Abstract
Open reading frame (ORF) Mm2058 of the methanogenic archaeon Methanosarcina mazei strain Gö1 was shown in vivo and in vitro to encode the nonorthologous replacement of the α-ribazole-phosphate phosphatase (CobC; EC 3.1.3.73) enzyme of Salmonella enterica serovar Typhimurium LT2. Bioinformatics analysis of sequences available in databases tentatively identified ORF Mm2058, which was cloned under the control of an inducible promoter and was used to support growth of an S. enterica strain under conditions that demanded CobC-like activity. The Mm2058 protein was expressed with a decahistidine tag at its N terminus and was purified to homogeneity using nickel affinity chromatography. High-performance liquid chromatography followed by electrospray ionization mass spectrometry showed that the Mm2058 protein had phosphatase activity that converted α-ribazole-5′-phosphate to α-ribazole, as reported for the bacterial CobC enzyme. On the basis of the data reported here, we refer to ORF Mm2058 as cobZ. We tested the prediction by Rodionov et al. (D. A. Rodionov, A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand, J. Biol. Chem. 278:41148-41159, 2003) that ORF HSL01294 (also called Vng1577) encoded the nonorthologous replacement of the bacterial CobC enzyme in the extremely halophilic archaeon Halobacterium sp. strain NRC-1. A strain of the latter carrying an in-frame deletion of ORF Vng1577 was not a cobalamin auxotroph, suggesting that either there is redundancy of this function in Halobacterium or the gene was misannotated.
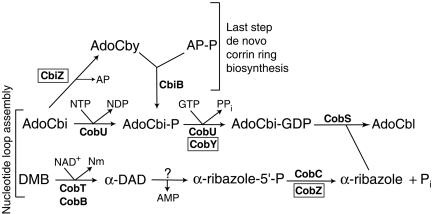
Cobalamin (Cbl; also called B12) is a cobalt-containing cyclic tetrapyrrole whose synthesis requires a great deal of genetic information (18, 20). Cbl is an essential nutrient for many organisms, including humans; however, its biosynthesis is restricted to bacteria and archaea (19, 22, 27, 29, 30). Although most of our current knowledge of how Cbl is assembled comes from research using bacterial systems (reviewed in references 6 and 27), important differences in corrinoid salvaging and in the late steps of the pathway in archaea have been reported (25, 28-30) (Fig. 1).
FIG. 1.
Late steps in corrin ring biosynthesis in Salmonella enterica. Shown is a model of the enzymatic reactions needed for nucleotide loop assembly in S. enterica. Shown are the enzymes, substrates, and intermediates in the pathway. Abbreviations: AdoCby, adenosylcobyric acid; AdoCbi, adenosylcobinamide; AdoCbl, adenosylcobalamin; α-DAD, α-5,6-dimethylbenzimidazole adenine dinucleotide; Nm, nicotinamide; AP, aminopropanol; AP-P, aminopropanol phosphate; CbiB, putative AdoCbi-phosphate synthase; CobU, AdoCbi kinase and guanylyltransferase; CobT, NaMN:5,6-dimethylbenzimidazole phosphoribosyltransferase; CobS, adenosylcobalamin synthase; CobC (CobZ in M. mazei), α-ribazole-phosphate phosphatase; CbiZ, amidohydrolase. Archaeal enzymes are boxed.
Bioinformatics analysis of available genome sequences, such as the one performed by Rodionov et al. (18), serve as tools to predict, based on homology, which organisms may have the capacity to synthesize Cbl and what the underpinning biochemistry may be. In some cases, however, when no orthologous genes are identified, the existence of new enzymes catalyzing similar reactions (25, 29) or new enzymes comprising new pathways is plausible (28, 30). The availability of genetic systems like the one in Salmonella enterica greatly facilitates the search for nonorthologous replacement functions using in vivo genetic screens or selections aimed at restoring growth by plasmid-encoded archaeal enzymes.
