Abstract
Vibrio cholerae is the causative agent of the severe diarrheal disease cholera. A number of environmental stimuli regulate virulence gene expression in V. cholerae, including quorum-sensing signals. At high cell densities, quorum sensing in V. cholerae invokes a series of signal transduction pathways in order to activate the expression of the master regulator HapR, which then represses the virulence regulon and biofilm-related genes and activates protease production. In this study, we identified a transcriptional regulator, VqmA (VCA1078), that activates hapR expression at low cell densities. Under in vitro inducing conditions, constitutive expression of VqmA represses the virulence regulon in a HapR-dependent manner. VqmA increases hapR transcription as measured by the activity of the hapR-lacZ reporter, and it increases HapR production as measured by Western blotting. Using a heterogenous luxCDABE cosmid, we found that VqmA stimulates quorum-sensing regulation at lower cell densities and that this stimulation bypasses the known LuxO-small-RNA regulatory circuits. Furthermore, we showed that VqmA regulates hapR transcription directly by binding to its promoter region and that expression of vqmA is cell density dependent and autoregulated. The physiological role of VqmA is also discussed.
The gram-negative bacterium Vibrio cholerae is the causative agent of cholera, an acute dehydrating diarrhea still endemic in many parts of the developing world (6). The ability of V. cholerae strains to cause severe enteric infection in humans is dependent on the expression of cholera toxin (CT) and a pilus colonization factor known as the toxin-coregulated pilus (TCP). The genes involved in CT and TCP production are controlled by a series of signal transduction cascades. Two membrane-localized complexes thought to respond to environmental signals, ToxRS and TcpPH, activate the transcription of toxT. ToxT, in turn, activates the transcription of the virulence regulon (ToxR regulon), including the cholera toxin genes ctxA and ctxB and the tcp genes responsible for TCP biosynthesis. In addition, Skorupski and coworkers have identified AphA as an activator of tcpPH expression (18, 29). The environmental cues influencing the expression of virulence genes in vivo are poorly characterized, but extensive studies have indicated the importance of a variety of stimuli, including temperature, pH, bile salts, and quorum-sensing signals (15, 19).
Single-celled bacteria are able to produce and respond to small diffusible molecules called autoinducers. These molecules accumulate as cell density increases and regulate the expression of a range of genes to control a variety of physiological functions, in a process known as quorum sensing (10, 33). Many gram-negative bacteria use a set of diffusible N-acyl homoserine lactones (AHLs) that generally serve as cell-to-cell communication signals. The key regulatory components of these signaling systems are LuxI-type proteins, which act as AHL synthases, and LuxR-type proteins, which serve as AHL receptors and AHL-dependent transcription factors (9). The quorum-sensing system in V. cholerae, which does not possess AHL-dependent signal transduction pathways, has been shown to respond to at least two autoinducer molecules: CAI-1, whose structure has yet to be solved, and AI-2, a furanosyl borate diester also produced by Vibrio harveyi and many other bacteria.
In V. cholerae, quorum sensing has been shown to negatively regulate virulence gene expression (26, 38). Accumulation of these autoinducers modulates the activity of a central regulator, LuxO, via membrane receptors CqsS and LuxPQ (26). At low cell densities, LuxO actively represses the expression of another key quorum-sensing regulator, HapR, by activating the expression of a set of small RNAs (21). At high cell densities, LuxO is inactivated, and thus hapR expression is activated. In V. cholerae, quorum sensing has been shown to negatively regulate virulence gene expression (26, 38). HapR decreases tcpPH transcription indirectly by repressing transcription of aphA (18). HapR also represses the vps (Vibrio polysaccharide synthesis) operon, thus negatively regulating biofilm formation (13, 35, 37). In addition, HapR directly up-regulates the expression of hapA, which produces secreted hemagglutinin (HA)/protease, responsible for detachment of the vibrios from the intestinal epithelium (7, 17, 23).
Recent studies have suggested that quorum-sensing regulation in V. cholerae is far more complicated than previously thought. The VarS/VarA two-component sensory system comprises an additional quorum-sensing-dependent regulatory input, which uses an additional set of small RNAs together with the global regulatory protein CsrA to modulate gene expression in V. cholerae (20). Furthermore, hapR expression is repressed at high cell densities by HapR itself, although the significance of this autorepression is unknown (22). In this study, we identified an additional regulator, VCA1078, that modulates the V. cholerae quorum-sensing regulon by increasing the expression of hapR. Genetic and biochemical studies suggest that this regulator binds directly to the hapR promoter. We thus name VCA1078 VqmA (Vibrio quorum modulator A).
