Abstract
Marinocine is a broad-spectrum antibacterial protein synthesized by the melanogenic marine bacterium Marinomonas mediterranea. This work describes the basis for the antibacterial activity of marinocine and the identification of the gene coding for this protein. The antibacterial activity is inhibited under anaerobic conditions and by the presence of catalase under aerobic conditions. Marinocine is active only in culture media containing l-lysine. In the presence of this amino acid, marinocine generates hydrogen peroxide, which causes cell death as confirmed by the increased sensitivity to marinocine of Escherichia coli strains mutated in catalase activity. The gene coding for this novel enzyme was cloned using degenerate PCR with primers designed based on conserved regions in the antimicrobial protein AlpP, synthesized by Pseudoalteromonas tunicata, and some hypothetical proteins. The gene coding for marinocine has been named lodA, standing for lysine oxidase, and it seems to form part of an operon with a second gene, lodB, that codes for a putative dehydrogenase flavoprotein. The identity of marinocine as LodA has been demonstrated by N-terminal sequencing of purified marinocine and generation of lodA mutants that lose their antimicrobial activity. This is the first report on a bacterial lysine oxidase activity and the first time that a gene encoding this activity has been cloned.
Antimicrobial peptides and proteins are synthesized by different organisms throughout the phylogenetic scale, from bacteria to mammals. In higher organisms they participate in the defense against pathogens (22). In microorganisms, this class of compounds includes bacteriocins and antimicrobial peptides (15). It has been proposed that these antimicrobials are used in competition between microorganisms, offering an advantage to the producer strains (2). Although most of these antimicrobial agents are peptides of a relatively low molecular weight, others are proteins such as the pore colicins produced by Escherichia coli (21). Regarding gram-positive bacteria, there are examples of large proteins such as lysostaphin, an endopeptidase produced by Staphylococcus simulans, of interest for the treatment of infections caused by Staphylococcus spp. (19).
Among these proteins, the antimicrobial properties of enzymes generating hydrogen peroxide have also been described, and in fact this is one of the mechanisms used by lactic acid bacteria to control the growth of competitors (23). l-Amino acid oxidases (l-AAOs) (EC 1.4.3.2) are flavoenzymes found in different organisms, although the best characterized are the snake venom l-AAOs (9). These flavoenzymes catalyze the oxidative deamination of l-amino acids to the respective α-keto acids with the release of ammonium and hydrogen peroxide, which determines their antimicrobial properties (41). Most l-AAOs oxidize l-lysine slightly or not at all. However, l-lysine oxidases are a subclass of these enzymes. l-Lysine oxidases have been isolated from fungi, particularly of the genus Trichoderma (20). This fungal enzyme has been characterized at the biochemical level and shows antibacterial effects (29), but the cloning of the gene encoding the activity has not been reported.
Some marine bacteria also produce antimicrobial proteins. Pseudoalteromonas tunicata produces an autolytic protein that is key to biofilm development in this microorganism (30). Pseudoalteromonas (Alteromonas) luteoviolacea strain 9K-V10 also produces an antimicrobial protein active against gram-positive and gram-negative bacteria (32). Some gliding bacteria synthesize glycoproteins that participate in microbial competition in biofilms. In the last scenario, it was proposed that the high molecular weight of the glycoprotein should help to prevent its diffusion to the surrounding environment (4).
Marinomonas mediterranea is a melanogenic marine bacterium that displays a rich secondary metabolism. It expresses two different growth-phase-regulated polyphenol oxidases (PPOs), a tyrosinase and a laccase (26). The tyrosinase is involved in melanin synthesis using tyrosine as substrate (25). The genus Marinomonas bears many phenotypic similarities to other aerobic marine γ-proteobacteria with polar flagellation and a G+C content below 50 mol% (36). In fact, many Marinomonas and Pseudoalteromonas species were originally included in the genus Alteromonas. Like some Pseudoalteromonas strains, M. mediterranea also synthesizes an antibacterial protein, named marinocine, with activity against both gram-positive and gram-negative bacteria (28). Marinocine is detected in the supernatants of the culture medium and shows remarkable resistance against many hydrolytic enzymes (28). In this study, the basis of its antibacterial properties has been characterized and the gene coding for the antibacterial protein has been cloned. It is shown that marinocine shows lysine oxidase activity and that the antibacterial effect is a consequence of the hydrogen peroxide generated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. The protocol for marinocine extraction from M. mediterranea cultures was previously described (28). E. coli strains were grown in LB or M9 medium. Catalase, l-lysine α-oxidase from Trichoderma viride, l-lysine, amino acids, and related amines were purchased from Sigma.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| M. mediterranea | ||
| MMB-1 | Wild-type strain, Rifs Gms | 39 |
| MMB-1R | MMB-1, spontaneous Rifr | 40 |
| T101 | MMB-1R, ppoA::Tn10 Gmr [DMPO−] | 40 |
| T105 | MMB-1R, Ω Tn10 Kmr [TIR− MEL−] | 25 |
| ng56 | MMB-1, NG [TIR− MEL−] | 39 |
| T103 | MMB-1R, ppoS::Tn10 Kmr [PPO+/−, MEL+/−] | 26 |
| MUTMAR2 | MMB-1R, lodA::pFSVK-mar8 | This study |
| SB1 | MMB-1R, lodA::pFSVK-SS [MAR−] | This study |
| E. coli | ||
| MP180 | thi-1 HrfH | 24 |
| UM120 | MP180 katE::Tn10 | 24 |
| UM122 | MP180 katF::Tn10 | 24 |
| UM202 | MP180 katG::Tn10 | 24 |
| S17-1(λpir) | thi pro hsd(r− m+) recA::RP4-2-Tcr::Mu Kmr::Tn7 Tpr Smr, lysogenized by λpir phage | 8 |
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM5) hsdR17 recA endA gyrA96 thi-1 relA | 14 |
| K-12 | Commercially available | |
| Plasmids | ||
| pLOFKm | ori R6K mob RP4 Apr mini-Tn10 Kmr, VM | 17 |
| pFSV | Cmr Tcrori R6K mob RP4 | 3 |
| pFSVK | ori R6K mob RP4 mini-Tn10 Kmr, 1-kb NcoI-SacI ‘ppoA’ fragment | 27 |
| pGEM-T | Commercially available | |
| pMAR28 | pGEM-T containing a 1.3-kb inner lodA fragment amplified by degenerate PCR | This study |
| pFSVK-mar8 | pFSVK containing a ca. 1.1-kb fragment comprising the lodA fragment from pMAR28 excised at NcoI-SacI flanking sites of pGEM-T | This study |
| pFSVK-SS | pFSVK-mar8 lacking a 404-bp inner lodA fragment of that present in pMAR28 excised by ScaI-StuI digestion and religation | This study |
| pRECMAR4 | pFSVK-mar8 rescued from genomic DNA of MUTMAR2 by XbaI excision so that it contains genomic regions flanking lodA | This study |
Descriptions of the strains include their genotypes and most relevant phenotypes in brackets. The parent strain is mentioned first. Abbreviations used: NG, strain obtained by chemical mutagenesis with nitrosoguanidine; Ω, strain obtained by transposon mutagenesis with the transposon next mentioned; ::, the insertion locus of the transposon or other element has been characterized; VM, mutagenesis vector. Phenotypes: DMPO−, loss of laccase activity; TIR−, loss of SDS-activated tyrosinase activity; PPO+/−, decrease in all PPO activities; MEL−, amelanogenic; MEL+/−, decrease in melanin synthesis; MAR−, nonproducer of marinocine.
