Abstract
Agrobacterium tumefaciens wild-type strains have a unique quorum-sensing (QS)-dependent Ti plasmid conjugative transfer phenotype in which QS signaling is activated by corresponding conjugative opine inducers. Strain K588, with a nopaline-type chromosomal background harboring an octopine-type Ti plasmid, however, is a spontaneous mutant displaying a constitutive phenotype in QS. In this study, we show that a single amino acid mutation (L54P) in the QS antiactivator TraM encoded by the traM gene of Ti plasmid is responsible for the constitutive phenotype of strain K588. Introduction of the L54P point mutation to the TraM of wild-type strain A6 by allelic replacement, however, failed to generate the expected constitutive phenotype in this octopine-type strain. Intriguingly, the QS-constitutive phenotype appeared when the pTiA6 carrying the mutated traM was placed in the chromosomal background of the nopaline-type strain C58C1RS, suggesting an unknown inhibitory factor(s) encoded by the chromosomal background of strain A6 but not by C58C1RS. Low-stringency Southern blotting analysis showed that strain A6, but not strain C58 and its derivatives, contains a second traM homologue. The homologue, designated traM2, has 64% and 65% identities with traM at the DNA and peptide levels, respectively. Similar to TraM, TraM2 is a potent antiactivator that functions by blocking TraR, the QS activator, from specific binding to the tra gene promoters. Deletion of traM2 in strain A6 harboring the mutated traM confers a constitutive QS phenotype. The results demonstrate that the QS system in strain A6 is subjected to the dual control of TraM and TraM2.
Quorum sensing (QS) is a bacterial community genetic regulatory mechanism that controls diverse biological functions in different bacterial species. Among the several bacterial QS systems reported (for reviews, see references 2, 32, and 38), the most characterized one is the acylhomoserine lactone (AHL)-based QS system. AHL signals are known to regulate many interesting microbial activities, including bioluminescence in Vibrio species (7), Ti plasmid conjugative transfer in Agrobacterium tumefaciens (23, 36), antibiotics production and virulence in Erwinia carotovora (25), and virulence and biofilm formation in Pseudomonas aeruginosa (5, 20). The central components of AHL-based QS systems are LuxI (I)- and LuxR (R)-type proteins. The former is the AHL synthase and the latter is the AHL-dependent transcription factor, both of which are conserved in different bacterial species containing AHL-based QS systems (11). In most cases, at low bacterial population density, the I-type enzyme produces a basal level of AHL signals, which accumulate as bacterial cells proliferate and interact with the R-type transcription factors. Subsequently, the R-AHL complexes induce higher-level expression of I-type enzymes, which boosts AHL production (AHL signals are also known as autoinducers), and activate the transcriptional expression of other QS-dependent genes (37).
Differing from the above generally conserved QS model, the QS system in the wild-type strains of A. tumefaciens, which regulates Ti plasmid conjugative transfer, is normally not active until the bacterial cells detect conjugative opines produced by the crown gall tumors incited by the A. tumefaciens strain harboring this Ti plasmid (9). The conjugative opines regulate QS and, hence, Ti plasmid conjugative transfer by controlling the expression of traR, which encodes the R-type protein of A. tumefaciens. More specifically, in A. tumefaciens octopine-type strains, the QS-dependent Ti plasmid conjugative transfer is induced by octopine (21), while in nopaline-type strains, it is induced by agrocinopines A and B (8). In nopaline strain C58, traR is a member of a five-gene operon of pTiC58, which is expressed from a promoter regulated by the transcriptional repressor AccR. Repression by AccR is relieved in the presence of agrocinopine A or B, and the operon, including traR, is expressed (22), whereas in octopine strains, traR is located in a 14-member operon that is regulated by the transcription factor OccR (12). OccR acts either as a repressor or an activator depending on the absence or presence of octopine, respectively, by binding to different positions of the promoter of the traR operon (31).
TraR activity is negatively modulated by antiactivator TraM. TraM interferes with TraR function by formation of a stable antiactivation complex that prevents TraR from recognizing its target DNA-binding sites (4, 17, 29, 30). It was shown that TraM is critical for setting the QS threshold in A. tumefaciens and for prevention of TraR from initiating transcription of the tra regulon in the absence of opine inducer (24). In octopine-mannityl opine-type strains, a Ti plasmid-encoded TrlR, which is a truncated version of TraR with the DNA-binding domain at its carboxyl terminus eliminated due to a frameshift mutation, could also interfere with the function of TraR (19, 41). Purified TrlR formed inactive heterodimers with TraR in the presence of AHL signals and blocked TraR for both specific DNA binding and transcription of a tra promoter (3).
In A. tumefaciens, production of the AHL signals, including N-(3-oxo-octanoyl)-l-homoserine lactone and other minor derivatives, is encoded by traI (15, 36), which is under the control of the transcription factor TraR. The findings that TraR is subject to several levels of regulations and modulations may explain our early observation that production of the AHL signal is hardly detectable in the wild-type A. tumefaciens strains in the absence of opine inducers (39). Nevertheless, spontaneous mutants of A. tumefaciens, which are constitutive in AHL signal production and Ti plasmid conjugative transfer, have been identified (1, 16, 39). Genetic analysis of a spontaneous Trac (transfer constitutive) mutant of nopaline strain C58 showed that mutation of the repressor AccR is responsible for the constitutive phenotype (1). Transposon mutagenesis studies revealed that knockout of traM can also lead to constitutive AHL production and Ti plasmid conjugative transfer in nopaline strain C58 and octopine strain R10 (10, 14).
We have previously reported a spontaneous Trac mutant K588 of A. tumefaciens (34); the mutant, carrying a Trac derivative of the octopine-type plasmid pTiB6S3 in the genetic background of nopaline-type strain C58C1RS, is constitutive in AHL production. Although the mechanism is unknown, the constitutive nature of this mutant has greatly facilitated the in vivo investigations of genetic regulation of QS signal degradation in A. tumefaciens, as a wild-type strain such as A6 does not produce a detectable level of AHL signals in liquid medium even in the presence of octopine (33, 34). In this study, we have investigated the QS constitutive nature of strain K588. By DNA sequencing and genetic analysis, we identified a single nucleotide mutation in the traM of pTiK588 that accounts for the QS constitutive phenotype of strain K588. Based on this finding, we questioned whether introduction of the same mutation in the traM of pTiA6 could also render a QS constitutive phenotype in strain A6. The analysis led to identification of a traM homologue in octopine-type strains A6 and Ach5C3. We further demonstrated that the traM homologue, designated traM2, also encodes a potent QS antiactivator.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. A. tumefaciens strains were grown at 28°C in LB medium (per liter contains 10 g Bacto tryptone, 5 g yeast extract, and 10 g NaCl, pH 7.0) or in BM minimal medium (basic minimum nutrient with 0.2% mannitol added as the sole carbon source and 0.2% ammonia sulfate as the sole nitrogen source unless otherwise indicated) (33, 39). Escherichia coli strains were grown at 37°C in LB medium. Antibiotics were added at the following concentrations when required: kanamycin, 50 μg/ml (A. tumefaciens) or 100 μg/ml (E. coli); tetracycline, 5 μg/ml (A. tumefaciens) or 10 μg/ml (E. coli); rifampin, 100 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotypes and/or characteristicsa | Source or reference |
|---|---|---|
| A. tumefaciens | ||
| NT1(traR tra::lacZ749) | C58C1RS harboring pJM749 and pSVB33, AHL bioassay indicator strain | 25 |