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Escherichia coli and V. cholerae strains used in this study are listed in Table 1 and were propagated in LB containing appropriate antibiotics at 37°C, unless otherwise noted. The plasmid harboring Ptac-controlled vqmA was constructed by PCR amplifying the vqmA (VCA1078) coding sequence and cloning it into the pMal-c2x vector (New England Biolabs), resulting in pZL3 (no malE) or pJZ179 (in frame with malE). A vqmA in-frame deletion was constructed by overlapping PCR of the vqmA flanking region and cloning into pWM91 (24). The resulting plasmid was then introduced into V. cholerae strains and selected for double homologous recombination events. Plasmids containing vqmA lacking its helix-turn-helix DNA binding domain sequences (ΔHTH) were constructed by digesting pJZ179 with NruI and XbaI, filling in with Klenow fragment, and religating. The hapR-lacZ reporter plasmid was constructed by cloning the PCR-amplified hapR promoter region into the transcriptional LacZ reporter vector pKP302 (28). The vqmA-lacZ transcriptional reporter fusions were constructed by cloning of either an internal fragment of vqmA or intact 5′ vqmA sequences into a suicide plasmid containing promoterless lacZ (pJZ244) on pGP704 (27). The resulting plasmids were then introduced into V. cholerae strains lacking lacZ and screened for homologous recombination events.
TABLE 1.
Strains and plasmids used in this study
| Strains or plasmids | Relevant characteristic(s) | Source or reference |
|---|---|---|
| V. cholerae strains | ||
| C6706 | El Tor, wild type | 31 |
| C6706lacZ | lacZ deletion | 38 |
| C6706mpc | luxO constitutive allele (L104Q) | 32 |
| MM194 | hapR deletion | 38 |
| ZLV101 | vqmA deletion in C6706lacZ | This study |
| ZLV102 | pMal-c2x in ZLV101 | This study |
| ZLV103 | pMal-c2x in C6706lacZ | This study |
| ZLV104 | pZL3 in ZLV101 | This study |
| ZLV105 | pZL3 in C6706lacZ | This study |
| ZLV106 | hapR-lacZ in ZLV102 | This study |
| ZLV107 | hapR-lacZ in ZLV103 | This study |
| ZLV108 | hapR-lacZ in ZLV104 | This study |
| AHV301 | vqmA-lacZ in C6706lacZ, vqmA disrupted | This study |
| AHV302 | vqmA-lacZ in C6706lacZ, with intact vqmA | This study |
| Plasmids | ||
| pMal-c2x | Cloning vector to make MBP fusions | New England Biolabs |
| pKP302 | lacZ transcriptional fusion vector | 28 |
| pGP704 | R6K origin vector, mob RP4 | 27 |
| pWM91 | R6K origin vector with sacB | 24 |
| pJZ142 | hapR-lacZ on pVIK112 for integrating into the V. cholerae genome | 38 |
| pBB1 | luxCDABE of V. harveyi on a cosmid | 1 |
| pJZ244 | Promoterless lacZ of V. cholerae in pGP704 | This study |
| pJZ177 | vqmA flanking region cloned into pWM91 | This study |
| pJZ179 | vqmA coding sequence in pMal-c2x, in frame with malE | This study |
| pZL3 | Ptac-vqmA, vqmA coding sequence in pMal-c2x | This study |
| pZL4 | Same as pJZ179 with vqmA DNA binding sequence deleted | This study |
| pZL5 | hapR promoter region in pKP302 | This study |
| pAH301 | Internal fragment of vqmA cloned into pJZ244 | This study |
| pAH302 | Intact 5′-end fragment of vqmA cloned into pJZ244 | This study |
Microarray analysis.
The primers for PCR amplification of all V. cholerae full-length open reading frames (ORFs) were synthesized as previously published (5). All PCR products were spotted onto GAPSII slides (Corning) using OminiGrid 100 (Gene Machines, Inc) (performed by Penn Microarray Core Facilities). Strains ZLV102 and ZLV105 were grown to mid-log phase in AKI medium, which is optimized for virulence gene expression (16), containing appropriate antibiotics and 0.1 μM isopropyl-β-d-thiogalactopyranoside (IPTG). The RNA was isolated using TRIzol reagent (Invitrogen) and cleaned with an RNeasy kit (QIAGEN). Fluorescently labeled cDNA was prepared by direct incorporation of fluorescent nucleotide analogues (Cy3-dCTP and Cy5-dCTP) during a first-strand randomly primed reverse transcription reaction. The differentially labeled cDNAs were combined and subsequently applied to the array surface under conditions that favor hybridization. Microarray slides were scanned using a GenePix 4000B (Axon, Inc). For every ORF-specific spot, the resulting fluorescence intensity of each of the labels was measured, and intensities were compared using the GenePix Pro 6.0 software system (Axon, Inc).