Oxygen consumption measurements.
For oxygen consumption measurements, a Clark type electrode coupled to a Hansatech CD1B oxygraph unit was used. A saturated KCl solution provided the necessary conductance between both electrodes for a constant input voltage of 700 mV. The output-generated current was amplified and drawn by an OmniScribe graphic detector from Houston Instruments. To establish the zero level of emitted current, we used a sodium dithionite solution that reduces all free molecular oxygen in the chamber.
E. coli K-12 grown at 25°C in LB up to exponential phase was used to test the effect of marinocine on oxygen consumption. Cells were washed with phosphate buffer (0.05 M, pH 7) and resuspended to an optical density at 600 nm of 0.4. After stabilization of endogenous oxygen consumption, the compounds to be tested (glucose or marinocine) were injected into the chamber.
N-terminal amino acid sequencing.
The N-terminal amino acid sequence of marinocine was analyzed by automatic Edman degradation on an Applied Biosystems Procise Sequencer. A purified sample of marinocine was run in 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under denaturing conditions (28). The proteins were blotted onto an Immobilon polyvinylidene difluoride membrane (Millipore) and stained with amido black. The bands were sequenced at the CIB-CSIC (Madrid).
Lysine oxidase activity.
In order to test the enzymatic production of hydrogen peroxide by marinocine samples, the Amplex Red kit (Molecular Probes) was used. The fluorimetric assay is based on the detection of H2O2 using 10-acetyl-7-hydroxyphenoxazine (Amplex Red reagent). In the presence of horseradish peroxidase, Amplex Red reacts with H2O2 at a 1:1 stoichiometry, producing a highly fluorescent product, resorufin, which can be detected by excitation and emission wavelengths of 550 and 590 nm, respectively. The assays were performed in 96-well plates at a final volume of 100 μl. Lysine was added to a final concentration of 50 mM. All tests were made in 50 mM (pH 7.4) phosphate buffer containing 50 μM Amplex Red reagent and 0.1 units/ml horseradish peroxidase. Background fluorescence was corrected by subtracting the values derived from all the reagents except for the l-lysine oxidase. Appropriate controls, including catalase at 0.1 mg/ml, were also included to ensure that the fluorescence signal was due to H2O2. For each sample, all measurements were made in duplicate.
Cloning of the marinocine locus.
Cloning of the marinocine locus was achieved by PCR amplification using degenerate deoxyoligonucleotides designed according to the alignment of the P. tunicata AlpP gene to homologous genes in other bacteria (Table 2). The three designed primers were as follows: MARDIR1, for the forward direction, and MAREV1 and MAREV2, for the reverse direction. PCR was performed with genomic DNA of M. mediterranea MMB-1R as template. Two consecutive PCRs were performed, varying the annealing temperature with 10 cycles at 44°C followed by 33 cycles at 48°C in each PCR. For the first reaction, primers MARDIR1-MAREV2 were used. The PCR product of this reaction, a faint band of 1,500 bp, was used as template for the second PCR, using MARDIR1 and MAREV1. The 1,300-bp fragment obtained was purified and cloned into pGEM-T (Promega), and two of the plasmids obtained, pMAR9 and pMAR28, were sequenced in both directions. The sequencing of the DNA fragment inserted in both plasmids gave the same results; therefore, pMAR28 was selected for further analysis.
TABLE 2.
Conserved regions 3, 7, and 8 (the others are not shown) in proteins similar to AlpP used to clone the gene coding for marinocine by degenerate PCR
| Organism | Accession no. (size [amino acids]) | Sequences ofa:
|
||
|---|---|---|---|---|
| Conserved region 3 | Conserved region 7 | Conserved region 8 | ||
| P. tunicata | AAP73876 (727) | 82IVWTVHLANKKAAWY98 | 488RMAOPWQADFFQCT503 | 595YYSYWWPPQSPWDVL610 |
| C. violaceum | AAQ60932 (606) | 82IRWTAHLANKKPSWY98 | 476YMAIPWQADFNECS491 | 496RRLWWWPAQRPEFVY512 |
| Magnetococcus sp. strain MC-1 | ZP_00289207 (608) | 90IEWTVHLANKKSIWY106 | 486FMAIPWQADFNLCS501 | 506RTLWWWPAQRPLNVY522 |
| C. crescentus | NP_419374 (687) | 116IVWTVHLANKKANSY132 | 556FMAVPWHTDYNACA571 | 582ALYWSWPAQRPVQVH598 |
| M. degradans 2-40 | ZP_00318138 (767) | 119IEWSVYLANKKSSWF135 | 649FSALPWQADFNECS664 | 692SETLWWPAYRPMQVY708 |
| R. palustris | NP_947813 (684) | 111VTWSVHVANTKAAWY127 | 523WMGVPWQGDAFSCQ538 | 545PTPVWWPALLPVDVL561 |
| R. baltica | NP_866009 (527) | 79IQWRVSLANRKAAGD95 | 449GNALPWQADFLKCA464 | 461CAGSWWPAQRPDEVF477 |
| Degenerate deoxyoligonucleotidesb | 5′ GTNYAYYTNGCNAAYAARAA 3′ | 5′ RAARTCNGCYTGCCANGG 3′ | 5′ GGNNNYTGNGSNGGCCACC 3′ | |
| M. mediterraneac | AY968053 (726) | 82IEWTVHLANKKAAWY98 | 503RMACPWQADFFNCT518 | 576YSYWWPPQSPWDVL591 |
Boldface letters indicate the specific amino acids actually used in the design of the three degenerate deoxyoligonucleotides (one forward, MARDIR1; two reverse, MAREV1 and MAREV2).
