Abstract
Porphyromonas gingivalis is an anaerobic microorganism that inhabits the oral cavity, where oxidative stress represents a constant challenge. A putative transcriptional regulator associated with oxidative stress, an oxyR homologue, is known from the P. gingivalis W83 genome sequence. We used microarrays to characterize the response of P. gingivalis to H2O2 and examine the role of oxyR in the regulation of this response. Most organisms in which oxyR has been investigated are facultative anaerobes or aerobes. In contrast to the OxyR-regulated response of these microorganisms to H2O2, the main feature of the response in P. gingivalis was a concerted up-regulation of insertion sequence elements related to IS1 transposases. Common OxyR-regulated genes such as dps and ahpFC were not positively regulated in P. gingivalis in response to H2O2. However, their expression was dependent on the presence of a functional OxyR, as revealed by microarray comparison of an oxyR mutant to the wild type. Phenotypic characterization of the oxyR mutant showed that OxyR plays a role in both the resistance to H2O2 and the aerotolerance of P. gingivalis. Escherichia coli and other bacteria with more complex respiratory requirements use OxyR for regulating resistance to H2O2 and use a separate regulator for aerotolerance. In P. gingivalis, the presence of a single protein combining the two functions might be related to the comparatively smaller genome size of this anaerobic microorganism. In conclusion, these results suggest that OxyR does not act as a sensor of H2O2 in P. gingivalis but constitutively activates transcription of oxidative-stress-related genes under anaerobic growth.
Porphyromonas gingivalis is a gram-negative, nonmotile, pleomorphic rod and obligate anaerobe (35). Several studies based on microbiological and immunological findings have classified P. gingivalis as one of the causative agents in periodontitis (25, 36). However, P. gingivalis is also found as part of the indigenous supra- and subgingival microflora in healthy individuals of all ages (40, 47, 48). During colonization of the oral tissues, P. gingivalis is exposed to various oxidative stress conditions (e.g., during survival in saliva) in which the presence of an unfavorable redox potential and the damaging effects of reactive oxygen species (ROS) might challenge its survival before finding the appropriate anaerobic microenvironment in which to establish itself and proliferate. Hydrogen peroxide, in particular, poses a problem for microorganisms in dental biofilms as it is produced by other community members such as streptococci (6) and can freely permeate the cell envelopes of adjacent bacteria. Once P. gingivalis forms part of a subgingival biofilm, lower redox potentials (17, 24) might favor its proliferation, but the microorganism would still encounter other sources of oxidative stress originating from the host defenses (e.g., neutrophils).
ROS such as O2·−, HO·, and H2O2 are produced inside bacterial cells in aerobic environments (15, 29). ROS are toxic as they are highly reactive and can cleave nucleic acids and oxidize essential proteins and lipids (8, 12). Obligate anaerobes cannot grow in the presence of oxygen but can survive transient periods of aerobiosis (11, 42). In contrast to the increasing knowledge of oxygen toxicity and antioxidant systems in aerobes, the basis for anaerobiosis and how anaerobes react to oxidative stress are poorly understood. It is believed that anaerobes cannot grow in the presence of oxygen due to the inactivation of key metabolic enzymes (28) and the absence of the adequate oxidative-stress defense systems (23). However, some anaerobes have been shown to possess antioxidant enzymes and regulatory networks similar to those in their aerobic counterparts (4, 31).
In aerobes and facultative anaerobic bacteria, the expression of antioxidant-related genes is usually regulated by transcriptional modulators that sense oxidative-stress-generating agents (30, 38). The SoxR/SoxS and the OxyR systems are examples of these regulators that respond, respectively, to superoxide-generating compounds and H2O2. OxyR is a redox-sensitive protein in the LysR family of DNA-binding transcriptional modulators (38). In Escherichia coli, OxyR is not activated (oxidized) during aerobic growth. Rather, it requires the addition of exogenous H2O2. The levels of OxyR, however, remain constant after treatment with H2O2 (37). Under aerobic growth of E. coli low amounts of reduced OxyR molecules are present in the cells, bound to the promoters of the OxyR-regulated genes. When H2O2 is produced inside or diffuses into the cell, direct oxidation of the OxyR protein induces disulfide bond formation between two cysteine residues, C199 and C208 (19, 50), an event that changes OxyR DNA-binding specificity and allows recruitment of RNA polymerase, leading to the induction of a variety of target genes (41, 43). Several approaches have contributed to the identification of the E. coli OxyR regulon, which includes dps (a nonspecific, protective, DNA-binding protein), ahpF and ahpC (alkylhydroperoxide reductase subunits F and C, respectively), katG (catalase), gor (glutathione reductase), grxA (glutaredoxin A), and trxC (thioredoxin 2), among other genes (51). An OxyR homologue has also been identified in the aerotolerant anaerobe Bacteroides fragilis (4, 13, 31, 33). The OxyR regulon of B. fragilis includes katB (catalase), ahpFC, dps, tpx (thioredoxin peroxidase), rbpA (RNA binding protein), ftnA (ferritin), and rbr (rubrerythrin) (13, 33). In B. fragilis, several OxyR-regulated genes are induced not only after H2O2 addition, as occurs in E. coli, but also after exposure to air. However, oxyR is necessary only for resistance to H2O2; its inactivation does not affect the aerotolerance of B. fragilis, perhaps because of compensatory mechanisms that are not OxyR dependent (32).
P. gingivalis is a catalase-negative organism (4); however, several studies have shown that it possesses alternate antioxidant defenses. These include superoxide dismutase, which appears to be protective against atmospheric oxygen (20), as well as rubrerythrin, Dps, and AhpFC, all of which have been demonstrated to be protective against exogenously added H2O2 (16, 39, 45). Furthermore, an oxyR homologue has been identified in the P. gingivalis genome sequence (26). The purpose of the present study was to evaluate the role of oxyR in the P. gingivalis response to H2O2. We report here that putative OxyR-controlled genes, identified by microarray analysis, are not inducible after H2O2 treatment. However, their expression during anaerobic growth requires the presence of a functional OxyR. We also report that P. gingivalis oxyR is important not only for resistance to H2O2 but also for the aerotolerance of the microorganism.
MATERIALS AND METHODS
Microorganisms and growth conditions.
P. gingivalis W83 (a kind gift of M. J. Duncan, Department of Molecular Genetics, The Forsyth Institute, Boston, MA) and W50 (ATCC 53978) were maintained short-term on anaerobic blood agar plates, incubated at 37°C in a Shel Lab Bactron IV anaerobic chamber (Sheldon Manufacturing, Inc., Cornelius, OR) with an atmosphere of H2-CO2-N2 (5/5/90 ratio). All strains were grown in brain heart infusion (BHI) medium (Becton Dickinson and Company, Sparks, MD) supplemented, after autoclaving, with 5 mg of hemin liter−1 and 0.5 g of cysteine liter−1. For all experiments, supplemented BHI medium was prereduced in the anaerobic chamber for a minimum of 4 h prior to inoculation with P. gingivalis.
