on the formation of complex I than the insertion of 10 nucleotides (Fig. 5; data not shown). NADPH still increased the affinity of CbbR for the DNA template with an insertion of 10 nucleotides, but not with an insertion of 5 nucleotides.
FIG. 5.
(A) Gel retardation assays of mutant cbb promoters with either 5 (+5) or 10 (+10) nucleotides inserted between CbbR binding sites IR1 and IR2. F, I, and II and the symbols for the concentrations of CbbR and NADPH are as defined in the legend for Fig. 2. (B) Percentages of bound DNA in complex I and complex II for wild-type (wt) and mutated cbb promoters.
Fusions between lacZ and mutant cbb promoters with an insertion of 5, 6, 10, 12, or 14 nucleotides between IR1 and IR2 were constructed to determine the effects of helical phasing on the activity of the cbb promoter. cbb promoters with an increase in phasing of less (5 or 6 nucleotides) or more (12 or 14 nucleotides) than one helical turn of DNA were inactive during both heterotrophic and autotrophic growth of X. flavus (Fig. 4A). However, the insertion of one helical turn of DNA (10 nucleotides) between IR1 and IR2 did not abolish the activity of the cbb promoter; the β-galactosidase activity in X. flavus harboring a fusion between this mutant cbb promoter and lacZ was 65% of that of the wild type following autotrophic growth (Fig. 4B). Interestingly, similar β-galactosidase activities were observed following heterotrophic growth, which shows that introduction of one helical turn between IR1 and IR2 resulted in a constitutive cbb promoter. The +10 insertion mutant promoter was not active in X. flavus R22, which lacks a functional CbbR (Fig. 4C). This shows that the activity of this mutant promoter was completely dependent on CbbR and not due to the activity of a cryptic promoter introduced by the insertion of 10 nucleotides.
Autoregulation of cbbR.
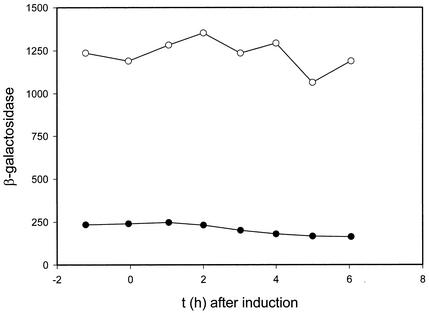
We have previously shown that CbbR binds to a region containing IR1 which is immediately adjacent to cbbR (27). It is therefore likely that the binding of CbbR to IR1 represses the transcription of cbbR, resulting in an autoregulatory circuit. To test this assumption, a fusion between cbbR and lacZ which includes the cbbR-cbbL intergenic region which contains the cbbR promoter was constructed. The Calvin cycle was induced in wild-type X. flavus strain H4-14 harboring the cbbR-lacZ fusion by the addition of formate to cells growing on gluconate-containing medium (Fig. 6). β-Galactosidase was present at a constant level before autotrophic growth induction, and its level decreased to two-thirds of the initial level (Fig. 6) following induction of the Calvin cycle, as indicated by the appearance of RuBisCO (data not shown), which is encoded by the first two genes of the cbb operon (11). A fivefold-higher level of β-galactosidase activity was seen when the experiment was repeated with X. flavus R22 lacking CbbR (Fig. 6). However, in contrast to what was found for the wild-type strain, RuBisCO activities did not appear (data not shown) and the expression level of the cbbR-lacZ fusion remained constant. These results show that CbbR negatively regulates its own expression.
FIG. 6.
Activities of β-galactosidase in extracts of X. flavus H4-14 (wild-type; •) and R22 (cbbR; ○) containing a cbbR-lacZ fusion and growing on 5 mM gluconate. The results before and following the induction of the Calvin cycle by the addition of 20 mM formate and automatic titration with formic acid (25% [vol/vol] at 0 h) are shown. Enzyme amounts are expressed as nanomoles per minute per milligram of protein.
DISCUSSION
LysR-type proteins bind to inverted repeats which contain a conserved thymidine and adenine (T-N11-A) separated by 11 nucleotides (5). The promoter of the cbb operon contains three of these motifs, which are separated by one, two, and three turns of the DNA helix (Fig. 1). We previously noted that these motifs are conserved in the cbb promoters of other chemoautotrophic bacteria, which led to the proposal of the CbbR binding motif TNA-N7-TNA (Fig. 1) (19). The data presented in this paper show that all three binding sites are functional but play different roles in DNA-protein interaction and activation of the cbb promoter.
