Abstract
The location of Helicobacter pylori in the gastric mucosa of mammals is defined by natural pH gradients within the gastric mucus, which are more alkaline proximal to the mucosal epithelial cells and more acidic toward the lumen. We have used a microscope slide-based pH gradient assay and video data collection system to document pH-tactic behavior. In response to hydrochloric acid (HCl), H. pylori changes its swimming pattern from straight-line random swimming to arcing or circular patterns that move the motile population away from the strong acid. Bacteria in more-alkaline regions did not swim toward the acid, suggesting the pH taxis is a form of negative chemotaxis. To identify the chemoreceptor(s) responsible for the transduction of pH-tactic signals, a vector-free allelic replacement strategy was used to construct mutations in each of the four annotated chemoreceptor genes (tlpA, tlpB, tlpC, and tlpD) in H. pylori strain SS1 and a motile variant of strain KE26695. All deletion mutants were motile and displayed normal chemotaxis in brucella soft agar, but only tlpB mutants were defective for pH taxis. tlpD mutants exhibited more tumbling and arcing swimming, while tlpC mutants were hypermotile and responsive to acid. While tlpA, tlpC, and tlpD mutants colonized mice to near wild-type levels, tlpB mutants were defective for colonization of highly permissive C57BL/6 interleukin-12 (IL-12) (p40−/−)-deficient mice. Complementation of the tlpB mutant (tlpB expressed from the rdxA locus) restored pH taxis and infectivity for mice. pH taxis, like motility and urease activity, is essential for colonization and persistence in the gastric mucosa, and thus TlpB function might represent a novel target in the development of therapeutics that blind tactic behavior.
Helicobacter pylori is a highly motile, urease-positive, gram-negative pathogen that establishes lifelong infections of the gastric mucosa of much of the world's population (8, 29, 35). These bacteria live deep in the mucus layer, near the underlying epithelial cells and away from the highly acidic lumen. H. pylori must also survive direct exposure to stomach acid during the initial colonization process or perhaps during repopulation following failed eradication therapy. Mutational studies have established both motility and functional urease activity as essential for colonization in animal models of infection (4, 14, 26, 38). Infections with H. pylori tend to be antral predominant, but little is known regarding how bacteria ultimately select the site for colonization. It has been suggested that gastric acidity dictates the site of colonization (1, 17, 34), and in areas of endemicity, constant ingestion of bacteria, particularly by young children, exposes all areas of the stomach to colonization, allowing infections to predominate in the more permissive regions such as the less acidic antrum.
Most H. pylori bacteria are located deep within the gastric mucus layer, being distributed within ∼30 μm of the mucosal epithelial cells, and on histology can be observed within the gastric crypts (13, 25). Persistence in the gastric mucus must depend on tactic behavior and motility to avoid being eliminated by the constant turnover of gastric mucus. Direct studies by Schreiber et al. in H. pylori-infected Mongolian gerbils demonstrated that bacteria oriented within pH gradients, and in the absence of these gradients, bacteria were found throughout the gastric mucus (25). Their studies also showed that inversion of bicarbonate/CO2 or urea/ammonia gradients did not alter orientation of bacteria within the pH gradient, suggesting that pH taxis may be dominant over other chemotactic responses. These observations seem to suggest that H. pylori can sense and respond to temporal changes in local acidity by coordinating tactic signal transduction to the flagellar motors to remain in or seek out an optimal pH zone. Remarkably, the chemoreceptors and signal transduction networks associated with pH taxis have not been identified.
In Escherichia coli, pH-tactic responses to permeant weak acids and bases are modulated by chemoreceptors Tsr and Tar, respectively, which monitor changes in cytoplasmic pH (15, 32). In contrast, the cytoplasmic pH of H. pylori does not vary more than 1 pH unit in response to strong acids over a pH range from 1 to 6 when urea is present (28). Under these conditions, the pH-gated urease activity contributes to buffering of the cytoplasm, pH stasis, and acid survival (26, 28, 38). It is conceivable that behavioral responses to strong acids differ from weak-acid responses or even involve energy-tactic mechanisms that respond to the ΔpH component of the proton motive force. Energy taxis is best exampled by behavioral responses of bacteria to molecular oxygen (aerotaxis), for which aerotaxis receptor Aer transduces chemotactic signals based on the redox status of a bound flavin (19, 24). While Campylobacter jejuni, a close relative of H. pylori, contains an aerotactic chemoreceptor system (11), these genes are not present in the Helicobacter sequenced genomes (2, 31). The H. pylori genome encodes three canonical membrane-spanning chemoreceptor proteins (TlpA, TlpB, and TlpC) and one soluble chemoreceptor protein (TlpD) (3, 5, 9, 23, 30). While several groups have demonstrated chemotactic responses to a variety of substrates including urea, bicarbonate, and amino acids (3, 5, 22, 40), the substrate specificities of these chemoreceptors have not been determined.
In the present study, we have developed an assay system to track and measure pH-tactic behavior of H. pylori, and through systematic mutational analysis of each of the chemoreceptor genes, we identified tlpB as necessary for pH-tactic responses. Using a highly permissive interleukin-12 (IL-12)-deficient mouse infection model (13), we show that only tlpB mutants are defective for colonization (restored by allelic complementation in the rdxA locus), suggesting that pH taxis, like urease activity and motility, is essential for colonization and persistence in the gastric mucosa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strains SS1 and motile KE26695 (KE88-3887) were grown under humid microaerobic conditions at 37°C on brucella-based medium supplemented with 7.5% newborn calf serum (Sigma) and antimicrobials as previously described (10, 18, 21). Escherichia coli strains were cultured in Luria-Bertani medium and supplemented as required with appropriate antibiotics.
Generation of tlp allelic-replacement mutants.
