Abstract
Whole-genome shotgun sequencing was used to study the sequence variation of three Pseudomonas aeruginosa isolates, two from clonal infections of cystic fibrosis patients and one from an aquatic environment, relative to the genomic sequence of reference strain PAO1. The majority of the PAO1 genome is represented in these strains; however, at least three prominent islands of PAO1-specific sequence are apparent. Conversely, ∼10% of the sequencing reads derived from each isolate fail to align with the PAO1 backbone. While average sequence variation among all strains is roughly 0.5%, regions of pronounced differences were evident in whole-genome scans of nucleotide diversity. We analyzed two such divergent loci, the pyoverdine and O-antigen biosynthesis regions, by complete resequencing. A thorough analysis of isolates collected over time from one of the cystic fibrosis patients revealed independent mutations resulting in the loss of O-antigen synthesis alternating with a mucoid phenotype. Overall, we conclude that most of the PAO1 genome represents a core P. aeruginosa backbone sequence while the strains addressed in this study possess additional genetic material that accounts for at least 10% of their genomes. Approximately half of these additional sequences are novel.
Pseudomonas aeruginosa is a ubiquitous environmental bacterium that is of increasing importance as an opportunistic human pathogen. P. aeruginosa infections are common in patients with compromised antibacterial defenses. A particular niche for infections that has been created by advances in pediatric care is the lungs of cystic fibrosis (CF) patients. As effective management of other clinical complications has been introduced and life expectancy has increased, chronic pulmonary infections with P. aeruginosa have emerged as the leading cause of morbidity and mortality in CF (11, 17). Despite steady improvements in treatment, CF pulmonary infections resist eradication with antibiotics and lead, over a period of many years, to irreversible tissue destruction (11).
Available data suggest that most CF patients are infected by unique strains of P. aeruginosa acquired from environmental reservoirs (15, 30, 31) and that the infections are frequently clonal (5, 29, 36, 39). During the time that a particular strain is resident in the airways of a CF patient, considerable genetic adaptation occurs. A well-known example of this process is the conversion of strains from nonmucoid to mucoid phenotypes due to mutations in mucA (11, 21, 22), a gene whose product is a negative regulator of the biosynthesis of the secreted polysaccharide alginate. Another example is the frequent loss of O antigen (8, 14, 18, 28, 29), a change that also appears to be due to mutation.
Restriction mapping of P. aeruginosa strains from diverse backgrounds indicates that a significant proportion of the variation among isolates is due to insertions and deletions of up to 500 kbp of genomic material (30, 34). In this respect, P. aeruginosa is superficially similar to Escherichia coli. Comparison of the pathogenic O157 and the nonpathogenic K-12 E. coli strains has revealed that much of their difference can be attributed to blocks of DNA that are strain specific (26). These so-called K islands and O islands comprise 12% of the genome of K-12 and 26% of the genome of O157, respectively, and are interspersed in a conserved backbone sequence with relatively little polymorphism (26). In P. aeruginosa, few strain-specific regions have been well characterized. One example is the 50-kbp P. aeruginosa genomic island 1, PAGI-1, found in many clinical isolates of P. aeruginosa in place of the 7-kbp of PAO1 sequence and hypothesized to play a role in evading the host immune response (20). Other examples include a 20-kbp island found in P. aeruginosa strain PAK, containing genes involved in glycosylation of a-type flagellin (2), and the recently described 11 groups of gene clusters at the O-antigen biosynthetic locus (27). However, despite the evidence indicating that genomic islands may play an important role in P. aeruginosa biology, little is known about the sequence and location of other islands.
Sequence-based studies of genetic variation in P. aeruginosa appear to support the presence of conserved backbone sequences, similar to the E. coli model. Sequencing of six housekeeping genes in 19 environmental and clinical isolates revealed levels of genetic diversity even lower than in the E. coli comparisons, with average pairwise nucleotide substitution rates on the order of a few tenths of a percentage point (15). These data, along with those from restriction-mapping experiments, suggest that P. aeruginosa genomes have numerous strain-specific regions interspersed in a well-conserved backbone. More detailed studies of DNA sequence variation in P. aeruginosa are needed to define these backbone and strain-specific sequences, which may promote our understanding of the genetic determinants of pathogenicity in CF lung infections.
The recent sequencing of the genome of the standard laboratory strain of P. aeruginosa, PAO1 (37), has laid the groundwork for more extensive studies of genomic variation in clinical and environmental isolates of this bacterium. We have carried out whole-genome-sample sequencing of two P. aeruginosa strains isolated from late-stage CF-related infections and a strain from an environmental source. These data provide a detailed view of the pattern of sequence variation among the three new strains relative to the PAO1 reference.
MATERIALS AND METHODS
Isolation of clinical strains.
All samples were processed at Children's Hospital and Regional Medical Center in Seattle. Oropharyngeal specimens were obtained using a cotton-tipped swab to collect the specimen from the posterior oropharyngeal wall and tonsillar pillars. Oropharyngeal samples were inoculated directly onto selective media. Bronchoalveolar lavage fluid and sputum were cultured using selective media as previously described (5). The media from which P. aeruginosa were isolated were MacConkey and cetrimide agars. Organisms were identified by standard techniques including the use of a biochemical panel for the identification of lactose nonfermenters (5). All isolates were taken from the primary culture plate and frozen at −80°C in sterile skim milk. The Institutional Review Board at Children's Hospital and Regional Medical Center, Seattle, Wash., approved this protocol.
Whole-genome fingerprinting.
Chromosomal DNA of each isolate was extracted from overnight 20-ml Luria broth cultures by sodium dodecyl sulfate lysis and phenol-chloroform extraction followed by ethanol precipitation and spooling with a glass rod (20). Restriction enzyme digestions of 2.5 to 3.5 μg of DNA using 20 to 28 U of the enzyme SfoI (New England Biolabs, Beverly, Mass.) were performed at 37°C for 2 to 4 hs. Reactions were run on 1% agarose gels at 4°C for 18 to 22 h at 150 V, and the products were stained with SYBR Green (Molecular Probes Inc., Eugene, Oreg.). Images were captured using a Fluorimager (Molecular Dynamics, Foster City, Calif.).
Strain selection and whole-genome-sequence sampling.
P. aeruginosa clinical strains 1-60 and 2-164 were selected as typical isolates present in the lungs of two CF patients in an advanced stage of a clonal infection, as determined by whole-genome fingerprints. Strain MSH, donated by Stephen Lory, was collected near Mount St. Helens and selected to represent a typical environmental isolate. Genomic libraries were made by sonication of chromosomal DNA and subsequent ligation into either M13 or pBluescript vectors. Sequence traces from each library were generated using primarily dye-terminator sequencing chemistry. For artificial sampling of PAO1, 11,609 traces were randomly selected from the PAO1 sequencing project (37). All four sets of individual traces were analyzed using the phred/phrap software package (9, 10).
PAO1 coverage analysis.