Luminescence assay.
Cosmid pBB1, carrying the V. harveyi lux operon (1), was introduced into V. cholerae strains by electroporation. The resulting strains were grown in LB with appropriate antibiotics at 30°C overnight, diluted to a concentration of 1:1,000 in fresh LB, and incubated at 30°C. Luminescence was read at 30-min intervals for 7 h using a Bio-Tek Synergy HT spectrophotometer. Cultures were diluted, and cell density was monitored by counting CFU. Relative light units are defined as 103 light units/CFU ml−1.
Western blotting (immunoblotting) of HapR.
Strains were grown in LB overnight and inoculated at 1:100 into LB broth. Samples were withdrawn at the time points indicated. Cell pellets were lysed, and total protein was measured by a Bio-Rad protein assay kit. Portions (0.1 mg) of total protein were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and immunoblotted with affinity-purified polyclonal anti-HapR rabbit antiserum.
Detection of cholera toxin.
GM1 ganglioside enzyme-linked immunosorbent CT assays (12) were carried out following overnight incubations of V. cholerae strains with shaking at 37°C in AKI medium with appropriate antibiotics and IPTG (0.1 μM).
β-Galactosidase activity assays.
E. coli or V. cholerae strains containing LacZ reporter fusions were grown at 37°C in LB with IPTG (0.1 μM). Samples were withdrawn at the time points indicated and then assayed for β-galactosidase activity as previously described (25).
Gel retardation assays.
Maltose-binding protein (MBP)-VqmA fusions were purified through amylose columns according to the manufacturer's instructions (New England Biolabs). PCR products containing different lengths of the hapR promoter region were digested with XbaI and end labeled using [α-32P]dCTP and the Klenow fragment of DNA polymerase I. Binding reaction mixtures contained 0.1 ng DNA and 0.5 μg wild-type or ΔHTH VqmA proteins in a buffer consisting of 10 mM Tris-HCl (pH 7.9), 1 mM EDTA, 1 mM dithiothreitol, 60 mM KCl, 30 mg/ml calf thymus DNA, 20 mg/ml bovine serum albumin, and 10% glycerol. After 20 min of incubation at room temperature, samples were size fractionated using 5% polyacrylamide gels in 0.5× TAE buffer (20 mM Tris-acetate, 1 mM EDTA [pH 8.5]). The radioactivity of free DNA and VqmA-DNA complexes was visualized by using a StormB840 PhosphorImager (Molecular Dynamics).
Infant mouse colonization assay.
The infant mouse colonization assay has been described previously (11). Briefly, V. cholerae mutant strains were mixed with the wild-type strain, and approximately 105 cells were inoculated into 6-day-old CD-1 suckling mice. After a 20-h period of colonization, intestinal homogenates were collected, and the ratio of mutant to wild-type bacteria was determined.
RESULTS
Overexpression of the LuxR family protein VqmA represses virulence gene expression in vitro.
In bacteria, gene expression is often highly regulated by various transcription factors. This is supported by the large numbers of conserved transcriptional regulators revealed by microbial genome sequencing. For example, in the V. cholerae genome, there are eight open reading frames annotated as LuxR family proteins (14). LuxR family proteins are thought to share an AHL interaction region and DNA binding motif (9). However, while all annotated LuxR family proteins in V. cholerae share a conserved helix-turn-helix DNA binding motif (3), they show very weak homology to the conserved AHL binding domains (data not shown). Furthermore, no AHL synthase genes are present in the V. cholerae genome, and no AHL signals can be detected under laboratory culture conditions using a sensitive AHL bioassay strain (36) (data not shown). Thus, these LuxR-type proteins in V. cholerae may not be involved in the process of interaction with self-produced AHL signals. To investigate the genes possibly regulated by these LuxR family proteins, we performed microarray analysis to compare the transcriptional profiles of strains with chromosomal in-frame deletions of these genes to those of strains with plasmids overexpressing the corresponding genes. Artificially activating these regulatory genes enabled us to detect target genes that otherwise might not be affected under the experimental conditions we normally use. Interestingly, overexpression of one of the annotated LuxR family genes, VCA1078, which is predicted to encode a 319-amino-acid protein sharing a conserved carboxyl-terminal DNA binding motif with only the LuxR protein of Vibrio fischeri, leads to the repression of the entire ToxR virulence regulon (Table 2). For example, genes in the TCP island are repressed 2.3- to 5.2-fold in the VCA1078-overexpressing strain. Similarly, the ctxA and ctxB genes, which encode the two subunits of cholera toxin, are also inhibited (threefold) in such a strain. The inhibition of virulence gene expression is not due to a general inhibitory effect of protein overproduction, since no virulence gene repression was observed when other LuxR family proteins were overexpressed (data not shown) or with expression of the control protein MalE. We renamed VCA1078 vqmA (Vibrio quorum-sensing modulator A), because its gene product regulates the quorum-sensing system of V. cholerae (see below).