Conserved region 3, MARDIR1; conserved region 7, MAREV1; conserved region 8, MAREV2.
Marinocine sequence.
To isolate a genomic region encompassing the complete gene coding for marinocine in M. mediterranea, as well as upstream and downstream flanking DNA, the fragment cloned into pMAR28 was excised by digestion with SacI, eliminating ca. 250 bp from the 5′ end of the PCR product, and NcoI, which cuts the pGEM sequence, and ligated into similarly digested pFSVK. pFSVK is a suicide vector made up with selected characteristics from other described vectors. It contains oriR6K and mob RP4 from pFSV (3), the mini-Tn10 (Kmr) from pLOFKm (17), and an NcoI-SacI inner fragment from the ppoA gene (35). The replacement of the NcoI-SacI fragment in pFSVK with the fragment cloned in pMAR28 generated the suicide vector pFSVK-mar8. This vector was subsequently used to transform E. coli S17-1λpir, which was later conjugated with M. mediterranea MMB-1R as previously described (40). Genomic DNA of two of the kanamycin-resistant mutants, MUTMAR2 and MUTMAR3, was then digested using XbaI, a restriction endonuclease that did not cut the pFSVK-mar8 vector, and the fragments containing the regions of interest were selected after ligation, transformation into S17-1λpir, kanamycin marker selection on plates, and plasmid extraction. Eventually, two different plasmids, pRECMAR4 and pRECMAR6, obtained from MUTMAR2 and MUTMAR3, respectively, and therefore from different conjugation plates, were sequenced and no differences between them were detected.
Construction of lodA mutants.
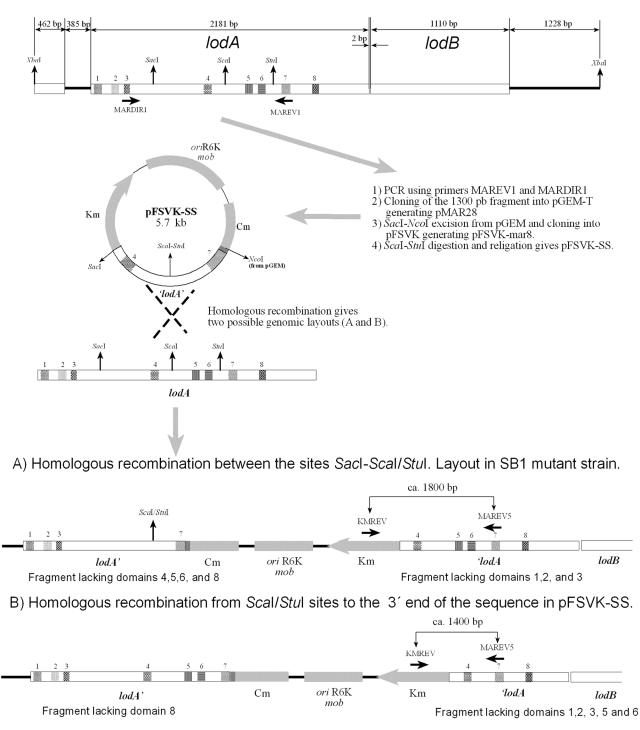
In order to generate lodA mutants, the protocol outlined in Fig. 1 was followed. pFSVK-mar8 was digested by ScaI-StuI. These enzymes cut a fragment of 400 bp inside the cloned region of lodA in pFSVK-mar8, which contained the fifth and sixth conserved domains, out of a total of eight, between homologous proteins. In other words, the insert that initially comprised nucleotides 518 to 1542 was excised in the middle, from 1027 to 1429. Thus, we obtained a suicide vector, pFSVK-SS, that gave two types of mutants when inserted by homologous recombination into the wild-type genome: producers and nonproducers of marinocine, such as strain SB1, according to the results obtained in the antibiogram assay against E. coli DH5α. Genomic DNA was extracted from both types of mutants and by PCR with the appropriate primers (KMREV, 5′-GTAACATCATTGGCAACG-3′; MAREV5, 5′-TGCCAAGGGCATGCCATGCGC-3′); it was checked that the phenotype depended on the region in the vector where the recombination process had taken place.
FIG. 1.
Construction of lodA mutant strains. Plasmid pFSVK-SS was created as shown. After integration of this plasmid into the genome of M. mediterranea by homologous recombination, two different genomic layouts were possible (A and B). They were distinguished by PCR using primers KMREV and MAREV5, obtaining PCR products of ca. 1,800 bp and 1,400 bp, respectively. Mutant strains with construction A, such as SB1, are nonproducers of marinocine. In contrast, mutant strains with construction B are producers of marinocine.
Nucleotide sequence accession number.
The DNA sequences of the lodA and lodB genes have been deposited in the GenBank database under accession number AY968053.
RESULTS
Oxygen and catalase effects on marinocine activity.
Studies of the environmental conditions required for detection of the antimicrobial effects of marinocine revealed that it is not active under anaerobic conditions. The inhibition halo in the antibiogram assay was not observed at all when the plates were incubated anaerobically.