Sequence data from strains 1-60, 2-164, MSH, and the PAO1 trace simulation were compared to the known PAO1 genome with cross_match (http://bozeman.mbt.washington.edu/phrap.docs/phrap.html), using its default parameters. The midpoint of the best hit for each trace and phred quality scores for aligned bases were used to determine Q20 (sequencing base calls with an error rate of less than 1%) coverage of the PAO1 genome in 5,000-bp nonoverlapping windows. The PAO1 simulation data were adjusted to approximate the sequence data from the three sampled strains. The number of matching simulation traces was reduced to equal the average number of traces from the sampled strains that could be aligned with the PAO1 genome (7,821 traces). The numbers of Q20 bases in simulation traces were multiplied by a factor to reduce the average number of Q20 bases per trace for the entire simulation set to a value approximately equal to the average number of Q20 bases per trace in the sequence-sampling data sets (385 bp). To create a completely random coverage model, a random-number generator was used to select 7,821 positions in the PAO1 genome around which 385-bp hypothetical traces were centered. Coverage in Q20 bases was calculated using the same nonoverlapping-window approach with these simulated data. Total genome coverage for all four calculations was estimated by averaging the coverage values for each of the 1,253 windows. Starting and ending coordinates of each match in all cases were used to calculate coverage gaps.
Annotation of strain-specific sequences.
Individual sequence traces with no match to the complete PAO1 genome by the above methods were extracted from the data sets of the three sampled strains. These traces were subjected to a filtering process which included removing those with the following attributes; (i) fewer than 140 bases of quality Q20 or greater, (ii) more than 75% vector-masked bases, (iii) hits to the E. coli K-12 genome with less than 5% mismatches, and (iv) any hits to eukaryotic repetitive elements found by Repeatmasker. The remaining sequences were submitted to the NR protein database via the BLASTX query translation program. Only the single hit with the lowest expect value for each entry was kept for further analysis.
Identification and analysis of SNPs from pairwise strain comparisons.
The traces for each strain and the PAO1 simulation were assembled with phrap. The resulting contigs and single traces were compared to the PAO1 genome, and to each other, using cross_match. The default values were used for all of the cross_match parameters. Single-nucleotide polymorphism (SNPs) were identified from mismatches in the alignments produced by cross_match. To avoid false-positive results due to sequencing errors, mismatches were called as SNPs only if they satisfied the following criteria. (i) Each nucleotide within a 5-bp window centered at the mismatch must have a quality value equal to or greater than Q25 (sequencing base calls with an error rate of less than 0.3%). The quality value may be a phrap quality in the case of contigs or a phred quality value for single traces. (ii) There can be no insertion or deletion within the 5-bp window. (iii) There can be no additional mismatches within the 5-bp window.
To verify the accuracy of this SNP identification method, the algorithm was tested in two ways. First, the complete PAO1 genome was compared to the randomly selected set of PAO1 traces. The substitution rate between these sequences was expected to approach 0% as the quality threshold and window size were increased. With a window size of 5 bp and a quality threshold of Q25, 655 mismatches were found in 3,082,441 bases analyzed, a 0.0204% error rate. Second, mutations were randomly introduced into the known PAO1 sequence to produce an artificial genome with a SNP rate of 0.5% relative to PAO1. The SNP finding algorithm was applied to a comparison between this mutated genome and the partial coverage supplied by the randomly selected PAO1 traces to recover the introduced SNPs. The simulation yielded 3,021,197 bases that met the predetermined quality criteria (Q25 quality and a window size of 5 bp). Mismatches were found at 15,640 of these positions, which is a SNP rate of 0.518%. On verification with the correct PAO1 sequence, 639 of these mismatches were errors and 15,001 were true SNPs (i.e., 96% of detected SNPs were real). Each SNP was entered into a database and classified according to the specific base pair difference. These data were used to quantify the frequency of transition versus transversion base pair substitutions. A complete listing of the SNPs that were found is available on the internet at www.genome.washington.edu/UWGC. Histograms of percent nucleotide substitution of comparisons between the complete PAO1 genome and the three sampled strains were created using the above algorithm applied to 5,000- bp windows that overlap by 2,500 bp along the PAO1 genome.
The analysis used to identify regions of high sequence variation was derived from a SNP analysis of 5-kbp windows of the PAO1 genome, offset by 2.5 kbp, compared to the three partially sequenced strains. Windows with at least 500 bp of alignable sequence and a SNP value of more than 3 standard deviations from the mean (≥4.35%) are reported, as are the exact number of SNPs, alignable bases, and percent alignable bases with SNPs. The G+C content is reported for the entire 5,000-bp window. The genome-wide G+C content for PAO1 is 66.56%, with a minimum 5-kbp window of 47.64%, a maximum of 72.96%, and a 95% confidence interval of 60.73 to 72.38%. The GC skew was calculated as described by Grigoriev (12). All annotated open reading frames (ORFs) that overlap regions of high variation are listed, and more detail on these ORFs is available at www.pseudomonas.com.
Yeast recombinational cloning.
The O-antigen gene clusters from clinical strains 1-60 and 2- 164 were isolated with plasmid pEHS4, as described previously (27). The pyoverdine clones were retrieved in a similar manner, as well as, in some cases, by fosmid cloning. For the yeast recombinational cloning, the targeting sequences used were from PAO1 sequence coordinates 2638501 to 2639000 and from 2689501 to 2690000. Recombinant clones were detected using primers 5′ CGAGCTCATCGCTAATAACTTCGTA 3′ and 5′ CTCAGCGACACCCTGCTGTCGGTGC 3′ for the upstream side and primers 5′ TATAGCACGTGATGAAAAGGACCGC 3′ and 5′ CTTCAAGCGTCCCGACGGCGAGTTC 3′ for the downstream side.
Phenotypic and genotypic analysis of O antigen in patient 1 isolates.
The presence of the insertional element in the clinical strain 1-60 O-antigen biosynthetic region was detected using PCR with primers 5′CGGCATAGCCTTGTTGACTT 3′ and 5′GCGACCAAACCTTTTGGATT 3′. PCR reagents used were from the Advantage GC kit for high-G+C templates (Clontech Laboratories, Palo Alto, Calif.). Products were visualized using standard gel electrophoresis techniques. Serotyping of clinical strains was performed with an O1-specific monoclonal antibody (ERFA, Westmount, Quebec, Canada) as described previously (27).
PCR amplification and DNA sequencing of mucA, and examination of isolates for a mucoid phenotype.
The primers used for mucA amplification were 5′CTCGTGAAGCAATCGACAAA 3′ and 5′ AAAAGCAACAGGGAGGTGGT 3′. Reagents used for these reactions were the same as above. Products were visualized using standard gel electrophoresis techniques and then treated with shrimp alkaline phosphatase and exonuclease enzymes (USB, Cleveland, Ohio) for DNA sequencing with the above primers, using dye terminator chemistry. Sequencing reactions were purified via ethanol precipitation and run on an ABI PRISM 377XL automated sequencer (Applied Biosystems, Foster City, Calif.). All of the isolates were examined for a mucoid phenotype by the same experienced clinical microbiologist at Children's Hospital and Regional Medical Center after overnight growth on cetrimide agar.