| K588 | C58C1 harboring octopine type Ti plasmid pTiB6S3Trac, Eryr, Chlr | A. Kerr |
| A6 | Wild-type octopine strain containing pTiA6, Trai | A. Kerr |
| Ach5 | Wild-type octopine strain containing pTiAch5, Trai | C. Fuqua |
| R10 | Wild-type octopine strain containing pTiR10, Trai | C. Fuqua |
| B6 | Wild-type octopine strain containing pTiB6, Trai | C. Fuqua |
| B6S3 | Wild-type octopine strain containing pTiB6S3, Trai | C. Fuqua |
| ATCC 15955 | Wild-type octopine strain containing pTi15955, Trai | C. Fuqua |
| C58 | Wild-type nopaline strain containing pTiC58, Trai | S. Q. Pan |
| C58C1RS | Derivative of strain C58, pTi−, Rifr, recipient strain for Ti plasmid conjugative transfer | S. Q. Pan |
| K749 | Derivative of strain C58, pTi−, pAT−, Rifr | A. Kerr |
| Ach5C3 | Derivative of strain Ach5, Ti plasmid cured, Rifr | S. K. Farrand |
| A6(pTiA6traMK588) | Derivative of strain A6 where the traM gene was replaced with the traM from strain K588 | This study |
| C58C1RS(pTiA6traMK588) | Derivative of strain C58C1RS with the Ti plasmid donated by A6(pTiA6traMK588) | This study |
| C58C1RS(pTiK588) | Derivative of strain C58C1RS harboring pTiK588 of K588 | This study |
| Ach5C3(pTiA6traMK588) | Derivative of strain Ach5C3 with the Ti plasmid donated by A6(pTiA6traMK588) | This study |
| Ach5C3(pTiK588) | Derivative of strain Ach5C3 harboring pTiK588 of K588 | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 λpir | Laboratory collection |
| S17-1 | pro res− mod+ integrated copy of RP4, mob+ | A. Kerr |
| Plasmids | ||
| pLAFR3 | IncP, broad-host-range cosmid vector, Tetr | 28 |
| pAW19 | f1(+)ori lacZα of pBluescript II (SK+), sacB, Sucr | S. K. Farrand |
| pLA-traMA6-PT | pLAFR3 harboring the coding region of traM amplified from pTiA6; the gene is under the control of the lac promoter of the vector, Tetr | This study |
| pLA-traM2 | pLAFR3 harboring the coding region of traM2 that is placed under the control of the lac promoter of the vector, Tetr | This study |
| pJZ358 | Expression construct for TraRR10 expression | 41 |
| pGC-traM2 | Expression construct for TraM2 expression | This study |
| pBK129 | Cosmid clone containing about 26-kb tra region from pTiK588 | This study |
| pAW19-traMK588 | pAW19 containing the traM coding sequences and its flanking region from pTiK588 | This study |
| pAW-traM2d | pAW19 containing the flanking regions of traM2 from strain A6 | This study |
Abbreviations: Trac, constitutive in AHL production and Ti plasmid conjugative transfer; Trai, AHL production and Ti plasmid conjugative transfer are inducible by opine inducer; Ampr, ampicilin resistant; Kanr, kanamycin resistant; Tetr, tetracycline resistant; Rifr, rifampicin resistant; Eryr, erythromycin resistant; Chlr, chloramphenicol resistant; Sucr, sucrose resistant.
DNA manipulation and plasmid construction.
Plasmids were purified using a plasmid miniprep kit as recommended by the manufacturer (QIAGEN). PCR product purification and DNA recovery from agarose gel were carried out with the QIAquick PCR purification kit and QIAquick gel extraction kit (QIAGEN), respectively. For construction of pAW-traMK588, the DNA fragment containing traM was amplified from A. tumefaciens strain K588 with forward primer 5′-CCCAAGCTTGATCCTAACTATTTCTCCTG and reverse primer 5′-CGGGATCCGTTTCGAGGCTCATCAAAAG, end-blunted using Klenow enzyme before ligation to vector pAW19 (18), which was cut by NotI and subsequently filled in with Klenow enzyme. The resulting plasmid was screened by PCR and confirmed by DNA sequencing. The construct pLA-traMA6-PT was assembled as follows: the allele containing the TraM-encoding region and its putative promoter was cloned from A. tumefaciens A6 with the PCR primers 5′-CCCAAGCTTGATCCTAACTATTTCTCCTG and 5′-CGGGATCCGTTTCGAGGCTCATCAAAAG (enzymatic cut sites underlined), enzyme digested, and introduced between the BamHI and HindIII sites of pLAFR3. The resulting construct was further analyzed to ensure that the insert was placed in the orientation opposite that of the lac promoter of the vector so that the traMA6 expression was under the control of its cognate promoter.
Replacement of traMA6 with traMK588 in A. tumefaciens A6.
The allelic replacement of traM in strain A6 (traMA6) with that from strain K588 (traMK588) was carried out following the procedure described previously (18) with minor modifications. Briefly, the plasmid pAW-traMK588 was introduced into E. coli S17-1 by electroporation, and then kanamycin-resistant colonies of E. coli S17-1(pAW-traMK588) were selected and mated with A. tumefaciens A6. The transconjugants were grown on BM medium supplemented with 50 μg/ml kanamycin for 4 days; a single colony was then picked and streaked for further purification. Subsequently, allelic replacement candidates were selected on BM agar plates containing 5% sucrose at 28°C (13). These candidates were then selected for loss of their kanamycin resistance on LB agar plates and confirmed by DNA sequencing.
Deletion of traM2 in strain A6(pTiA6traMK588) and complementation.
Deletion of the traM2 gene in strain A6(pTiA6traMK588) was performed following the previously described method (18) with minor modifications. In brief, two PCR fragments flanking traM2 were amplified from the genomic DNA sample of A. tumefaciens A6 with two pairs of PCR primers (5′-GAAGGTGTGGCGGTGTTC and 5′-CGGGATCCGACACTACTGAATCTTTC; 5′-CGGGATCCGCTGGTTGGACTTCTGG and 5′-CAGGAATCACTATCCCCA). The PCR products were digested with BamHI and ligated with T4 DNA ligase. The fusion fragment was amplified by PCR using ligation mixture as a template and a pair of primers (forward, 5′-GAAGGTGTGGCGGTGTTC; reverse, 5′-CAGGAATCACTATCCCCA). The product was purified using the QIAquick gel extraction kit (QIAGEN) after agarose gel separation. The fragment, filled in with Klenow enzyme, was cloned into the pAW19 vector which had been cut with NotI and blunted by Klenow enzyme; the resultant construct was designated pAW-traM2d. After DNA sequencing confirmation, pAW-traM2d was introduced into E. coli S17-1 by electroporation. The kanamycin-resistant colonies of E. coli S17-1(pAW-traM2d) were selected, confirmed by PCR, and mated with A. tumefaciens A6(pTiA6traMK588) by biparental mating. Selection of kanamycin-resistant transconjugants and subsequent identification of the traM2 deletion mutant were conducted as described above.
For complementation, the coding region of the native traM2 gene was amplified with a primer pair (5′-CGGGATCCATGGATTTGAAAGATTCAG and 5′-CGGGATCCGCAGGCTTCAGTTGACC). The PCR products were digested with BamHI, purified with the QIAquick PCR purification kit (QIAGEN), and then ligated into the vector pLAFR3 cut with the same enzyme. The construct, in which traM2 was placed under the control of the lac promoter from the vector, was selected after PCR and sequencing verification and introduced into A. tumefaciens strains by following the procedures described above.
Quantitative determination of AHL.
The amount of AHL signals produced by bacterial cells was quantitatively determined as described previously (6). Briefly, 20 ml BM agar plates supplemented with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (50 μg/ml) were cut into separated slices (1 cm in width) and then used for AHL bioassay. To measure the concentration of AHL in bacterial supernatants, the cell culture was sampled at given time points and centrifuged at 14,000 rpm for 30 min at 4°C. Supernatants (5 μl) were added to one end of an agar slice, and AHL indicator strain NT1 (traR tra::lacZ749) was spotted at progressively further distances from the loaded samples. The plates were incubated at 28°C overnight, and the distance from the last induced blue colony to the origin of the AHL sample in each agar slice was measured for quantification of the amount of AHL, as described previously (6). To measure AHL production on solid agar plates, 10 μl of fresh culture with an optical density at 600 nm (OD600) of 0.6 was spotted on bioassay plates and incubated at 28°C for 12 h before the indicator strain was loaded. Based on the signal diffusion distance, the amount of AHL produced was calculated and expressed as the equivalent of N-(3-oxo-octanoyl)-l-homoserine lactone, as described previously (6, 34). Each assay was repeated at least three times with two replicates.
