Abstract
Transcriptional profiling of Pseudomonas aeruginosa grown under steady-state hyperosmotic stress conditions showed an up-regulation of genes associated with osmoprotectant synthesis, putative hydrophilins, and the type III secretion system with associated cytotoxins. A large number of regulatory genes, including several two-component systems not previously known to be influenced by osmolarity, were differentially expressed by P. aeruginosa in immediate response to hyperosmotic shock.
Changes in osmolarity are likely an inescapable reality for microbes colonizing any environment (22), and bacteria must be able to both respond immediately to osmotic shock, which might occur during transitions between growth environments, and sustain growth under lasting conditions of osmotic stress. As an opportunistic pathogen of humans, Pseudomonas aeruginosa employs adaptive responses that originally evolved for survival in diverse and often stressful environmental conditions (21, 22). Hence, genes differentially expressed in response to osmotic stress may play a key role in the prevalence and persistence of P. aeruginosa in osmotically stressful infection sites (9). Aside from the accumulation of osmoprotectants, very little is known about osmoadaptive processes in P. aeruginosa (3, 5). In this study we used Affymetrix GeneChip microarrays to investigate the transcriptional responses of P. aeruginosa to both osmotic up-shock and growth under steady-state osmotic stress conditions.
Bacterial growth conditions and genetic manipulations.
P. aeruginosa PA14 was grown in modified minimal A medium (14) containing 60 mM K2HPO4, 30 mM KH2PO4, 7.5 mM (NH4)2SO4, 1 mM MgSO4, 10 μM FeSO4, and 17 mM glucose, with or without 0.3 M NaCl or 0.7 M sucrose, pH 7.1. A 40-ml volume of medium was inoculated with 0.3 ml of washed cells from an overnight culture to yield a starting optical density at 600 nm (OD600) of ∼0.03. Log-phase cells used for DNA microarray analysis under steady-state conditions were harvested at an OD600 of 0.25. For the osmotic up-shock experiments, 3.9 ml of prewarmed minimal medium containing 3 M NaCl (pH 7.1) was added to 35 ml of log-phase culture (OD600, 0.25) to yield a final NaCl concentration of 0.3 M. As a control, 3.9 ml of prewarmed minimal medium without NaCl was added to 35 ml of log-phase culture. Cells were harvested for gene expression analysis 15 to 60 min after up-shock. All incubations were performed in a 37°C water bath with shaking at 200 rpm.
RNA isolation, removal of contaminating DNA, cDNA synthesis, and fragmentation of cDNA were performed as previously described (16). Processing of the P. aeruginosa GeneChips (Affymetrix) was performed at the University of Tulsa Microarray Facility per Affymetrix protocols. Data analysis was performed using Affymetrix Microarray Suite software; only those genes differentially regulated at least threefold were considered. In general, 70% to 75% of the ∼5,500 open reading frames (ORFs) in P. aeruginosa (19) were detectable on the GeneChips.
The steady-state osmotic stress regulon.
The expression levels of 116 and 81 genes were changed at least threefold in cells grown in the presence of 0.3 M NaCl and 0.7 M sucrose, respectively. However, 66 genes showed an expression level change of at least threefold in response to both stressors (Table 1), indicating a response to osmotic stress in general rather than a response to a specific compound or ionic strength. Although P. aeruginosa GeneChips have proven reliable for evaluating mRNA levels (16), reverse transcription-PCR experiments confirmed GeneChip data for two up-regulated genes (pscC and PA2152) and one down-regulated gene (PA2260) (data not shown). These 66 genes are referred to here as the steady-state osmotic stress regulon.
TABLE 1.