TABLE 2.
Differential gene expression in vqmA mutant and vqmA(Con) strains in AKI medium
| ORF | ID | vqmA(Con)/vqmA mutant expression ratioa | SDb |
|---|---|---|---|
| Pathogenesis | |||
| TagD protein | VC0824 | −2.7 | 0.0 |
| Toxin-coregulated pilus biosynthesis protein P | VC0826 | −3.2 | 0.0 |
| Toxin-coregulated pilus biosynthesis protein H | VC0827 | −2.6 | 0.0 |
| Toxin-coregulated pilin TcpA | VC0828 | −3.3 | 0.1 |
| Toxin-coregulated pilus biosynthesis protein B | VC0829 | −2.6 | 0.0 |
| Toxin-coregulated pilus biosynthesis protein Q | VC0830 | −3.2 | 0.1 |
| Toxin-coregulated pilus biosynthesis protein C | VC0831 | −2.3 | 0.1 |
| Toxin-coregulated pilus biosynthesis protein R | VC0832 | −2.3 | 0.0 |
| Toxin-coregulated pilus biosynthesis protein D | VC0833 | −3.6 | 0.1 |
| Toxin-coregulated pilus biosynthesis protein S | VC0834 | −2.7 | 0.0 |
| Toxin-coregulated pilus biosynthesis protein T | VC0835 | −4.0 | 0.1 |
| Toxin-coregulated pilus biosynthesis protein E | VC0836 | −2.2 | 0.0 |
| Toxin-coregulated pilus biosynthesis protein F | VC0837 | −2.7 | 0.2 |
| TCP virulence regulatory protein ToxT | VC0838 | −2.9 | 0.1 |
| Leader peptidase TcpJ | VC0839 | −3.6 | 0.0 |
| Accessory colonization factor AcfB | VC0840 | −2.3 | 0.0 |
| Accessory colonization factor AcfC | VC0841 | −5.2 | 0.0 |
| Cholera enterotoxin beta subunit | VC1456 | −3.4 | 0.1 |
| Cholera enterotoxin A subunit | VC1457 | −2.6 | 0.1 |
| Hemolysin | VCA0219 | −8.8 | 0.0 |
| Regulation | |||
| LuxR family transcriptional regulator VqmA | VCA1078 | 40.0 | 8.0 |
| Hemagglutinin/protease regulatory protein HapR | VC0583 | 4.3 | 0.3 |
| LuxR family transcriptional regulator | VCA0888 | 20.8 | 5.5 |
| Metabolism | |||
| Malate dehydrogenase | VC0432 | −3.7 | 0.0 |
| Aconitate hydratase 2 | VC0604 | −2.9 | 0.0 |
| Carbon starvation protein A (putative) | VC0687 | −4.5 | 0.0 |
| Glutaredoxin | VC1146 | 5.2 | 0.5 |
| Alpha-acetolactate decarboxylase | VC1589 | 4.4 | 0.4 |
| Acetolactate synthase | VC1590 | 2.5 | 0.1 |
| Glyceraldehyde 3-phosphate dehydrogenase | VC2000 | 3.3 | 0.4 |
| 2-Oxoglutarate dehydrogenase E1 component | VC2087 | −3.3 | 0.1 |
| GTP-binding protein TypA | VC2744 | 2.4 | 0.2 |
| Glutamate-ammonia ligase | VC2746 | 2.9 | 0.4 |
| Glycerophosphoryl diester phosphodiesterase | VCA0136 | −30.3 | 0.0 |
| GMP reductase | VCA0197 | 4.5 | 1.8 |
| Acrobic glycerol-3-phosphate dehydrogenase | VCA0657 | −24.9 | 0.0 |
| Glycerol kinase | VCA0744 | −47.3 | 0.0 |
| Glycerol uptake facilitator protein | VCA0745 | −18.3 | 0.0 |
| 3-Oxoacyl-(acyl carrier protein) synthase III | VCA0751 | −3.1 | 0.1 |
| Proline dehydrogenase | VCA1073 | −4.6 | 0.0 |
| Protein processing/transport | |||
| 16-kDa heat shock protein A | VC0018 | 4.3 | 0.3 |
| Peptide ABC transporter ATP-binding protein | VC0170 | −2.7 | 0.0 |
| Peptide ABC transporter periplasmic peptide-binding protein | VC0171 | −2.7 | 0.0 |
| Na+/H+ antiporter (putative) | VC0389 | 2.8 | 0.2 |
| PTS system glucose-specific IIBC componentc | VC2013 | 3.6 | 0.1 |
| Glycerol-3-phosphate transporter | VCA0137 | −63.9 | 0.0 |
| Maltose ABC transporter permease protein | VCA0943 | 2.7 | 0.5 |
| Maltose ABC transporter permease protein | VCA0944 | 3.7 | 0.8 |
| Unknown function | |||
| VC0134, VC0491, VC0669, VC0770, VC1492, VC2155, VCA0324, VCA0367, VCA0483, VCA0587, VCA0887 | >2 | ||
| VC0669, VC1492, VC2155, VCA1097 | <−2 |
ZLV102 and ZLV104 were grown in AKI medium in the presence of IPTG (0.1 μM). Positive values represent activation in ZLV104 [vqmA(Con)]; negative values represent repression. Average ratios from two independent experiments are shown.