Conjugative transfer efficiency assay.
The Conjugation assay was carried out by modification of the method of drop mating described previously (39). The recipient strains used in this study were the rifampin-resistant A. tumefaciens nopaline strain C58C1RS and octopine strain Ach5C3 (Table 1). Donor strains were grown for 24 h at 28°C in BM minimal agar plates (when required, octopine was included at a final concentration of 0.8 g per liter). The bacterial cells were resuspended in BM medium without any carbon source and adjusted to an OD600 of 1.0, and a 10-fold dilution series was prepared. From each dilution, 5 μl of bacterial cells was spotted onto a lawn of the recipient strain growing on selective plates, which were supplemented with antibiotic rifampin and octopine as the only carbon source to support the growth of transconjugants only. To determine the number of viable donor cells, 10 μl of cells of each dilution was spread onto LB plates. Plates were incubated at 28°C. The numbers of donors and transconjugants were counted 1 week after mating. Each assay was repeated two times with three replicates. The transfer efficiency of the donor strain was calculated as the number of transconjugants observed per donor cell applied.
RNA preparation and real-time RT-PCR.
Bacterial strains were cultivated in minimal medium with or without octopine, as indicated. When the OD600 reached 1.2, cells were collected by centrifugation at 4°C. Total RNA was subsequently isolated using the RNeasy Mini kit (QIAGEN) according to the manufacturer's protocol. Residual DNA present in the RNA samples was removed by the RNase-free DNase I. The quality and quantity of the RNA samples were determined with a Nanodrop ND-1000 (Nanodrop Technologies). An aliquot of 0.2 μg total RNA was used as a template for normal reverse transcription (RT)-PCR to amplify the traI coding region using forward primer 5′-CGACCAATACCAACACCAG-3′ and reverse primer 5′-GACGGTGTTGCCAATCGC-3′. A fragment of 16S rRNA was also amplified under the same conditions as the internal control using forward primer 5′-TGACGAGTGGCGGACGGGTG and reverse primer 5′-ATGCAGTTCCCAGGTTGAGC.
The real-time RT-PCR was conducted on a Lightcycler from Roche Diagnostics with the QuantiTect SYBR green RT-PCR kit (QIAGEN) according to the manufacturer's protocol. The traI cDNA fragments were amplified using primers 5′-CGACCAATACCAACACCAG-3′ and 5′-GACGGTGTTGCCAATCGC-3′. The 16S rRNA transcripts were included as an endogenous control for each sample using primers 5′-GAGGAAGGTGGGGATGACG and 5′-CCAACTCCCATGGTGTGACG. The data were analyzed using the LightCycler 3.5.3 software (Roche Diagnostics Corporation).
Southern blotting analysis and cloning of traM2.
Total DNA samples were prepared from different strains using the MasterPure DNA purification kit (Epicenter Technologies, Madison, WI). Probes were generated by PCR from strain A6 with the PCR digoxigenin (DIG) labeling kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's recommendations. Primers for traM and traM2 amplifications were 5′-CGTGACGAAAAAGGTCGAG (forward) and 5′-GGGTATGAAGCCAAGAATG (reverse) and 5′-GATTCAGTAGTGTCGGAC (forward) and 5′-GATGAAACCCAGAAGTCC (reverse), respectively. Total DNA samples from different strains were completely digested by BamHI and separated on a 1.2% agarose gel, and Southern blotting analyses were performed with DIG-labeled probes according to the manufacturer's protocols (Roche Molecular Biochemicals, Mannheim, Germany). For blotting using traM probe, low-stringency hybridization was performed at 37°C and washed consecutively at 60°C twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer containing 0.1% sodium dodecyl sulfate (SDS) and twice with 0.5× SSC containing 0.1% SDS. For traM2 blotting, high-stringency hybridization and washing were done at 42°C and 68°C, respectively, with the same initial washing buffer as above, but the washing buffer in the last two rounds was changed to 0.1× SSC containing 0.1% SDS. Signals were detected with the DIG luminescent detection kit (Roche Molecular Biochemicals, Mannheim, Germany).
For cloning of traM2, several pairs of PCR primers were designed based on the coding and flanking sequences of traM of strain A6. The primer pair that produced the expected DNA fragments was 5′-CGGATTCTAACCCCCGGC (forward) and 5′-TACCGCGCTCGCCATCCGG (reverse). The DNA fragments amplified from the total DNAs of A. tumefaciens strains were separated on 1.2% agarose gels and purified with the QIAquick gel extraction kit (QIAGEN) for DNA sequencing.
Gel mobility assay.
The wild-type TraM and TraR, encoded by pTiR10, were purified as previously described (4). The coding region of traM2 was cloned into pET15b (Novagen) at the NdeI and BamHI restriction sites, respectively, to generate plasmid pGC-traM2. TraM2 was expressed in E. coli BL21(DE3) harboring pGC-traM2. A gel mobility assay was performed as described before (29). An infrared dye (IR)-labeled tra box-containing DNA fragment was amplified from pJZ304 (42) by PCR using two synthetic oligonucleotides, 5′-AATTAACCCTCACTAAAG (IRD700 labeled; LI-COR) and 5′-GTTTTCCCAGTCACGAC (Integrated DNA Technologies). TraR (0.1 μM) was incubated with an IR-labeled tra box in reaction buffer (12 mM HEPES-NaOH, 4 mM Tris-HCl, 60 mM potassium glutamate, 1 mM EDTA, 1 mM dithiothreitol, 12% glycerol). TraM2 or TraM at the indicated concentrations was added to the above reaction mixture and incubated for 30 min at room temperature. The mixture was directly subject to 8% nondenaturing polyacrylamide (80:1 acrylamide:bis-acrylamide) gels. The IR signal was visualized in the Odyssey infrared image system (LI-COR).
RESULTS
Phenotype comparison of QS-constitutive strain K588 and QS-inducible strain A6.
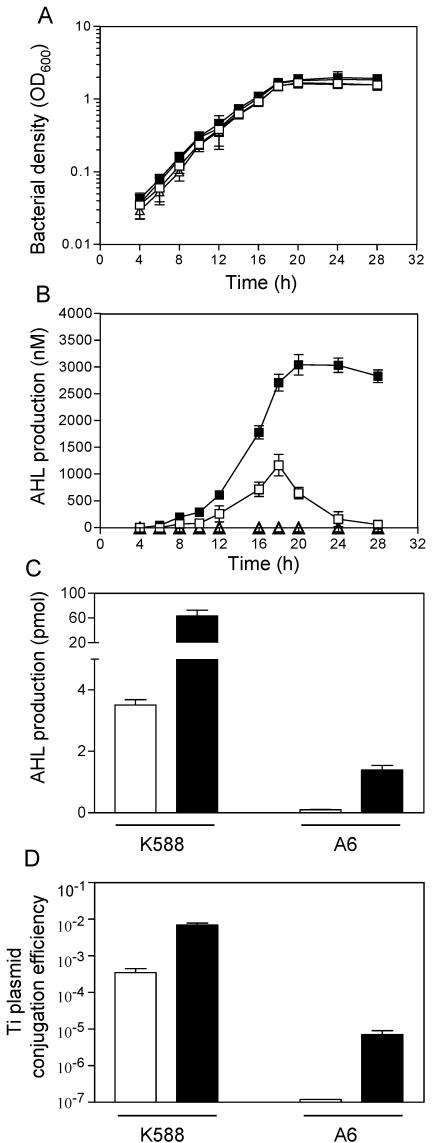
In an effort to elucidate the cause that rendered K588 the QS-constitutive phenotype, we made careful analyses of bacterial growth, AHL production, and Ti plasmid conjugative transfer efficiency by comparison with A. tumefaciens strain A6. A6 is a wild-type octopine-type strain carrying pTiA6, whose QS-dependent conjugative transfer is activated by induction of octopine (39). The results showed that K588 and A6 exhibited similar growth patterns; bacterial cells grew exponentially in minimal medium until about 18 h after inoculation, when they moved into stationary phase (Fig. 1A). Addition of octopine at the concentration of 0.8 mg/ml in BM minimal medium did not have a significant effect on bacterial growth. In the absence of octopine, AHL was detectable in the supernatant of K588 when the bacterial population density (OD600) reached around 0.2. The signal molecules accumulated rapidly along with the exponential growth of the bacterial population and peaked at the transition to stationary phase, with a maximal concentration reaching about 1,200 nM, and then decreased quickly (Fig. 1B). In the presence of octopine, AHL could be detected in the K588 supernatant at an earlier stage (OD600 = ∼0.1). Furthermore, the concentration was significantly higher than that without octopine induction throughout the growth phases, approximately threefold at the peak. The results suggest that even though the tight control of QS was released in K588, octopine could still significantly boost AHL accumulation. Strain A6, however, produced no detectable AHL in liquid culture even in the presence of octopine (Fig. 1B), indicating that its concentration in the supernatant was lower than 0.05 pmol, which is the detection threshold of the AHL reporter strain used in this study.