Construction of P. gingivalis W50 oxyR isogenic mutant and complemented mutant.
The P. gingivalis genome sequence (26) was accessed at http://www.tigr.org, and all gene designations correspond to TIGR gene identification (ID) numbers. Table 1 shows the oligonucleotides used in this study. Construction of the P. gingivalis oxyR isogenic mutant was based on methodology previously described (10). Primers oxyR1 and oxyR2 (Table 1) were designed from PG0270 encoding a putative OxyR in P. gingivalis W83. A 902-bp fragment was amplified by PCR of P. gingivalis W50 genomic DNA using primers oxyR1 and oxyR2, and the product was ligated into pGEM-T Easy (Promega, Madison, WI) to generate pOX1. Transformants were selected on Luria-Bertani plates supplemented with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 80 μg X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) ml−1, and 100 μg ampicillin ml−1. A 2.65-kb EcoICRI TetQ fragment from Bacteroides thetaiotaomicron, conferring tetracycline resistance to P. gingivalis, was removed from pNJR12 (22) and ligated into a unique BsaMI site on pOX1 to generate pOX2. The resulting pOX2 vector was transformed into E. coli JM109 by electroporation using standard procedures. Purified, linearized (ScaI) pOX2 was then electroporated into P. gingivalis W50 as previously described (10). Electroporated cells were allowed to recover in 1 ml BHI medium for 2 h under anaerobic conditions and plated on blood agar containing 1 μg of tetracycline ml−1. Colonies were recovered after 5 to 6 days. Southern blot analysis was performed to confirm the construction of the oxyR mutant.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence 5′ to 3′a |
|---|---|
| Construction of oxyR mutants | |
| oxyR1 | TCGAATACATAGCCGCATTG |
| oxyR2 | CCTGTCTGCAACTTGTGCAT |
| oxyR3 | CGAATAAAAGGTGCCGACAT |
| oxyR4 | ATTCGTTCAAGCCAAATGCT |
| Northern blot hybridization | |
| dps (PG0090) | GCGTATCGGTCACATTCTTCAGC GTACTTG |
| ahpC (PG0618) | GGCCGGACATACCTGACCATCGTG AGCA |
| Real-time PCR | |
| dps (PG0090) | F: CGGTGAGGCAGGCGATGAGGTA |
| R: CTTGGCAGCGTAGGCAGACAGC | |
| ahpC (PG0618) | F: GGCTTACCGTGGCTCTTTCGTGA |
| R: GGACATACCTGACCATCGTGAGCA | |
| Hypothetical protein (PG0421) | F: AATGCTGCAGTGCGAGTTATC |
| R: AGGGTGGGCTGGTTGAA | |
| sod (PG1545) | F: GCACGGAATTTGAAAACGCTGAC |
| R: CTTGCCCGGACGGAACTGAG | |
| Ferritin (PG1286) | F: CAAATCAAGGCCGAAATGTGGTCT |
| R: TTTCCTCGAGGCTCTGCTTTTTGA | |
| Thioredoxin (PG0275) | F: TTGGCAGGGATTTTCGGTGTCAGA |
| R: GCCCATCGTTTGCGTCGGTATTC | |
| Formate-tetrahydrofolate ligase (PG1321) | F: AGGAAATATACAGCGCAGGGAGTG |
| R: TGGCAATACAAACGGGGAGATGAT | |
| 16S rRNA gene | F: AGGCGTGAGGAAGGTGTGGATGAC |
| R: CGCCCGGTAGCTGCCCTTTGT | |
| Glucokinase (PG1737) | F: ATGAATCCGATCCGCCACCAC |
| R: GCCTCCCATCCCAAAGCACT | |
| Excinuclease (PG2210) | F: CGGAAGGAACGGTGGAGGAAC |
| R: GGCATGCCCCGATAGGATTG |
F, forward; R, reverse.
To construct a complemented oxyR mutant (designated comp), primers oxyR3 and oxyR4 (Table 1) were used to amplify oxyR and flanking regions by PCR using W50 genomic DNA as template. The PCR product was ligated into pGEM-T Easy to generate pOX3. EcoRI inserts were excised from pOX3 and inserted into the EcoRI site of the P. gingivalis/E. coli shuttle vector pYH411 (unpublished), a 12.3-kb vector derived from pYH400 (12.8 kb) (49) that confers ampicillin resistance in E. coli and erythromycin resistance in P. gingivalis. Purified recombinant plasmid DNA was transferred into the electrocompetent P. gingivalis oxyR mutant strain as previously described (27). Colonies were selected on blood agar containing 1 μg of tetracycline ml−1 and 10 μg erythromycin ml−1 after 7 to 10 days of growth. The recombinant insert from plasmid DNA isolated from the comp strain was sequenced with primers oxyR3 and oxyR4 and shown to be 100% identical to wild-type oxyR.
Determination of resistance to H2O2 treatment and tolerance to air.
One milliliter of an overnight culture was used to inoculate 100 ml of BHI medium preincubated anaerobically at 37°C. Culture densities, expressed in Klett units (KU), were registered every hour using a Klett-Summerson photoelectric colorimeter (Arthur H. Thomas Co., Philadelphia, PA) until stationary phase was reached (KU 210). Hydrogen peroxide (250 μM) was added at early exponential phase (typically 9 h after inoculation).
To compare the levels of resistance of cells to killing by atmospheric oxygen, P. gingivalis wild type and mutants were grown in BHI medium until late logarithmic phase (KU 180). Serial 10-fold dilutions were performed in the same prereduced medium, and 0.1 ml was spread on prereduced blood agar plates, which were then exposed to air for different periods of time, followed by anaerobic incubation for 4 to 6 days. The CFU appearing on plates exposed to air divided by the CFU on the control plates (not exposed to air) times 100 was equal to the survival percentage.
RNA isolation, Northern blot hybridization, and real-time PCR.
P. gingivalis cultures, grown to mid-exponential phase, were divided in half. One aliquot was left untreated while the other was treated with H2O2, anaerobically in most cases, or aerobically, when indicated. RNA was isolated from treated and untreated cultures by mixing 10 ml of culture with an equal volume of hot phenol saturated with 0.1 M citrate buffer (pH 4.3), following conventional protocols (34). Genomic DNA was removed from RNA samples by treatment with RNase-free DNase I (Promega). For Northern blot analysis, 10 μg of RNA was electrophoresed in 1× MOPS (morpholinepropanesulfonic acid) buffer, transferred to a Hybond N+ nylon membrane with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer, and hybridized with the [γ-32P]dATP-labeled oligonucleotide probe. Oligonucleotides used to detect RNA transcripts are listed in Table 1. Membranes were exposed to a phosphoimager detection screen, and the values were normalized to that of the respective 16S rRNA band detected on the ethidium bromide-stained agarose gel to correct for any loading differences.