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in GenBank under accession numbers AF540990, AF540991, AF540992, and AF540993.
RESULTS
Whole-genome-sequence sampling of late-stage P. aeruginosa isolates.
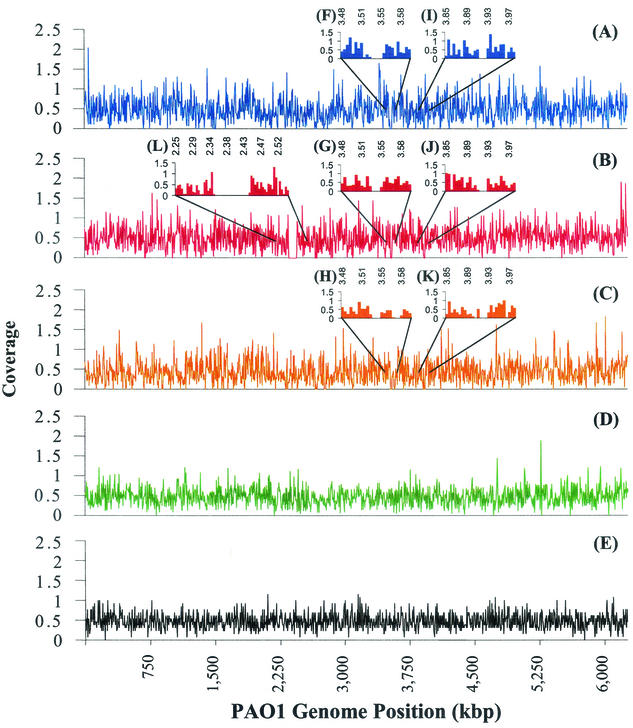
Two P. aeruginosa clinical isolates from CF patients (strain 1-60 from patient 1 and strain 2-164 from patient 2) and an environmental isolate MSH, were chosen for whole-genome-sequence sampling (Table 1). The average coverage of the three sampled genomes was approximately 0.5×, a value that implies a 40% chance of observing any particular base pair at least once. Figure 1 shows alignment histograms of the coverage across the PAO1 genome for each strain. These histograms conform reasonably well to expectations based on a random-sampling model, as illustrated by comparing Fig. 1A to C with the idealized histograms shown in Fig. 1D and E. Figure 1D illustrates variations in coverage of the PAO1 genome by a randomly selected set of PAO1 sequencing traces (37), while Fig. 1E illustrates variations in the coverage of the PAO1 genome in a random simulation.
TABLE 1.
Strains analyzed in this study
| Strain | Source | Reference | No. of sequencing traces | Genomic coverage in high-quality dataa |
|---|---|---|---|---|
| 1-60 | Isolated from CF patient 1 sputum at age 60 mo | 5 | 10,088 | 0.57× |
| 2-164 | Isolated from CF patient 2 sputum at age 164 mo | J. L. Burns, personal communication | 11,328 | 0.67× |
| MSH | Isolated from a lake on Mount St. Helens following the 1980 eruption | J. Staley, personal communication | 10,567 | 0.49× |
High-quality data are defined as base calls with a phred quality score of >20 (i.e., Q20 bases), which corresponds to a <1% error rate (9). Genomic coverage is estimated by dividing the total number of Q20 bases acquired by the 6.3- Mbp size of the PAO1 genome.
FIG. 1.
Histograms of the coverage of the PAO1 genome in high-quality data that could be aligned with the PAO1 reference sequence. (A to C) Data are from strains 1-60 (A), 2-164 (B), and MSH (C). Coverage was estimated by calculating the number of bases with a phred quality score of 20 or higher that could be aligned within 5-kbp windows across the PAO1 genome. (D to L) Inset plots indicate fine-grained views of coverage gaps at the O-antigen biosynthetic locus at PAO1 coordinates 3.53 to 3.55 Mbp (F to H), a 17-kbp deletion common to all three sampled strains relative to PAO1 at coordinate 3.91-mbp (I to K), and a 100-kbp gap in strain 2-164 at coordinate 2.4-mbp (L). (D) Coverage simulation using a random subset of sequencing traces from the PAO1 sequencing project. (E) Coverage histogram obtained in a pure simulation of random sampling to an average coverage of 0.48×.
Genomic regions unique to PAO1.
Large gaps in the coverage histograms of Fig. 1A to C are likely to represent segments of the PAO1 genome that lack alignable counterparts in one or more of the other strains. There are two sizeable segments of the PAO1 genome to which no sequencing traces could be aligned for any of the sampled strains. One of these coverage gaps encodes a set of enzymes required for O-antigen biosynthesis (Fig. 1F to H). An additional coverage gap of approximately 17 kbp occurs at the PAO1 coordinate at 3.9 Mbp in all three sampled strains (Fig. 1I to K). Through further experimental analysis via PCR, we established that this gap in coverage corresponds to a 16,635-bp deletion common to all three strains, relative to PAO1 (data not shown). None of the PAO1-specific genes in this region (PAO1 ORFs 3497 to 3514) are well characterized. In addition, we found a 100-kbp coverage gap in strain 2-164 at the PAO1 coordinate at 2.4 Mbp (Fig. 1L). The DNA missing from strain 2-164 includes a diverse set of 92 genes (PAO1 ORFs 2130 to 2221). The absence of this DNA was confirmed by PCR analysis of genomic DNA from strain 2-164 and all other isolates available for patient 2 (data not shown). One end of the gap corresponds to the insertion location of the previously described 50-kbp P. aeruginosa genomic island 1, PAGI-1 (20); however, none of the sequencing traces from strain 2-164 match the sequence of PAGI-1.
Genomic regions unique to the clinical and environmental isolates.
Genomic segments present in the genomes of the three sampled strains but not in PAO1 can be detected by analyzing the sequencing traces that do not align with the PAO1 genome. We found 910 traces from strain 1-60 (11.6% of all high-quality bases generated), 1,671 traces from strain 2-164 (17.8% of all high-quality bases) and 844 traces from strain MSH (10.2% of all high-quality bases) that fail to align, indicating that a substantial portion of the genome of each strain is composed of sequence tracts of at least a few hundred base pairs with no PAO1 counterparts. Strain 2-164, which carries a large and unique deletion relative to PAO1, has the highest proportion of unalignable reads. We attempted to annotate these unalignable sequences by comparing their translation products to the nonredundant protein database using BLASTX. Results from these similarity searches are summarized in Table 2.
TABLE 2.