FIG. 1.
Phenotype analysis of A. tumefaciens strains K588 (square) and A6 (triangle). The assays were performed with (solid symbol or bar) or without (open symbol or bar) octopine. (A) Time course of growth of the two strains in BM minimal medium. Bacterial overnight culture was diluted in BM medium to same population density (OD600 = 1.0), inoculated into BM medium at a 1:100 ratio, and incubated with shaking (200 rpm) at 28°C. (B) AHL production by A. tumefaciens strains in liquid cell culture. The culture conditions were the same as above. Note, the data for A6 with and without octopine were identical, and as a result, solid triangles overlapped with open triangles. (C) AHL production by A. tumefaciens strains on BM medium agar plate. The procedures for inoculation and bioassay were described in Materials and Methods. (D) Ti plasmid conjugative transfer efficiency of A. tumefaciens strains. Induction of donor cells was done on solid agar medium as described in Materials and Methods. Values are presented as means ± standard deviations of results from at least three independent repeats.
To provide more accurate comparison of the relative AHL production between the two strains, we measured the signal production by adding and growing same amount of K588 and A6 cells on solid BM agar plates. As shown in Fig. 1C, A6 produced a basal level of 0.15 ± 0.04 pmol when grown overnight on octopine-free plates, whereas K588 produced about 3.5 ± 0.29 pmol, which was about 23-fold higher than A6. In the presence of octopine, the AHL production by A6 was increased up to 1.7 ± 0.8 pmol, showing a typical octopine-inducible phenotype. As expected, similar to what had been observed in liquid culture, K588 also produced substantially more AHL (60 ± 3.75 pmol) on plates containing octopine. It is noteworthy that the amount of AHL produced by A6 with octopine induction was still less than that of constitutively produced by K588, reflecting very strict regulatory mechanisms in strain A6.
Furthermore, we determined the Ti plasmid conjugative transfer efficiencies of strains K588 and A6 with and without octopine induction. The results (Fig. 1D) showed that the transfer efficiency of A6 was not detectable (<10−9) in the absence of octopine but increased by at least 1,000-fold with octopine induction. In contrast, the QS-constitutive strain K588 transferred its Ti plasmid at a high frequency (4 × 10−4), and octopine induction moderately increased the efficiency by about 10 times. Given the significant difference of the two strains in AHL production, we further tested whether the addition of AHL to strain A6 could bypass the requirement of octopine induction. However, no significant difference in Ti plasmid transfer efficiency of strain A6 was observed with or without external addition of the QS signals (1, 5, 10 μM). Taken together, these data suggest that the enhanced AHL production in strain K588 is not the primary cause responsible for the constitutive Ti plasmid conjugative transfer phenotype.
Sequence analysis of the key genes implicated in QS signal production and regulation in strains K588 and A6.
It is known that the majority of the genes involved in QS signal production and Ti plasmid conjugative transfer are carried by Ti plasmid. To check if there was any significant insertion or deletion in pTiK588, we purified pTiK588 and pTiA6 from strains K588 and A6, respectively, using a previously established simple method (35). Digestion of the two plasmids with restriction enzymes EcoRI and HindIII, respectively, however, appeared to produce the same digestion pattern (data not shown) as that previously reported for pTiA6 (35), implying that point mutations or small deletions may be more likely the cause.
To identify the mutated gene(s) responsible for the QS- and Ti plasmid transfer-constitutive phenotype in strain K588, we constructed a cosmid library of pTiK588 by ligating the HindIII partially digested plasmid to the cosmid vector pLAFR3. The library was introduced to strain A6, and the transconjugants constitutively producing the QS signal were screened. One transconjugant, which contained the cosmid clone pBK129, was found to produce a similar level of AHL signals as strain K588 in the absence of octopine. The insert of pBK129 was sequenced, and analysis of the DNA sequence (GenBank accession no. DQ195264) showed that it contained about 26 kb of nucleotides, including 24 intact genes ranging from traD to mclB as well as a part of traG (27). These genes were positioned in the same order as their counterparts of pTiR10, a wild-type Ti plasmid of octopine strain R10 (40). Moreover, as expected, the whole insert region showed a high degree of homology (99%) at the nucleotide level with the same region in pTiR10. Nevertheless, nucleotide variations were identified in 11 of the 24 genes, including traA, traF, traB, traM, ophEDCBA, msh, and ooxA. Interestingly, among the several key genes known to be involved in regulation of both QS signal production and Ti plasmid conjugative transfer, including occR, traR, and traM, only the traM of pTiK588 contained three nucleotide variations (T161C, T268A, A269T), which resulted in two amino acid substitutions (L54P, Y90I), in comparison with the traM of pTiR10 (10). We also amplified and sequenced the corresponding genes from strain A6 and found that the occR, traR, and trlR genes are identical between strains K588 and A6. However, the traM gene of A6 also contained the T268A and A259T variations, indicating that only one single nucleotide mutation (T161C), which caused leucine-to-proline substitution at the 54th amino acid (L54P) of the 102-amino acid TraM, was worthy of further investigation. The data seem to be agreeable with the previous finding that substitution of Leu54 with alanine of the TraM of pTiR10 by site-directed mutagenesis could result in partial reduction in inhibitory activity against the TraR-mediated reporter gene expression (29). It is important that the cosmid pBK129 carries both traM and traR. Previous study showed that mutations in traM lead to constitutive expression of the TraR-AHL-dependent tra genes in the absence of conjugative opines (10, 14), indicating that the strains lacking a functional TraM could produce a critical amount of TraR and AHL for tra gene expression. The DNA sequence analysis suggested that pBK129 may encode a nonfunctional TraM but produce a functional TraR, which may well explain why introduction of the cosmid into strain A6 resulted in constitutive production of AHL signals.
Expression of the traM from strain A6 rescued the QS-constitutive phenotype of strain K588.