IR1 is the promoter-distal CbbR motif. In contrast to IR2 and IR3, it is a perfect inverted repeat, to which CbbR binds with high affinity (27). In sharp contrast, DNA binding to the A site harboring IR2 and IR3 does not occur in the absence of IR1. In addition, disruption of the conserved nucleotides in IR1 abolished DNA binding by CbbR to the cbb promoter, including binding to the unaltered A site of the cbb promoter (Fig. 2). This indicates that the binding of a CbbR dimer to IR1 is essential for the binding of CbbR to the adjacent low-affinity site. Increasing the spacing between IR1 and IR2 had a strong impact on formation of DNA-protein complex I, which is the result of the interaction between two CbbR dimers and the cbb promoter. However, formation of complex II, which represents interaction between a single CbbR dimer and the high-affinity binding site IR1, is not affected. These data are consistent with cooperative binding between two CbbR dimers, in which the primary role of IR1 is to recruit a CbbR dimer to the cbb promoter, which subsequently facilitates the binding of a second CbbR dimer to the A site. Cooperative binding between LysR-type proteins at R and A sites has been observed for other LysR-type proteins (3, 6, 16, 29). A second function of IR1 is to control expression of cbbR. Since IR1 is located adjacent to the initiation codon of cbbR, it is likely that the binding of CbbR to IR1 results in repression of cbbR transcription. Disruption of the cbbR gene indeed results in increased activity of the cbbR promoter (Fig. 6), which is consistent with this model. Negative autoregulation of gene expression by LysR-type proteins is common among this class of transcriptional regulators (18).
The A site of the cbb promoter contains two partially overlapping CbbR motifs, IR2 and IR3, to which CbbR binds with low affinity (27). Single point mutations disrupting the LysR motif of IR3 have no effect on the DNA binding affinity of CbbR. However, mutations in both conserved nucleotides of IR3 (TA(−39/−27)CG) have a severe negative effect on the formation of complex I (Fig. 2). In contrast, mutations in IR2 do not affect the binding of CbbR to the low- and high-affinity sites. These data strongly suggest that CbbR binds to IR1 in the high-affinity site and to IR3 in the low-affinity site. We have previously shown that NADPH enhances the formation of complex I, which indicates that NADPH binding by CbbR increases the affinity of the protein for the promoter-proximal binding site (27). The double mutation in IR3 not only reduces the binding of CbbR to the promoter-proximal site but also abolishes the NADPH effect. This indicates that NADPH enhances the affinity of CbbR for IR3, resulting in an overall increase in DNA binding by CbbR. The data presented here therefore indicate that IR3 functions as the primary low-affinity binding site of CbbR.
Mutations in IR2 do not affect DNA binding of CbbR in vitro but have a severe negative effect on the activity of the cbb promoter in vivo (Fig. 4). The data suggest that interaction between CbbR and IR2 is required for transcriptional activation of the cbb promoter. This hypothesis is supported by the effects of an increased spacing between IR1 and IR2. When the distance between the two repeats is increased by one helical turn, the cbb promoter becomes constitutive. The constitutive nature of this mutant promoter is completely dependent on CbbR, since the mutant promoter is not active in an X. flavus strain which is devoid of CbbR. Due to the increased spacing in the mutant promoter, the distance between IR1 and IR2 in the mutant is the same as that between IR1 and IR3 in the wild type. As a result, it is likely that CbbR bound to IR1 recruits a second CbbR dimer to IR2, which, due to the additional helical turn, is now in the position of IR3, to which CbbR normally binds. These data show that IR2 is required for transcriptional activation of the cbb promoter.
Despite the fact that mutations in IR3 reduced the binding of CbbR to the low-affinity site, the activity of the cbb promoter was increased during both heterotrophic and autotrophic growth. This was not because these mutations created a constitutive promoter, since expression of the cbb-lacZ fusion was completely dependent on the presence of CbbR. It seems therefore likely that the binding of CbbR to IR3 has a repressive effect on the wild-type cbb promoter. Since IR3 partially overlaps the −35 region of the cbb promoter, it is possible that CbbR bound to IR3 prevents the binding of RNA polymerase to the cbb promoter, preventing initiation of transcription.