Real-time PCR relative quantification was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Four micrograms of RNA was reverse transcribed in a reaction mixture containing 2 μl random hexamers (3 mg/ml; Invitrogen, Carlsbad, CA), 1.2 μl of a 12.5 mM deoxynucleoside triphosphate mix, 100 U RNase inhibitor (Ambion, Austin, TX), 3 μl 0.1 M dithiothreitol (DTT), 6 μl 5× Superscript buffer, 2 μl Superscript II (Invitrogen), and diethyl pyrocarbonate-treated water to a 30-μl final volume. Reaction mixtures were incubated at 42°C for 16 h. Primer sequences for real-time PCR are listed in Table 1. The optimal primer concentration for all genes was determined to be 300 nM. The absence of nonspecific amplification was determined by generating dissociation curves after PCR was complete. Amplification efficiency was determined in a reaction mixture containing 0 to 20 ng cDNA template. For real-time relative quantification, all genes were amplified for 40 cycles in a 50-μl reaction mixture containing 1× SYBR Green PCR Master Mix, 300 nM (each) primer, and 5 ng cDNA template μl−1 with an annealing temperature dependent on the primer pair used. The P. gingivalis 16S rRNA gene and the open reading frames (ORFs) PG1737, encoding a glucokinase, and PG2210, encoding an excinuclease, were selected as endogenous controls. The expression of PG1737 and PG2210 was shown to remain unchanged by microarray analysis (data not shown). Target gene expression was normalized to that of the endogenous control gene which had the amplification efficiency closest to that of the target. The comparative cycle threshold (CT) method was used for relative quantification according to Applied Biosystems ABI Prism 7700 Sequence Detection System User bulletin no. 2. Briefly, the ΔCT was determined by subtracting the average CT value of the housekeeping gene from the average CT value of the target gene. Then the ΔΔCT for each condition was calculated by subtracting the ΔCT of the calibrator condition (untreated wild type) from the ΔCT of the condition evaluated (H2O2-treated wild type, untreated oxyR mutant, or H2O2-treated oxyR mutant). The range for each condition relative to the calibrator was determined by evaluating the expression 2−ΔΔCT with ΔΔCT + S and ΔΔCT − S, where S is the standard deviation of the ΔΔCT value.
Microarray experiments.
P. gingivalis 70-mer oligonucleotide-based microarrays were fabricated at TIGR and provided by the NIDCR through the NIAID PFGRC facility. Arrays were based on ORFs annotated in the W83 genome sequence with each oligonucleotide printed four times on the glass slide. Samples for hybridization on each array were prepared in the following manner: cDNA was generated by reverse transcription of 16 μg of total RNA in a reaction containing 2 μl random hexamers (3 mg/ml; Invitrogen), 1.2 μl amino allyl-deoxynucleotide triphosphate mix (12.5 mM dATP, dCTP, and dGTP; 4.16 mM dTTP; and 8.33 mM amino allyl-dUTP), 100 U RNase inhibitor (Ambion), 3 μl 0.1 M DTT, 6 μl 5× Superscript buffer, 2 μl Superscript II (Invitrogen), and diethylpyrocarbonate-treated water to a 30-μl final volume. Reaction mixtures were incubated at 42°C for 16 h. RNA template was hydrolyzed at 65°C for 15 min with the addition of 10 μl 1 M NaOH and 10 μl 0.5 M EDTA. The reaction was then neutralized by the addition of 25 μl 1 M Tris (pH 7.4), and the aminoallyl-cDNA was cleaned using a Qiaquick PCR purification Kit (QIAGEN). Speed-Vac-dried cDNA was resuspended in 4.5 μl 0.1 M Na2CO3 buffer, pH 9.0, and mixed with 4.5 μl N-hydroxysuccinimide-Cy5 or N-hydroxysuccinimide-Cy3 dye (Amersham Biosciences, Piscataway, NJ) dissolved in dimethyl sulfoxide. The coupling reaction was allowed to proceed for 1 h at room temperature in the dark. Uncoupled Cy dyes were removed with a Qiaquick PCR purification kit. The Speed-Vac-dried labeled samples were resuspended in 20 μl hybridization buffer and mixed for hybridization. Slides were hybridized overnight at 42°C. Hybridization buffers and washing procedures are described at http://www.tigr.org/tdb/microarray/conciseguide.html. Microarrays were scanned using a GenePix 4000B (Axon, Union City, CA) scanner and analyzed using GenePix Pro 6.0. RNA samples from three biological replicates were analyzed with dye swapping (to avoid any differences in Cy3 and Cy5 labeling efficiency), resulting in a total of six microarray slides for each comparison. The three comparisons studied were (i) untreated wild-type samples compared to wild-type samples treated with 125 μM H2O2 for 5 min, (ii) untreated oxyR mutant compared to H2O2-treated oxyR mutant, and (iii) untreated wild type compared to untreated oxyR mutant. Statistical analysis was performed by calculating the P values in a two-tailed t test.
Nucleotide sequence accession number.
The GenBank accession number for the P. gingivalis W50 oxyR homologue sequence is DQ098106.
RESULTS
Insertional inactivation of oxyR decreases P. gingivalis aerotolerance and resistance to H2O2.
The P. gingivalis W50 oxyR homologue was sequenced (GenBank accession no. DQ098106) and found to be 100% identical to the oxyR homologue (PG0270) from the P. gingivalis W83 genome sequence (26). A PSI BLAST analysis (2) revealed that PG0270 exhibited 58% identity to B. fragilis oxyR (accession no. AAG02619) and 34% identity to E. coli oxyR (accession no. P11721). Importantly, the helix-turn-helix motif region for DNA binding and promoter recognition, present at the N-terminal domain of LysR-type regulators (18), is highly conserved, as are the cysteine residues in positions 199 and 208, which have been shown to be critical for the ability of the transcription factor to sense H2O2 in vivo and in vitro (50).
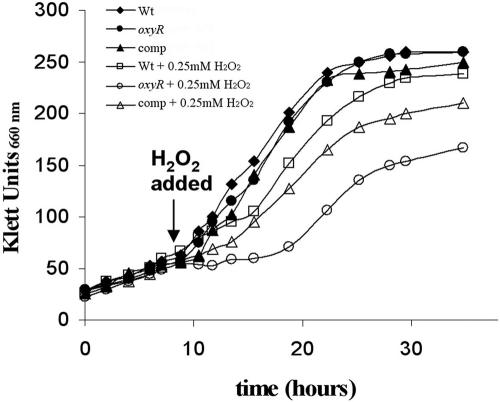
Insertional inactivation of P. gingivalis oxyR had no significant effect on the growth rate of the mutant under anaerobic conditions. However, as shown in Fig. 1, inactivation of oxyR reduced the ability of the oxyR mutant to recover after treatment (anaerobically) with 250 μM H2O2. Complementation of the oxyR mutant with oxyR expressed from pYH411 partially restored the wild-type phenotype. Insert sequencing in pYH411 showed no discrepancies with P. gingivalis genomic DNA sequence. Furthermore, approximately 150 bp upstream of the oxyR starting site were included to ensure complete coverage of the promoter region. These results suggest, however, that DNA sequence elements outside the cloned region may be required for efficient transcription of oxyR.