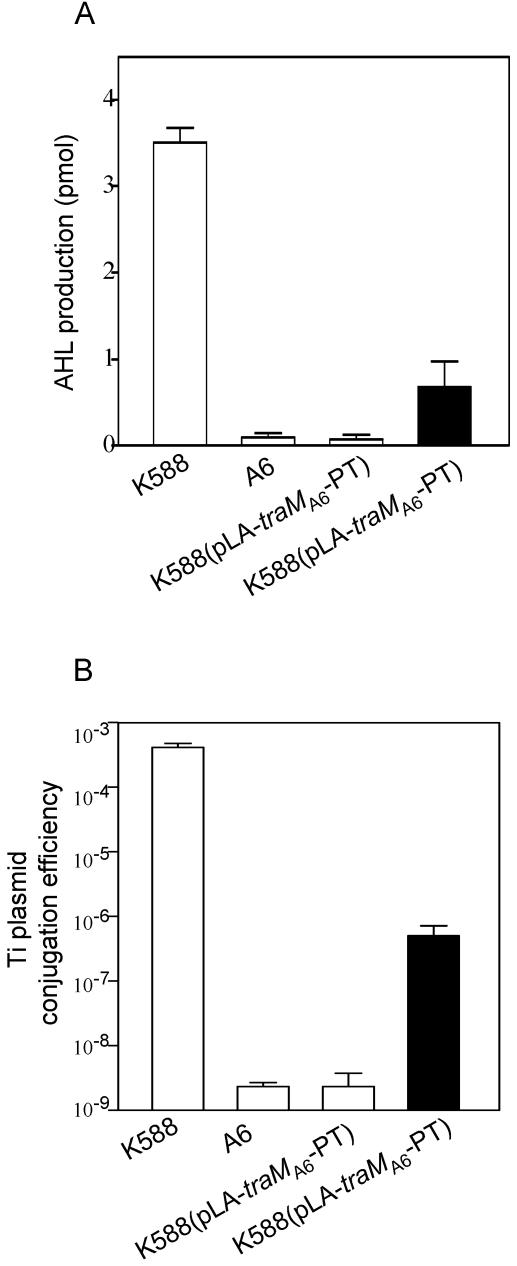
To test whether the L54P mutation in homogenously expressed TraM was responsible for the QS-constitutive phenotype in strain K588, we cloned the wild-type traM gene from A. tumefaciens strain A6 (traMA6) and tested whether it could effectively rescue the QS-constitutive phenotype of K588. The construct pLA-traMA6-PT, in which the traMA6 gene was controlled under its cognate promoter, was introduced into strain K588, and then AHL production was measured. As shown in Fig. 2A, in the absence of octopine induction, the amount of AHL produced on the solid agar plate by K588(pLA-traMA6-PT) was dramatically decreased to almost the same level as that of wild-type strain A6. Consistently, we were also unable to detect AHL in the supernatant of K588(pLA-traMA6-PT) cell culture. Moreover, expression of traMA6 abolished the constitutive Ti plasmid conjugative transfer of K588 (Fig. 2B). As expected, the inhibition of AHL production and Ti plasmid transfer provided by TraMA6 was significantly alleviated by supplementation of octopine (Fig. 2A and B).
FIG. 2.
The wild-type TraM from A. tumefaciens strain A6 rescued the QS-constitutive phenotype of strain K588. The assays were conducted in the absence (open bar) or presence (solid bar) of octopine. (A) AHL production by strain K588 and its derivatives on solid agar plate. (B) Ti plasmid conjugative transfer efficiency of the above strains. The donor cells were grown on a solid agar plate as described. Strain A6 was included as a control.
Replacement of traMA6 with traMK588 in strain A6 did not generate the expected QS-constitutive phenotype.
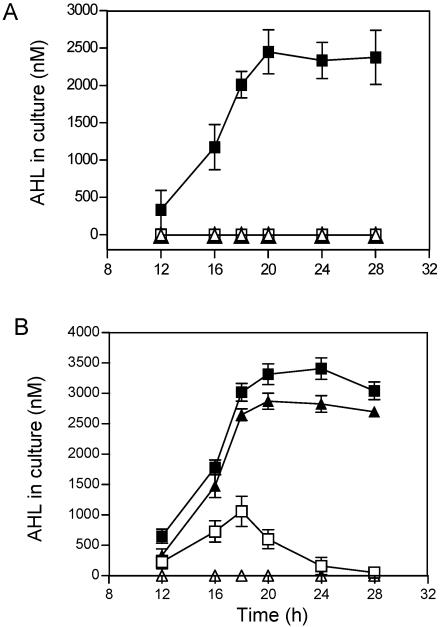
To further examine the consequence of L54P mutation in TraM, we cloned the traM gene from K588 (designated traMK588) and used it to replace the A6 counterpart by gene replacement. The allele-exchanged strain, verified by DNA sequencing analysis, was designated A6(pTiA6TraMK588) and subjected to phenotype analyses. Most surprisingly, however, strain A6(pTiA6traMK588) failed to accumulate detectable AHL in the culture supernatant in the absence of octopine induction (Fig. 3A). When analyzed on the solid agar plates, we found that strain A6(pTiA6traMK588) produced a basal level of QS signal (0.15 ± 0.02 pmol) similar to that of wild-type strain A6 but much less than that of K588 (Fig. 1B).
FIG. 3.
Differential impact of TraM mutation on QS signal production by different A. tumefaciens strains in liquid culture. The assays were done with (solid symbol) or without (open symbol) octopine. (A) Effect of TraM point mutation (L54P) on AHL production by strain A6. Squares, A6(pTiA6traMK588); triangles, A6. Note, data for A6 with and without octopine were identical, and as a result, solid triangles overlapped with open triangles. (B) Plasmid pTiA6traMK588 displayed different AHL production phenotypes in A. tumefaciens strains C58C1RS and Ach5C3. Squares, C58C1RS(pTiA6traMK588); triangles, Ach5C3(pTiA6traMK588).
When induced by octopine, however, the amount of AHL signals accumulated in the culture supernatant of strain A6(pTiA6traMK588) reached about 2,500 nM at 20 h after inoculation (Fig. 3A), which was slightly lower than that of strain K588 (Fig. 1B). On solid agar plates supplemented with octopine, we found that strain A6(pTiA6traMK588) produced a >40-times-higher amount of AHL signal than its parental strain A6 (75 ± 0.3 pmol versus 1.7 ± 0.1 pmol), which was at the same level as that of strain K588 (75 ± 0.6 pmol). It becomes highly intriguing that the L54P mutation in TraM, which results in the QS-constitutive phenotype in strain K588, is not able to change the QS-inducible phenotype of strain A6. The data strongly suggest the existence of critical genetic variations between strains K588 and A6 other than the difference in traM.
The differential impact of TraM mutation on QS in strains C58C1RS and A6 could be attributed to the chromosomal variations between the two strains.
To probe whether the critical genetic variations are in pTiA6 or in the chromosomal background of strain A6, we transferred pTiA6traMk588 from strain A6(pTiA6traMK588) into the Ti plasmid-free nopaline-type strain C58C1RS by conjugative transfer, as described in Materials and Methods. The resultant transconjugant was designated C58C1RS(pTiA6traMK588). The bioassay results showed that, unlike its donor strain A6(pTiA6traMK588), C58C1RS(pTiA6traMK588) displayed typical QS-constitutive phenotypes like K588, proficiently producing the AHL signals in liquid culture without octopine (Fig. 3B) and transferring Ti plasmid with high efficiency (data not shown), which were comparable to strain K588 (Fig. 1B and D). Similarly, the AHL amount produced by C58C1RS(pTiA6traMK588) could be further increased by octopine induction (Fig. 3B).
The above data strongly imply that an unknown factor(s) encoded by the A6 chromosomal background, which is missing in strains K588 and C58C1RS, is responsible for the differential impact of the L54P mutant in TraM on QS in different strains, such as A6 and K588. To test whether this phenomenon could also occur in other strains, we then transferred the plasmid pTiA6traMK588 from A6 to Ach5C3, which is a Ti plasmid-free octopine-type A. tumefaciens strain. The transconjugants, designated Ach5C3(pTiA6traMK588), were assayed for AHL production. The results in Fig. 3B show, interestingly, a pattern similar to that of pTiA6traMK588 in the genetic background of strains A6, with no QS signal production in the absence of octopine (Fig. 3A). Based on these data, we concluded that octopine strains, at least A6 and Ach5C3, but not the nopaline strain C58, might contain a gene(s) that encodes an unidentified factor(s) suppressing QS signal production, directly or indirectly, in the absence of opine inducer.
The unknown factor in octopine strain A6 regulates traI expression at the transcriptional level.