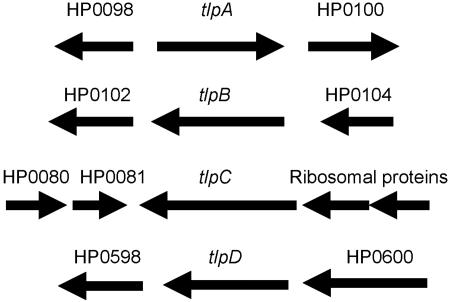
Figure 1 depicts the genetic organization of the four tlp genes of H. pylori KE26695, and primer sets used to generate mutants are presented in Table 1. The vector-free allelic-replacement mutagenesis strategy used to generate tlp-deficient mutants is outlined in Fig. 2 (6). Either the kanamycin resistance (Kmr) cassette aphA3 from pDH37 (10) or chloramphenicol resistance (Cmr) cassette cat originating from Campylobacter coli (37) was amplified with the appropriate primers listed in Table 1. H. pylori genomic DNA was isolated as previously described (13). Two pairs of gene-specific primers designated P1/P2 and P3/P4 for each tlp gene were used to PCR amplify the up- and downstream regions, respectively, of each target gene in order to produce fragments of ∼500 base pairs (bp) flanking the region to be deleted. The P2 and P3 primers contained 5′ leaders complementary to the E1 and E2 primers, respectively, for the appropriate antibiotic cassette and 20 to 21 bp of gene-specific sequence. The standard PCR in 50 μl contained 5 μl 10× PCR buffer, 1 μl of SS1 DNA (250 to 500 ng) or plasmid containing antibiotic cassette (1 to 2 ng), 200 μM (each) deoxynucleoside triphosphates, 300 nM of each primer, and 0.75 μl (2.5 U) of high-fidelity DNA polymerase (Roche Diagnostics). The PCR conditions were as follows: 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 54°C, and 1 min at 72°C, and a final extension of 7 min at 72°C. The amplicons were gel purified with a QIAquick gel extraction kit (QIAGEN), and the flanking amplicons and the drug resistance cassette amplicon were amplified in a second PCR with P1 and P4 primers for each tlp gene as originally described by Chalker et al. (6), with a modification of four precycles at 54°C. The resulting long amplicons were either directly transformed into H. pylori (36) or cloned into the pBC(ks) vector (Stratagene). The vector-free protocol was used to directly construct tlpA (HP0099), tlpB (HP0103), and tlpD (HP0599) gene replacements of SS1 and KE88-3887. The tlpC (HP0082) construct generated by the vector-free method was first cloned into plasmid pBC(ks) by blunt-end ligation at the EcoRV site, after filling in the A overhang of the amplicon. After transformation of E. coli DH5α, plasmid DNA was isolated and used to transform SS1 and KE88-3887. All tlp knockout constructs were verified by diagnostic PCR techniques to confirm loss of the tlp gene and replacement with the appropriate cassette. The diagnostic PCRs comprised sets of DNA primers designed to hybridize within the aphA3 or cat cassette paired with primers hybridizing to distal chromosomal sequences, and to confirm deletion of the respective tlp gene, primers were designed to internal sequences of the respective gene. All amplicons from validations of gene replacements were cloned into pBC or pBS vectors and sequenced in our lab (Center for Functional Microbial Genomics and Host Defense core facility) on a Beckman CEQ8000 automated DNA sequencer (Beckman Coulter).
FIG. 1.
Genetic organization of the tlp genes of H. pylori strain KE26695. For allelic replacement mutagenesis, cat or aphA3 cassette constructs were directionally designed to minimize polar effects on downstream genes. Note that the membrane-spanning tlp genes are clustered (HP0082 to HP0103), though not linked with other chemotaxis genes.
TABLE 1.
DNA primers
| Primer | Sequencea |
|---|---|
| TLPA-P1 | 5′-AACAAGCTCGCTAAAGGCTG |
| TLPA-P2K | 5′-TTGGCGTATAACATAGTATCGTCA CAATCAACGCCACGCAC |
| TLPA-P3K | 5′-TAGTACCTAGATTTAGATGTCATC GGTTACTGAGGGCAATC |
| TLPA-P4 | 5′-AAAGCTCGTTTTCTCTCGCC |
| TLPB-P1 | 5′-TGGTTACAGACGCTGATAGG |
| TLPB-P2K | 5′-TTGGCGTATAACATAGTATCGAAC ACGACCAGCATGATACG |
| TLPB-P3K | 5′-TAGTACCTAGATTTAGATGTCATG TGAGTGGAACGACCATG |
| TLPB-P4 | 5′-CAAGCGGATGATCATCTC |
| TLPC-P1 | 5′-ATCGCTAAAGTGTGGCTCAC |
| TLPC-P2K | 5′-TTGGCGTATAACATAGTATCGTAA CAACCGCACACACCATC |
| TLPC-P3K | 5′-TAGTACCTAGATTTAGATGTCGAA GATGTGAGCAGGAAG |
| TLPC-P4 | 5′-TGAACGACGATATGGGAGAC |
| TLPD-P1 | 5′-GTGTGCATCCATGAATAAGA |
| TLPD-P2C | 5′-CCCAGTTTGTCGCACTGATAAGGA AGATAGTGCTAAACATG |
| TLPD-P3C | 5′-ATCCACTTTTCAATCTATATCTGT TCAAGCTTGCAAGATTCC |
| TLPD-P4 | 5′-CTTTAATCATTACACACCGC |
| CATF-E1 | 5′-GATATAGATTGAAAAGTGGAT (cat) |
| CATR-E2 | 5′-TTATCAGTGCGACAAACTGGG (cat) |
| KAMF-E1 | 5′-CGATACTATGTTATACGCCAA (aph3-A gene) |
| KAMR-E2 | 5′-GACATCTAAATCTAGGTACTA (aph3-A gene) |
| RDXFSac | 5′-CACGAGCTCTGGTAATTGTTTCGTTAGGG (5′ rdxA) |
| RDXRXba | 5′-CACTCTAGACTTTATAAGACTCCGGATAG (5′ rdxA) |
| RDXFXho | 5′-TTGCTCGAGTGCTTGGCG (3′ rdxA) |
| RDXRKpn | 5′-ATCGGTACCAAGTAATCGCATC (3′ rdxA) |
| TLPBCompF | 5′-GCAGGATCCTCAAAGGATGGGAGGACTT |
| TLPCompR | 5′-GCAGAATTCACGGAAAGAATGGTGTCTTC |
Restriction endonuclease cleavage sites are underlined.
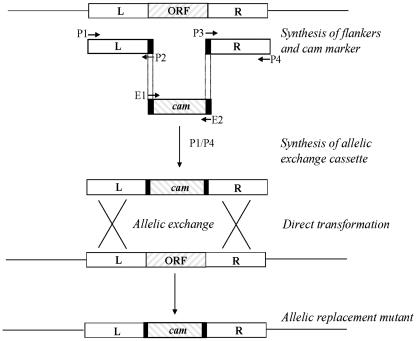
FIG. 2.
Outline of the vector-free allelic replacement mutagenesis strategy as originally described by Chalker et al. (6). DNA sequences flanking the target gene (∼500 bp) were amplified with primer sets P1/P2 (L) and P3/P4 (R), with primers P2 and P3 containing leader sequences complementary to primers E1 and E2 used to amplify the respective drug resistance cassette (cam = cat). Amplicons generated with each primer pair (L, R, and cam) were amplified in a second PCR with P1 and P4 primers to generate the long PCR product. This product was used for natural transformation of H. pylori, followed by selection on the appropriate antibiotic. ORF, open reading frame.
Construction of pRDX-C vector and complementation of the ΔtlpB mutant.