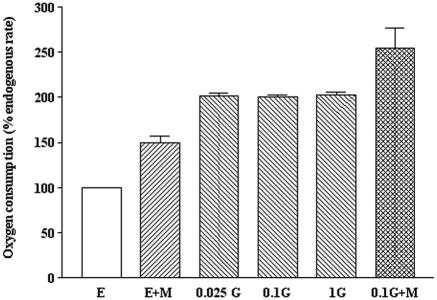
Some marine bacteria belonging to the genus Alteromonas synthesize macromolecular antimicrobial compounds which modify bacterial respiration (12). Due to this precedent, the implication of oxygen in marinocine activity was examined by measuring its consumption by susceptible cells. In order to avoid bactericidal effects, marinocine was assayed at concentrations below the minimal bactericidal concentration, 200 U/ml for E. coli K-12, the strain used in these experiments. Under these conditions, marinocine increased the oxygen consumption compared to the endogenous rate of control cells (Fig. 2). The effect of marinocine was not related to a potential increase in the carbon source, since the addition of this compound to glucose-saturated cells provoked an additional increase in the respiration rate (Fig. 2).
FIG.2.
Effect of marinocine on E. coli K-12 respiration. E, endogenous rate; E+M, 65 U/ml marinocine added; 0.025G, 0.025% glucose; 0.1G, 0.1% glucose; 1G, 1% glucose; 0.1G+M, 0.1% glucose plus 65 U/ml marinocine.
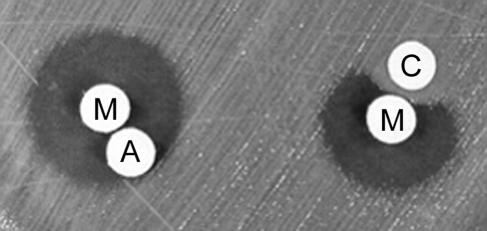
In order to check whether reactive oxygen species were generated at the time the antibacterial effect was observed, the activity of marinocine against several susceptible strains was assayed in the presence of catalase. The results indicated that the antibacterial activity was completely abolished under these conditions (Fig. 3), suggesting that the inhibitory effect was mediated by the action of hydrogen peroxide.
FIG. 3.
Catalase inhibition of the antibacterial effect of marinocine on E. coli K-12. M, disk loaded with 17 U of marinocine; A, disk with distilled water; C, disk with 0.2 mg of catalase.
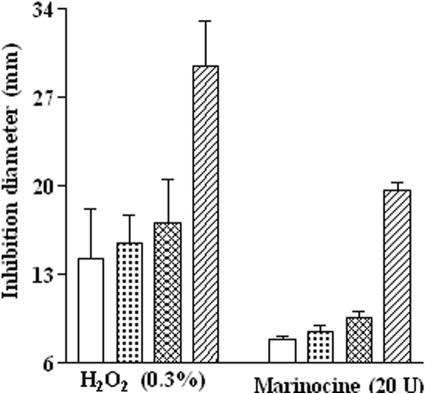
To verify the role of hydrogen peroxide in the antibacterial activity of marinocine, several antibiogram tests were performed against three different E. coli strains derived from the wild-type strain MP180: strain UM202 (a katG mutant), mutated in the production of hydroperoxidase (catalase) HPI, and strains UM120 (a katE mutant) and UM122 (a katF mutant), mutated in the production of HPII (24). These strains showed similar patterns of response to hydrogen peroxide and marinocine. It was observed that strain UM202 is the most sensitive to both hydrogen peroxide and marinocine (Fig. 4). This result is in agreement with the fact that the bifunctional catalase-peroxidase HPI, encoded by the katG gene mutated in strain UM202, is inducible by H2O2 and is the main catalase involved in the resistance to this agent (24, 38). Because of its increased sensitivity to marinocine, this strain was selected for further studies.
FIG. 4.
Antibacterial effects of marinocine and hydrogen peroxide on different E. coli strains mutated in catalase activity. Open bars, MP180 (wild-type strain); checked bars, UM120 (katE mutant); cross-hatched bars, UM122 (katF mutant); striped bars, UM202 (katG mutant).
Effect of growth medium used in the antibiogram on susceptibility to marinocine.
The generation of hydrogen peroxide by marinocine suggested some kind of enzymatic activity for this protein. Two possibilities were considered. Hydrogen peroxide could be generated after marinocine interaction with some cellular component, or it could be produced from some ingredient in the growth medium. To test the latter hypothesis, antibiograms were performed under different culture conditions. It was observed that marinocine was active only in complex media, not in defined medium such as M9. This fact could have been the result of the different sensitivities of E. coli in rich and defined media to the hydrogen peroxide produced. However, the sensitivity of E. coli to commercially available hydrogen peroxide was tested and no differences were observed (data not shown).
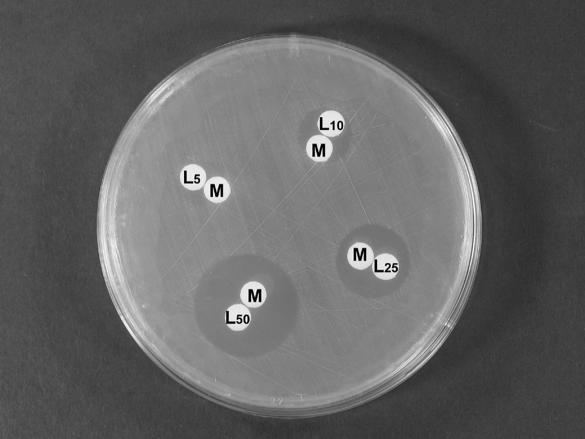
Next, the effects of the addition of different protein hydrolysates and biological extracts (bacteriological peptone, soya peptone, tryptone, casein, yeast extract, or Casamino Acids) to a basal M9 medium suggested that an amino acid present in complex media could be involved in the generation of hydrogen peroxide. The screening of the different protein amino acids revealed that l-lysine was the only amino acid required for marinocine to show its antibacterial effect. This effect was dependent on the amount of l-lysine added to the medium (Fig. 5). Other compounds related to l-lysine, such as l-arginine, l-ornithine, d-lysine, and polyamines (putrescine, cadaverine, spermine, spermidine, 1,6-hexanodiamine) were assayed under the same conditions with disks loaded with 20 μl at a 50 mM concentration. However, none of these compounds were able to induce the antimicrobial effect of marinocine.