FIG. 1.
Effect of the addition of 250 μM H2O2 (open symbols) on the growth of P. gingivalis W50 wild type (Wt), oxyR mutant (oxyR), and complemented oxyR mutant (comp) compared to nontreated controls (closed symbols). All cultures were grown anaerobically in BHI medium except for the oxyR complemented mutant, which also contained 10 μg erythromycin ml−1.
Table 2 compares the abilities of the wild type, the oxyR mutant, and the complemented mutant to tolerate exposure to atmospheric oxygen for different periods of time. It was demonstrated that in P. gingivalis OxyR seems to play a role in aerotolerance, as the oxyR mutant was more sensitive to air than the wild type and the complementation partially restored aerotolerance.
TABLE 2.
Air sensitivity of P. gingivalis wild type (wt), oxyR mutant (oxyR), and complemented oxyR mutant (comp)
| Time of exposure to air (hours) | % CFUa
|
||
|---|---|---|---|
| wt | oxyR | Comp | |
| 1 | 97 ± 1 | 92 ± 2 | 96 ± 2 |
| 2.5 | 83 ± 3 | 64 ± 3 | 68 ± 3 |
| 5 | 59 ± 1 | 34 ± 1 | 42 ± 1 |
| 10 | 39 ± 3 | 4 ± 1 | 17 ± 4 |
| 20 | 34 ± 1 | <0.05 | 11 ± 3 |
| 25 | 16 ± 4 | <0.05 | 2 ± 3 |
Results represent the percentage of colony-forming units appearing on plates exposed to air compared to the control (time of exposure = 0 h) and are presented as the mean survival percentage ± standard deviation of duplicate experiments with duplicate samples per experiment.
Expression of dps and ahpFC in P. gingivalis requires OxyR but does not increase in response to H2O2.
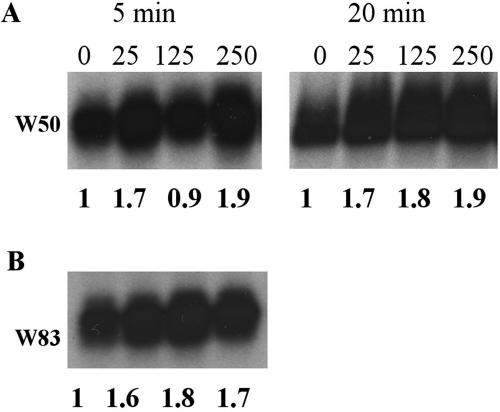
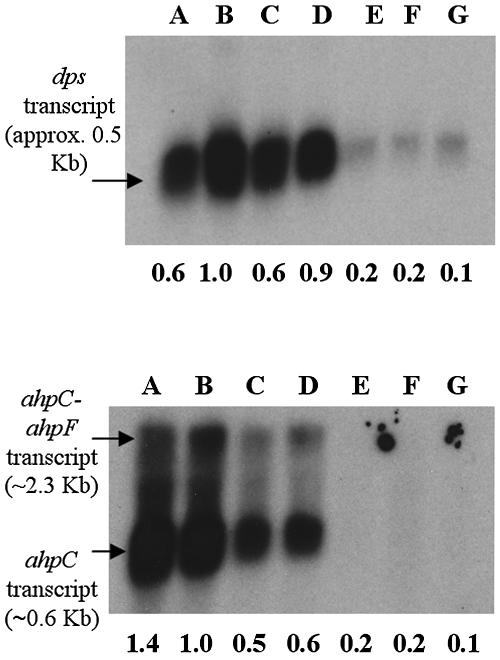
The dps gene has been shown to play a role in protection of E. coli from peroxide stress, and its expression is regulated by OxyR (3). Dps is significantly up-regulated in E. coli and B. fragilis after H2O2 exposure, increasing 180- and 37-fold, respectively, compared to untreated cultures (31, 51). P. gingivalis possesses a Dps homologue which has been shown to be protective against H2O2 and possibly regulated by OxyR (45). To evaluate dps transcriptional levels in response to H2O2, P. gingivalis W50 cultures were exposed to different H2O2 concentrations for 5 or 20 min. Results showed that H2O2 did not have a significant effect on the expression of dps (Fig. 2). A similar result was obtained with strain W83 after treatment with different H2O2 concentrations for 5 min (Fig. 2). If it is assumed that in P. gingivalis dps transcription is also regulated by OxyR, these results would suggest that OxyR is in a constitutively active state since the dps transcript was detected in wild-type cultures grown anaerobically without H2O2 treatment, and it did not increase after treatment with H2O2. In order to investigate whether BHI medium contained any H2O2, catalase was added to the medium 4 h before inoculation with P. gingivalis; the activity of the enzyme was verified, and it was demonstrated that 5 μg of catalase ml−1 was able to remove 20 μM H2O2 to undetectable amounts (data not shown). The effect of the addition of catalase on the expression of dps, ahpC, and ahpF was evaluated and compared to that for cultures grown in growth media without catalase. Figure 3 shows that removal of trace H2O2 did not significantly affect the expression of dps, ahpC, and ahpF, suggesting that the presence of H2O2 in the medium is not the cause of the high levels of dps transcript seen without H2O2 treatment. To test if exposure to oxygen would further induce the expression of dps, ahpC, and ahpF, P. gingivalis was treated with H2O2 under normal atmospheric conditions. Exposure to air had no effect on the expression of these genes compared to H2O2 treatment of anaerobically grown cultures (Fig. 3). This finding also suggested that OxyR is at its maximum level of activation during anaerobic growth. Furthermore, in contrast to the wild type, the expression of dps, ahpC, and ahpF was greatly reduced in the oxyR mutant under all the conditions tested, before and after H2O2 treatment (Fig. 3), suggesting that OxyR is indeed necessary for the expression of these genes under anaerobic growth.
FIG. 2.
Northern blot analysis of the expression of dps in P. gingivalis W50 and W83 after treatment with different H2O2 concentrations. Panel A shows dps expression in W50 treated for 5 and 20 min. Panel B shows expression of dps in W83 after treatment for 5 min. The probe used was dps (Table 1). The number above each lane represents the H2O2 μM concentration. The number below each lane represents the relative intensity of the bands compared to the untreated sample (no H2O2 added). All bands were normalized to the intensity of the corresponding 16S rRNA band in the ethidium bromide-stained gel photographed prior to transfer.