From two independent experiments.
PTS, phosphotransferase.
Repression of virulence genes by constitutively expressed VqmA is HapR dependent.
To verify the microarray data, we measured the production of CT, a major virulence determinant, after growth in AKI medium, which is known to induce virulence factor production (16). The strains with constitutively expressed VqmA produced 93% less cholera toxin than a wild-type strain (Fig. 1). Deletion of vqmA did not affect CT production.
FIG. 1.
Constitutive expression of vqmA inhibits cholera toxin production in a HapR-dependent manner. V. cholerae strains (vqmA, wild type [wt], or hapR) with pMal-c2x (vector control) (white bars) or pZL3 (constitutively expressed vqmA) (gray bars) were grown in AKI medium in the presence of 0.1 μM IPTG. CT in supernatants was measured using a CT enzyme-linked immunosorbent assay. Data are expressed as percentages of the level of CT produced by wild-type C6706 grown under the same conditions. Results are representative of three experiments ± standard deviations.
Microarray analysis indicated that the expression of two regulators, the LuxR family protein VCA0888 and HapR, was up-regulated when VqmA was constitutively expressed (Table 2). Overexpression of VCA0888 did not affect CT production (data not shown), indicating that VCA0888 is not involved in virulence gene regulation. However, previous studies have demonstrated that HapR represses virulence gene expression by repressing aphA transcription (18, 38). Remarkably, hapR expression is increased more than fourfold in the strain constitutively expressing VqmA [VqmA(Con)]. This suggests that VqmA may act on virulence gene expression through HapR. To test this possibility, we examined CT production in a hapR mutant. In contrast to hapR+ strains harboring the Ptac-vqmA plasmid (producing only 7% of the CT made in wild-type strains), overexpression of VqmA in the hapR mutant had only a minimal effect on CT production (80% of the CT produced by the wild type) (Fig. 1), while a plasmid constitutively expressing hapR in such a strain restored the repression of CT production (data not shown). These results indicated that the inhibitory effect of VqmA on the virulence regulon required functional HapR.
VqmA activates hapR transcription at low cell densities.
Our microarray experiment revealed that VqmA regulates hapR transcription. To further confirm the microarray data and to investigate how VqmA affects hapR transcription, we monitored the expression of a chromosomal hapR-lacZ transcriptional fusion in a vqmA deletion mutant, a wild-type vqmA strain, and a strain with multiple copies of constitutively expressed vqmA as a function of cell density. Figure 2A shows that hapR expression is higher at low cell densities (optical density at 600 nm [OD600], <0.6) in strains with a single copy of vqmA (wild type) and strains constitutively expressing vqmA than in vqmA deletion mutants. However, at high cell densities (OD600, ≥1.5), the expression of hapR is similar in all three strains tested.
FIG. 2.
VqmA increases hapR transcription at low cell densities. (A) Chromosomal hapR-lacZ expression in the vqmA deletion strain (−) (white bars), wild-type vqmA (wt) (gray bars), and the strain with constitutively expressed vqmA (C) (black bars) at the cell densities indicated. Results are representative of three experiments ± standard deviations. (B) Western blot analysis of HapR production in the same strains at the cell densities indicated. Strains with different VqmA levels are indicated as explained in the legend to panel A. The vqmA deletion mutant and the wt strain contained a vector control, and the strain constitutively expressing vqmA contained plasmid pZL3). IPTG (0.1 μM) was added to all cultures. Results are representative of three experiments.