Annotation of strain-specific sequencing traces
| Protein BLASTX hit category | No. (%) of traces for strain:
|
|||
|---|---|---|---|---|
| 1-60 | 2-164 | MSH | Combined | |
| P. aeruginosa, PAO1 genome | 32 (3.5) | 67 (4) | 34 (4) | 133 (3.9) |
| Elements from other P. aeruginosa strains | ||||
| PAGI-1 | 44 (4.8) | 5 (0.6) | 49 (1.4) | |
| O antigena | 164 (18) | 155 (9.3) | 12 (1.4) | 331 (9.7) |
| Flagellin glycosylation island | 17 (1.9) | 17 (0.5) | ||
| Other | 6 (0.7) | 51 (3.1) | 10 (1.2) | 67 (2.0) |
| Other Pseudomonas species | 20 (2.2) | 96 (5.7) | 27 (3.2) | 143 (4.2) |
| Xylella fastidiosa | 50 (5.5) | 122 (7.3) | 31 (3.7) | 203 (5.9) |
| Mezorhizobium loti | 25 (2.7) | 32 (1.9) | 15 (1.8) | 72 (2.1) |
| E. coli | 9 (1) | 42 (2.5) | 19 (2.3) | 70 (2) |
| Other bacteria | 76 (8.4) | 236 (14.1) | 43 (5.1) | 355 (10.4) |
| Mobile genetic elements (phage, plasmid) | 11 (1.2) | 60 (3.6) | 25 (3.0) | 96 (2.8) |
| Other | 2 (0.2) | 1 (0.1) | 3 (0.1) | |
| Traces with no BLASTX hits | 454 (49.9) | 810 (48.5) | 622 (73.7) | 1,886 (55.1) |
| Total no. of high quality traces unaligned with PAO1 | 910 | 1,671 | 844 | 3,425 |
Traces in this category correspond to the O-antigen type of each strain; type O1 for strains 1-60 and MSH, and type O6 for strain 2-164.
Of the combined total of 3,425 traces from the three sampled strains that did not align with the PAO1 genome, only 1,886 (55%) had identifiable similarity to known gene products. Hits to products of genes in the PAO1 genome account for 4% of the total for each strain; hence, there appear to be genes in these strains that have diverged significantly at the nucleic acid level from related PAO1 genes while still coding for similar proteins. Another prominent category of sequences encode proteins similar to those already reported in P. aeruginosa strains other than PAO1. This category includes reads that hit proteins involved in O-antigen synthesis. Also included in this category are hits to genes in the previously described 50-kbp P. aeruginosa genomic island 1 PAGI-1 (20) and a 20-kbp flagellin-related glycosylation island (2), the cytotoxin-converting phage phi CTX (24), and alternate forms of flagellar (3, 35) and pyocin (7, 33) genes. Hits to proteins encoded by other bacteria comprise a third category, dominated by proteins encoded by the plant pathogen Xylella fastidiosa and the nitrogen-fixing symbiont Mesorhizobium loti. The remaining traces, which comprise almost half of all traces that are unalignable with the PAO1 genome, do not encode peptides with matches to known proteins. This result suggests the presence of substantial novel genetic material in the three strains.
Levels of genetic variation within shared segments of the genome.
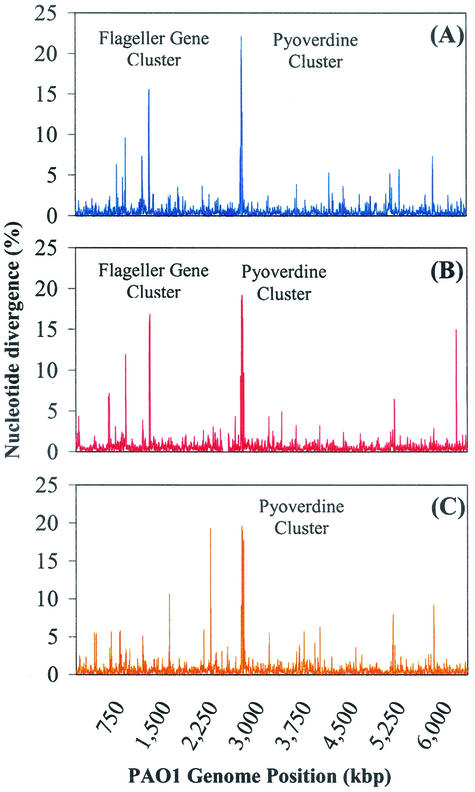
The data sets described in Table 1 provide insights into the genetic variation between shared regions of PAO1 and each of the comparison strains. Sequence analysis was performed between the PAO1 reference and high-quality comparison strain sequences that had an error rate of less than 1%. An overall assessment of SNPs revealed a ratio of transition to transversion mutations of 2.5:1. This is similar to the 3:1 ratio observed in strain comparisons of E. coli (26). With respect to transversions, 50% are G↔T or A↔C, 41% are G↔C, and 9% are A↔T. The apparent bias is presumably a reflection of the overall high G+C content of P. aeruginosa (66.56%) but may underscore an active mechanism that preserves the high G+C content in this organism. Figure 2 shows a whole-genome scan of genetic variation between PAO1 and each of the comparison strains. These histograms show the percent sequence divergence in 5-kbp windows that overlap by 2.5 kbp. Average sequence variation between PAO1 and strains 1-60, 2-164, and MSH was 0.49, 0.50, and 0.47%, respectively. We also attempted pairwise comparisons between 1-60, 2-164, and MSH. These estimates are more problematical because there is uncertainty about the precise boundaries of an alignable region. However, our best estimates suggest that no two of these strains are more closely related to each other than any of them is to PAO1.
FIG. 2.
Percent nucleotide mismatches of aligned sequence between PAO1 and strains 1-60 (A), 2-164 (B), and MSH (C) in sliding 5-kbp windows that overlap by 2.5 kbp.
Large peaks in the histograms in Fig. 2 represent regions in the PAO1 genome that are much more variable than the background level of 0.5% variation. The dramatic peak at the coordinate at 2.7 Mbp is particularly striking. This highly variable genome segment contains genes whose products are involved in the biosynthesis of the siderophore pyoverdine and the receptor for ferripyoverdine (e.g., pvdD, pvdE, pvdF, pvdI, pvdJ, pvdK, and fpva) (19, 38). The pyoverdine locus was studied in greater depth, as presented below. The variation peak at coordinate 1.2 Mbp in both clinical isolates includes a large number of genes whose products are involved in flagellar synthesis. These genes have been previously implicated in the biosynthesis of the P. aeruginosa serologically distinct a- and b-type flagellae (1, 35). Both the pyoverdine and flagellar peaks involve sequence divergences of approximately 20%, a value that approaches the threshold for sequence alignability. Hence, they represent a “gray zone” between normal levels of interstrain variation on the one hand and the complete loss of alignability on the other.