The pRDX-C vector was constructed in pBlueScript KS(+) as essentially described by Smeets et al. for cloning rdxA sequences into pBC (27). The 5′ and 3′ portions of rdxA were PCR amplified from H. pylori KE26695 chromosomal DNA with the primers listed in Table 1. Amplicons were sequentially cloned into pBlueScript following restriction digestion (SacI and XbaI for the 5′ rdxA and XhoI and KpnI for the 3′ rdxA), creating pRDX. The cat cassette was PCR amplified using the primers listed in Table 1 and, following treatment with T4 polymerase, the blunt-ended DNA was cloned into the EcoRV site of pRDX, yielding pRDX-C. The forward orientation (same orientation as rdxA) of the cat cassette was confirmed by restriction analysis and DNA sequencing.
Wild-type (WT) tlpB, with its flanking regions, was PCR amplified from H. pylori SS1 chromosomal DNA with primers TLPBCompF and TLPBCompR listed in Table 1. The ∼2.1-kb fragment was digested with BamHI and EcoRI and cloned into similarly restricted pRDX-C, yielding pRDX-C::tlpB (see Fig. 6A for genetic organization). Following transformation into E. coli DH5α (ampicillin resistance/Cmr selection), the plasmid was naturally transformed into SS1 ΔtlpB and plated on brucella agar supplemented with 20 μg/ml each of Km and Cm (no selection on metronidazole). The resulting SS1 ΔtlpB/tlpB+ strain was screened for correct chromosomal insertion of tlpB in the rdxA locus by PCR.
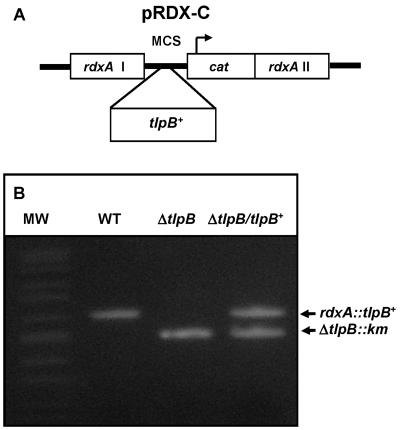
FIG. 6.
Complementation of ΔtlpB mutant. (A) The pRDX-C vector was constructed in pBlueScript as described in the text. A multiple cloning sequence (MCS) from the vector is flanked by the 5′ coding sequences of rdxA (I) and 3′ rdxA sequences (II). rdxA encodes an oxygen-insensitive NADPH nitroreductase that activates the prodrug metronidazole to toxic hydroxylamine adducts (10). (B) PCR validation of the TlpB complement. Amplicons were generated by PCR with primers designed to flanking sequences of tlpB (outside the promoter region and at the 3′ end) which were present in all constructs. WT = SS1 (2 kb); SS1 ΔtlpB::aphA3 (Km) (1.5 kb) and ΔtlpB/tlpB+ complement (doublet). MW, molecular weight.
pH taxis assay.
pH-tactic assays in 50- or 100-μl capillary tubes were modeled on prior work with enteric bacteria and H. pylori (9, 39). Capillary tubes were filled with suspensions of H. pylori strains (SS1 or KE88-3887) in phosphate-buffered saline, pH 7.0, containing 1 mM MgCl2 or in brucella broth. The ends, one of which contained HCl (pH 0.1 to 2), were plugged with 2% agarose, and the tactic behavior was visualized under low power with a Zeiss light microscope (160× to 400×). Strains of E. coli and Proteus mirabilis (defective for pH and negative chemotaxis) (39) served as controls. A wet-mount microscope slide assay was developed in which a drop of bacterial suspension (optical density at 660 nm [OD660] of ∼0.1) was covered by a no. 1 coverslip (22 by 50 mm) that was sealed around the perimeter (three sides) with high-vacuum grease (Dow Corning). The slide was visualized with the 40× objective of an inverted phase-contrast microscope (Olympus IX71) equipped with a video capture system (Evolution QEi; Media Cybernetics) and image analysis software package (ImagePro Plus; Media Cybernetics) that enabled continuous monitoring in still or video modes of data collection. The software allowed tracking of individual bacteria, permitting time-based measures of tactic behavior. Once baseline bacterial motility had been recorded, a drop of diluted HCl (pH 0.1 to 3.0 or medium control) or HCl supplemented with 5 mM urea was placed along the open side of the coverslip and, following a few seconds to enable slight mixing to cease, tactic behavior was recorded. All recordings were performed in triplicate to ensure reproducibility and all studies were performed with different batches of bacteria grown to a common OD, to ensure reproducibility of measurements. Individual bacteria were tracked (five each) to obtain a composite of swimming and arcing behavior, since arcing behavior correlated with tactic responses.
Chemotaxis and aerotaxis assays.
Motility of all strains was examined microscopically by wet-mount assay. Nonmotile KE26695 (TIGR strain) served as a visual control. General chemotactic behavior (qualitative) was assessed in semisolid medium prepared with brucella broth containing 0.3 to 0.35% agar in standard petri plates. The plates were inoculated with 1 μl of matched bacterial suspensions (OD600 of 0.1) by stabbing the plastic tip of a pipette into the soft agar. The plates were then incubated for 5 days at 37°C under microaerobic conditions. The outward migration of chemotactic bacteria was measured (diameter) every 24 h. Aerotaxis was observed in semisolid brucella broth (5 ml in 13-ml screw-cap test tubes) by inoculating the medium below the surface with 100 μl of 0.1-OD suspension and then visually tracking the migration of an aerotactic band as it migrated up to the surface of the medium. As a control for these experiments, aerotactic Campylobacter jejuni strain H840 was used as previously described (16).
Mouse infection protocols.
IL-12-deficient mice (C57BL/6 p40−/−) were bred in-house at the Carleton Animal Care Facility, Dalhousie University, Halifax, Nova Scotia, Canada (13). C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) were maintained in the University of Virginia School of Medicine animal quarters. Animals were bred under isolator conditions and placed in conventional housing for at least 4 weeks prior to experimentation. All animals were given access to water and commercial chow throughout the course of the experiments. Animals were sacrificed using CO2 asphyxiation. Animal protocols were approved by the animal ethics committee of the Dalhousie Medical School animal care facility and the University of Virginia.
To ensure that mouse-colonizing H. pylori strain SS1 was fully infectious, the strain was passaged two times through mice (3 weeks each), and then all four tlp mutations were created newly in this strain by the allelic replacement methods described earlier. Tactic phenotypes for the newly constructed mutants were verified prior to mouse infections. Mice (groups of five) were infected with 0.2 ml of tlpB, tlpC, and tlpD mutants, WT SS1, 0.2 ml saline (mock infection), and a non-mouse-colonizing strain G27 by oral gavage (three times over a 5-day period). The SS1 ΔtlpB/tlpB+ complement and wild-type SS1 strains were used to infect C57BL/6J mice as described above, except that two infections over 3 days were used. All animals were sacrificed at 3 weeks postinfection, and sections of stomach tissue were weighed, homogenized, diluted, and plated on Columbia blood agar plates supplemented with Dent's antibiotics for microbiological counts (13, 18). Infectivity is reported as the number of animals colonized of the total infected, and the microbial load in the stomach was determined from the means and standard deviations of the counts of CFU/g stomach material (triplicate plating) from the infected animals of each group.