FIG. 3.
Northern blot analysis of the expression of dps (top panel) and ahpFC (bottom panel) in P. gingivalis W50 wild type and oxyR mutant. Lanes correspond to wild type grown in the presence of 5 μg/ml catalase (A), wild type grown without catalase (B), wild type grown without catalase and treated with 125 μM H2O2 in the anaerobic chamber (C), wild type grown without catalase and treated with 125 μM H2O2 under aeration (D), oxyR mutant grown in the presence of 5 μg/ml catalase (E), oxyR mutant grown without catalase (F), and oxyR mutant grown without catalase and treated with 125 μM H2O2 in the anaerobic chamber (G). The probes used were dps and ahpC. The ahpC probe recognizes polycistronic expression of ahpF and ahpC and monocistronic expression of ahpC. The number below each lane represents the relative intensity of the bands compared to lane B (untreated wild type). All bands were normalized to the intensity of the corresponding 16S rRNA band in the ethidium bromide-stained gel photographed prior to transfer.
Microarray analysis of P. gingivalis response to H2O2 treatment.
Table 3 shows the genes found to be up-regulated greater than 2.5-fold in P. gingivalis W50 after treatment with H2O2. Eight out of nine up-regulated genes were transposase-related insertion sequence elements belonging to the IS1 family. P. gingivalis DNA microarrays contain 10 different oligonucleotides that correspond to IS1 transposases. The P. gingivalis genome, however, contains 32 ORFs identified as IS1 transposases (according to TIGR annotation). The reason for this discrepancy is probably the high similarity among genes, as each of the oligonucleotide sequences present in the microarray slide matches up to 16 ORFs for transposases in the genome, covering the 32 ORFs with minimal redundancy. The two remaining oligonucleotide sequences for IS1 transposases in the microarray slides that did not yield up-regulated results with our cutoff of more than 2.5-fold, and consequently do not appear in Table 3, were PG0852, which increased 2.2-fold in expression in the H2O2-treated strain, and PG0988, which increased 1.73-fold. These results could suggest that all transposases from the IS1 family showed some degree of up-regulation. Microarray analysis, however, does not allow discrimination among specific ORFs. Hydrogen peroxide treatment did not result in down-regulation by more than 2.5-fold of any genes.
TABLE 3.
Microarray analysis of the effect of H2O2 (125 μM) treatment on Porphyromonas gingivalis W50
| Genea | Gene name/function | Other matchesb | Fold inductionc | P value |
|---|---|---|---|---|
| PG0460 | ISPg1, transposase | PG2169, PG1244, PG0549 | 4.28 | 0.0016 |
| PG0051 | ISPg1, transposase, degenerate | PG1384, PG0760 | 4.06 | 0.0049 |
| PG0813 | ISPg1, transposase, truncation | 4.01 | 0.0008 | |
| PG0944 | ISPg1, transposase, truncation | 3.66 | 0.0022 | |
| PG2195 | ISPg1, transposase, truncation | 3.26 | 0.0083 | |
| PG0294 | Glycosyl transferase, group 2 family protein | 2.76 | 0.0172 | |
| PG0988 | Transposase, ISPg1-related, authentic frameshift | 2.69 | 0.0116 | |
| PG0184 | ISPg1, transposase | PG1906, PG1448, PG0184, PG1624, PG1197, PG117, PG1031, PG0825 | 2.57 | 0.0036 |
| PG2169 | ISPg1, transposase, degenerate | 2.55 | 0.0268 |
Gene number corresponds to TIGR ID.
Matching ORF information is from The Bioinformatics Resources for Oral Pathogens (BROP) facility (http://www.brop.org). All matching ORFs correspond to IS1-related transposases.
Genes exhibiting a greater-than-2.5-fold induction in transcription only. No genes were down-regulated after exposure to H2O2.
Microarray comparison of P. gingivalis W50 wild type and oxyR mutant.
Northern blot analysis of the expression of ahpFC and dps in P. gingivalis wild type and oxyR mutant suggested that these OxyR-dependent genes are expressed at their highest levels during anaerobic growth. Therefore, in order to identify other OxyR-dependent genes we carried out a microarray comparison of the wild type and the oxyR mutant grown under anaerobic conditions and without H2O2 treatment. Table 4 shows the 28 genes with decreased expression (more than 2.5-fold) in the oxyR mutant. The identification of ahpC, dps, and ahpF as the genes with the most decreased levels of expression suggested that this methodology was useful in identifying OxyR-dependent genes. On the other hand, only four genes were expressed at a higher level in the oxyR mutant than in the wild type (Table 5). Three of these genes were transposase related, and two of them were also induced in the wild type after H2O2 treatment (Table 3), suggesting that a response seen for oxidative stress occurs in the oxyR mutant even when grown under anaerobic conditions.
TABLE 4.
Genes with a decreased level of expression in the oxyR mutant compared to Porphyromonas gingivalis W50 wild type (during anaerobic growth)
| Genea | Gene name/function | Fold decreaseb | P value |
|---|---|---|---|
| PG0618 | Alkyl hydroperoxide reductase, C subunit | 16.3 | 0.00565 |
| PG0090 | Dps family protein | 6.70 | 0.01006 |
| PG0619 | Alkyl hydroperoxide reductase, F subunit | 6.53 | 0.00711 |
| PG0421 | Hypothetical protein | 6.34 | 0.00897 |
| PG1545 | Superoxide dismutase, Fe-Mn | 5.73 | 0.01323 |
| PG1286 | Ferritin | 5.57 | 0.03179 |
| PG0275 | Thioredoxin | 4.56 | 0.01633 |
| PG1321 | Formate-tetrahydrofolate ligase | 4.46 | 0.10428 |
| PG1116 | Methylenetetrahydrofolate dehydrogenase | 3.86 | 0.01301 |
| PG0686 | Conserved hypothetical protein | 3.80 | 0.09954 |
| PG1540 | S-Adenosylmethionine:tRNA ribosyltransferase-isomerase | 3.59 | 0.00707 |
| PG1124 | DUF80 domain protein | 3.54 | 0.00134 |
| PG0257 | Conserved hypothetical protein | 3.27 | 0.03744 |
| PG0888 | Hypothetical protein | 3.20 | 0.02188 |
| PG1089 | DNA-binding response regulator RprY | 3.04 | 0.00349 |
| PG1960 | Ribosomal protein L28 | 2.95 | 0.01197 |
| PG0385 | Ribosomal protein S21 | 2.91 | 0.09041 |
| PG0707 | Hypothetical protein | 2.90 | 0.03812 |
| PG1076 | Acyl coenzyme A dehydrogenase, short chain specific | 2.84 | 0.01287 |
| PG0037 | Ribosomal protein L19 | 2.83 | 0.03776 |
| PG0595 | Ribosomal protein S6 | 2.83 | 0.03506 |
| PG2117 | Ribosomal protein S16 | 2.82 | 0.11780 |
| PG1134 | Thioredoxin reductase | 2.81 | 0.09180 |
| PG1108 | Hypothetical protein | 2.62 | 0.01818 |
| PG0594 | RNA polymerase sigma-70 factor | 2.62 | 0.02485 |
| PG0193 | Cationic outer membrane protein OmpH | 2.56 | 0.03085 |
| PG1542 | Collagenase | 2.53 | 0.02568 |
| PG0434 | Hypothetical protein | 2.53 | 0.01712 |
Gene number corresponds to TIGR ID.