The absence of an ortholog of the S. enterica cobC gene in sequenced archaeal genomes suggested that either archaea use an enzyme of distinct evolutionary origin to dephosphorylate α-ribazole-5′-phosphate (α-RP) or the latter is not the substrate for the archaeal enzyme. In S. enterica, the product of the CobC reaction (EC 3.1.3.73) (α-ribazole [α-R]) is the cosubstrate of the cobalamin synthase (CobS; EC 2.7.8.26) enzyme. The bioinformatics analysis by Rodionov et al. identified a putative gene (referred to as cobZ) which they proposed encoded the nonorthologous replacement of the bacterial CobC enzyme in all archaeal genomes except Sulfolobus solfataricus and Thermoplasma spp., which have an apparent cobC ortholog, and Archaeoglobus fulgidus and Halobacterium spp., which do not have a cobC ortholog (18). It was suggested that in Halobacterium sp. strain NRC-1, open reading frame (ORF) HSL01294 (Vng1577) encoded the nonorthologous replacement of CobC. To date, no experimental evidence supporting the involvement of the hypothetical CobZ or Vng1577 protein in cobamide synthesis has been reported.
A role for the Halobacterium sp. strain NRC-1 ORF Vng1577 gene product in cobalamin synthesis (if any) is not discernible.
We tested whether a function encoded by ORF Vng1577 of Halobacterium sp. was required for Cbl synthesis in this archaeon. For this purpose, an in-frame deletion of ORF Vng1577 was constructed using reported methodology and verified by DNA sequencing using BigDye (ABI PRISM) protocols (Biotechnology Center, University of Wisconsin—Madison) (17, 30). Deletion of ORF Vng1577 (strain JE6693; Table 1) did not cause any discernible Cbl phenotype (data not shown), suggesting either that the protein encoded by ORF Vng1577 was not involved in de novo Cbl biosynthesis or that redundant functions exist in Halobacterium sp. strain NRC-1. We also investigated whether the putative Vng1577 protein was involved in salvaging the precursor cobyric acid (Cby) or cobinamide (Cbi) (30). JE6792, a strain of Halobacterium sp. strain NRC-1 carrying a block in de novo corrin ring biosynthesis and a chromosomal in-frame deletion of ORF Vng1577 (ΔVng1577-1 ΔcbiP1), grew when the medium was supplemented with Cby or Cbi, indicating that the putative Vng1577 gene product did not have a role in the conversion of either one of these precursors to cobalamin or that redundant precursor-salvaging functions exist in this archaeon.
TABLE 1.
Strains and plasmidsa
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Salmonella enterica | ||
| TR6583, formerly SA2929 | metE205 ara-9 | K. Sanderson via J. Roth |
| Derivatives of TR6583 | ||
| JE2192 | cobT109::MudI1734b (lacZ+kan+) cobC1175::Tn10d16d17 (tet+) | Laboratory collection |
| JE7493 | JE2192/pCOBZ2 | |
| JE7404 | JE2192/pJO46 | |
| JE7403 | JE2192/pT7-7 | |
| JE7492 | JE2192/pT7-5 | |
| Escherichia coli | ||
| BL21 λDE3 | PlacUV5-T7 RNA polymerase | Novagen |
| Plasmids | ||
| pCOBZ1 | M. mazei Gö1 cobZ+ in pET-16b | |
| pCOBZ2 | M. mazei Gö1 cobZ+ in pT7-7 bla+ | |
| pJO46 | cobC+ in pT7-5 bla+ | 15 |
| pT7-5, pT7-7 | Cloning vectors bla+ | 23 |
| pET-16b | Cloning vector bla+ | Novagen |
| Halobacterium sp. strains | ||
| JE6693 | Δura-3 ΔVng1577-1 | |
| JE6792 | Δura-3 ΔVng1577-1 ΔcbiP1 |
All S. enterica strains used in these studies carry a null allele of the metE gene (16), making growth of the cell dependent on the activity of the Cbl-dependent methionine synthase (MetH) enzyme (9, 10, 24). No-carbon E (NCE) medium (2) was used as minimal medium. When added, the following supplements were at the indicated concentrations: glucose, 11 mM; MgSO4, 1 mM; corrinoids (cobyric acid, cobinamide, cobalamin), 10 nM; 5,6-dimethylbenzimidazole (DMB), 0.3 mM; and trace minerals, 10 ml/liter (1). All cobyric acid (Cby), Cbi, and Cbl (Sigma) were added in their cyano forms; DMB was purchased from Aldrich. Nutrient broth (NB; Difco Laboratories) (0.8%, wt/vol) containing NaCl (85 mM) was used as rich medium to culture S. enterica strains; lysogeny broth (LB) (3, 4) was used as rich medium to culture E. coli strains. Strains and plasmids were constructed during the course of this work unless stated otherwise.