Phylogenetic and protein structure analyses.
Motif searches utilized SMART software (http://smart.embl-heidelberg.de), homology searches used a BLAST search of microbial genomes (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), and alignments of closely related sequences were done with CLUSTALW (http://www.ebi.ac.uk/clustalw/). Protein-protein interactions were evaluated using the Hybrigenics PimRider, and gene organizations as protein structure analyses were examined on the Pedant web site (http://pedant.gsf.de/).
RESULTS
Development of pH taxis assays.
We initially set up pH taxis assays in capillary tubes, modeled on prior work with enteric bacteria (39). In these assays, E. coli strains formed a chemotactic band within a few millimeters of the HCl-agar-plugged end, and this band migrated away from the acid. In contrast, Proteus mirabilis, which is defective for negative chemotaxis (39), did not respond to the acid gradient and quickly became nonmotile as the acid diffused down the capillary tube. However, due to light distortion with round capillary tubes, we were unable to utilize the more quantitative video analysis system to track tactic responses of control bacteria or H. pylori. We next investigated the possibility of using a simple wet-mount assay on a standard microscope slide to visualize pH-tactic behavior. The advantage of this assay is that it provides a much wider field of view to fully assess population behavior. For this assay, bacterial suspensions of H. pylori in brucella broth were seeded onto the microscope slide and covered with a coverslip sealed on three sides with vacuum grease. Brucella broth cultures were used directly, as collection of bacteria by centrifugation and resuspension resulted in a population of poorly motile bacteria as previously noted (9).
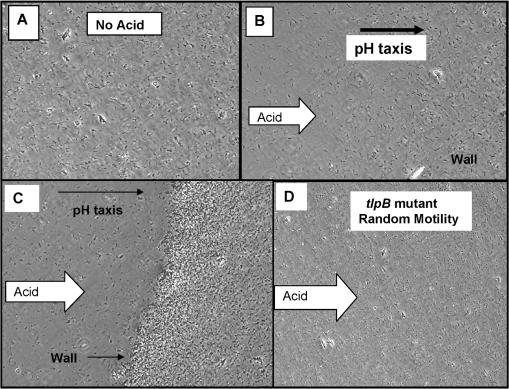
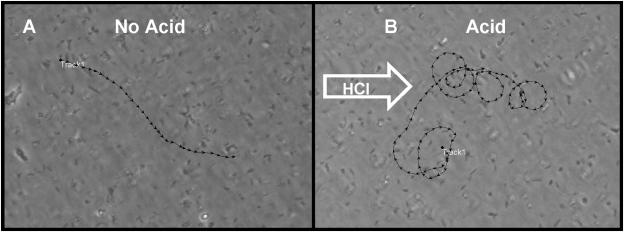
Prior to acid exposure, baseline motility was video recorded. Most of the bacteria were motile but clearly sluggish, and swimming patterns were essentially random (Fig. 3A) (see Video S1 [http://intmed.med.virginia.edu/hoffmanlab/]). In response to acid, bacterial motility increased, and the majority of bacteria could be observed swimming in large clockwise arcs that were particularly sharp in the most concentrated area of acid, such that the net population movement was away from the acid (Fig. 3B) (see Video S2 [http://intmed.med.virginia.edu/hoffmanlab/]). Arcing or swimming in wide circles has been noted by others for H. pylori (9, 23). The arcing behavior differed from the typical straight-line swimming of enteric bacteria. Flagella are bundled (one to five) at one pole, but it is not known how rotational activity promotes arcing. Tumbling behavior was not appreciably observed with H. pylori; rather, the bacteria would stop and move a short distance in a straight line, then resume arcing behavior, but there was no reversal of direction of the arc. The net result of this arcing behavior was a collective movement of the bacterial population away from the strong acid, and within minutes, there was an absence of motile bacteria within the now acidic field of view (see Video S2). As the bacteria moved away from the acid and, presumably, to a more neutral region, bacteria concentrated into a dense band (wall) of organisms, and motility became sluggish to nonexistent as depicted in Fig. 3C (see Video S3 [http://intmed.med.virginia.edu/hoffmanlab/]). In response to solutions of higher pH (>3 or medium alone), no tactic behavior was noted, eliminating fluid streaming as a cause of the tactic behavior. To test whether the wall of bacteria was nonviable, we added another drop of 0.1 N HCl, and in response to the additional acid, the bacteria within the wall became highly motile once again and engaged in another round of frantic arcing taxis away from the acid source and formed a new wall some distance from the previous one. This pH-tactic response was highly reproducible and was observed with a variety of H. pylori strains in our collection. In contrast to enteric bacteria (39), H. pylori did not swim toward the acid (moving up the pH gradient to a slightly acidic optimal pH zone), so the tactic response observed appears more consistent with negative chemotactic (avoidance) behavior. The addition of 5 mM urea to the 0.1 M HCl did not alter the pH-tactic behavior of H. pylori but did protect those bacteria directly exposed to the highest concentration of acid (ordinarily rendered nonmotile in HCl alone) (data not presented).
FIG. 3.
pH-tactic behavior of H. pylori strains and mutants. (A) No acid. Random distribution of WT SS1 bacteria in the microscopic field. (B) 0.1 N HCl. Directed movement of bacterial population from left (less concentrated) to right (more concentrated) in response to acid (∼2 min). (C) 0.1 N HCl. Formation of wall of bacteria distal to the approaching acid (note that most of the bacterial population has moved from left to right) (5 min). (D) tlpB mutant response to 0.1 N HCl. Bacterial motility is random (appears like no-acid depiction of panel A), and there is no migration of bacteria from left to right (2 min). Not depicted: loss of motility and coccoid body formation are observed at 5 min post-acid challenge.
Using a software feature, we tracked the movement of individual bacteria in the presence or absence of acid. In the absence of acid (Fig. 4A), bacteria exhibited sluggish straight-line swimming (linear velocity of 11.4 ± 1.7 μm per s) that was essentially random (Table 2). In response to acid, bacterial motility increased dramatically, and individual bacteria swam in arcs whose net activity was away from the acid source, as depicted in Fig. 4B. Table 2 provides data on motility, tactic behavior, and arcing behavior (based on tracking five organisms per sample). Note that the relative increase in linear velocity is influenced by the degree of arcing.
FIG. 4.
Computer-analyzed tracking motility patterns. (A) No acid, motile SS1 bacteria swim in linear fashion and do not display arcing patterns; (B) 0.1 N HCl, motile SS1 bacteria swim in a series of arcs that move each bacterium from left to right over a period of 30 s. Multiple trackings under each condition yielded similar results (summarized in Table 2): no arcing and random swimming patterns (no acid) and arcing tactic patterns in response to acid.
TABLE 2.