Genes exhibiting a greater-than-2.5-fold decrease in transcription only.
TABLE 5.
Genes with increased level of expression in the oxyR mutant compared to Porphyromonas gingivalis W50 wild type (during anaerobic growth)
| Genea | Gene name/function | Other matchesb | Fold increasec | P value |
|---|---|---|---|---|
| PG0184 | ISPg1, transposase | PG1906, PG1448, PG0184, PG1624, PG1197, PG117, PG1031, PG0825 | 3.28 | 0.00839 |
| PG0852 | ISPg1, transposase, authentic frameshift | PG2169, PG2059, PG1845, PG1384, PG0764, PG0051, PG2193, PG2011, PG1909, PG1399, PG1349, PG1228, PG0939, PG0845, PG0760 | 3.24 | 0.01317 |
| PG0265 | Hypothetical protein | 2.82 | 0.16103 | |
| PG0051 | ISPg1, transposase, degenerate | PG1384, PG0760 | 2.57 | 0.03737 |
Gene number corresponds to TIGR ID.
Matching ORF information from The Bioinformatics Resources for Oral Pathogens (BROP) facility (http://www.brop.org). All matching ORFs correspond to IS1-related transposases.
Genes exhibiting a greater-than-2.5-fold induction in transcription only.
Confirmation of levels of expression of putative OxyR-dependent genes by real-time PCR.
Real-time PCR was used to evaluate expression levels of the eight genes most affected by the insertional inactivation of oxyR, as determined by microarray analysis. Relative quantification confirmed that the expression of these genes was decreased in the oxyR mutant (Table 6). Consistent with studies that suggest that DNA microarray analysis may underestimate changes in gene expression (46), the decrease as measured with real-time PCR was in some cases greater than the result obtained by microarray analysis. Real-time PCR was also used to analyze the patterns of expression of the putative OxyR-regulated genes in the wild type after treatment with H2O2. A decrease in expression in response to H2O2 was observed for all the genes analyzed. Microarray results also showed a decrease in expression of these genes but less than 2.5-fold (data not shown). The only exception was dps, where expression was slightly up-regulated (1.56-fold) when analyzed by microarrays.
TABLE 6.
Real-time PCR relative quantification of the expression of OxyR-dependent genesa
| Gene ID/name | wt | wt + H2O2 | oxyR mutant | oxyR mutant + H2O2 |
|---|---|---|---|---|
| PG0618/alkyl hydroperoxide reductase, C subunit | 1 (0.7-1.3) | ↓4.27 (2.0-9.1) | ↓92.00 (83.1-101.8) | ↓59.70 (54.1-65.78) |
| PG0090/Dps family protein | 1 (0.9-1.1) | ↓1.25 (1.0-1.5) | ↓11.7 (8.2-16.5) | ↓6.95 (6.0-8.1) |
| PG0421/hypothetical protein | 1 (0.8-1.4) | ↓3.64 (2.2-6.2) | ↓50.57 (41.1-62.2) | ↓70.4 (57.1-86.7) |
| PG1545/superoxide dismutase, Fe-Mn | 1 (0.7-1.4) | ↓24.25 (16.0-36.7) | ↓959.41 (640.4-1,437.1) | ↓749.2 (467.1-1,201.9) |
| PG1286/ferritin | 1 (0.7-1.4) | ↓2.95 (2.4-3.5) | ↓7.7 (5.0-11.9) | ↓17.7 (11.3-27.7) |
| PG0275/thioredoxin | 1 (0.7-1.4) | ↓8.41 (4.5-15.8) | ↓6.21 (5.4-7.1) | ↓9.2 (5.8-14.6) |
| PG1321/formate-tetrahydrofolate ligase | 1 (0.7-1.4) | ↓3.16 (2.0-5.1) | ↓4.91 (3.2-7.4) | ↓3.92 (2.18-7.03) |
All numbers represent the range of expression relative to the wild-type strain not treated with H2O2 (wt). Arrows indicate a decrease in expression. Average CT values for each gene were normalized to an endogenous control with similar amplification efficiency. Range (in parentheses) was determined by evaluation of the expression 2−ΔΔCT as described in Materials and Methods.
DISCUSSION
Our results show that the transcription of certain P. gingivalis antioxidant-related genes requires the presence of a functional OxyR within the cells. OxyR seems to operate differently in P. gingivalis compared to facultative anaerobic or aerobic microorganisms in which the regulator has been studied (9, 21, 44, 51). Furthermore, our results indicate that OxyR from P. gingivalis differs also from that of the anaerobe B. fragilis (31, 32). In P. gingivalis, under the conditions tested, the expression of the OxyR-dependent genes occurs during anaerobic growth and not in response to H2O2. The genes dependent on OxyR, however, seem important for the resistance of the microorganism to H2O2 exposure and aerotolerance. The ability to maintain constitutive expression of antioxidant genes might be in fact an advantage in the oral cavity, where oxidative stress is ubiquitous, and might represent an evolutionary adaptation to the oral environment. We cannot rule out, however, the possibility that other environmental conditions might further increase the expression of this set of genes, perhaps through different regulatory mechanisms. For example, a recent study has found that nine genes for which we report decreased levels of expression in the oxyR mutant (Table 4) were up-regulated in P. gingivalis after contact with epithelial cells (14). It was unexpected to find that treatment with H2O2 slightly decreased the levels of expression of the OxyR-dependent genes. This observation confirms that OxyR does not act as a sensor for H2O2 in P. gingivalis. The explanation for this effect, however, requires further investigation.