The MudI1734 element is described elsewhere (8).
ORF Mm2058 of the methanogenic archaeon Methanosarcina mazei strain Gö1 encodes a protein with α-ribazole-5′-phosphate phosphatase activity.
We investigated the possibility that the putative cobZ gene encoded the archaeal nonorthologous replacement of the bacterial CobC enzyme. We chose the putative cobZ gene (ORF Mm2058) of M. mazei Gö1 because in the past we successfully expressed Cbl biosynthetic genes from this archaeon in Salmonella enterica (28).
i. In vivo studies.
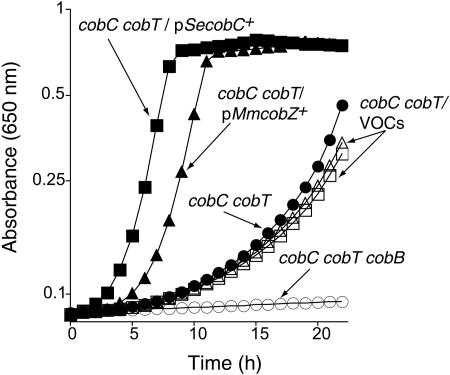
Using standard recombinant DNA techniques, we cloned the wild-type allele of ORF Mm2058 (cobZ) from M. mazei into plasmid pT7-7 and used the resulting plasmid (pCOBZ2 MmcobZ+) in complementation studies. We introduced plasmid pCOBZ2 into a cobC cobT S. enterica strain to ask whether CobZ would restore Cbl biosynthesis from Cbi and 5,6-dimethylbenimidazole (DMB) in this strain. The effect of the lack of CobC activity on nucleotide loop assembly from Cbi and DMB is best assessed in a genetic background that lacks NaMN:DMB phosphoribosyltransferase (CobT; EC 2.4.2.21) activity (15). In such a genetic background, the ability of the cobC cobT strain (JE2192; Table 1) to assemble the nucleotide loop is substantially impaired relative to the cobC cobTpSecobC+ strain (Fig. 2, open versus solid squares; Table 1, JE7404). The residual ability of strain JE2192 to assemble the nucleotide loop was due to CobB sirtuin function in the background (Fig. 2, open versus closed circles). As expected, a cobC cobT strain harboring an empty vector (JE7403 and JE7492; Table 1) made Cbl from Cbi and DMB, but the growth kinetics was very different from that of cultures of the cobC cobT strain harboring a plasmid carrying the wild-type allele of the S. enterica cobC gene or the M. mazei cobZ gene (Fig. 2, open versus solid squares and open versus solid triangles). This was strong in vivo evidence supporting the conclusion that the archaeal CobZ function compensated for the lack of CobC during the assembly of the nucleotide loop in the cobC S. enterica strain.
FIG. 2.