Measures of pH tactic behavior of H. pylori strains and tlp mutantsa
| Strain | Acid (+/−) | Swimming pattern (random or arc) | Linear velocity (μm/s) | Mean arc radius (μm) |
|---|---|---|---|---|
| SS1 (5) | − | Random | 11.4 ± 1.7 | None |
| SS1 (5) | + | Arc | 14.1 ± 1.4 | 3.53 ± 0.49 |
| tlpD (5) | − | Arc/tumble | 17.8 ± 1.6 | None |
| tlpD (5) | + | Arc/tumble | 9.3 ± 2.6 | 2.67 ± 0.71 |
| tlpB (5) | − | Random | 13.6 ± 1.8 | None |
| tlpB (5) | + | Random | 16.3 ± 2.9 | None |
| tlpC (5) | − | Random | 57.4 ± 4.5 | Linear |
| tlpC (5) | + | Linear/arc | No change | ND |
Helicobacter pylori strain SS1 and mutants were exposed to HCl (+/−) 0.1 N, and five individual bacteria were tracked for tactic behavior. Random, absence of tactic behavior with bacteria displaying mostly linear swimming, but arcing and tumbling are also observed in the population; arc, much of the population is engaged in this behavior. The radius of the recorded arcs for each bacterium was computed and the mean and standard deviation are presented. ND, not determined.
To determine whether H. pylori also displayed aerotactic behavior, we examined semisolid medium in a test tube for appearance of an aerotactic band as has been described with other bacteria (19). We could not demonstrate aerotaxis with motile wild-type strains of H. pylori in soft agar (data not presented). Aerotactic behavior was observed with C. jejuni strain H840 under identical conditions. Microscopic examination of wet mounts can often reveal aerotactic behavior, especially around small air bubbles (commonly observed with Campylobacter strains), and in the case of H. pylori strains, we never observed such behavior. Therefore, in the absence of clear evidence for aerotactic behavior by H. pylori, aerotaxis was not further pursued in this study.
Isolation of pH taxis mutants.
In other bacteria displaying tactic responses to changes in pH or oxygen (aerotaxis), methyl-accepting chemoreceptors (MCPs) with similarities to Tsr or Aer proteins are often involved (19, 24). Examination of the two annotated genomes of H. pylori revealed four orthologs of MCPs, tlpA, tlpB, tlpC, and tlpD (2, 31), none of which share any appreciable homology in the N-terminal regions with similar sequences of Tsr or Aer. Furthermore, none of the Tlp proteins shares any homology with aerotactic proteins (CetA and CetB) of Campylobacter jejuni that were previously shown to mediate aerotaxis (11). To analyze the roles of MCP genes in pH taxis, knockout mutations were created in each of the four tlp genes. The advantage of this high-throughput vector-free technology is that amplicons go unaltered directly into H. pylori by natural transformation (6). PCR-based analysis was performed on each of the mutants, and since the replaced cassette (aphA3 or cat) is substantially smaller than the tlp genes, mutant constructs were readily distinguishable from the respective intact genes based on the size of the amplicons generated. Based on the observation that none of the clones picked contained doublets (mutant and WT amplicons), together with additional PCR-based validations, we concluded that all tlp knockouts were authentic.
Assessment of motility and chemotaxis.
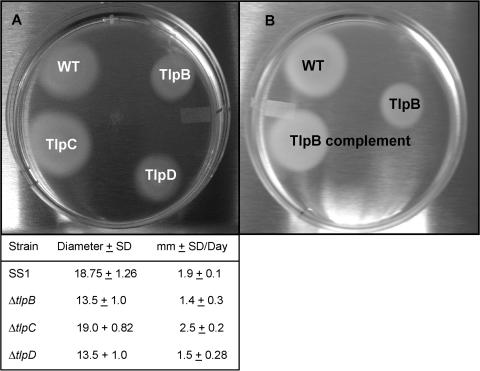
All tlp knockout mutants of SS1 and motile KE26695 were motile, as visualized by wet mounts, and were video recorded (data not presented). Since previous studies had shown that tlpA, tlpB, and tlpC mutants were chemotactic as measured in brucella-based soft agar (3), we examined all four tlp knockout mutants for similar tactic behavior by this assay. As seen in Fig. 5A, all mutants (the tlpA mutant is not depicted) exhibited chemotactic behavior, as shown by their ability to migrate outward from the central inoculum. The results presented are for the SS1 strain and mutants. Note that the tlpB and tlpD mutants seemed to be slightly less chemotactic than the WT and a tlpC mutant as measured at 5 days.
FIG. 5.
Qualitative chemotaxis assay in soft agar. (A) Wild-type SS1 and isogenic tlpC, tlpD, and tlpB mutants were inoculated into the semisolid brucella medium, and the outward migration of motile bacteria was observed over a period of 5 days. Not depicted: nonmotile KE26695 (TIGR-sequenced strain) forms a central colony but shows no spreading in soft agar. Diameters of the total distance migrated in 5 days are the means and standard deviations of four determinations; means and standard deviations for distance traveled per day are for three determinations. (B) Comparison of WT SS1, ΔtlpB mutant (TlpB), and the SS1 ΔtlpB/rdxA::tlpB+ (TlpB complemented) mutant. Note that full chemotactic capability was restored to the complemented mutant. SD, standard deviation.
Defects in pH taxis.
Each of the tlp mutants was screened for pH-tactic responses by the wet-mount slide assay and was visualized by phase contrast microscopy and video recorded. Our studies established that tlpA, tlpC, and tlpD mutants displayed pH-tactic behavior, though there were some noted differences compared with WT SS1. Video tracking of motility in the presence or absence of HCl showed that tlpC mutants were hypermotile and tended to swim in straight lines regardless of acidity. The linear velocity of this mutant was ∼57 μm per second (Table 2), and these bacteria rapidly formed a wall of less- motile bacteria. In contrast, tlpD mutants tended to swim in arcs in the absence of acid and to tumble more frequently. In response to acid, tlpD mutants were less robust in negative chemotaxis and wall formation. The linear velocity was decreased by nearly half by exposure to acid (Table 2). Motility measurements were not collected for the tlpA mutant. In contrast to the pH-tactic behavior of the other mutants, tlpB mutants, while displaying increased motility in response to acid (Table 2), showed no tactic response (Fig. 3D) (see Video S4 [http://intmed.med.virginia.edu/hoffmanlab/]). The behavior of tlpB mutants was similar to that observed with P. mirabilis, a bacterium that does not display pH taxis or negative chemotaxis (39). As the acid moved across the slide, large numbers of bacteria became nonmotile, and the typical clearing of the field observed with pH-tactic strains was not observed with the tlpB mutant. The noted random motility also included some arcing and tumbling behavior, but no taxis. If 5 mM urea is added to the HCl, bacterial motility is not inhibited by acid, presumably due to the protective action of urea hydrolysis. These results suggest that TlpB function is required for pH taxis of H. pylori.
Allelic complementation.