The data presented in this paper are consistent with the “sliding-dimer model” proposed for the LysR-type regulators OccR and OxyR (23, 28). In this model (Fig. 7), LysR-type proteins initially bind to the promoter-distal or R site and subsequently recruit a second dimer to the A site at a binding site equivalent to IR3. Following activation of the regulator by the binding of a ligand, the second dimer repositions itself from IR3 to IR2, resulting in the binding of the regulator adjacent to the binding site of RNA polymerase. The regulator is now properly positioned to make productive contacts with the alpha or other subunits of RNA polymerase, resulting in transcription initiation (21, 28). Like OccR and OxyR, CbbR introduces a bend in the DNA, which is relaxed following the binding of the ligand (27). It was proposed for OxyR and OccR that the relaxation of the DNA bending angle is due to a conformational change following the binding of the ligand, which causes the “sliding” of the dimer by one helical turn to the adjacent major groove, i.e., a repositioning from IR3 to IR2. The data presented in this paper on CbbR support this model.
FIG. 7.
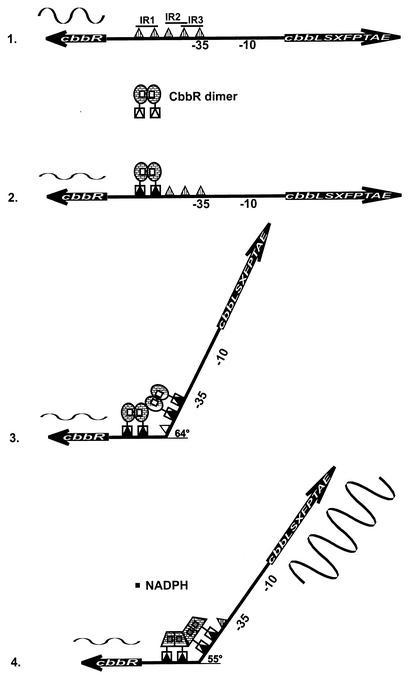
Proposed model of CbbR-mediated positive and negative regulation of the cbb operon and the cbbR gene and the response to the inducer NADPH. (1) No CbbR bound to the cbbR-cbbL intergenic region. cbbR is expressed highly, and the cbb operon is silent. (2) One CbbR dimer bound to IR1. (3) Two CbbR dimers bound to IR1 and IR3, resulting in the bending of DNA at an angle of 64°. cbbR expression is downregulated, and the cbb operon remains silent. (3) Two CbbR dimers bound to sites IR1 and IR2, relaxing the DNA bend by 9° to 55° in the presence of the CbbR inducer NADPH. cbbR expression is further downregulated, and the cbb operon is expressed highly. The sizes of wave patterns above cbbR or below the cbb operon are an indication of the expression of the respective genes.