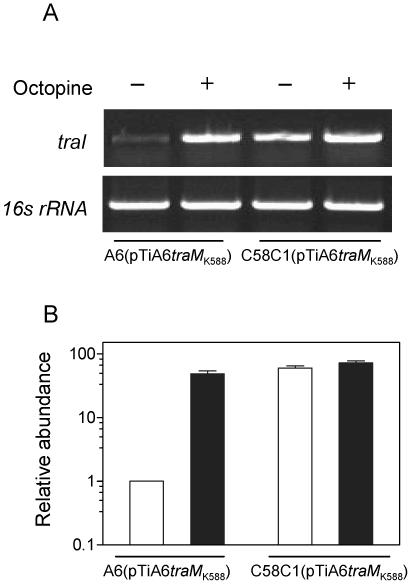
It remained to be determined whether the inhibitory effect of the unknown factor on QS signal production and conjugation in octopine strains was exerted at the transcriptional or translational level. For this purpose, we used RT-PCR to analyze the transcriptional expression patterns of traI, which is located on the Ti plasmid and encodes synthesis of the QS signal AHL in A. tumefaciens (15, 36), in different chromosomal genetic backgrounds and with or without octopine. As shown in Fig. 4A, the traI transcriptional expression in strain A6(pTiA6traMK588) was low without octopine but was significantly promoted by the addition of the opine inducer. While in the case of C58C1RS(pTiA6traMK588), the traI transcription maintained a high level regardless of whether there was octopine in the medium or not. The RT-PCR results were further confirmed by real-time RT-PCR analysis (Fig. 4B). By normalizing the abundance of traI transcripts of the bacterial cells grown with or without octopine to the amount of 16S rRNA in the same set of sample, we showed no significant difference in traI transcription by strain C58C1RS(pTiA6traMK588) under both conditions. However, in strain A6(pTiA6traMK588), we found that octopine induction enhanced traI transcription by about 60-fold, reaching a level similar to that of strain C58C1RS(pTiA6traMK588). The data confirm that the unknown factor in strain A6 inhibits traI transcriptional expression, either directly or indirectly, but that repression can be released by the conjugative opine.
FIG. 4.
Analysis of traI transcriptional expression in A. tumefaciens. (A) RT-PCR detection of traI transcripts in A. tumefaciens strains A6(pTiA6traMK588) and C58C1RS(pTiA6traMK588) with (+) and without (−) octopine induction. The 16S rRNA gene fragment was amplified under the same conditions as the internal control. (B) Real-time RT-PCR quantification analysis of the traI transcripts with (solid bar) or without (open bar) octopine. The bacterial strains were grown in BM liquid medium, and total cellular RNAs were extracted from bacterial cultures when the OD600 reached 1.2 and subjected to RT-PCR and real-time RT-PCR analysis as described in Materials and Methods. The relative amount of the traI transcripts was normalized to the 16S rRNA content within each reaction, and the analysis was repeated twice. For the convenience of comparison, the relative traI transcript abundance in strain A6(pTiA6traMK588) without octopine induction was arbitrarily assigned a value of 1.
Identification of traM2 in A. tumefaciens A6.
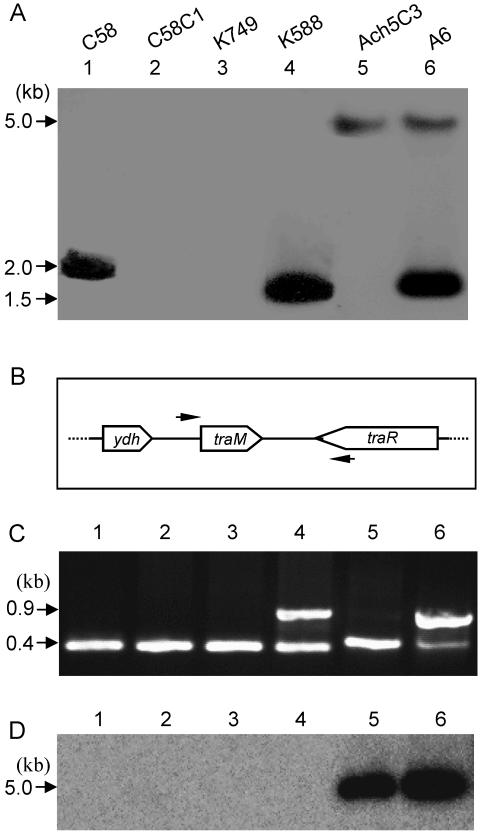
Interestingly, several phenotypes of this unknown factor are reminiscent of those of TraM. First, similar to the traM mutant of strains C58 and R10 (10, 14), overexpression of wild-type traMA6 in strain K588 restored the QS-inducible phenotype (Fig. 2). Second, the unknown factor suppressed the transcriptional expression of traI in the absence of opine inducer (Fig. 4), which is again similar to that of TraM (14). It was, hence, highly pertinent to determine whether strains A6 and Ach5C3 had a traM homologue. Total DNA samples were prepared from several related A. tumefaciens strains and digested with BamHI. After separation by gel electrophoresis, the DNA samples were hybridized at low stringency with traMA6 probe. Strains C58 and K588 produced one hybridization band about 2 or 1.5 kb in size (Fig. 5A), which was the expected DNA band size of traM in nopaline and octopine strains, respectively. Strain A6 contained two hybridization bands: one was the traM band about 1.5 kb in size and the second band was about 5 kb. Coincidently, strain Ach5C3, which contained the unknown factor as well (Fig. 3B), also produced the 5-kb band (Fig. 5A). The weak hybridization signal of the 5-kb band suggests that strains A6 and Ach5C3 may contain a second copy of traM (designated traM2) in the chromosomal DNA, which is homologous, but not identical, to the traM carried by the Ti plasmid.
FIG. 5.
Southern blot and PCR analysis of traM homologues in various A. tumefaciens strains. (A) Southern blot hybridization of nopaline-type strains (C58, C58C1RS, K749, and K588) and octopine-type strains (Ach5C3 and A6) using traM probe under low-stringency conditions (Materials and Methods). (B) Designing PCR primers to amplify the traM homologue from chromosomal DNA based on the traM flanking sequences in pTiA6. The relative locations of the final PCR primer pair used for amplification in panel C are indicated by arrows. (C) PCR fragments amplified from different strains as indicated in panel A. The 0.9-kb band was the expected DNA fragment amplified from individual octopine-type Ti plasmid, which was absent from three Ti plasmid-free strains, i.e., C58C1RS, K749, and Ach5C3. The band could not be amplified from Ti plasmid-containing nopaline strain C58 because of low homology in the region. The about 0.4-kb bands from strain A6 and Ach5C3 were shown to contain traM homologues. The similarly sized nonspecific bands were also amplified from the 4 nopaline-type derivatives (see text). (D) High-stringency Southern blot analysis using traM2 as probe. In panels C and D, the samples used were the same as in panel A.
Assuming that traM2 shares a reasonable degree of homology with the traM gene of Ti plasmid origin, several pairs of PCR primers were designed and tested in amplification of traM2, using the total DNA sample of strain A6 as a template. While the majority of primer pairs produced only one band with the expected size of traM amplification (data not shown), one pair of primers, which were designed based on the flanking sequence of traM (Fig. 5B), produced two PCR fragments of about 900 and 400 bp, respectively (Fig. 5C). We then used the same primer pair to test the total DNA samples of other strains and found that the 400-bp band was amplified from all the strains tested, including octopine-type A6 and Ach5C3 and nopaline-type C58, C58C1RS, K749, and K588 (Fig. 5C). Sequence analysis confirmed that the 400-bp PCR products from strain A6 and Ach5C3 contained an identical traM2 gene (GenBank accession no. DQ192640), which showed 64% and 65% homologies with traM of strain A6 at the nucleotide and peptide levels, respectively. In contrast, the similarly sized PCR product from C58 and its derivatives C58C1RS, K749, and K588 corresponded to an intergenic region on the linear chromosome of C58. The region contains two truncated open reading frames (Atu3178, Atu3179) (http://www.ncbi.nlm.nih.gov/), but none of them showed homology to traM (data not shown). High-stringency Southern blotting analysis using traM2 as probe further confirmed that, among the six strains tested, only A6 and Ach5C3 contained the traM2 gene (Fig. 5D). We further determined the presence of traM2 in other octopine strains, including Ach5, R10, ATCC 15955, B6, and B6S3, under the same high-stringency Southern blotting conditions. However, no traM2-specific hybridization signal was detected in all the tested strains except Ach5 (data no shown). The existence of traM2 in strain Ach5 is expected, as strain Ach5C3 is a Ti plasmid-free derivative of strain Ach5 (Table 1).