To determine whether TlpB function was necessary for pH taxis, the SS1 ΔtlpB mutant was complemented by cloning a wild-type tlpB gene into the rdxA locus. This was accomplished by first cloning the tlpB gene into the multiple cloning sequence region flanked by rdxA sequences as shown in Fig. 6A and was validated by PCR as demonstrated in Fig. 6B (note two amplicons in SS1 ΔtlpB/tlpB+ complement representing the mutant allele and complement allele). Expression of tlpB under its own promoter in the rdxA locus of SS1 ΔtlpB/tlpB+ restored pH-tactic behavior, as demonstrated by migration of the bacteria away from the acid and the accumulation of bacteria into a wall as observed with wild-type SS1. The bacteria regained the arcing motility in response to strong acid and displayed pH-tactic behavior as described for WT SS1 (data not presented). Moreover, as shown in Fig. 5B, the ΔtlpB complement regained full chemotaxis in the soft agar assay, suggesting that in addition to defects in pH taxis, the tlpB mutation also affects other chemotactic behaviors or possibly full motility.
The tlpB mutants do not colonize mice.
Based on the work of Schreiber et al. (25), one would predict that defects in pH taxis should manifest as deficiencies in mouse colonization. To test this possibility, we infected highly permissive IL-12 (p40−/−)-deficient mice with the tlpB, tlpC, and tlpD mutants and determined the colonization efficiency and CFU/g of stomach material for each set of mice at 3 weeks postinfection (Table 3). We had tested tlpA mutants in an earlier mouse experiment and obtained results similar to those reported by Andermann et al. (3). As seen in Table 3, bacterial densities for tlpC and tlpD mutants were comparable to those obtained with the WT SS1 strain (∼107 CFU/g). In contrast, tlpB mutants did not colonize IL-12-deficient mice (two of five animals), and bacterial counts for tlpB mutants were >5 log units lower than for the other tlp mutants, similar to the colonization efficiency determined for strain G27 (<100 CFU/g), which does not colonize mice (3, 7). To determine whether restoration of pH-tactic behavior to the ΔtlpB mutant could also restore mouse infectivity, we infected four C57BL/6J mice with the ΔtlpB/tlpB+ complement and compared infectivity to WT SS1. As seen in Table 3, complementation of the ΔtlpB mutant restored infectivity to within 1 log unit of that obtained with WT SS1 (4.6 × 104 for the complement versus 3.3 × 105 for SS1). It should be noted that the colonization density of SS1 strains and mutants for C57BL/6J mice is ∼2 log units lower than for the IL-12-deficient mouse (13). These studies indicate that pH-tactic behavior mediated by TlpB is essential for mouse colonization by the SS1 strain.
TABLE 3.
Mouse colonization densities for H. pylori strain SS1 and mutantsa
| Strain or mutant | Infected animals/total | CFU/g stomach ± SD |
|---|---|---|
| Mock infected (saline) | 0/5 | 0 |
| SS1 WT | 5/5 | 3.87 × 107 ± 1.26 × 107 |
| SS1 tlpB | 3/5 | 81 ± 61 |
| SS1 tlpC | 5/5 | 2.59 × 107 ± 1.1 × 107 |
| SS1 tlpD | 5/5 | 3.15 × 107 ± 1.4 × 107 |
| G27 | 2/5 | 81 ± 74 |
| SS1 WTb | 4/4 | 3.3 × 105 ± 2.1 × 105 |
| SS1 tlpB/tlpB+b | 4/4 | 4.6 × 104 ± 2.0 × 104 |
Five animals each were infected with SS1 and tlp mutants and 3 weeks post infection, bacterial load was determined (CFU/g stomach) as described in the text.
Four C57BL/6J mice each were infected with SS1 WT and complemented SS1 ΔtlpB mutant (SS1 ΔtlpB/tlpB+). The colonization efficiency of SS1 for IL-12-deficient mice is ca. 100-fold greater than for C57BL/6J mice as previously described (13).
Protein analysis.
The TlpB protein contains two membrane-spanning domains in the N-terminal half of the protein (amino acids 9 to 32 and 211 to 231) and methyl-accepting chemotaxis domains in the C-terminal cytoplasmic half. A HAMP domain (a domain commonly found in histidine kinases, adenylyl cyclases, MCPs, and phosphatases) is located in the cytoplasmic domain between amino acids 213 and 284. However, the HAMP domain does not include a PAS (signaling) domain which in aerotaxis protein Aer contains bound flavin (19). The HAMP domain does not share amino acid similarity with the HAMP domains of the Tsr protein of E. coli, but the locations of basic amino acids and acidic amino acids seem to conserve the charge distribution that is required for cytoplasmic pH sensing in E. coli (19, 32) (Fig. 7). BLAST searches show that the N-terminal 250 amino acids are unique to H. pylori strains J99 and KE26695. There is homology in this region with a Helicobacter mustelae tlp mutant (35% identity and 54% similarity), which also colonizes the stomachs of ferrets. There was no similarity with proteins from Helicobacter hepaticus or with other species of epsilon proteobacteria (Campylobacter spp. and Wolinella succinogenes). Moreover, BLAST searches of the entire protein database identified no significant homology to any other protein that might provide clues as to how TlpB senses changes in pH. The 250-amino-acid region predicted to be exposed to the periplasm contained no obvious motifs detected by SMART or similar protein analysis software. Protein interaction information on HP0103 on the Hybrigenics web site did not identify any proteins interacting with the N-terminal region of TlpB.
FIG. 7.
Charge distribution within the HAMP domains of TlpB and Tsr. HAMP domains are cytoplasmic linker regions found in bacterial chemoreceptors. The HAMP sequence of Tsr senses changes in cytoplasmic pH (32). By convention, basic amino acids are given a “+” charge and acidic amino acids are given a “−” charge. The HAMP domains were identified as indicated in the text for TlpB from H. pylori strains KE26695 and J99 and a Tlp ortholog (Hmus) of H. mustelae. Tsr is the serine-sensing chemoreceptor of the E. coli K12 strain.
DISCUSSION
We developed a simple wet-mount-based pH taxis assay in which H. pylori displays pH-tactic behavior that can best be described as directed arcing motility away from the acid source. Using this assay system, we screened the four tlp chemoreceptor mutants for any defects in pH taxis and identified chemoreceptor tlpB as required for pH taxis. Moreover, only tlpB mutants were defective for mouse colonization. Allelic complementation of the ΔtlpB mutant by expression of the WT tlpB gene, cloned in single copy into the rdxA locus, restored both pH-tactic behavior and infectivity for mice, indicating that pH taxis, like motility and urease activity, is an essential factor for colonization of the gastric mucosa. The addition of urea to HCl did not abolish pH taxis by H. pylori, consistent with the well-established role of the pH-gated urease in protecting bacteria while they swim out of the extreme acid. Our findings are also consistent with those of Schreiber et al., who provided convincing in vivo evidence that the orientation of H. pylori in the gastric mucus of gerbils requires a pH gradient (25). Our studies support the notion that pH taxis is dominant over other tactic behaviors and enables colonizing bacteria entering the gastric mucus to travel down natural gradients of hydrogen ions to areas more hospitable for bacterial growth. Moreover, temporal pH sensing and millisecond tactic responses help confine the bacteria within a narrowly pH-defined zone which prevents them from swimming up into the more acidic regions of gastric mucus, where bacteria would be eliminated in the normal turnover of mucus.