P. aeruginosa genes differentially regulated by osmotic stress
| Functional classa | Genea | Fold regulationb
|
Functional classa | Genea | Fold regulationb
|
|||
|---|---|---|---|---|---|---|---|---|
| 0.3 M NaCl | 0.7 M sucrose | 0.3 M NaCl | 0.7 M sucrose | |||||
| Secreted factors | Export protein | pscK | 5.2 | 7.0 | ||||
| ADP-ribosyltransferase | exoT | 5.3 | 7.3 | Export protein | pscL | 5.8 | 6.8 | |
| Adenylate cyclase | exoY | 6.1 | 7.6 | Chaperone | spcU | 6.0 | 6.3 | |
| Probable chaperone | PA3842 | 6.6 | 7.9 | |||||
| Protein secretion (type III secretion system) | ||||||||
| Translocation protein | pscU | 4.4 | 3.6 | Adaptation/protection | ||||
| Translocation protein | pscT | 4.9 | 5.4 | Osmotically inducible protein | osmC | 3.6 | 4.7 | |
| Translocation protein | pscS | 5.1 | 6.2 | |||||
| Translocation protein | pscR | 5.5 | 5.8 | Posttranslational modification/degradation | ||||
| Translocation protein | pscQ | 5.1 | 6.4 | Ribosome modulation factor | rmf | 7.8 | 6.2 | |
| Translocation protein | pscP | 5.2 | 5.4 | Leucine aminopeptidase | pepA | 5.3 | 6.5 | |
| Translocation protein | pscO | 6.2 | 7.1 | Probable protease | PA4171 | 5.2 | 6.9 | |
| ATP synthase | pscN | 5.6 | 6.6 | |||||
| Outer membrane protein | popN | 5.5 | 6.0 | Metabolism/enzymes | ||||
| Secretion protein | pcr1 | 5.5 | 6.7 | Protein activator | pra | −3.4 | −3.7 | |
| Secretion protein | pcr2 | 5.2 | 6.3 | Probable aldose-epimerase | PA2260 (kguE) | −7.3 | −3.9 | |
| Secretion protein | pcr3 | 5.6 | 7.1 | Probable 2-ketogluconate kinase | PA2261 (kguK) | −9.0 | −3.8 | |
| Secretion protein | pcr4 | 5.0 | 6.4 | Probable 2-ketogluconate transporter | PA2262 (kguT) | −9.2 | −4.5 | |
| Apparatus protein | pcrD | 4.8 | 6.1 | 2-Hydroxyacid dehydrogenase | PA2263 (kguD) | −6.2 | −3.8 | |
| Transcriptional regulator | pcrR | 5.0 | 5.7 | Probable gluconate dehydrogenase subunit | PA2264 | −5.3 | −4.5 | |
| Regulator protein | pcrG | 9.5 | 8.0 | Gluconate dehydrogenase | PA2265 | −6.2 | −4.4 | |
| Secretion protein | pcrV | 7.1 | 8.2 | Probable cytochrome c precursor | PA2266 | −6.3 | −4.0 | |
| Regulatory protein | pcrH | 6.5 | 7.5 | Probable trehalose synthase | PA2152 | 3.4 | 3.6 | |
| Translocation protein | popB | 4.2 | 5.3 | Probable glutamine amidotransferase | PA3459 | 5.7 | 6.3 | |
| Translocation protein | popD | 5.6 | 6.3 | Probable acetyltransferase | PA3460 | 5.8 | 6.3 | |
| Regulator protein | exsC | 4.8 | 5.7 | Probable peptidase | PA3461 | 4.9 | 4.8 | |
| Hypothetical protein | PA1711 | 7.2 | 6.7 | Probable bacterioferritin | PA4880 | 5.2 | 5.8 | |
| Regulator protein | exsB | 8.4 | 6.0 | |||||
| Transcriptional regulator | exsA | 6.6 | 6.9 | Unknown | ||||
| Regulator protein | exsD | 5.5 | 6.3 | Hypothetical protein | PA0988 | 5.0 | 5.6 | |
| Export protein | pscB | 7.6 | 8.7 | Hypothetical protein | PA1323 | 4.5 | 5.4 | |
| Export protein | pscC | 6.0 | 6.6 | Hypothetical protein | PA1324 | 3.8 | 4.9 | |
| Export protein | pscD | 5.6 | 6.9 | Conserved hypothetical protein | PA2146 | 14.4 | 10.6 | |
| Export protein | pscE | 5.8 | 6.8 | Hypothetical protein | PA2747 | 3.0 | 3.9 | |
| Export protein | pscF | 5.7 | 6.8 | Hypothetical protein | PA3691 | 3.7 | 5.1 | |
| Export protein | pscG | 5.5 | 6.8 | Hypothetical protein | PA3843 | 7.5 | 7.1 | |
| Export protein | pscH | 5.6 | 6.6 | Hypothetical protein | PA4139 | −5.9 | −4.8 | |
| Export protein | pscI | 5.6 | 6.9 | Hypothetical protein | PA5482 | 3.2 | 4.3 | |
| Export protein | pscJ | 6.7 | 7.4 | |||||
From the P. aeruginosa genome project website (www.pseudomonas.com). Proposed gene names for the 2-ketogluconate utilization operon are in parentheses (20).
Severalfold regulation of P. aeruginosa genes differentially expressed under high-osmolarity conditions (0.3 M NaCl or 0.7 M sucrose) as compared to low-osmolarity conditions; a positive number indicates an up-regulation of the gene under high-osmolarity conditions. Values are the averages of three trials.
Interestingly, 40 of those 66 genes are associated with virulence factor expression, encoding proteins of a type III secretion system (TTSS), the type III cytotoxins ExoT and ExoY, and two ancillary chaperones (7, 10) (Table 1). The significance of an osmoinducible TTSS in cystic fibrosis disease pathogenesis is debatable (2, 4, 12), but osmoinduction of this system may have significance for P. aeruginosa living in other settings. For instance, a functional TTSS (and cytotoxin ExoU) has been shown to protect P. aeruginosa from predation by the amoeba Dictyostelium discoideum (13).