CobZ compensates for the lack of CobC during Cbl-dependent growth of a S. enterica cobC strain. Plasmid pMmcobZ+ (pCOBZ2) contained a wild-type allele of M. mazei strain Göl cobZ+ gene (ORF Mm2058; hypothetical protein, accession no. NP_634082) under the control of the T7 promoter and ribosome-binding site present in vector pT7-7 (23). The 5′ primer MM2058-5′-NdeI (5′-CATATGCATATGAAGTTATCAGATATTGAGGA-3′; restriction enzyme site is underlined) and the reverse primer MM2058-3′-BamHI (5′-CATTATGTACCGCCGGATCCTCAGTCTTTTCTTT-3′) were used to amplify a 569-bp fragment from M. mazei genomic DNA. The resulting product contained an ATG initiation codon instead of the original TTG start site. The amplified fragment was cut with NdeI/BamHI restriction enzymes, gel purified, and cloned into the NdeI/BamHI restriction site of the cloning vector pT7-7. Cbl-dependent growth was assessed in minimal medium supplemented with glucose, Cbi, and DMB. The following plasmids were introduced into strain JE2192 (cobC cobT): VOCs (vector-only controls), plasmids pT7-7 and pT7-5; plasmid pSecobC+ (pJO46); and plasmid pMmcobZ+ (pCOBZ2). Growth at 37°C with continuous shaking (19 Hz) was monitored using an EL808 UltraMicroplate Reader (BioTek Instruments). Plasmids were introduced into S. enterica by electroporation (14). S. enterica strains were grown to full density (2 × 109 CFU/ml) in nutrient broth supplemented with ampicillin (Ap; 100 μg/ml) to ensure that plasmids were maintained.
ii. In vitro studies.
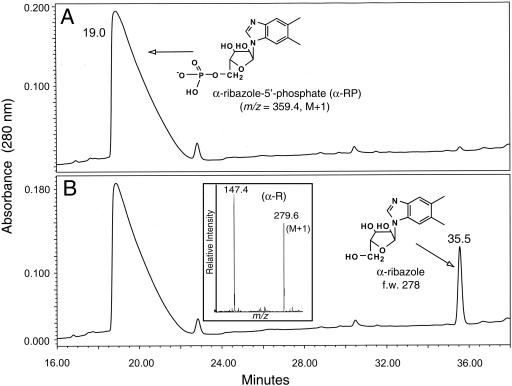
To provide in vitro support for the complementation studies described above, the cobZ+ allele of M. mazei was cloned into vector pET-16b to direct the synthesis of a CobZ protein with a decahistidine tag fused to its N terminus (H10CobZ; plasmid pCOBZ1). We purified H10CobZ protein using an ÄKTA explorer (Amersham Pharmacia Biotech) fast protein liquid chromatography system equipped with a prepacked nickel-affinity chromatography HiTrap chelating column (Amersham Pharmacia Biotech). The column was developed with a linear gradient of 1× binding buffer (20 mM phosphate, 0.5 M NaCl, pH 7.4 to 7.6) containing increasing amounts of imidazole (from 10 to 500 mM). After purification, H10CobZ protein was dialyzed using snakeskin-pleated dialysis tubing with a molecular weight cutoff of 10,000 (Pierce) at 4°C against 1 liter of 50 mM PIPES buffer, pH 6.8, containing EDTA (5 mM). Purification of the H10CobZ protein was monitored using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (11), and protein was visualized by staining gels with Coomassie blue (21); protein concentration was determined as described previously (5). Highly purified H10CobZ protein (>98% homogeneity; data not shown) was incubated with α-RP, and product formation was monitored by reverse-phase high-performance liquid chromatography. As shown in Fig. 3, CobZ dephosphorylated α-RP into α-R, whose identity was confirmed using electrospray ionization mass spectrometry (Fig. 3B, inset, m/z = 279.6), by its UV-visible spectrum (data not shown) (7, 13), and by the retention time of α-R (35.5 min) generated by incubating α-RP with alkaline phosphatase (data not shown).
FIG. 3.