FIG. 5.
Positive effect of l-lysine presence on marinocine activity against E. coli UM202 in antibiograms in M9 medium. Disks M contained 4.5 U of marinocine, while disks L contained 20 μl of l-lysine at the indicated millimolar concentrations.
l-Lysine oxidase activity of purified marinocine.
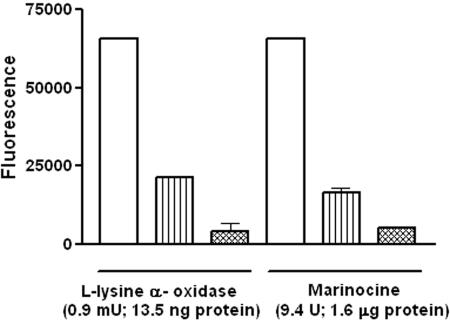
The above-mentioned results, and the fact that the antibacterial effect of marinocine was inhibited by catalase, suggested that marinocine is able to oxidize l-lysine to generate hydrogen peroxide. To test this possibility, l-lysine oxidase activity was determined using a fluorimetric assay in which the commercially available l-lysine α-oxidase from T. viride was included as a positive control. It was observed that in the presence of l-lysine, both marinocine and l-lysine oxidase from T. viride generated hydrogen peroxide (Fig. 6). This result confirms the l-lysine oxidase activity of marinocine and accounts for the mechanism of generation of hydrogen peroxide mediating its antibacterial effect. Comparison of the experimental data indicates that l-lysine α-oxidase from T. viride showed a much higher specific activity for the generation of hydrogen peroxide than did the M. mediterranea enzyme. In contrast, determination of the Km has given a value of 2 μM for marinocine, lower than the value of 40 μM reported for the Trichoderma enzyme (20). In additional experiments, it was observed that, like marinocine, the commercial l-lysine α-oxidase showed antibacterial activity in minimal medium only in the presence of l-lysine (data not shown).
FIG. 6.
Production of hydrogen peroxide by commercially available l-lysine α-oxidase and marinocine. Open bars represent assays in which 50 mM l-lysine was added as substrate. For vertical-striped bars, 0.1 mg/ml catalase was also added to the assay mixture. Cross-hatched bars represent control reactions with no l-lysine added. Since the definitions of enzymatic units are different for the two proteins (20, 28), the amounts of protein are provided to facilitate comparison between them.
Cloning of the gene coding for marinocine.
Thus far, there has been no report of the cloning of genes coding for l-lysine oxidase activity. However, the autolytic protein AlpP, synthesized by the marine bacterium P. tunicata (18), shows electrophoretic properties similar to those of marinocine (28). AlpP has been recently cloned and sequenced (30). Using the AlpP sequence as a query in a BLAST search (1) allowed us to detect several hypothetical proteins, including one from Saccharophagus (Microbulbifer) degradans strain 2-40 (accession number ZP_00318137). Interestingly, in this strain this gene is close to another gene that putatively encodes a laccase similar to PpoA from M. mediterranea (35). In order to explore the existence of a protein homologous to AlpP in M. mediterranea, a PCR-based protocol was used. Degenerate PCR primer mixtures were designed based on the sequences of some consensus regions detected in proteins similar to AlpP. After alignment (5), a total of eight conserved regions were found in all of those proteins, and it was considered that the sequences of regions 3, 7, and 8, shown in Table 2, provided the best conserved domains upon which to base our PCR primer designs.
After two rounds of PCR using a different set of primers (see Materials and Methods), a DNA fragment in the range of the expected size, ca. 1,300 bp, was successfully amplified; cloned in pGEM vector, generating plasmid pMAR28; and sequenced. Sequence analysis of this fragment showed that it encodes a protein with high similarity to the proteins used to design the primers, providing confidence that the fragment of the appropriate gene had been amplified.
In order to clone the complete gene coding for marinocine, plasmid pMAR28 was digested with SacI, eliminating ca. 250 nucleotides from the 5′ region of the PCR product, and NcoI, which cuts the pGEM sequence, and cloned into pFSVK, generating pFSVK-mar8. This vector was mobilized into M. mediterranea and, since it is a suicide vector unable to replicate in this microorganism, became integrated in the target gene by homologous recombination between the chromosome and the fragment generated by PCR. Transconjugants were selected by the kanamycin resistance encoded by the plasmid. The suicide vector inserted in the chromosome of strain MUTMAR2, one of those mutants, was rescued from the genomic DNA by digestion with the enzyme XbaI, which does not cut in the plasmid, followed by religation and transformation of E. coli S17-1λpir. The plasmid obtained, pRECMAR4, contained a chromosomal region of 5,368 bp.
Sequence analysis of lodA and lodB.
The complete sequencing of the genomic fragment in pRECMAR4 revealed the presence of two adjacent open reading frames (ORFs) that appear to be organized in an operon-like fashion. The operon has been named lod, for l-lysine oxidase activity, the enzymatic activity shown by marinocine.
The first ORF, named lodA, is 2,181 bases in length. Translation analysis predicts a protein of 726 amino acids, with an expected molecular mass of 80.9 kDa. Neither a putative signal peptide nor a transmembrane region have been detected with, respectively, the Signal P program (version 3.0; Center for Biological Sequence Analysis, Technical University of Denmark [http://www.cbs.dtu.dk]) (10) and the Dense Alignment Surface method (Institute for Molecular Pathology, Vienna, Austria [http://mendel.imp.univie.ac.at]) (6, 7). These theoretical predictions suggest either that marinocine is released after cell death or that it is released by a nonclassical mechanism. lodA shows strong similarity to the genes used as patterns for degenerated primer design (Table 2). The highest identity scores were found with the antibacterial protein AlpP of P. tunicata (protein AAP73876, 52% identity) and with the predicted protein AAQ60932 of Chromobacterium violaceum (33% identity).