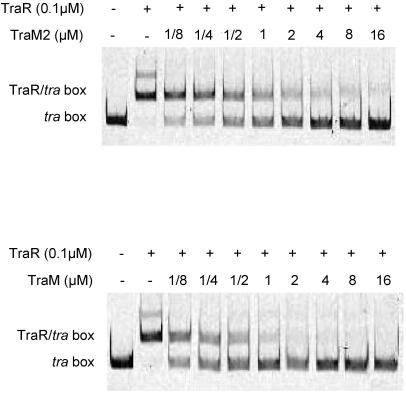
TraM2 is a potent antiactivator in vitro.
Sequence alignment showed that basically all the key amino acid residues, which significantly contribute to antiactivation activity against TraR (29), were conserved identically between TraM and TraM2 (Fig. 6). The data suggest that TraM2 might have a potency similar to that of TraM in the formation of an antiactivation complex that prevents TraR from binding to its target promoter. As expected, our gel mobility assay (Fig. 7) showed that TraM2 of A6 was able to block the binding of TraR to the tra box almost as effectively as the cognate antiactivator TraM. Thus, like TraM, TraM2 is a potent antiactivator to TraR under in vitro conditions.
FIG. 6.
Sequence alignment of three TraM homologues used in this study. Alignment of amino acid sequences was performed using the computational program Clustalw (http://sbcr.bii.a-star.edu.sg/clustalw/). The key amino acid residues showing strong influence on inhibition activity against TraR-mediated gene activation, identified in a previous study (29), were indicated by a triangle above the sequence. TraMK588 differs from TraM in only one amino acid (L54P).
FIG. 7.
Inhibitory effect of TraM2 on DNA binding to TraR by gel mobility assay. The inhibition potency of TraM2 from strain A6 (A) is compared with that of TraMR10 (B). Concentrations of proteins (TraR, TraM, and TraM2) in the reaction mixtures are indicated above the nondenaturing gels (8%). Positions of the IR-labeled tra box in both free and complexed with TraR forms are indicated. At 16 μM, TraM2 displaced TraR completely from the tra box, compared to a lower concentration (∼2 μM) of TraM. −, not included in the reaction mixture.
TraM2 is a functional quorum-sensing modulator in vivo.
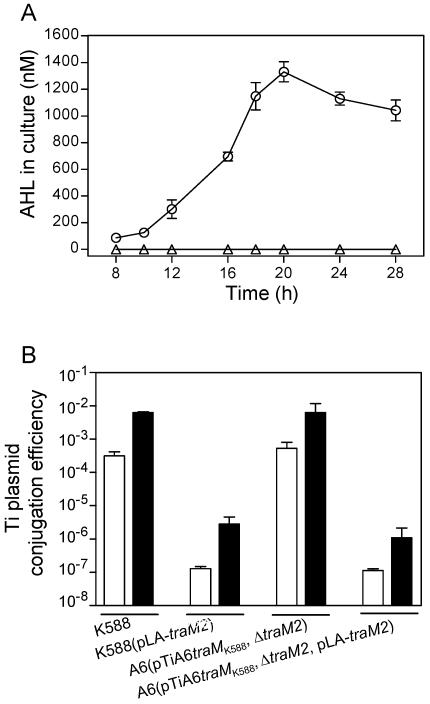
To determine the physiological effect of traM2 in strain A6, we generated a traM2 deletion mutant in the genetic background of strain A6(pTiA6traMk588), in which a central region of 234 nucleotides (corresponding to 78 amino acids) of traM2 was deleted [designated A6(ΔtraM2, pTiA6traMk588)]. As described above, strain A6(pTiA6traMk588) was not able to produce detectable AHL signals; however, deletion of traM2 from the strain resulted in constitutive production of AHL in the absence of octopine inducer (Fig. 8A). Moreover, this traM and traM2 double mutant A6(ΔtraM2, pTiA6traMk588) displayed a constitutive phenotype in Ti plasmid conjugal transfer, similar to that of strain K588 (Fig. 8B). We further showed that heterogeneous expression of traM2 under the control of the constitutive lac promoter in both strains K588 and A6(ΔtraM2, pTiA6traMk588) abrogated the transfer-constitutive phenotype (Fig. 8B). The data demonstrate that TraM2 is involved in modulation of quorum sensing in strain A6.
FIG. 8.
Effect of null mutation of traM2 on quorum sensing in A. tumefaciens strain A6. (A) AHL accumulation by A. tumefaciens strains A6(pTiA6traMK588) (triangle) and A6(ΔtraM2, pTiA6traMK588) (circle) in the supernatant of bacterial culture without octopine induction. (B) Ti plasmid conjugative transfer efficiency of A. tumefaciens strains with (solid bar) or without (open bar) octopine induction on solid medium. Value are presented as means ± standard deviations of the results from at least three independent repeats.
DISCUSSION
The QS antiactivator TraM encoded by the Ti plasmid was discovered about a decade ago in A. tumefaciens nopaline-type strain C58 and octopine-type strain R10 consecutively by two laboratories (10, 14). These researchers found that transposon knockout of traM in both strains resulted in the constitutive production of AHL signals and high-efficiency transfer of Ti plasmid even in the absence of corresponding opine inducers. In this study, we showed that a single amino acid mutation in TraM, in which leucine 54 was replaced with proline, could fully account for the constitutive phenotypes of strain K588, a spontaneous mutant that produces AHL signals and transfers Ti plasmid efficiently without the need for octopine inducer (34). Our findings are highly consistent with the alanine scanning mutagenesis analysis of the TraM from pTiR10, which showed that an L54A substitution in TraM basically abolished its inhibitory activity against TraR-mediated gene expression when TraR was expressed at a low level, although the impact of L54A mutation on anti-TraR activity was reduced when TraR was expressed at a high level (29). The crystal structure of TraM revealed that the L54 residue is located at the end of an α-helix and lies on a region potentially interacting with TraR (4, 30). Replacement of L54 with proline may cause more severe conformational changes than with alanine, as proline is conformationally restricted in a peptide and consequently could have severe effects on the secondary and tertiary structure of the protein. Our results have verified that under the native TraR expression status, the L54P substitution in TraM is sufficient to switch the Ti plasmid from QS-inducible mode to QS-constitutive mode in the chromosomal background of strain C58C1RS.
Surprisingly, however, we failed to generate the expected QS-constitutive phenotypes in octopine strain A6 after we replaced its wild-type traM with the mutated traM from pTiK588, which contained the L54P substitution (Fig. 3A). The same allele-exchanged Ti plasmid, when residing in the genetic background of nopaline strain C58C1RS, however, produced the QS-constitutive phenotype (Fig. 3B), similar to strain K588 (Fig. 1B). Subsequent genetic and biological analysis revealed that there is a second copy of traM carried by the chromosomal DNA of two octopine-type strains of A. tumefaciens, i.e., A6 and Ach5C3. The homologue, designated traM2, shares 64% and 65% identity with traM at the nucleotide and peptide levels, respectively. Although the identity between TraM and TraM2 is not particularly high, the key amino acid residues contributing to the antiactivation activity of TraM as revealed by alanine scanning analysis, including L29, A36, I37, H40, R41, L43, L54, A81, Q82, L93, G94, and P97 (29), are all conserved in TraM2 (Fig. 6). These data suggest that TraM2, similar to TraM, could also function as a potent QS antiactivator.