We demonstrated that the constitutive expression of the OxyR regulon was not a consequence of residual H2O2 in the medium, as addition of catalase did not significantly change expression of several OxyR-dependent genes. The finding that OxyR is constitutively active, despite anaerobic conditions, could be explained by the possibility that P. gingivalis possesses an OxyR molecule with a locked-oxidized conformation. Indeed, it has been demonstrated that certain mutations in oxyR are capable of inducing constitutive phenotypes in B. fragilis and E. coli (18, 32). Some of these strains have been isolated as spontaneous mutants that showed increased tolerance to H2O2. However, sequence analysis of P. gingivalis OxyR could not identify any amino acid substitutions that could correspond to those of the constitutive mutants. Since P. gingivalis oxyR shares only about 30% and 50% identity to oxyR from E. coli and B. fragilis, respectively, other substitutions/mutations in the nonconserved region of the sequence may be responsible for the constitutive phenotype. Another possible explanation for OxyR activation under anaerobic conditions is the lack of an effective system in P. gingivalis to maintain OxyR in its reduced form. OxyR is activated in E. coli by two mechanisms that include direct reaction with H2O2 and a change in the thiol-disulfide redox status of the cells (5). The latter is maintained by small proteins such as glutaredoxin 1 (grxA) and thioredoxin (trxA), which are able to reduce OxyR in vitro. However, it seems that glutaredoxin 1 (grxA) is preferred in vivo as the reductant of the disulfide bonds that lead to the deactivation of OxyR (5). The importance of these two thiol-disulfide-reducing systems in maintaining OxyR in its reduced state in E. coli has been confirmed by the observation that double mutants lacking these disulfide-reducing systems have a constitutively active phenotype whereby OxyR is activated without H2O2 treatment (5). In the P. gingivalis genome sequence, no homologue of glutaredoxin is present; however, a homologue of thioredoxin is found (50% sequence identity). Perhaps the reason why constitutive activation occurs in this anaerobe is the inability of the thioredoxin system to maintain the reduced status of OxyR. Also, the induction of transposase-related genes in the oxyR mutant under anaerobic conditions (Table 5) might indicate that the lack of expression of the OxyR-dependent genes creates an “oxidative-stress”-like response (as in Table 3), perhaps because of a change in the intracellular redox status.
Microarray analysis of the response to H2O2 in P. gingivalis revealed a limited ability to induce genes related to oxidative stress compared to that in a facultative anaerobe such as E. coli. A microarray analysis of E. coli gene expression in response to H2O2 showed induction of 140 genes more than fourfold (51). In contrast, P. gingivalis does not seem to possess such transcriptome versatility and a concerted up-regulation of transposase-related insertion elements was the only feature of the response to H2O2. Although the function of these transposase-encoding genes in P. gingivalis remains largely unknown, an increase in transposase activity in response to stress could be a way of increasing genomic plasticity and therefore diversity of the population, generating variants with better chances of surviving the unstable environmental conditions (7). Microarray analysis, however, does not allow determination of which specific transposase-related ORFs are up-regulated after H2O2 addition; therefore, the nature of this response requires further investigation.
Our results demonstrate that the P. gingivalis oxyR mutant is less resistant to oxygen and H2O2 exposure than is the wild type. However, it is interesting that both strains had the ability to recover and resume growth when treated with a sublethal concentration of H2O2 (Fig. 1). This observation suggests that P. gingivalis possesses OxyR-independent mechanisms for the detoxification of H2O2. One of these mechanisms could be rubrerythrin, which has been shown to be important for the H2O2 resistance of P. gingivalis (39) but was not identified as OxyR dependent in the present study.
Investigation of the transcriptome of the oxyR mutant by microarray analysis identified 28 genes that showed decreased expression after oxyR inactivation. It is not expected that all of these genes are directly OxyR regulated, as some are likely to be down-regulated due to the absence of the OxyR-dependent genes. It was nevertheless reassuring to find ahpC, dps, and ahpF as the genes most affected by deletion of oxyR. These three transcripts seem to be present at relatively high levels in bacterial cells (51), and perhaps the majority of the effect of oxyR inactivation could be attributed to their decrease. The following genes have also been affected by oxyR inactivation: PG0421, a hypothetical protein with no apparent homology to other oxidative-stress-related genes; superoxide dismutase, which has a clear role in oxidative stress protection but has not been demonstrated to date to be part of the OxyR regulon in other organisms; ferritin, partially regulated by OxyR in B. fragilis (33), possibly acting as an iron storage protein that decreases available intracellular iron and the production of ROS through the Fenton reaction (1); and thioredoxin, which has been shown to be part of the OxyR regulon in E. coli (51) and B. fragilis (31). Further studies to confirm OxyR binding ability to the promoter region of these genes are necessary.
OxyR has a role in H2O2 resistance as well as in aerotolerance in P. gingivalis. This might be a consequence of the fact that the OxyR regulon in this microorganism includes genes such as superoxide dismutase, regulated by SoxR in other bacteria. No homologous equivalent of SoxR was found in P. gingivalis. The small size of the genome of P. gingivalis (2.3 Mb) compared to other organisms such as E. coli (4.6 Mb) and B. fragilis (5.3 Mb) could perhaps be a result of the combination of various functions (H2O2 and O2·− protection in this case) in the same molecule. As opposed to anaerobic bacteria, the amplification of control mechanisms in aerobes might reflect the need to deal with more complex environments. Further comparative studies of the transcriptional switches operating in different anaerobic microorganisms might help us to understand the evolution of their antioxidant defenses.
Acknowledgments
S. R. Gill is acknowledged for his advice on the microarray experiments; A. Kingman and T. Wu for their help with statistical analysis; and G. Storz, J. Imlay, R. J. Palmer, Jr., N. S. Jakubovics, A. H. Rickard, and C. Seers for helpful discussions. pYH411 was a kind gift from Hisashi Yoshimoto, Department of Oral Biology, Kanagawa Dental College, Japan.