CobZ protein dephosphporylates α-RP. Shown are the high-pressure liquid chromatograms of CobZ reactions. A. Reaction mixture containing heat-inactivated CobZ enzyme; B. CobZ reaction. α-RP and α-ribazole eluted 19 and 35.5 min after injection, respectively. The inset in panel B shows the electrospray ionization mass spectrometry spectrum (positive ionization mode) of the reaction product. The signals with m/z values of 279.6 (M + 1) and 147.4 (M + 1) were consistent with the molecular masses of α-ribazole and DMB, respectively. Substrate and products of the CobZ reaction were filtered using a Spin X centrifuge tube filter (Corning) and analyzed by reverse-phase high-pressure liquid chromatography as described previously (12, 26). Phosphatase activity was measured in reaction mixtures (100 μl) containing α-RP (40 nmol), in 30 mM Tris-HCl buffer (pH 8) containing MgCl2 (9 mM) and KCl (0.1 M), and in homogeneous H10CobZ protein (5 μg [0.25 nmol]). Reaction mixtures were prepared on ice, transferred, and kept in a 37°C heater for 1 h and then heated to 80°C for 10 min to terminate the reaction. Heat-inactivated H10CobZ protein was used as a negative control, and shrimp alkaline phosphatase (Promega) was used as a positive control. High-performance liquid chromatography-purified product of the CobZ reaction was dried under vacuum using a SpeedVac concentrator (Savant) prior to mass spectrometry analysis at the MS Facility at the University of Wisconsin-Madison Biotechnology Center. Mass spectra were obtained using a Perkin-Elmer Sciex API365 mass spectrometer. f.w., formula weight.
Conclusions.
We have identified the archaeal gene encoding the nonorthologous replacement of the bacterial CobC enzyme (EC 3.1.3.73), which dephosphorylates α-ribazole-5′-phosphate (α-RP) to yield α-ribazole (α-R) during the late steps of cobalamin biosynthesis. The gene encoding the archaeal α-ribazole-5′-phosphate phosphatase is cobZ. These studies illustrate the positives and negatives of gene annotation without experimental support. The experimental data do not support the assignment of ORF HSL01294 in Halobacterium spp. as the gene encoding the nonorthologous replacement of CobC. In contrast, such a prediction is supported by the data in the case of ORF Mm2058 in Methanosarcina mazei Gö1. We suggest that, to avoid confusion, genes be named only when experimental support for the function of the encoded protein exists.
Acknowledgments
This work was supported by NIH grant GM40313 to J.C.E.-S. C.L.Z. was supported in part by a Ruth L. Kirschstein National Research Service Award (F31-GM64009). J.D.W. was supported in part by the Ira Baldwin and the Jerome Stefaniak fellowships awarded by the Department of Bacteriology, University of Wisconsin—Madison.
We thank P. Renz for his gift of CNCby and G. Gottschalk for his gift of M. mazei DNA.
REFERENCES
- 1.Balch, W. E., and R. S. Wolfe. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-255. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. L. 2005. Chemistry and enzymology of vitamin B12. Chem. Rev. 105:2075-2149. [DOI] [PubMed] [Google Scholar]
- 7.Brown, K. L., J. M. Hakimi, D. M. Nuss, Y. D. Montejano, and D. W. Jacobsen. 1984. Acid-base properties of α-ribazole and the thermodynamics of dimethylbenzimidazole association in alkylcobalamins. Inorg. Chem. 23:1463-1471. [Google Scholar]
- 8.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusions with mini-Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drennan, C. L., S. Huang, J. T. Drummond, R. G. Matthews, and M. L. Ludwig. 1994. How a protein binds B12: a 3.0A X-ray structure of B12-binding domains of methionine synthase. Science 266:1660-1674. [DOI] [PubMed] [Google Scholar]
- 10.Hall, D. A., C. W. Vander Kooi, C. N. Stasik, S. Y. Stevens, E. R. Zuiderweg, and R. G. Matthews. 2001. Mapping the interactions between flavodoxin and its physiological partners flavodoxin reductase and cobalamin-dependent methionine synthase. Proc. Natl. Acad. Sci. USA 98:9521-9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage and structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 1999. In vitro synthesis of the nucleotide loop of cobalamin by Salmonella typhimurium enzymes. Proc. Natl. Acad. Sci. USA 96:11798-11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Toole, G. A. 1994. Biochemistry and genetics of cobalamin nucleotide loop assembly in Salmonella typhimurium. Ph.D. dissertation. University of Wisconsin—Madison.