The second ORF, lodB, with a length of 1,110 bp, appears to start immediately downstream of lodA, at an ATG codon just 2 bp from the lodA stop codon. The deduced product of lodB is a protein of 369 amino acids with a predicted molecular mass of 41.4 kDa. LodB shows the putative conserved domain FixC, described in flavin-dependent dehydrogenases (COG0644.1) (31).
No other genes seem to form part of the same operon. In the 5′ end of the fragment cloned in pRECMAR4, an incomplete ORF coding for a hypothetical protein is detected. However, this ORF is 385 bp apart from lodA. On the other hand, 18 bp downstream from lodB we found a palindromic sequence (TAAACGAAGAGAGCCGACGATTGCCGGAGGCTCCTTTCGTTTA) that might act as putative transcriptional termina-tor. In addition, no other ORF was detected in the 1,228-bp region cloned downstream of lodB.
As for the genetic features controlling transcription and translation of this operon, a typical ribosome binding site, AGGAG, is located 6 bp upstream of the initial codon of lodA. The promoter region is less clearly identified, although a putative −35 region, TTGCTC, and a −10 region, TATAAA, are detected. The AT content from the putative −10 region to the ribosome binding site is high (71.4%), as expected for this region.
Identity of marinocine with the product of lodA.
The correlation between lodA and marinocine in M. mediterranea was achieved by sequencing the N-terminal end of the protein responsible for the inhibitory effect. The active protein was purified as previously described (28) and run in 8% SDS-PAGE under denaturing conditions. Under these conditions, two bands, at 97 and 185 kDa, were observed; it was postulated that the 185-kDa band was a dimer of the smaller species (28). The sample was then blotted onto a polyvinylidene difluoride membrane, and the proteins were subjected to N-terminal sequencing. The amino acid sequence of the 97-kDa band was LALSVHPS. Comparison of this sequence with the translation of lodA, MALSVHPS, indicated the correspondence between the antibacterial protein and LodA. Regarding the codon AUG being translated as Leu for the beginning of translation, to our knowledge there is no report of this feature for other proteins. The discrepancy between the experimental and predicted sequ-ences was repeatedly confirmed by sequencing, but the cause of this result is unknown.
To confirm that lodA encodes the marinocine protein, strain SB1, mutated in this gene, was created by insertional mutagenesis (see Materials and Methods and Fig. 1). The pFSVK-SS vector was inserted into the MMB-1R genome by homologous recombination. In the mutant SB1, lodA was split into two fragments, each of which lacked at least three of the eight conserved regions in this protein, as identified by BLAST with homologous proteins. Next, lysine oxidase activity and antimicrobial activity were determined for the SB1 strain. It was observed that this strain shows neither lysine oxidase activity nor antimicrobial activity, confirming the identity of LodA as marinocine.
Lysine oxidase activity was also determined in different strains with mutations in structural genes coding for PPO activities. Strain T101 is affected in laccase activity (40), and strains T105 and ng56 are affected in tyrosinase activity (25, 39). It was observed that, in agreement with previous reports indicating that these strains produce marinocine (28), they also showed lysine oxidase activity. In contrast, regulatory gene mutants such as strain T103, affected in PpoS, a sensor histidine kinase regulating PPO activities (26), and other strains with uncharacterized mutations affecting the regulation of PPO activities, such as ngC1, T102, MIT1, and MIT2 (16), showed neither lysine oxidase nor marinocine activity.
DISCUSSION
M. mediterranea synthesizes a broad-spectrum antimicrobial protein named marinocine. Some characteristics of this protein have been recently reported (28). In this study, we describe the molecular properties responsible for its antimicrobial activity and the cloning of the gene coding for that protein.
The inhibitory effect of marinocine was detectable only when antibiograms were performed on media containing l-lysine and under aerobic conditions. It has been shown that marinocine has l-lysine oxidase activity, generating hydrogen peroxide as a product of the reaction. The antimicrobial activity of marinocine is due to this hydrogen peroxide, as judged by the protective role of catalase and the increased sensitivity toward marinocine and hydrogen peroxide of catalase mutant strains. The generation of hydrogen peroxide as a mechanism of microbial competition has been studied mainly in lactic acid bacteria (23). It is a mechanism employed to inhibit the growth of other microorganisms, not only by those involved in food fermentation but also by pathogenic microorganisms such as Streptococcus pneumoniae when growing in the upper respiratory tract (34). Hydrogen peroxide is a small molecule that diffuses rapidly, has high membrane permeability, and is able to affect a wide variety of microorganisms. This mechanism of action could explain the wide range of activity observed for marinocine (28), and it can be an advantage in environments such as biofilms, in which a complex microbial community competes for nutrients. In fact, the protein with highest similarity to marinocine, AlpP from P. tunicata, is involved in biofilm development by the producer strain (30).
The cloning of the gene responsible for marinocine was achieved by degenerated PCR using primers designed based on the published sequences of AlpP from P. tunicata and similar hypothetical proteins. A gene in M. mediterranea coding for a protein, LodA, with high similarity to those proteins was detected. The N-terminal sequence of the translation product of lodA coincides with the results obtained from the sequencing of purified marinocine in SDS-PAGE. This result strongly indicates the identity of LodA with marinocine. Moreover, the directed mutation of lodA by homologous recombination also suppressed marinocine and lysine oxidase activities. lodA is followed by a second gene, lodB, that putatively codes for a flavin-dependent dehydrogenase. The separation of both genes by only 2 bp strongly suggests that they are in the same transcriptional unit. This is also supported by the fact that, according to the published sequences of the genomes of the microorganisms in Table 2, proteins similar to lodA and lodB are always contiguous in the genome.