To further confirm that VqmA regulates hapR, we used a Western blot assay to measure HapR production in strains with plasmids with different forms of vqmA. Equal amounts of total protein were loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. In the wild-type strain, HapR was detectable when mid-log phase was reached, although little HapR was found in vqmA mutants at these cell densities (Fig. 2B). In contrast, when VqmA was constitutively expressed, HapR was detected at lower cell densities (OD600, ≈0.2). At high cell densities, all strains produced similar amounts of HapR protein. We interpret these data as indicating that VqmA contributes to hapR regulation primarily at low cell densities.
VqmA increases HapR-activated gene expression.
In addition to repressing virulence gene expression, the quorum-sensing regulator HapR also activates the heterologous promoter of the V. harveyi luxCDABE operon (26). To study the relationship between VqmA and quorum-sensing regulation, we introduced the V. harveyi luciferase genes into strains with different vqmA constructs and used light production as an indicator of quorum-sensing-dependent gene expression. Cultures were diluted, and light production per cell was measured during subsequent growth. Figure 3A shows that the wild-type strain produced light in a cell density-dependent pattern. This U-shaped curve is due to the dilution of the culture and thus a drop in the extracellular autoinducer levels to below the threshold concentration required for stimulation of lux expression. As cell density increases, lux is induced over time due to the accumulation of new autoinducers (26). Constitutive VqmA expression in either vqmA mutants or wild-type strains caused earlier induction of Lux activity than that observed for the wild type with a control vector. On the other hand, Lux induction was delayed in the vqmA deletion strain containing only a vector plasmid. In addition, overproduction of VqmA in either luxO mutants (displaying constitutive lux expression) or hapR mutants (no light produced) did not affect light production (data not shown). These data indicate that the quorum-sensing system is activated at lower cell densities when VqmA is present. Furthermore, VqmA proteins do not affect autoinducer production, as evidenced by the fact that all four strains produced similar amounts of both CAI-1 and AI-2 under these conditions (data not shown).
FIG. 3.
VqmA activates quorum-sensing-regulated luminescence at lower cell densities and can bypass the LuxO-sRNA regulatory circuits. (A) Light production per cell in strains containing the V. harveyi lux operon cosmid pBB1. The wild-type strain containing either the pMal-c2x vector (wt) (open squares) or pZL3, constitutively expressing vqmA (solid squares), and vqmA mutants containing either pMal-c2x (open triangles) or pZL3 (solid triangles) were grown in LB containing 0.1 μM IPTG at 30°C. At various time points, light production and densities of cultures (in CFU) were measured. Relative luminescence units were calculated as 103 light units/CFU ml−1. Results are representative of three experiments. (B) Effect of VqmA on light production in a mutant with constitutively active LuxO. Strains containing either pMal-c2x (vector control) (white bars) or pZL3 [vqmA(Con)] (gray bars) were grown overnight at 30°C in LB containing 0.1 μM IPTG, and luminescence was measured. Results are representative of three experiments ± standard deviations.
Recent work from Bassler and colleagues reveals that when V. cholerae is at a low cell density, phosphorylated LuxO (LuxO-P) activates a set of small regulatory RNAs, which work together with Hfq to destabilize hapR mRNA (21). At high cell densities, LuxO is dephosphorylated and thus inactive. Constitutive mutations in the luxO allele, such as D47E (8, 13) and L104Q (32), “lock” the LuxO protein into a state mimicking LuxO-P. These constitutive LuxO mutants therefore inactivate the quorum-sensing system by constitutively repressing hapR expression. To investigate the interaction between VqmA regulation of hapR and the LuxO-small-RNA (LuxO-sRNA) regulatory circuits described above, we introduced pMal-c2x (vector control) or pZL3 (Ptac-vqmA) into the strain containing a constitutive luxO (L104Q) allele and measured Lux expression. Figure 3B shows that Lux was not expressed in the luxO(Con) mutant containing a vector control, as previously reported (32), but overexpression of VqmA induced light production more than 100-fold greater than that with the vector control. However, compared to that in wild-type strains, VqmA cannot fully recover light production in luxO constitutive mutants. These data suggest that VqmA can partially overcome the LuxO-sRNA inhibitory effect on hapR transcription, possibly by activating the production of more hapR mRNA. No light was detected in the hapR mutants, suggesting that VqmA itself cannot activate the lux operon.
VqmA regulates hapR transcription directly.