The previous results showed that TraM exerts its effect by direct interaction with the transcriptional factor TraR, which serves as the receptor of AHL QS signal and the key activator of the QS-dependent tra regulon (26, 42). The complex is highly stable, with a dissociation constant in the nanomolar range (29). DNA gel retardation analysis confirmed that TraM2, similar to the TraM encoded by the Ti plasmid, could effectively prevent TraR from binding to a tra promoter (Fig. 7), indicating that in vivo TraM2 might repress the QS-dependent expression of tra genes by interacting with the TraR activator encoded by pTiA6. The physiological effect of TraM2 has been demonstrated by deletion analysis. While the traM single mutant A6(pTiA6traMk588) was dependent on octopine inducer for activation of AHL production in liquid culture (Fig. 3A), the double mutant A6(ΔtraM2, pTiA6traMk588) displayed a QS-constitutive phenotype in AHL production and Ti plasmid conjugal transfer (Fig. 8). The genetic data are consistent with the DNA gel retardation analysis that TraM2 is a strong antiactivator.
Even though mutation of the traM of pTiA6 along failed to change the QS-inducible phenotype (Fig. 3A), the role of its product in modulation of QS signaling in strain A6 is beyond any doubt. The wild-type strain A6 was unable to produce detectable AHL signals in liquid culture supplemented with octopine inducer; however, introduction of the L54P mutation in TraM enabled the strain to produce abundant AHL signals when induced by the conjugative opine (Fig. 3A). This is consistent with the demonstrated role of TraM in other bacterial strains (10, 14). All things considered, we can conclude that, together with TraM, TraM2 provides additional control to modulate the TraR-AHL-dependent activation of tra regulon in A. tumefaciens strain A6.
However, TraM2 does not seem to be a widely conserved quorum-sensing component in A. tumefaciens. From a limited A. tumefaciens strain collection, we found that the traM2 gene exists only in octopine-type strains A6, Ach5, and its Ti plasmid-free derivative Ach5C3 but neither in the other tested octopine-type strains, i.e., R10, ATCC 15955, B6, and B6S3, nor in the nopaline-type strain C58. The absence of traM2 in strain R10 is consistent with the previous finding that a null mutation of the traM carried by pTiR10 in octopine strain R10 resulted in constitutive Ti plasmid conjugative transfer (14), a phenotype typical to the A. tumefaciens strains that contain only one copy of traM (10).
Acknowledgments
We thank S. K. Farrand, S. Q. Pan, and C. Fuqua for providing A. tumefaciens strains and plasmids, respectively.
This work was funded by the Agency of Science, Technology, and Research (A*Star), Singapore.
REFERENCES
- 1.Beck von Bodman, S., G. T. Hayman, and S. K. Farrand. 1992. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. USA 89:643-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck von Bodman, S., W. D. Bauer, and D. L. Coplin. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455-482. [DOI] [PubMed] [Google Scholar]
- 3.Chai, Y., J. Zhu, and S. C. Winans. 2001. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40:414-421. [DOI] [PubMed] [Google Scholar]
- 4.Chen, G., J. W. Malenkos, M. R. Cha, C. Fuqua, and L. Chen. 2004. Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR. Mol. Microbiol. 52:1641-1651. [DOI] [PubMed] [Google Scholar]
- 5.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 6.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, J. G., A. Kerr, A. Petit, and J. Tempe. 1982. Conjugal transfer of nopaline and agropine Ti-plasmids-the role of agrocinopines. Mol. Gen. Genet. 186:269-274. [Google Scholar]
- 9.Farrand, S. K. 1998. Conjugation in the Rhizobiaceae, p. 199-233. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae: molecular biology of model plant-associated bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 10.Fuqua, C., M. Burbea, and S. C. Winans. 1995. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J. Bacteriol. 177:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signaling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua, C., and S. C. Winans. 1996. Localization of OccR-activated and TraR-activated promoters that express two ABC-type permeases and the traR gene of Ti plasmid pTiR10. Mol. Microbiol. 20:1199-1210. [DOI] [PubMed] [Google Scholar]
- 13.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang, I., D. M. Cook, and S. K. Farrand. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang, I., P. L. Li, L. H. Zhang, K. R. Piper, D. M. Cook, M. E. Tate, and S. K. Farrand. 1994. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. USA 91:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klapwijk, P. M., and R. A. Schilperoort. 1979. Negative control of octopine degradation and transfer genes of octopine Ti plasmids in Agrobacterium tumefaciens. J. Bacteriol. 139:424-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, Z. Q., Y. Qin, and S. K. Farrand. 2000. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275:7713-7722. [DOI] [PubMed] [Google Scholar]
- 18.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Oger, P., K. S. Kim, R. L. Sackett, K. R. Piper, and S. K. Farrand. 1998. Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27:277-288. [DOI] [PubMed] [Google Scholar]
- 20.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 21.Petit, A., J. Tempe, A. Kerr, M. Holsters, M. van Montagu, and J. Schell. 1978. Substrate induction of conjugative activity of Agrobacterium tumefaciens Ti plasmids. Nature 271:570-572. [Google Scholar]
- 22.Piper, K. R., S. Beck von Bodman, I. Hwang, and S. K. Farrand. 1999. Hierarchical gene regulatory systems arising from fortuitous gene associations: Controlling quorum sensing by the opine regulon in Agrobacterium. Mol. Microbiol. 32:1077-1089. [DOI] [PubMed] [Google Scholar]
- 23.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 24.Piper, K. R., and S. K. Farrand. 2000. Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the activator TraM. J. Bacteriol. 182:1080-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin, Y., Z. Q. Luo, A. J. Smyth, P. Gao, S. Beck von Bodman, and S. K. Farrand. 2000. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 19:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shmatkov, A. M., A. A. Melikyan, F. L. Chernousko, and M. Borodovsky. 1999. Finding prokaryotic genes by the ‘frame-by-frame’ algorithm: targeting gene starts and overlapping genes. Bioinformatics 15:874-886. [DOI] [PubMed] [Google Scholar]
- 28.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swiderska, A., A. K. Berndtson, M. R. Cha, L. Li, G. M. Beaudoin III, J. Zhu, and C. Fuqua. 2001. Inhibition of the Agrobacterium tumefaciens TraR quorum-sensing regulator. Interactions with the TraM anti-activator. J. Biol. Chem. 276:49449-49458. [DOI] [PubMed] [Google Scholar]
- 30.Vannini, A., C. Volpari, and S. Di Marco. 2004. Crystal structure of the quorum-sensing protein TraM and its interaction with the transcriptional regulator TraR. J. Biol. Chem. 279:24291-24296. [DOI] [PubMed] [Google Scholar]
- 31.Wang, L., J. D. Helmann, and S. C. Winans. 1992. The A. tumefaciens transcriptional activator OccR causes a bend at a target promoter, which is partially relaxed by a plant tumor metabolite. Cell 69:659-667. [DOI] [PubMed] [Google Scholar]
- 32.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, H. B., C. Wang, and L. H. Zhang. 2004. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol. Microbiol. 52:1389-1401. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, L. H., and A. Kerr. 1993. Rapid purification of Ti plasmids from Agrobacterium by ethidium bromide treatment and phenol extraction. Lett. Appl. Microbiol. 16:265-268. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, L. H., P. J. Murphy, A. Kerr, and M. E. Tate. 1993. Agrobacterium conjugation and gene regulation by N-acyl-L-homoserine lactones. Nature 362:446-448. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, L. H. 2003. Quorum quenching and proactive host defense. Trends Plant Sci. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, L. H., and Y. H. Dong. 2004. Quorum sensing and signal interference: diverse implications. Mol. Microbiol. 53:1563-1571. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, L. H., and A. Kerr. 1991. A diffusible compound can enhance conjugal transfer of the Ti plasmid in Agrobacterium tumefaciens. J. Bacteriol. 173:1867-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, J., and S. C. Winans. 1998. Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol. Microbiol. 27:289-297. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837.10220379 [Google Scholar]