REFERENCES
- 1.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Chang, M., and I. P. Crawford. 1990. The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 18:979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson, J. L., and F. R. Tabita. 1977. Different molecular forms of D-ribulose-1,5-bisphosphate carboxylase from Rhodopseudomonas sphaeroides. J. Biol. Chem. 252:943-949. [PubMed] [Google Scholar]
- 5.Goethals, K., M. van Montagu, and M. Holsters. 1992. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc. Natl. Acad. Sci. USA 89:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang, J., and M. A. Schell. 1991. In vivo interactions of the NahR transcriptional activator with its target sequences. J. Biol. Chem. 266:10830-10838. [PubMed] [Google Scholar]
- 7.Kusian, B., and B. Bowien. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135-155. [DOI] [PubMed] [Google Scholar]
- 8.Lehmicke, L. G., and M. E. Lidstrom. 1985. Organization of genes necessary for growth of the hydrogen-methanol autotroph Xanthobacter sp. strain H4-14 on hydrogen and carbon dioxide. J. Bacteriol. 162:1244-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lidstrom-O'Conner, M. E., G. L. Fulton, and A. E. Wopat. 1983. ‘Methylobacterium methanolicum’: a syntrophic association of two methylotrophic bacteria. J. Gen. Microbiol. 129:3139-3148. [Google Scholar]
- 10.Meijer, W. G. 1994. The Calvin cycle enzyme phosphoglycerate kinase of Xanthobacter flavus required for autotrophic CO2 fixation is not encoded by the cbb operon. J. Bacteriol. 176:6120-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijer, W. G., A. C. Arnberg, H. G. Enequist, P. Terpstra, M. E. Lidstrom, and L. Dijkhuizen. 1991. Identification and organization of carbon dioxide fixation genes in Xanthobacter flavus H4-14. Mol. Gen. Genet. 225:320-330. [DOI] [PubMed] [Google Scholar]
- 12.Meijer, W. G., L. M. Croes, B. Jenni, L. G. Lehmicke, M. E. Lidstrom, and L. Dijkhuizen. 1990. Characterization of Xanthobacter strains H4-14 and 25a and enzyme profiles after growth under autotrophic and heterotrophic growth conditions. Arch. Microbiol. 153:360-367. [DOI] [PubMed] [Google Scholar]
- 13.Meijer, W. G., P. de Boer, and G. van Keulen. 1997. Xanthobacter flavus employs a single triosephosphate isomerase for heterotrophic and autotrophic metabolism. Microbiology 143:1925-1931. [DOI] [PubMed] [Google Scholar]
- 14.Meijer, W. G., E. R. E. van den Bergh, and L. M. Smith. 1996. Induction of the gap-pgk operon encoding glyceraldehyde-3-phosphate dehydrogenase and 3-phosphoglycerate kinase of Xanthobacter flavus requires the LysR-type transcriptional activator CbbR. J. Bacteriol. 178:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Parsek, M. R., D. L. Shinabarger, R. K. Rothmel, and A. M. Chakrabarty. 1992. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 174:7798-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penninga, D., B. A. van der Veen, R. M. Knegtel, S. A. van Hijum, H. J. Rozeboom, K. H. Kalk, B. W. Dijkstra, and L. Dijkhuizen. 1996. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J. Biol. Chem. 271:32777-32784. [DOI] [PubMed] [Google Scholar]
- 18.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 19.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 20.Simon, R., M. O'Connell, M. Labes, and A. Pühler. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 21.Tao, K., C. Zou, N. Fujita, and A. Ishihama. 1995. Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase alpha subunit. J. Bacteriol. 177:6740-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terazono, K., N. R. Hayashi, and Y. Igarashi. 2001. CbbR, a LysR-type transcriptional regulator from Hydrogenophilus thermoluteolus, binds two cbb promoter regions. FEMS Microbiol. Lett. 198:151-157. [DOI] [PubMed] [Google Scholar]
- 23.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 24.van den Bergh, E. R. E., S. C. Baker, R. J. Raggers, P. Terpstra, E. C. Woudstra, L. Dijkhuizen, and W. G. Meijer. 1996. Primary structure and phylogeny of the Calvin cycle enzymes transketolase and fructosebisphosphate aldolase of Xanthobacter flavus. J. Bacteriol. 178:888-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Bergh, E. R. E., L. Dijkhuizen, and W. G. Meijer. 1993. CbbR, a LysR-type transcriptional activator, is required for expression of the autotrophic CO2 fixation enzymes of Xanthobacter flavus. J. Bacteriol. 175:6097-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Keulen, G., L. Dijkhuizen, and W. G. Meijer. 2000. Effects of the Calvin cycle on the nicotinamide adenine dinucleotide concentrations and redox balances of Xanthobacter flavus. J. Bacteriol. 182:4637-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Keulen, G., L. Girbal, E. R. E. van den Bergh, L. Dijkhuizen, and W. G. Meijer. 1998. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J. Bacteriol. 180:1411-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, L., and S. C. Winans. 1995. The sixty nucleotide OccR operator contains a subsite essential and sufficient for OccR binding and a second subsite required for ligand-responsive DNA bending. J. Mol. Biol. 253:691-702. [DOI] [PubMed] [Google Scholar]
- 29.Wek, R. C., and G. W. Hatfield. 1988. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J. Mol. Biol. 203:643-663. [DOI] [PubMed] [Google Scholar]