The results presented in this work confirm our previous observations that PPOs expressed by M. mediterranea are not directly involved in antimicrobial activity (28), since mutans of the structural genes coding for those enzymes maintain lysine oxidase activity. However, all five strains isolated by our group that have been previously characterized as being affected in the regulation of PPO activities and melanogenesis (16, 26) are also affected in the synthesis of marinocine. It has also been shown that expression of the antimicrobial protein and pigmentation are coregulated in P. tunicata (11). The molecular mechanisms coordinating the expression of these characteristics in M. mediterranea remain to be determined, but at least a two-component regulatory system may be involved since one of these strains, T103, is mutated in the gene coding for PpoS, a sensor histidine kinase (26). The common regulation by PpoS of PPO activities and marinocine activity could offer a competitive advantage, since melanins are known free radical scavengers and could protect the producer strain from the oxidative stress caused by synthesis of the autotoxic compound.
A BLAST search performed with the sequence of marinocine as a query reveals that, as previously indicated, the most similar protein is AlpP from P. tunicata, whose antimicrobial activity has been demonstrated previously (18). All other proteins detected and shown in Table 2 are hypothetical. The degree of distribution of this kind of antimicrobial protein in other microorganisms is not known, but some antimicrobial macromolecular compounds produced by other marine bacteria closely resemble the properties of marinocine in terms of increasing oxygen consumption and the inhibitory effect of catalase (12, 13).
The substrate of marinocine is the amino acid l-lysine. This raises the question of whether the enzymatic activity of marinocine is involved in the metabolism of l-lysine in M. mediterranea. However, strain SB1, mutated in marinocine activity, is able to grow in minimal medium, indicating its capacity to synthesize l-lysine. It has also been observed that neither the wild type nor the mutant is able to use l-lysine as the sole carbon and energy source, but both of them can use it as the sole nitrogen source (unpublished results). These results suggest that marinocine is not involved in l-lysine metabolism, although this possibility cannot be completely ruled out since alternative pathways may exist in a single microorganism.
Our study is the first in which the gene coding for an enzyme with l-lysine oxidase activity has been cloned. l-Lysine α-oxidase activity has been widely detected, together with other l-amino acid oxidases, in the venoms of several snakes (43). However, most of the studies of this activity have been performed with enzymes produced by fungi of the genus Trichoderma. The first of these studies reported the antineoplastic properties of the enzyme from T. viride (20). Subsequent works on enzymes with similar catalytic properties in other Trichoderma species describe a wide range of biological properties, mainly antibacterial, antiviral, immunomodulating, and antitumor effects (29). Technical uses to detect l-lysine in biological samples have also been proposed (37). In mammals, this enzymatic activity has been described in the mouse brain, where lysine catabolism occurs via the pipecolic acid pathway (33). However, as far as we know, the genes coding for those activities have never been reported, and hence it is not possible to assess the degree of similarity to lodA. As previously mentioned, when the sequence of marinocine was used as a BLAST query, only some bacterial proteins were detected. l-Lysine α-oxidase from Trichoderma and marinocine are similar in terms of the generation of hydrogen peroxide, which determines their antimicrobial activity. However, there are some differences in the catalytic properties between them. It has been observed that lysine oxidase from Marinomonas shows lower specific activity but a higher affinity for the substrate than the fungal enzyme. This could be related to the different environments that the microorganisms occupy. In marine waters, the estimated concentration of free amino acids falls in the range from 0.1 to 50 nM (42), and hence the catalytic properties of marinocine seem to be adapted to those conditions of low substrate availability. An additional difference is that l-amino acid oxidases and l-lysine α-oxidase from Trichoderma are enzymes containing flavin adenine dinucleotide as the coenzyme. In previous studies, marinocine did not show any absorbing band in its UV-visual spectrum peak that could indicate the presence of those aromatic coenzymes in the molecule (28). Further studies are being carried out to compare the physicochemical properties and enzymatic activities of both proteins.
This study shows the novel molecular basis of the antibacterial activity of marinocine and the cloning of the gene coding for this protein with l-lysine oxidase activity. Marinocine has homology with proteins distributed among different bacterial genera. For one of the organisms, P. tunicata, a role in biofilm dispersal has been proved (30). In addition, the antibacterial activity of the protein could play a central role in bacterial interactions and competition in natural environments. Additional research into these kinds of proteins is needed to elucidate their physiological roles and to explore possible applications in relation to their antimicrobial activities.
Acknowledgments
This work was supported by grant BIO2004-4803 from the Spanish MEC.