Identification of osmoprotectant genes required for growth under osmotic stress conditions.
In similarity to other bacteria, P. aeruginosa has been shown to accumulate K+, glutamate, trehalose, and N-acetylglutaminylglutamine amide (NAGGN) as cytoplasmic osmoprotectants in response to osmotic stress (5). As expected, cells grown under osmotic stress conditions showed elevated expression of genes that putatively encode proteins involved in the synthesis of the osmoprotectants trehalose (PA2152) and NAGGN (PA3459 to PA3461) (Table 1). Furthermore, the expression profile and genetic arrangement of ORFs PA3459 to PA3461 suggest that they constitute an operon (www.pseudomonas.com).
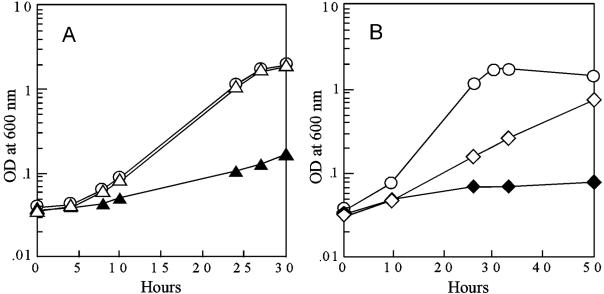
Recent evidence suggests that NAGGN is the major organic osmoprotectant in P. aeruginosa (5). Given this observation, mutants harboring a transposon insertion in PA3459 or PA3460 (11) (www.genome.washington.edu) were examined for growth in the presence of 0.5 M NaCl. Transposon insertion locations were confirmed by PCR (data not shown). The PA3459 and PA3460 mutants grew as well as the wild type in media that did not contain NaCl but were impaired for growth under osmotic stress conditions (Fig. 1A and B). The reversed orientation of the transposon in PA3460 (11) (see Mutant Search at www.genome.washington.edu) may have created a polar effect, inhibiting downstream transcription of PA3461 and thus preventing full complementation in the mutant (Fig. 1B). Similar results were seen with growth in 0.7 M sucrose (data not shown).
FIG. 1.
Disruption of putative osmoprotectant synthesis genes impairs growth of P. aeruginosa PAO1 in a high-osmolarity medium. Cells were grown at 37°C in a glucose minimal medium containing 0.5 M NaCl. For complementation, a wild-type copy of the interrupted gene containing the native ribosome binding site was PCR generated and cloned (1) as an EcoRI-HindIII or EcoRI-BamHI fragment downstream of the lac promoter in pUCP20 (17). Plasmid constructs were introduced into P. aeruginosa by use of electroporation (18), and transformants were selected using Trypticase soy agar containing 100 μg/ml carbenicillin. FA015, PA3459::ISphoA/hah; FA011, PA3460::ISlacZ/hah. PA3459 and PA3460 encode a putative glutamine amidotransferase and an acetyltransferase, respectively. (A) Open circles, PAO1/pUCP20; open triangles, FA015/pUCP20::PA3459; closed triangles, FA015/pUCP20. (B) Open circles, PAO1/pUCP20; open diamonds, FA011/pUCP20::PA3460; closed diamonds, FA011/pUCP20. Values are representative of at least three independent trials.
The osmoprotectant betaine can functionally complement NAGGN mutants.
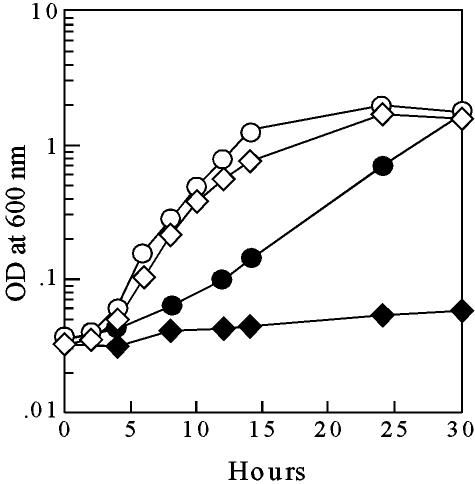
Betaine is a powerful osmoprotectant that accumulates to a relatively high concentration in the cytoplasm of osmotically stressed cells (3, 5). Exogenous betaine has been shown to increase the growth rate and to partially replace NAGGN in osmotically stressed wild-type P. aeruginosa cells (5). Hence, we reasoned that the NAGGN mutants would exhibit a wild-type growth rate under osmotic stress conditions when grown in the presence of betaine. Indeed, the doubling time for the PA3460 mutant in 0.5 M NaCl plus betaine was virtually identical to that seen in the wild type grown under the same conditions—i.e., doubling times were 117 and 120 min for the wild type and mutant, respectively (Fig. 2). These results suggest that betaine can substitute for NAGGN in osmotically stressed cells whereas trehalose and glutamate cannot. However, betaine does not exert transcriptional control over these osmoprotective genes because betaine had no significant effect on the expression of PA2152 or PA3459 to PA3461 in osmotically stressed cells (data not shown).