In comparison with other epsilon proteobacteria, H. pylori possesses a minimum set of chemoreceptors and lacks orthologs of cheB and cheR which are necessary for the methylation/demethylation adaptive response (2, 31). The TlpB of H. pylori shares no similarity with Tlp proteins from the epsilon proteobacteria in either the N-terminal region or in the putative cytoplasmic pH-sensing HAMP domain. The response of H. pylori to acid differs from that of Serratia marcescens and Pseudomonas aeruginosa, which use both positive and negative tactic responses to remain within an optimal pH zone in capillary tube assays (putatively optimizing the ΔpH component of the proton motive force) (39). It is significant to note that H. pylori bacteria did not swim toward the acid or away from the strong base, as observed with enteric bacteria, suggesting a one-dimensional “negative chemotactic” response. This phenotype also appears to resemble a tar (defective for negative taxis from alkali) mutant of E. coli for which Tsr transduces a one-dimensional pH-tactic response signaled by protonation within the cytoplasmic HAMP domain (32). While the HAMP domain of TlpB exhibits no amino acid similarity with the HAMP domain of Tsr, there is sufficient conservation in charge distribution (basic and acidic amino acids as indicated in Fig. 7) to suggest a cytoplasmic pH-sensing mechanism. This charge distribution is also noted in the TlpB ortholog of the H. mustelae TlpB but not in Tlp proteins of H. hepaticus. Mechanistic studies are in progress to determine how TlpB senses changes in hydrogen ion concentration.
Once highly motile H. pylori bacteria have escaped pH stress, motility decreases or ceases, and a wall of accumulating bacteria is observed in the in vitro assay. Interestingly, the bacterial density of the wall increases as highly motile tactic bacteria pile into the sedentary group comprising the wall. Wall formation is most likely an in vitro phenomenon, since in vivo, diffusion of hydrogen ions would be restricted by gastric mucus, and bacterial density would be lower. This version of pH displacement and recolonization might provide a survival advantage as erosion of epithelial cells due to inflammation and loss of gastric mucus (ulcer formation) would increase local acidity, causing the bacteria to move, but not too far from the nutrient source provided by inflammation. The ability of H. pylori to stop swimming in response to a more neutral pH environment might be relevant to the function of the Cag type IV secretion system, since it has been hard to conceptually reconcile how highly motile bacteria would be able to deploy the type IV pili into epithelial cells without first becoming sedentary. Our findings suggest that the sedentary state might optimize attachment to epithelial cells and activation of the Cag system to promote inflammation (8, 12). Thus, the chemotaxis system of H. pylori may be another example of the remarkable ability of Helicobacter spp. to adapt common physiological systems to optimize colonization and life-long persistence in the apparently hostile gastric mucosa of mammals.
Our findings that TlpB is required for pH taxis in H. pylori and is essential for mouse colonization are in stark contrast to the findings of McGee et al., who concluded that tlpB mutants were not defective for colonization of gerbils and who reported no phenotype associated with chemotactic behavior (20). In their study, animals infected with tlpB mutants exhibited measurable differences in inflammation and recruitment of lymphocytes (20). We obtained the tlpB mutant from David McGee, and it appeared to be pH tactic in our assay, though this strain and the supplied parental SS1 exhibited poor motility (data not presented). We also confirmed that the locus was genetically altered, and DNA sequence analysis indicated that the tlpB gene was indeed deleted as originally described. As pointed out in previous studies, gene duplications (mutant and functional allele) are not uncommon in this species (6) or are second-site mutations in genes that might produce an avirulent phenotype. In our studies, vector-free allelic replacement mutagenesis was used to create all mutants used, and in the case of tlpB, mutants produced in three independent experiments yielded a consistent phenotype. Therefore, allelic complementation of the ΔtlpB mutant by the single-copy insertion of a functional tlpB gene into the rdxA locus provided a critical test of the hypothesis and formally established the requirement of TlpB function for pH-tactic behavior and mouse infectivity. Studies are in progress to resolve pH-tactic differences between these strains.
The chemotaxis system of H. pylori is complex and includes a fused CheAY histidine-kinase/response regulator and three CheV paralogs, each of which contains an N-terminal CheW domain (23) and a response regulator domain of the CheY family (9). It has been suggested that the CheW portion of CheV proteins binds to the Tlp chemoreceptors and possibly modulates phosphorylation of CheA. However, none of the cheV genes can rescue a cheW mutant of H. pylori (23). Mutational analysis of the cheV genes indicated that cheV1 was essential for chemotactic behavior, while the other two mutants displayed WT chemotactic abilities as judged by soft agar assays (23). These findings do not exclude cheV2 and cheV3 in other specialized chemotactic responses. Several groups have reported chemotactic responses of H. pylori strains to bicarbonate, ammonia/urea, bile, and various amino acids (5, 9, 22, 40). Yet, other studies have not been able to reproduce some of these findings (3), underscoring the need for more mechanistic studies of this system. There is also controversy regarding the necessity for a functional chemotaxis system for infections in animal models. For example, initial studies showed that cheW, cheA, and cheY mutants failed to colonize mice (9, 23), whereas more-recent studies showed that these nonchemotactic mutants were able to colonize mice (3, 30). Clearly, much more study is required to identify both the substrates recognized by each chemoreceptor and the specific phosphorelay proteins coordinating each tactic response. Such studies will require development of more-robust nonnutrient assay systems in which single compounds can be tested. We found that H. pylori flagella are easily sheared (e.g., by centrifugation and resuspension of bacteria in other chemotaxis buffer systems), limiting pH-tactic studies to dilutions of bacteria in growth medium. Foynes et al. similarly diluted H. pylori bacteria in a phosphate-citrate buffer in their chemotaxis assays (9). Our studies also demonstrated variations in motility and pH-tactic behavior for tlpD and tlpC mutants, the former demonstrating more arcing and tumbling behavior and the latter displaying hypermotility that accelerated wall formation. Yet, these mutants behaved essentially as the wild type in terms of mouse colonization. Further study will be required to assess the interactions of these chemoreceptors in mediating or amplifying tactic behavior.
We do not know how pH taxis integrates into other acid-adaptive behaviors that include the effects of pH-gated urease (4, 26, 38) and the various transcription-based two-component regulators that control global gene expression, including adaptation to acid stress (7, 33). Studies by Delany et al. indicate that tlpB may be regulated by an orphan response regulator HP1043 (7), while one of the cheV genes appears to be regulated by the nickel response regulator NikR (33). Further study is needed to assess the role (if any) of tlpB in acid stress and, more importantly, to evaluate the extent to which pH sensing and tactic behavior minimize the effects of acid stress and the magnitude of acid-adaptive responses in general.