Overall, we detected 25 regions of significantly elevated sequence variation in pairwise comparisons between PAO1 and the three partially sequenced strains (Table 3). These regions are defined in units of 5,000-bp PAO1 segments that had at least 500 bp of alignable, high-quality reads (≥99% accuracy in base calls) from one of the three strains and where pairwise nucleotide differences exceeded 4.35% (more than 3 standard deviations above the median value). Interestingly, there were no obvious sequence features unique to these regions that flag them relative to the rest of the genomic sequence. The percentage of GC base pairs in these regions ranged from 63.56 to 66.87%, which is slightly lower but nonetheless similar to the genome- wide average of 66.56% GC base pairs. In addition, we did not detect any strand bias in G+C content (GC skew [12]). With respect to codon usage of ORFs in these regions, there appeared to be a lower G+C content in the synonymous third codon positions than the genome-wide average, as well as underutilization of optimal codons frequently used in highly expressed genes (13). Surprisingly, we did not identify exotoxin A (25) or homologs to the E. coli vgr/Rhs elements (6, 40) in our analysis. The highly polymorphic restriction fragment length polymorphisms upstream of the exotoxin A gene have been used for strain typing, and we found 1.5% nucleotide variation in this 10-kbp region. Similarly, the vgr/Rhs elements are associated with rearrangement hot spots in E. coli, yet we found unremarkable nucleotide substitution rates in the P. aeruginosa homologs that were examined.
TABLE 3.
PAO1 sequence coordinates for regions of high sequence diversitya
| Coordinates ofb | Length (bp)c | No. of SNPsd | No. of bases availablee | % SNPsf | PAO1 ORFsg | |
|---|---|---|---|---|---|---|
| PAO1 start | PAO1 end | |||||
| 290000 | 294999 | 5,000 | 83 | 1,488 | 5.58 | PA0259-PA0262 |
| 320000 | 324999 | 5,000 | 97 | 1,754 | 5.53 | PA0285-PA0289 |
| 515000 | 519999 | 5,000 | 143 | 2,103 | 6.80 | PA0457-PA0459 |
| 530000 | 534999 | 5,000 | 111 | 2,288 | 4.85 | PA0470-PA0473 |
| 555000 | 559999 | 5,000 | 66 | 1,252 | 5.27 | PA0495-PA0500 |
| 652500 | 657499 | 5,000 | 76 | 1,171 | 6.49 | PA0594-PA0596 |
| 685000 | 689999 | 5,000 | 44 | 789 | 5.58 | PA0625-PA0633 |
| 695000 | 699999 | 5,000 | 84 | 1,436 | 5.85 | PA0640-PA0643 |
| 790000 | 799999 | 10,000 | 274 | 2,535 | 10.81 | PA0719-PA0731 |
| 1060000 | 1064999 | 5,000 | 57 | 926 | 6.16 | PA0976-PA0982 |
| 1170000 | 1174999 | 5,000 | 224 | 1,881 | 11.91 | PA1084-PA1087 |
| 2150000 | 2154999 | 5,000 | 88 | 842 | 10.45 | PA1967-PA1972 |
| 2550000 | 2554999 | 5,000 | 59 | 1,346 | 4.38 | PA2312-PA2317 |
| 2635000 | 2654999 | 20,000 | 451 | 5,787 | 7.79 | PA2383-PA2397 |
| 2660000 | 2664999 | 5,000 | 115 | 551 | 20.87 | PA2399 |
| 2672500 | 2677499 | 5,000 | 186 | 1,132 | 16.43 | PA2402 |
| 2682500 | 2692499 | 10,000 | 139 | 1,723 | 8.07 | PA2402-PA2409 |
| 3647500 | 3652499 | 5,000 | 81 | 1,417 | 5.72 | PA3260-PA3264 |
| 4060000 | 4064999 | 5,000 | 85 | 1,695 | 5.01 | PA3624-PA3629 |
| 5037500 | 5042499 | 5,000 | 98 | 1,923 | 5.10 | PA4500-PA4503 |
| 5070000 | 5074999 | 5,000 | 162 | 2,030 | 7.98 | PA4526-PA4532 |
| 5097500 | 5102499 | 5,000 | 119 | 1,825 | 6.52 | PA4549-PA4554 |
| 5187500 | 5192499 | 5,000 | 91 | 1,561 | 5.83 | PA4625 |
| 5722500 | 5727499 | 5,000 | 162 | 1,966 | 8.24 | PA5084-PA5089 |
| 6090000 | 6094999 | 5,000 | 93 | 618 | 15.05 | PA5412-PA5415 |
Sequencing data for all three strains was compared to the PAO1 reference in 5,000-bp sliding windows that were sequentially offset by 2,500 bp. Regions with at least 500 bp of alignable sequence that exhibited nucleotide diversity values greater than three standard deviations (>4.35% sequence differences) from the mean value of 0.5% are shown.
Sequence coordinates of the PAO1 refence sequence, accessible at www.pseudomonas.com.
Overall span of the PAO1 region encompassed by high sequence variation.
Number of SNPs detected.
Total number of alignable bases in which SNPs were detected.
Percent SNPs among alignable bases.
Annotated ORFs within high-diversity regions. A more comprehensive description of these genes is available at www.genome.washington.edu/UWGC and at www.pseudomonas.com.
Clonal P. aeruginosa infections in two CF patients.
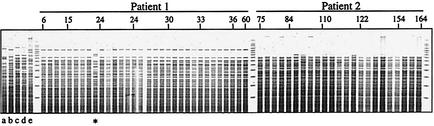
Sequential P. aeruginosa isolates were collected from the two CF patients for a period of years, starting at 6 months of age for patient 1 and 6 years 3 months of age for patient 2. Whole-genome restriction analysis was performed with the restriction enzyme SfoI (GGĈGCC). Figure 3 shows the restriction pattern of each isolate, together with strains MSH, PAO1, and three clinical isolates from separate CF individuals. With the exception of one strain from the patient 1 collection, all isolates from the same patient exhibit nearly identical fingerprints. Hence, it is likely that the infections in these patients are predominantly clonal, consistent with previous findings (5, 29, 36, 39). Close inspection reveals several restriction fragment length polymorphisms within isolates from a particular patient, a striking result consistent with our findings (reported below) that there is variation among patient isolates.
FIG. 3.
SfoI digestions of all P. aeruginosa strains from the patient 1 and patient 2 collections. For each patient, isolates are in historical sequence. Selected lanes are identified by the age (in months) of the patient at the time the isolates were cultured. The exceptional isolate from patient 1 is indicated by *. Additional digestions of PAO1 (a), MSH (b), and three clinical isolates from different CF patients (c to e) are indicated.
Cloning and resequencing of the pyoverdine and O-antigen biosynthetic loci.