- 14.O'Toole, G. A., M. R. Rondon, and J. C. Escalante-Semerena. 1993. Analysis of mutants of Salmonella typhimurium defective in the synthesis of the nucleotide loop of cobalamin. J. Bacteriol. 175:3317-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Toole, G. A., J. R. Trzebiatowski, and J. C. Escalante-Semerena. 1994. The cobC gene of Salmonella typhimurium codes for a novel phosphatase involved in the assembly of the nucleotide loop of cobalamin. J. Biol. Chem. 269:26503-26511. [PubMed] [Google Scholar]
- 16.Peariso, K., Z. S. Zhou, A. E. Smith, R. G. Matthews, and J. E. Penner-Hahn. 2001. Characterization of the zinc sites in cobalamin-independent and cobalamin-dependent methionine synthase using zinc and selenium X-ray absorption spectroscopy. Biochemistry 40:987-993. [DOI] [PubMed] [Google Scholar]
- 17.Peck, R. F., S. Dassarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 18.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278:41148-41159. [DOI] [PubMed] [Google Scholar]
- 19.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137-181. [DOI] [PubMed] [Google Scholar]
- 20.Roth, J. R., J. G. Lawrence, M. Rubenfield, S. Kieffer-Higgins, and G. M. Church. 1993. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 175:3303-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasse, J. 1991. Detection of proteins, p. 10.6.1-10.6.8. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. Wiley Interscience, New York, N.Y. [Google Scholar]
- 22.Scott, A. I. 2003. Discovering nature's diverse pathways to vitamin B12: a 35-year odyssey. J. Org. Chem. 68:2529-2539. [DOI] [PubMed] [Google Scholar]
- 23.Tabor, S. 1990. Expression using the T7 RNA polymerase/promoter system, p. 16.2.1.-16.2.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. Wiley Interscience, New York, N.Y. [Google Scholar]
- 24.Taylor, R. T., and H. Weissbach. 1973. N5-methylenetetrahydrofolate-homocysteine methyltransferases, p. 121-165. In P. D. Boyer (ed.), The enzymes, vol. 9. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 25.Thomas, M. G., and J. C. Escalante-Semerena. 2000. Identification of an alternative nucleoside triphosphate: 5′-deoxyadenosylcobinamide phosphate nucleotidyltransferase in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 182:4227-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trzebiatowski, J. R., and J. C. Escalante-Semerena. 1997. Purification and characterization of CobT, the nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J. Biol. Chem. 272:17662-17667. [DOI] [PubMed] [Google Scholar]
- 27.Warren, M. J., E. Raux, H. L. Schubert, and J. C. Escalante-Semerena. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 19:390-412. [DOI] [PubMed] [Google Scholar]
- 28.Woodson, J. D., and J. C. Escalante-Semerena. 2004. CbiZ, an amidohydrolase enzyme required for salvaging the coenzyme B12 precursor cobinamide in archaea. Proc. Natl. Acad. Sci. USA 101:3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodson, J. D., R. F. Peck, M. P. Krebs, and J. C. Escalante-Semerena. 2003. The cobY gene of the archaeon Halobacterium sp. strain NRC-1 is required for de novo cobamide synthesis. J. Bacteriol. 185:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodson, J. D., C. L. Zayas, and J. C. Escalante-Semerena. 2003. A new pathway for salvaging the coenzyme B12 precursor cobinamide in archaea requires cobinamide-phosphate synthase (CbiB) enzyme activity. J. Bacteriol. 185:7193-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]