In summary, we have demonstrated that chemoreceptor TlpB, whose N-terminal periplasmic and HAMP domains are conserved among gastric-colonizing strains of Helicobacter, is essential for pH taxis and for mouse infectivity. The fact that chemoreceptors tlpA, tlpC, and tlpD are dispensable for mouse colonization underscores the importance of acid in defining both the site of colonization by H. pylori and the ability of these bacteria to persist in the gastric mucosa.
Acknowledgments
We thank Rafael Garduno for assistance with the imaging system, Sander van Zanten for helpful discussions, and Donna Hutchison, Peter Ernst, and Elizabeth Wiznerowicz for the animal studies.
This work was supported by a grant from the Canadian Institutes for Health Research and startup funds from the University of Virginia Health Systems.
REFERENCES
- 1.Akada, J. K., K. Ogura, D. Dailidiene, G. Dailide, J. M. Cheverud, and D. E. Berg. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 149:1901-1909. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Andermann, T. M., Y. T. Chen, and K. M. Ottemann. 2002. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun. 70:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bury-Mone, S., S. Skouloubris, A. Labigne, and H. De Reuse. 2001. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Mol. Microbiol. 42:1021-1034. [DOI] [PubMed] [Google Scholar]
- 5.Cerda, O., A. Rivas, and H. Toledo. 2003. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224:175-181. [DOI] [PubMed] [Google Scholar]
- 6.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2002. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J. Bacteriol. 184:4800-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodwin, A., D. Kersulyte, G. Sisson, S. J. O. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 11.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, P. S. 2002. The bacteriology of Helicobacter pylori, p 1-17. In Y. Yamamoto, H. Friedman, and P. Hoffman (ed.), Immunology and pathogenesis of Helicobacter pylori. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 13.Hoffman, P. S., N. Vats, D. Hutchison, J. Butler, K. Chisholm, G. Sisson, A. Raudonikiene, J. S. Marshall, and S. J. O. Veldhuyzen van Zanten. 2003. Development of an interleukin-12-deficient mouse model that is permissive for colonization by a motile KE26695 strain of Helicobacter pylori. Infect. Immun. 71:2534-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Josenhans, C., K. A. Eaton, T. Thevenot, and S. Suerbaum. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 68:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kihara, M., and R. M. MacNab. 1981. Cytoplasmic pH mediates pH taxis and weak acid repellent taxis of bacteria. J. Bacteriol. 145:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krieg, N. R., and P. S. Hoffman. 1986. Microaerophily and oxygen toxicity. Annu. Rev. Microbiol. 40:107-130. [DOI] [PubMed] [Google Scholar]
- 17.Lee, A., M. F. Dixon, S. J. Danon, E. Kuipers, F. Megraud, H. Larsson, and B. Mellgard. 1995. Local acid production and Helicobacter pylori: a unifying hypothesis of gastroduodenal disease. Eur. J. Gastroenterol. Hepatol. 7:461-465. [PubMed] [Google Scholar]
- 18.Lee, A., J. O'Rourke, M. C. de Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 19.Ma, Q., M. S. Johnson, and B. L. Taylor. 2005. Genetic analysis of the HAMP domain of Aer aerotaxis sensor localizes flavin adenine dinucleotide-binding determinants to the AS-2 helix. J. Bacteriol. 187:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGee, D. J., M. L. Langford, E. L. Watson, J. E. Carter, Y. T. Chen, and K. M. Ottemann. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukhopadhyay, A. K., J.-Y. Jeong, D. Dailidiene, P. S. Hoffman, and D. E. Berg. 2003. The fdxA ferredoxin gene can down-regulate frxA nitroreductase gene expression and is essential in many strains of Helicobacter pylori. J. Bacteriol. 185:2927-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura, H., H. Yoshiyama, H. Takeuchi, T. Mizote, K. Okita, and T. Nakazawa. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect. Immun. 66:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pittman, M. S., M. Goodwin, and D. J. Kelly. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493-2504. [DOI] [PubMed] [Google Scholar]
- 24.Rebbapragada, A., M. S. Johnson, G. P. Harding, A. J. Zuccarelli, H. M. Fletcher, I. B. Zhulin, and B. L. Taylor. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber, S., M. Konradt, C. Groll, P. Scheid, G. Hanauer, H. O. Werling, C. Josenhans, and S. Suerbaum. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. USA 101:5024-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 115:628-641. [DOI] [PubMed] [Google Scholar]
- 27.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stingl, K., E. M. Uhlemann, R. Schmid, K. Altendorf, and E. P. Bakker. 2002. Energetics of Helicobacter pylori and its implications for the mechanism of urease-dependent acid tolerance at pH 1. J. Bacteriol. 184:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor, D. N., and J. Parsonnet. 1995. Epidemiology and natural history of H. pylori infections, p. 551-564. In M. J. Blaser, P. F. Smith, J. Ravdin, H. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 30.Terry, K., S. M. Williams, L. Connolly, and K. M. Ottemann. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 32.Umemura, T., Y. Matsumoto, K. Ohnishi, M. Homma, and I. Kawagishi. 2002. Sensing of cytoplasmic pH by bacterial chemoreceptors involves the linker region that connects the membrane-spanning and the signal-modulating helices. J. Biol. Chem. 277:1593-1598. [DOI] [PubMed] [Google Scholar]
- 33.van Vliet, A. H. M., F. D. Ernst, and J. G. Kusters. 2004. NikR-mediated regulation of Helicobacter pylori acid adaptation. Trends Microbiol. 12:489-494. [DOI] [PubMed] [Google Scholar]
- 34.Veldhuyzen van Zanten, S. J. O., T. Kolesnikow, V. Leung, J. L. O'Rourke, and A. Lee. 2003. Gastric transitional zones, areas where Helicobacter treatment fails: results of a treatment trial using the Sydney strain mouse model. Antimicrob. Agents Chemother. 47:2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veldhuyzen van Zanten, S. J. O., P. M. Sherman, and R. H. Hunt. 1997. Helicobacter pylori: new developments and treatments. Can. Med. Assoc. J. 156:1565-1574. [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, Y., P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 38.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 39.Williams, F. D., D. M. Anderson, P. S. Hoffman, R. H. Schwarzhoff, and S. Leonard. 1976. Evidence against the involvement of chemotaxis in the swarming of Proteus mirabilis. J. Bacteriol. 127:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worku, M. L., Q. N. Karim, J. Spencer, and R. L. Sidebotham. 2004. Chemotactic response of Helicobacter pylori to human plasma and bile. J. Med. Microbiol. 53:807-811. [DOI] [PubMed] [Google Scholar]