We are grateful to P. C. Loewen (University of Manitoba, Canada), who kindly supplied the E. coli strains mutated in different catalase genes, and to S. Kjelleberg (University of New South Wales, Australia), for critical reading of the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., and O. Schneewind. 1998. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 6:66-71. [DOI] [PubMed] [Google Scholar]
- 3.Bliska, J. B., K. L. Guan, J. E. Dixon, and S. Falkow. 1991. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc. Natl. Acad. Sci. USA 88:1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burchard, R. P., and M. L. Sorongon. 1998. A gliding bacterium strain inhibits adhesion and motility of another gliding bacterium strain in a marine biofilm. Appl. Environ. Microbiol. 64:4079-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corpet, F. 1988. Multiple sequence alignment with hierarchical-clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in procariotic membrane proteins: the Dense Alignment Surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 7.Cserzo, M., F. Eisenhaber, B. Eisenhaber, and I. Simon. 2002. On filtering false positive transmembrane protein predictions. Protein Eng. 15:745-752. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 9.Du, X. Y., and K. J. Clemetson. 2002. Snake venom l-amino acid oxidases. Toxicon 40:659-665. [DOI] [PubMed] [Google Scholar]
- 10.Dyrlov Bendtsen, J., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 11.Egan, S., S. James, and S. Kjelleberg. 2002. Identification and characterization of a putative transcriptional regulator controlling the expression of fouling inhibitors in Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 68:372-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauthier, M. J. 1976. Modification of bacterial respiration by a macromolecular polyanionic antibiotic produced by a marine Alteromonas. Antimicrob. Agents Chemother. 9:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier, M. J., and G. N. Flatau. 1976. Antibacterial activity of marine violet-pigmented Alteromonas with special reference to the production of brominated compounds. Can. J. Microbiol. 22:1612-1619. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E. W., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez-Romero, D., P. Lucas-Elio, D. Lopez-Serrano, F. Solano, and A. Sanchez-Amat. 2003. Marinomonas mediterranea is a lysogenic bacterium that synthesizes R-bodies. Microbiology 149:2679-2686. [DOI] [PubMed] [Google Scholar]
- 17.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James, S. G., C. Holmstrom, and S. Kjelleberg. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 62:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiri, N., G. Archer, and M. W. Climo. 2002. Combinations of lysostaphin with beta-lactams are synergistic against oxacillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 46:2017-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusakabe, H., K. Kodama, A. Kuninaka, H. Yoshino, H. Misono, and K. Soda. 1980. New anti-tumor enzyme, l-lysine alpha-oxidase from Trichoderma viride, purification and enzymological properties. J. Biol. Chem. 255:976-981. [PubMed] [Google Scholar]
- 21.Lazdunski, C. J., E. Bouveret, A. Rigal, L. Journet, R. Lloubes, and H. Benedetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy, O. 2004. Antimicrobial proteins and peptides: anti-infective molecules of mammalian leukocytes. J. Leukoc. Biol. 76:909-925. [DOI] [PubMed] [Google Scholar]
- 23.Lindgren, S. E., and W. J. Dobrogosz. 1990. Antagonistic activities of lactic-acid bacteria in food and feed fermentations. FEMS Microbiol. Rev. 87:149-163. [DOI] [PubMed] [Google Scholar]
- 24.Loewen, P. C., J. Switala, and B. L. Triggsraine. 1985. Catalases HpI and HpII in Escherichia coli are induced independently. Arch. Biochem. Biophys. 243:144-149. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Serrano, D., F. Solano, and A. Sanchez-Amat. 2004. Identification of an operon involved in tyrosinase activity and melanin synthesis in Marinomonas mediterranea. Gene 342:179-187. [DOI] [PubMed] [Google Scholar]
- 26.Lucas-Elío, P., F. Solano, and A. Sanchez-Amat. 2002. Regulation of polyphenol oxidase activities and melanin synthesis in Marinomonas mediterranea: identification of ppoS, a gene encoding a sensor histidine kinase. Microbiology 148:2457-2466. [DOI] [PubMed] [Google Scholar]
- 27.Lucas-Elío, P. 2003. Ph.D. thesis. Universidad de Murcia, Murcia, Spain.
- 28.Lucas-Elío, P., P. Hernandez, A. Sanchez-Amat, and F. Solano. 2005. Purification and partial characterization of marinocine, a new broad-spectrum antibacterial protein produced by Marinomonas mediterranea. Biochim. Biophys. Acta 1721:193-203. [DOI] [PubMed] [Google Scholar]
- 29.Lukasheva, E. V., and T. T. Berezov. 2002. l-Lysine alpha-oxidase: physicochemical and biological properties. Biochemistry (Moscow) 67:1152-1158. [DOI] [PubMed] [Google Scholar]
- 30.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. Q. He, D. I. Hurwitz, J. D. Jackson, Z. X. Ke, C. J. Lanczycki, C. A. Liebert, C. L. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. C. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy, S. A., R. M. Johnson, and D. Kakimoto. 1994. Characterization of an antibiotic produced by Alteromonas luteoviolacea Gauthier 1982, 85 Isolated from Kinko Bay, Japan. J. Appl. Bacteriol. 77:426-432. [DOI] [PubMed] [Google Scholar]
- 33.Murthy, S. N., and M. K. Janardanasarma. 1999. Identification of l-amino acid/l-lysine alpha-amino oxidase in mouse brain. Mol. Cell. Biochem. 197:13-23. [DOI] [PubMed] [Google Scholar]
- 34.Pericone, C. D., K. Overweg, P. W. M. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Amat, A., P. Lucas-Elío, E. Fernandez, J. C. Garcia-Borron, and F. Solano. 2001. Molecular cloning and functional characterization of a unique multipotent polyphenol oxidase from Marinomonas mediterranea. Biochim. Biophys. Acta 1547:104-116. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Amat, A., and F. Solano. 2005. Genus III. Marinomonas, p. 284-289. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York. N.Y.
- 37.Saurina, J., S. Hernandez-Cassou, S. Alegret, and E. Fabregas. 1999. Amperometric determination of lysine using a lysine oxidase biosensor based on rigid-conducting composites. Biosens. Bioelectron. 14:211-220. [DOI] [PubMed] [Google Scholar]
- 38.Schellhorn, H. 1994. Regulation of hydroperoxidase (catalase) expression in Escherichia coli. FEMS Microbiol. Lett. 131:113-119. [DOI] [PubMed] [Google Scholar]
- 39.Solano, F., E. Garcia, E. P. de Egea, and A. Sanchez-Amat. 1997. Isolation and characterization of strain MMB-1 (CECT 4803), a novel melanogenic marine bacterium. Appl. Environ. Microbiol. 63:3499-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solano, F., P. Lucas-Elío, E. Fernandez, and A. Sanchez-Amat. 2000. Marinomonas mediterranea MMB-1 transposon mutagenesis: isolation of a multipotent polyphenol oxidase mutant. J. Bacteriol. 182:3754-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stiles, B. G., F. W. Sexton, and S. A. Weinstein. 1991. Antibacterial effects of different snake-venoms—purification and characterization of antibacterial proteins from Pseudechis australis (Australian king brown or Mulga snake) venom. Toxicon 29:1129-1141. [DOI] [PubMed] [Google Scholar]
- 42.Williams, P. 2000. Heterotrophic bacteria and the dynamics of dissolved organic material, p. 153-200. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley, New York, N.Y.
- 43.Zhang, H. M., Q. Y. Yang, M. X. Sun, M. K. Teng, and L. W. Niu. 2004. Hydrogen peroxide produced by two amino acid oxidases mediates antibacterial actions. J. Microbiol. 42:336-339. [PubMed] [Google Scholar]