This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. M. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 4.Amano, A., H. Tamagawa, S. Shizukuishi, and A. Tsunemitsu. 1986. Superoxide dismutase, catalase and peroxidases in oral anaerobic bacteria. J. Osaka Univ. Dent. Sch. 26:187-192. [PubMed] [Google Scholar]
- 5.Aslund, F., M. Zheng, J. Beckwith, and G. Storz. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA 96:6161-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnard, J. P., and M. W. Stinson. 1999. Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect. Immun. 67:6558-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brawn, K., and I. Fridovich. 1981. DNA strand scission by enzymically generated oxygen radicals. Arch. Biochem. Biophys. 206:414-419. [DOI] [PubMed] [Google Scholar]
- 9.Chauvatcharin, N., S. Atichartpongkul, S. Utamapongchai, W. Whangsuk, P. Vattanaviboon, and S. Mongkolsuk. 2005. Genetic and physiological analysis of the major OxyR-regulated katA from Xanthomonas campestris pv. phaseoli. Microbiology 151:597-605. [DOI] [PubMed] [Google Scholar]
- 10.Dashper, S. G., L. Brownfield, N. Slakeski, P. S. Zilm, A. H. Rogers, and E. C. Reynolds. 2001. Sodium ion-driven serine/threonine transport in Porphyromonas gingivalis. J. Bacteriol. 183:4142-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fareleira, P., B. S. Santos, C. Antonio, P. Moradas-Ferreira, J. LeGall, A. V. Xavier, and H. Santos. 2003. Response of a strict anaerobe to oxygen: survival strategies in Desulfovibrio gigas. Microbiology 149:1513-1522. [DOI] [PubMed] [Google Scholar]
- 12.Harley, J. B., J. G. Flaks, H. Goldfine, M. E. Bayer, and H. Rasmussen. 1981. Hyperbaric oxygen toxicity and ribosome destruction in Escherichia coli K12. Can. J. Microbiol. 27:44-51. [DOI] [PubMed] [Google Scholar]
- 13.Herren, C. D., E. R. Rocha, and C. J. Smith. 2003. Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene 316:167-175. [DOI] [PubMed] [Google Scholar]
- 14.Hosogi, Y., and M. J. Duncan. 2005. Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect. Immun. 73:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imlay, J. A. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46:111-153. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, N. A., Y. Liu, and H. M. Fletcher. 2004. Alkyl hydroperoxide peroxidase subunit C (ahpC) protects against organic peroxides but does not affect the virulence of Porphyromonas gingivalis W83. Oral Microbiol. Immunol. 19:233-239. [DOI] [PubMed] [Google Scholar]
- 17.Kenny, E. B., and A. A. Ash, Jr. 1969. Oxidation reduction potential of developing plaque, periodontal pockets and gingival sulci. J. Periodontol. 40:630-633. [DOI] [PubMed] [Google Scholar]
- 18.Kullik, I., J. Stevens, M. B. Toledano, and G. Storz. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 177:1285-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C., S. M. Lee, P. Mukhopadhyay, S. J. Kim, S. C. Lee, W. S. Ahn, M. H. Yu, G. Storz, and S. E. Ryu. 2004. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11:1179-1185. [DOI] [PubMed] [Google Scholar]
- 20.Lynch, M. C., and H. K. Kuramitsu. 1999. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect. Immun. 67:3367-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maciver, I., and E. J. Hansen. 1996. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect. Immun. 64:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maley, J., N. Shoemaker, and I. S. Roberts. 1992. The introduction of colonic-Bacteroides shuttle plasmids into Porphyromonas gingivalis: identification of a putative P. gingivalis insertion-sequence element. FEMS Microbiol. Lett. 93:75-82. [DOI] [PubMed] [Google Scholar]
- 23.McCord, J. M., B. B. Keele, Jr., and I. Fridovich. 1971. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc. Natl. Acad. Sci. USA 68:1024-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mettraux, G. R., F. A. Gusberti, and H. Graf. 1984. Oxygen tension (pO2) in untreated human periodontal pockets. J. Periodontol. 55:516-521. [DOI] [PubMed] [Google Scholar]
- 25.Mouton, C., P. G. Hammond, J. Slots, and R. J. Genco. 1981. Serum antibodies to oral Bacteroides asaccharolyticus (Bacteroides gingivalis): relationship to age and periodontal disease. Infect. Immun. 31:182-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffman, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan, N., and J. A. Imlay. 2001. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol. Microbiol. 39:1562-1571. [DOI] [PubMed] [Google Scholar]
- 29.Park, M. K., R. A. Myers, and L. Marzella. 1992. Oxygen tensions and infections: modulation of microbial growth, activity of antimicrobial agents, and immunologic responses. Clin. Infect. Dis. 14:720-740. [DOI] [PubMed] [Google Scholar]
- 30.Pomposiello, P. J., and B. Demple. 2002. Global adjustment of microbial physiology during free radical stress. Adv. Microb. Physiol. 46:319-341. [DOI] [PubMed] [Google Scholar]
- 31.Rocha, E. R., C. D. Herren, D. J. Smalley, and C. J. Smith. 2003. The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe 9:165-173. [DOI] [PubMed] [Google Scholar]
- 32.Rocha, E. R., G. Owens, and C. J. Smith. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha, E. R., and C. J. Smith. 2004. Transcriptional regulation of the Bacteroides fragilis ferritin gene (ftnA) by redox stress. Microbiology 150:2125-2134. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Shah, H. N., and M. D. Collins. 1988. Proposal for the reclassification of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a new genus, Porphyromonas. Int. J. Syst. Bacteriol. 38:128-131. [Google Scholar]
- 36.Socransky, S. S., and A. D. Haffajee. 1994. Evidence of bacterial etiology: a historical perspective. Periodontol. 2000 5:7-25. [DOI] [PubMed] [Google Scholar]
- 37.Storz, G., L. A. Tartaglia, S. B. Farr, and B. N. Ames. 1990. Bacterial defences against oxidative stress. Trends Genet. 6:363-368. [DOI] [PubMed] [Google Scholar]
- 38.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 39.Sztukowska, M., M. Bugno, J. Potempa, J. Travis, and D. M. Kurtz. 2002. Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol. Microbiol. 44:479-488. [DOI] [PubMed] [Google Scholar]
- 40.Tanner, A. C., P. M. Milgrom, R. Kent, Jr., S. A. Mokeem, R. C. Page, C. A. Riedy, P. Weinstein, and J. Bruss. 2002. The microbiota of young children from tooth and tongue samples. J. Dent. Res. 81:53-57. [DOI] [PubMed] [Google Scholar]
- 41.Tao, K., N. Fujita, and A. Ishihama. 1993. Involvement of the RNA polymerase alpha subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol. Microbiol. 7:859-864. [DOI] [PubMed] [Google Scholar]
- 42.Thomas, S. J., and P. D. Eleazer. 2003. Aerotolerance of an endodontic pathogen. J. Endod. 29:644-645. [DOI] [PubMed] [Google Scholar]
- 43.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site—a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 44.Tseng, H. J., A. G. McEwan, M. A. Apicella, and M. P. Jennings. 2003. OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueshima, J., M. Shoji, D. B. Ratnayake, K. Abe, S. Yoshida, K. Yamamoto, and K. Nakayama. 2003. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 71:1170-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan, X. F., N. C. Verberkmoes, L. A. McCue, D. Stanek, H. Connelly, L. J. Hauser, L. Wu, X. Liu, T. Yan, A. Leaphart, R. L. Hettich, J. Zhou, and D. K. Thompson. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 186:8385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27:648-657. [DOI] [PubMed] [Google Scholar]
- 48.Yang, E. Y., A. C. Tanner, P. Milgrom, S. A. Mokeem, C. A. Riedy, A. T. Spadafora, R. C. Page, and J. Bruss. 2002. Periodontal pathogen detection in gingiva/tooth and tongue flora samples from 18- to 48-month-old children and periodontal status of their mothers. Oral Microbiol. Immunol. 17:55-59. [DOI] [PubMed] [Google Scholar]
- 49.Yoshimoto, H., Y. Takahasi, D. Kato, and T. Umemoto. 1997. Construction of a plasmid vector for transformation of Porphyromonas gingivalis. FEMS Microbiol. Lett. 152:175-181. [DOI] [PubMed] [Google Scholar]
- 50.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]
- 51.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]