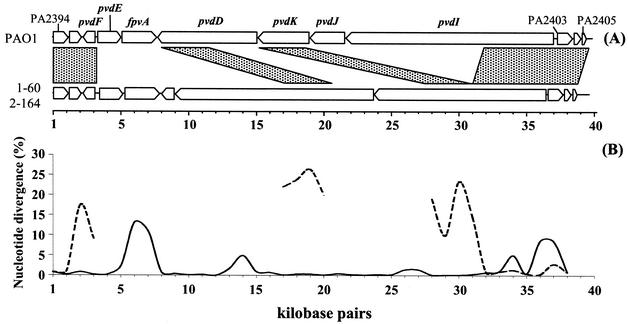
Our whole- genome variation scan led us to explore two highly variable regions among the P. aeruginosa genomes in more detail. The first was the pyoverdine segment, which stands out as the most prominent hypervariable feature in sequence alignments (Fig. 2). The majority of the pyoverdine locus from clinical strains 1-60 and 2-164 was cloned from fosmid libraries (a fosmid is related to a cosmid but has the single-copy F′ origin of replication [16]). Subsequently, we used targeted recombinational cloning in yeast (27) to isolate the ∼40-kbp pyoverdine regions from these isolates. Figure 4 shows the annotated sequences of the pyoverdine locus from PAO1 (ORFs PA2394 to PA2405) and from the two clinical strains. Surprisingly, the sequences of the two clinical strains are quite similar, with an overall nucleotide substitution rate of ∼2%. Most of the sequence variation between between these related clusters is found in the gene encoding the pyoverdine receptor, fpvA, and an ORF of unknown function, PA2403. Both clinical sequences differ by ≥25% from PAO1 across portions of the same region; sequence diversity is sufficiently high that alignments lose meaning in the central portions of the locus. Preliminary analysis using PCR suggest that the pyoverdine locus from the MSH environmental isolate is also similar to the that from the clinical strains (data not shown). Given that three major types of pyoverdine molecules are found among P. aeruginosa strains (reviewed in reference 23), our data suggest that PAO1 synthesizes one type of siderophore while the three strains used in this study all synthesize a different molecule and they have common sequences among their pyoverdine biosynthetic genes.
FIG. 4.
Gene structure of the pyoverdine locus. (A) PAO1 ORFs 2394 to 2405, and ORFs annotated from the pyoverdine regions of clinical strains 1-60 and 2-164. The shaded boxes indicate the alignable sequence conservation between the PAO1 reference and the pyoverdine synthesis region from strain 1-60. (B) Nucleotide divergence between strains 1- 60 and 2-164 (solid line) and between strains 1-60 and PAO1 (dashed line) in 1,000-bp nonoverlapping windows. Discontinuities in the 1-60/PAO1 comparison reflect the absence of alignable sequences throughout much of the region.
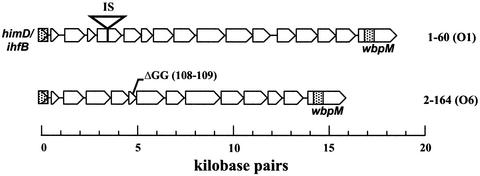
The second locus of interest, identified as a coverage gap in Fig. 1, is the O-antigen biosynthetic region. Bacterial lipopolysaccharide plays a critical role in the establishment of infection and subsequent interplay between the bacteria and the host immune system. Yeast recombinational cloning (27) was used to clone the O-antigen biosynthesis genes from clinical strains 1-60 and 2-164 (Fig. 5). DNA sequencing revealed that the cloned region from strain 1- 60 is nearly identical to the region corresponding to serotype O1 (27). The major difference is a 1.5-kbp insertion element present in the 1-60 sequence that disrupts the fourth ORF of O1, a gene encoding a member of the UDP-glucose/GDP-mannose dehydrogenase family (32, 41). Similarly, the region cloned from 2-164, which corresponds to serotype O6, contains a 2-nucleotide deletion in the fifth ORF of the cluster. This gene, which has previously been annotated as wbpQ, is of unknown function (4, 27). Neither strain 1-60 nor strain 2-164 possesses a discernable serotype on incubation with O1 and O6 monoclonal antibodies, respectively. Hence, it is probable that these mutations are actually responsible for loss of O-antigen synthesis. However, at least in the patient 1 isolates, the genotype-phenotype relationship may be more complex (see below). The indication that loss of O-antigen synthesis is due to specific mutations in the O-antigen gene clusters is supported by previous complementation studies (8).
FIG. 5.
O-antigen biosynthetic locus of clinical strains 1-60 and 2-164. Mutational events likely to disrupt lipopolysaccharide biosynthesis are indicated.
Analysis of O1 and mucA genes in strain isolates from patient 1.
We scored the mucoid phenotype and O1 serotype of sequential isolates from the patient 1 collection (Table 4). This analysis revealed marked heterogeneity among these isolates. Mutations in the mucA gene are known to cause the mucoid phenotype (11, 21, 22); therefore, we determined the DNA sequence of the mucA gene for the entire series of isolates. Two isolates collected at 24 months exhibited a mucoid phenotype at the time of isolation, although no corresponding mutation in mucA was detected. Mucoid isolates collected at later times harbor frameshift mutations in mucA. These data revealed three distinct mucA alleles, suggesting that multiple, independent mutational events gave rise to the mucoid phenotype.
TABLE 4.
Analysis of mucA O antigen for sequential patient 1 isolates
| Patient age at time of isolation (mo) | Serotype (+/−) | O1a | muc (+/−) | mucA |
|---|---|---|---|---|
| 6 | + | WT | − | WT |
| 9 | + | WT | − | WT |
| 12 | − | Insertion | − | WT |
| 12 | − | Insertion | − | WT |
| 15 | − | WT | − | WT |
| 18 | − | WT | − | WT |
| 21 | − | Insertion | − | WT |
| 21 | − | WT | − | WT |
| 24 | − | Insertion | − | WT |
| 24 | − | Insertion | − | WT |
| 24 | − | Insertion | + | WT |
| 24 | − | WT | − | WT |
| 24 | − | Insertion | − | WT |
| 24 | − | WT | + | WT |
| 27 | − | Insertion | − | WT |
| 27 | − | Insertion | − | WT |
| 27 | + | WT | + | ΔG430 |
| 30 | + | WT | + | ΔG430 |
| 30 | + | WT | + | ΔG357 |
| 30 | − | Insertion | − | WT |
| 30 | − | Insertion | − | WT |
| 33 | − | Insertion | − | WT |
| 33 | − | Insertion | − | WT |
| 33 | + | WT | + | ΔG357 |
| 36 | − | Insertion | − | WT |
| 36 | + | WT | + | ΔG430 |
| 36 | + | WT | + | ΔG357 |
| 36 | − | Insertion | − | WT |
| 36 | − | Insertion | − | WT |
| 36 | − | Insertion | − | WT |
O1-specific PCR products were scored either as wild type (wt) in size or as amplicons with an apparent insertion element (insertion).
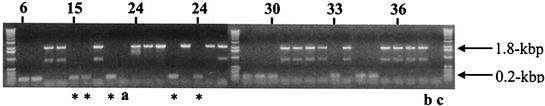
PCR was used to examine the presence or absence of the insertion element in the O1 gene cluster (Fig. 6). While the insertion element is present in most of the 23 untypeable isolates, there are 5 untypeable isolates obtained during the first 24 months of the patient's life that lack the insertion. At later times, all untypeable isolates possess the insertion. Isolates collected between 27 and 36 months were either mucoid or untypeable, suggesting that these genetically distinct strains coexisted in the patient's airways during this period. Finally, the mucoid and untypeable phenotypes coalesce in the 60-month isolate, 1-60, which was the focus of this study.