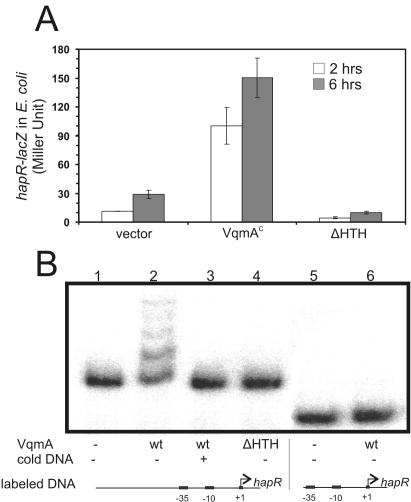
Our data suggest that VqmA plays a role in quorum-sensing regulation by increasing hapR expression. To investigate further whether VqmA activates hapR transcription directly or through other regulators, we constructed a plasmid containing the hapR promoter fused with a lacZ transcriptional reporter in E. coli and assayed for β-galactosidase activity. Compared to a vector control, VqmA increased hapR-lacZ expression (Fig. 4A). Deletion of the conserved helix-turn-helix DNA binding domain of VqmA completely abolished β-galactosidase activity. VqmA activation of hapR transcription in E. coli suggests that VqmA acts directly on the hapR promoter or, alternatively, activates a regulator gene that is conserved between V. cholerae and E. coli.
FIG. 4.
VqmA regulates the hapR promoter directly. (A) hapR-lacZ expression in E. coli. MC4100 with either a vector control, a plasmid constitutively expressing wild-type VqmA, or a plasmid expressing VqmA lacking its DNA binding domain (ΔHTH) was inoculated 1:100 into LB in the presence of 0.1 μM IPTG and grown at 37°C for 2 h (white bars) or 6 h (gray bars). Results are representative of three experiments. Error bars, standard deviations. (B) Gel retardation assays using purified MBP-VqmA proteins. Lanes 1 to 4, labeled hapR promoter region; lanes 5 and 6, labeled hapR promoter region without upstream sequences. Lanes 1 and 5, labeled DNA only; lanes 2 and 6, 0.5 μg MBP-VqmA; lane 3, 0.5 μg MBP-VqmA with 0.1 μg unlabeled DNA; lane 4, 0.5 μg MBP-VqmA(ΔHTH). wt, wild type.
We then purified the VqmA protein as MBP fusions and performed a gel retardation assay to test whether VqmA binds directly to the hapR promoter DNA. The MBP-VqmA fusion protein retained the same activity as the wild-type VqmA protein, as judged by the capacity of induction of hapR expression in E. coli and in V. cholerae (data not shown). Two kinds of DNA fragments were labeled. The shorter fragment comprised the region of the hapR promoter from the predicted −35 sequences (22) to the translational start site, while the second fragment extended an additional 200 bp upstream of the −35 region. Purified MBP-VqmA retarded the mobility of the DNA fragment containing the region upstream of −35 (Fig. 4B, lane 2) but not the shorter fragment (lane 6). Addition of unlabeled target DNA abolished the retardation (lane 3), indicating that the binding of VqmA is specific and is located upstream of the −35 region. Purified MBP-VqmA lacking the helix-turn-helix DNA binding domain did not shift the hapR promoter DNA (lane 4), further confirming that the DNA binding domain of VqmA is essential for its activity.
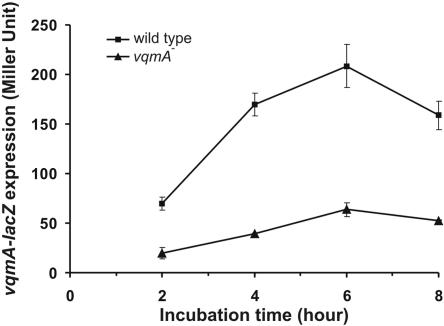
The expression of vqmA is autoregulated.
Our data demonstrated that VqmA regulates quorum sensing in V. cholerae by activating hapR expression. We were thus interested in how vqmA itself is expressed. To study this, we constructed two vqmA-lacZ chromosomal transcriptional fusions. One construct retains the intact vqmA gene, and the other has a disrupted vqmA gene (see Materials and Methods for details). Figure 5 shows that in the vqmA+ background, the expression of vqmA increased with cell density and reached maximal activity in late log phase. In contrast, vqmA expression in the vqmA disruptant background does not increase during cell growth. These data imply that VqmA autoregulates its own promoter.
FIG. 5.
VqmA is required for vqmA cell density-dependent expression. Wild-type (AHV302) (squares) and vqmA mutant (AHV301) (triangles) strains carrying a chromosomal vqmA-lacZ transcriptional reporter fusion were grown in LB at 37°C, and β-galactosidase activity was measured at the time point indicated. Results are representative of three experiments ± standard deviations.
Strains carrying mutations in vqmA have no colonization defect.
We have shown that constitutive expression of the vqmA genes repressed virulence gene expression in an in vitro inducing condition, while deletion of vqmA did not affect virulence gene expression. We examined whether VqmA affects V. cholerae pathogenesis by comparing the colonization capacities of the wild type and vqmA mutants using an infant mouse model. As expected, strains carrying a vqmA deletion can colonize infant mice as well as the wild-type strain (data not shown), indicating that vqmA is not involved in colonization, at least in the infant mouse model.