FIG. 6.
PCR assay to determine which isolates in the patient 1 collection have an insertion in the O-antigen B-band locus (presence of a 1.8-kbp band). Numbers indicate the approximate age in months of patient 1 at the time selected isolates were cultured. Isolates without the insertion that are not typeable are indicated by *. The outlying isolate, not clonally related to the predominant strain (a), strain 1-60 (b), and a no-DNA control (c) are indicated.
DISCUSSION
We investigated genetic variation in P. aeruginosa by using a collection of sequential isolates from the airways of two CF patients along with an isolate from an environmental source. Previous analyses of variation among CF isolates have established that infections within a particular patient are predominantly clonal (5, 29, 39). The relatedness of our sequential CF isolates was assessed using whole-genome restriction analysis, the results of which support this conclusion. It has also been observed that during long-term colonizations, strains from airways of CF patients tend to become mucoid (11, 17, 21, 22, 28) and lose the ability to produce O antigen (18, 28, 29). Both events were observed in this study. To investigate genomic variation among diverse strains of P. aeruginosa, the environmental isolate and two late-stage clinical isolates from our clonal collections were chosen for whole-genome-sample sequencing and comparison to the PAO1 reference genome. To date, the genomic structure of P. aeruginosa has been interrogated largely by restriction mapping, which has resulted in the conclusion that large rearrangements and acquisition and loss of large blocks of DNA are responsible for most of the genetic diversity among different strains (30, 34). At the nucleotide level, Kiewitz and Tümmler (15) reported that substitution rates in a limited set of conserved genes in the P. aeruginosa genome are lower than 0.5% In general, the results of our whole-genome-sample sequencing experiments are consistent with these observations.
Our high-resolution comparisons between sequence data from the sampled strains and the PAO1 reference genome allowed us make more detailed observations about the general character of P. aeruginosa genomes. First, the PAO1 genome is well conserved in the strains that we studied. Regions present in PAO1 but absent in other strains do exist, but they are mostly short (less than 20 -kbp), with the exception of one 100-kbp gap in clinical strain 2-164. There are two loci of significant size where no sequencing traces for any sampled strain could be aligned to the PAO1 genome. One of these involves a 17-kbp deletion, relative to PAO1, and the other involves genes at the O-antigen biosynthetic locus that are functionally related to the genes present in PAO1 but not conserved in sequence (27). The large fraction of the PAO1 genome that is common to all three strains leads us to believe that there is a backbone genome that is conserved in many P. aeruginosa strains, an observation supported by lower-resolution genome mapping of PAO1 and other strains (30, 34).
Second, our estimates indicate that the genome-wide average nucleotide substitution rate is 0.5%. This is almost an order of magnitude lower than the sequence diversity of genomic material shared between E. coli O157 and K-12 strains (26). Isolated hypervariable regions are apparent at specific loci. The most prominent examples include genes involved in synthesis and recognition of the siderophore pyoverdine and genes whose products synthesize serologically different flagellins.
Finally, strain-specific islands appear to be the primary mode of variation among different strains of P. aeruginosa. Although much of the sequence content of PAO1 is present in the three strains that were sampled, a significant amount of genetic material unique to the sampled strains was found. Approximately 10% of the sequencing traces that we gathered for each strain contain no sequence alignable with the PAO1 genome. This estimate is probably a conservative one because it does not account for chimeric or highly diverged fragments, which contain a significant amount of sequence that is not found in PAO1 but can still partially align with the PAO1 genome. Annotation of these strain-specific sequences resulted in identification of previously described islands, for example the PAGI-1 sequences from O-antigen biosynthetic genes and an island affecting flagellar glycosylation, but most are not homologous to any known genes. Few phage-related sequences were found. The G+C content of anonymous, unalignable fragments is significantly lower than the average for the PAO1 genome (50 to 54% versus 67%), which may indicate that much of this material was acquired via horizontal transfer from other bacterial species. One weakness of the present study is that it is not possible to group divergent sequences into islands that are not found in PAO1, nor is it possible to place these putative islands in the context of the PAO1 reference sequence. This type of analysis will be possible only with more complete sequence coverage of the genomes of clinical and environmental isolates.
The whole-genome-sample sequencing data motivated further investigation of specific regions. We recently used yeast recombinational cloning to isolate and characterize a diverse set of O- antigen biosynthesis genes (27). The same techniques revealed that clinical strains 1-60 and 2- 164 harbor O1 and O6 O-antigen gene clusters, respectively. These strains are untypeable, and sequencing revealed the probable causative mutations in both cases. We also used recombinational cloning to characterize the pyoverdine biosynthesis genes, and we found that all three strains possessed related sequences that are substantially diverged from those present in PAO1. Finally, we used phenotypic analysis coupled with sequence data to explore the heterogeneity of isolates derived from a clonal, CF-related infection. These studies point to sufficiently strong selective pressures for the overproduction of alginate to cause independent mutational events to occur in the same gene during the course of a single clonal infection. Both the mucA and O-antigen data also indicate that despite the strong selective pressure for mutations, selective sweeps resulting in fixation of favored mutations do not occur with any rapidity.
Our data suggest a relatively simple model for genetic variation in the 6-Mbp genome of P. aeruginosa. Much of the genome is relatively well conserved, with a level of variation, dominated by SNPs, that is not greatly higher than that found in many metazoan species such as humans. This conserved framework of the P. aeruginosa genome is interrupted by highly diverged segments that appear to reflect the effects of balancing selection on P. aeruginosa populations. Balancing selection leads to the maintenance in different strains of functionally diverged solutions to the same biological challenges (e.g., synthesis of the flagellum, the polysaccharide coating of the bacterium, and siderophores). More comprehensive analyses are required to determine the number of different functionally diverged “cassettes” that can plug into the conserved genomic framework; however, it is notable that systems that are already known and reasonably well understood account for a substantial proportion of the obvious sites of hypervariability.
We also obtained a first glimpse of the molecular evolution of P. aeruginosa during the course of a particular CF infection. The data emphasize the genetic heterogeneity that develops and is maintained for long periods within these clonal infections. Extensions of the work described here have the potential to produce a detailed model for genetic variation in P. aeruginosa both among CF patients and across time in individual patients. Since therapeutic interventions must be based on conserved features of these infections, such a model would provide a rational basis for evaluating the attractiveness of potential drug targets.
Acknowledgments
We thank the staff of the University of Washington Genome Center for their contributions to this work. We also thank Ben Voight, who generated the whole-genome fingerprints, and Michael Jacobs, who provided valuable suggestions for producing the manuscript.
This work was supported by a Cystic Fibrosis Foundation grant to Maynard Olson (Sam I. Miller, principal investigator), as well as a Center for Excellence in Genome Sciences grant (P50 HG02351) to Maynard Olson.
REFERENCES
- 1.Allison, J. S., M. Dawson, D. Drake, and T. C. Montie. 1985. Electrophoretic separation and molecular weight characterization of Pseudomonas aeruginosa H-antigen flagellins. Infect. Immun. 49:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, S. K., N. Dasgupta, S. Lory, and R. Ramphal. 2000. Identification of two distinct types of flagellar cap proteins, FliD, in Pseudomonas aeruginosa. Infect. Immun. 68:1474-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belanger, M., L. L. Burrows, and J. S. Lam. 1999. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145:3505-3521. [DOI] [PubMed] [Google Scholar]
- 5.Burns, J. L., R. L. Gibson, S. McNamara, D. Yim, J. Emerson, M. Rosenfeld, P. Hiatt, K. McCoy, R. Castile, A. L. Smith, and B. W. Ramsey. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444-452. [DOI] [PubMed] [Google Scholar]
- 6.Croft, L., S. A. Beatson, C. B. Whitchurch, B. Huang, R. L. Blakeley, and J. S. Mattick. 2000. An interactive web-based Pseudomonas aeruginosa genome database: discovery of new genes, pathways and structures. Microbiology 146:2351-2364. [DOI] [PubMed] [Google Scholar]
- 7.Duport, C., C. Baysse, and Y. Michel-Briand. 1995. Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J. Biol. Chem. 270:8920-8927. [DOI] [PubMed] [Google Scholar]
- 8.Evans, D. J., G. B. Pier, M. J. Coyne, Jr., and J. B. Goldberg. 1994. The rfb locus from Pseudomonas aeruginosa strain PA103 promotes the expression of O antigen by both LPS-rough and LPS-smooth isolates from cystic fibrosis patients. Mol. Microbiol. 13:427-434. [DOI] [PubMed] [Google Scholar]
- 9.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 10.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 11.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigoriev, A. 1999. Strand-specific compositional asymmetries in double-stranded DNA viruses. Virus Res. 60:1-19. [DOI] [PubMed] [Google Scholar]
- 13.Grocock, R. J., and P. M. Sharp. 2002. Synonymous codon usage in Pseudomonas aeruginosa PA01. Gene 289:131-139. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, R. E., L. M. Mutharia, L. Chan, R. P. Darveau, D. P. Speert, and G. B. Pier. 1983. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 42:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiewitz, C., and B. Tummler. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, U. J., H. Shizuya, P. J. de Jong, B. Birren, and M. I. Simon. 1992. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 20:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch, C., and N. Hoiby. 1993. Pathogenesis of cystic fibrosis. Lancet 341:1065-1069. [DOI] [PubMed] [Google Scholar]
- 18.Lam, M. Y., E. J. McGroarty, A. M. Kropinski, L. A. MacDonald, S. S. Pedersen, N. Hoiby, and J. S. Lam. 1989. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J. Clin. Microbiol. 27:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehoux, D. E., F. Sanschagrin, and R. C. Levesque. 2000. Genomics of the 35-kb pvd locus and analysis of novel pvdIJK genes implicated in pyoverdine biosynthesis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 190:141-146. [DOI] [PubMed] [Google Scholar]
- 20.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May, T. B., D. Shinabarger, R. Maharaj, J. Kato, L. Chu, J. D. DeVault, S. Roychoudhury, N. A. Zielinski, A. Berry, R. K. Rothmel, et al. 1991. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin. Microbiol. Rev. 4:191-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer, J. M. 2000. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 174:135-142. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama, K., S. Kanaya, M. Ohnishi, Y. Terawaki, and T. Hayashi. 1999. The complete nucleotide sequence of phi CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 31:399-419. [DOI] [PubMed] [Google Scholar]
- 25.Nociari, M. M., M. Catalano, D. Centron Garcia, S. C. Copenhaver, M. L. Vasil, and D. O. Sordelli. 1996. Comparative usefulness of ribotyping, exotoxin A genotyping, and SalI restriction fragment length polymorphism analysis for Pseudomonas aeruginosa lineage assessment. Diagn. Microbiol. Infect. Dis. 24:179-190. [DOI] [PubMed] [Google Scholar]
- 26.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-33. [DOI] [PubMed] [Google Scholar]
- 27.Raymond, C. K., E. H. Sims, A. Kas, D. H. Spencer, T. V. Kutyavin, R. G. Ivey, Y. Zhou, R. Kaul, J. B. Clendenning, and M. V. Olson. 2002. Genetic variation at the O- antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 184:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romling, U., B. Fiedler, J. Bosshammer, D. Grothues, J. Greipel, H. von der Hardt, and B. Tummler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 170:1616-1621. [DOI] [PubMed] [Google Scholar]
- 30.Romling, U., K. D. Schmidt, and B. Tummler. 1997. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J. Mol. Biol. 271:386-404. [DOI] [PubMed] [Google Scholar]
- 31.Romling, U., J. Wingender, H. Muller, and B. Tummler. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roychoudhury, S., T. B. May, J. F. Gill, S. K. Singh, D. S. Feingold, and A. M. Chakrabarty. 1989. Purification and characterization of guanosine diphospho-d- mannose dehydrogenase. A key enzyme in the biosynthesis of alginate by Pseudomonas aeruginosa. J. Biol. Chem. 264:9380-9385. [PubMed] [Google Scholar]
- 33.Sano, Y., H. Matsui, M. Kobayashi, and M. Kageyama. 1993. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J. Bacteriol. 175:2907-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt, K. D., B. Tummler, and U. Romling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spangenberg, C., T. Heuer, C. Burger, and B. Tummler. 1996. Genetic diversity of flagellins of Pseudomonas aeruginosa. FEBS Lett. 396:213-217. [DOI] [PubMed] [Google Scholar]
- 36.Speert, D. P., M. E. Campbell, S. W. Farmer, K. Volpel, A. M. Joffe, and W. Paranchych. 1989. Use of a pilin gene probe to study molecular epidemiology of Pseudomonas aeruginosa. J. Clin. Microbiol. 27:2589-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 38.Tsuda, M., H. Miyazaki, and T. Nakazawa. 1995. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J. Bacteriol. 177:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tummler, B., J. Bosshammer, S. Breitenstein, I. Brockhausen, P. Gudowius, C. Herrmann, S. Herrmann, T. Heuer, P. Kubesch, F. Mekus, U. Romling, K. D. Schmidt, C. Spangenberg, and S. Walter. 1997. Infections with Pseudomonas aeruginosa in patients with cystic fibrosis. Behring Inst. Mitt. 1997:249-255. [PubMed] [Google Scholar]
- 40.Wilderman, P. J., A. I. Vasil, Z. Johnson, and M. L. Vasil. 2001. Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol. 39:291-303. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, X., C. Creuzenet, M. Belanger, E. Egbosimba, J. Li, and J. S. Lam. 2000. WbpO, a UDP-N-acetyl-d-galactosamine dehydrogenase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 275:39802.. [DOI] [PubMed] [Google Scholar]