DISCUSSION
In this study, we have shown that the transcriptional regulator VqmA modulates V. cholerae quorum-sensing systems by directly increasing the expression of the quorum-sensing master regulator HapR. The gene encoding VqmA (VCA1078) is located on the small chromosome and may be divergently transcribed with a gene encoding pyridoxamine 5′-phosphate oxidase (VCA1079). Quorum sensing in V. cholerae has been shown to negatively regulate virulence gene expression in vitro by repressing the expression of aphA, whose gene products are required for the activation of the entire virulence regulon. Constitutive expression of VqmA induces quorum sensing early in growth and leads to repression of virulence genes. In the vqmA deletion mutant, quorum sensing displays a delay but is eventually fully induced. It is unclear what role VqmA plays during V. cholerae infection. vqmA is not expressed in V. cholerae isolated from cholera patients' stool samples (2) but is highly expressed in the rabbit ileal loop model of V. cholerae infection (34). Thus, we speculate that vqmA expression differs under different environmental conditions and affects quorum-sensing systems accordingly.
Although VqmA and other annotated LuxR family proteins may function independently of acyl homoserine lactone signals, used as quorum-sensing molecules in many other gram-negative bacteria, these proteins play regulatory roles in V. cholerae. In addition to VqmA, another protein in this group, VpsT (VCA0952), has been shown to serve as a positive transcriptional regulator of vps gene expression (4). Another noteworthy aspect of VqmA is that this protein has a conserved PAS domain (named after three proteins in which it occurs: Per, Arnt, and Sim), found widely among prokaryotes and eukaryotes (data not shown). PAS domains have been found to bind ligands and to act as sensors for light and oxygen in signal transduction (30). Although our in vitro MBP-VqmA DNA retardation assays indicated that the VqmA protein is able to bind its target promoter DNA, we cannot rule out the possibility that binding of additional ligands may enhance VqmA activity. We thus speculate that VqmA might be able to detect certain unknown environmental signals and modulate cell density-dependent cellular functions when needed.
Acknowledgments
We are grateful to John Mekalanos for providing insights and guidance to this study. We thank Jeffrey Weiser and Deborah Hung for helpful discussions and critical review of the manuscript. We also thank Bonnie Bassler for providing the HapR antiserum.
This study was supported by an NIH/NIAID K22 award (AI060715) and a Penn Genomics Institute seed grant. Z.L. is supported by the Felsen Diarrhea Research Fund.
REFERENCES
- 1.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 2.Bina, J., J. Zhu, M. Dziejman, S. Faruque, S. Calderwood, and J. Mekalanos. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. USA 100:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, R. G., and B. W. Matthews. 1989. The helix-turn-helix DNA binding motif. J. Biol. Chem. 264:1903-1906. [PubMed] [Google Scholar]
- 4.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finkelstein, R. A., M. Boesman-Finkelstein, Y. Chang, and C. C. Hase. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardel, C. L., and J. J. Mekalanos. 1994. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 235:517-526. [DOI] [PubMed] [Google Scholar]
- 13.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung, D. T., and J. J. Mekalanos. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. USA 102:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwanaga, M., K. Yamamoto, N. Higa, Y. Ichinose, N. Nakasone, and M. Tanabe. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30:1075-1083. [DOI] [PubMed] [Google Scholar]
- 17.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 18.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 19.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6:186-190. [DOI] [PubMed] [Google Scholar]
- 20.Lenz, D. H., M. B. Miller, J. Zhu, R. V. Kulkarni, and B. L. Bassler. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186-1203. [DOI] [PubMed] [Google Scholar]
- 21.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 22.Lin, W., G. Kovacikova, and K. Skorupski. 2005. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 187:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mel, S. F., K. J. Fullner, S. Wimer-Mackin, W. I. Lencer, and J. J. Mekalanos. 2000. Association of protease activity in Vibrio cholerae vaccine strains with decreases in transcellular epithelial resistance of polarized T84 intestinal epithelial cells. Infect. Immun. 68:6487-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 27.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pappas, K. M., and S. C. Winans. 2003. A LuxR-type regulator from Agrobacterium tumefaciens elevates Ti plasmid copy number by activating transcription of plasmid replication genes. Mol. Microbiol. 48:1059-1073. [DOI] [PubMed] [Google Scholar]
- 29.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 34.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 36.Zhu, J., Y. Chai, Z. Zhong, S. Li, and S. C. Winans. 2003. Agrobacterium bioassay strain for ultrasensitive detection of N-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl. Environ. Microbiol. 69:6949-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]