FIG. 2.
The osmoprotectant betaine recovers growth rate in an osmotic stress-susceptible mutant. Cells were grown in the presence of 0.5 M NaCl under the same conditions described for Fig. 1. Open symbols, cells grown with 0.5 mM betaine; closed symbols, cells grown without betaine. Circles, PAO1; diamonds, FA011 (PA3460::ISlacZ/hah). Values are representative of two independent trials.
Identification of putative hydrophilins.
The role of osmoprotectants in conferring resistance to osmotic stress has been relatively well characterized (3, 22). A lesser-known category of osmoprotective compounds is the hydrophilins, a family of relatively small basic proteins found in plants, Saccharomyces cerevisiae, and bacteria that are inducible by osmotic stress (8). The P. aeruginosa genome harbors (at least) three genes encoding proteins with similarity to Escherichia coli hydrophilins; i.e., ORFs PA3049, PA2146, and PA4738 encode proteins with 66%, 92%, and 71% similarity to E. coli Rmf, YciG, and YjbJ, respectively (www.pseudomonas.com). Moreover, the expression of ORFs PA3049 (rmf), PA2146 (yciG), and PA4738 (yjbJ) was up-regulated 5- to 16-fold in osmotically shocked P. aeruginosa cells. The precise function of hydrophilins in conferring osmoprotection in bacteria is not known (8), and the relative contribution of these proteins toward osmotolerance in P. aeruginosa has yet to be demonstrated.
Gene expression during osmotic up-shock.
The genes listed in Table 1 represent those that are differentially regulated in osmotically stressed cells growing under steady-state conditions several hours after initial exposure to hyperosmotic shock. To gain an understanding of the genetic events that occur during the early stages of osmoadaptation, we monitored transcription for 15 to 60 min in log-phase cells subjected to osmotic up-shock with 0.3 M NaCl.
Gene expression in osmotically shocked cells generally paralleled that seen in cells grown under steady-state osmotic stress, but in many cases the level of expression was enhanced. As would be expected, genes associated with osmoprotectant synthesis—i.e., PA2152 and PA3459 to PA3461—were among those showing enhanced levels of expression during the early stages of osmotic up-shock (Table 2). Interestingly, the expression of genes associated with the TTSS was substantially repressed in osmotically shocked cells (Table 2). This may be an energy-saving strategy for the cell, as resources that would otherwise be consumed by TTSS expression can be utilized for osmoadaptive purposes.
TABLE 2.
Effect of osmotic up-shock on expression of genes in the steady-state osmotic stress regulon of P. aeruginosa PA14
| Functional classa | Genea | Fold regulation
|
||
|---|---|---|---|---|
| Steady stateb | Up-shockc
|
|||
| 15 min | 60 min | |||
| Adaptation/protection | ||||
| Osmotically inducible protein | osmC | 3.6 | 10.6 | 1.7 |
| Posttranslational modification/degradation | ||||
| Ribosome modulation factor | rmf | 7.8 | −0.8 | 6.2 |
| Leucine aminopeptidase | pepA | 5.3 | −2.6 | −0.8 |
| Probable protease | PA4171 | 5.2 | 10.3 | 4.2 |
| Metabolism/putative enzymes | ||||
| Protein activator | pra | −3.4 | −6.6 | −1.6 |
| Probable aldose-epimerase | PA2260 (kguE) | −7.3 | −5.6 | −6.5 |
| Probable 2-ketogluconate kinase | PA2261 (kguK) | −9.0 | −4.7 | −5.5 |
| Probable 2-ketogluconate transporter | PA2262 (kguT) | −9.2 | −7.0 | −5.9 |
| 2-Hydroxyacid dehydrogenase | PA2263 (kguD) | −6.2 | −6.4 | −5.8 |
| Probable gluconate dehydrogenase subunit | PA2264 | −5.3 | −2.2 | −5.7 |
| Gluconate dehydrogenase | PA2265 | −6.2 | −2.8 | −5.7 |
| Probable cytochrome c precursor | PA2266 | −6.3 | −2.3 | −4.8 |
| Probable trehalose synthase | PA2152 | 3.4 | 10.6 | 2.8 |
| Probable glutamine amidotransferase | PA3459 | 5.7 | 11.5 | 5.6 |
| Probable acetyltransferase | PA3460 | 5.8 | 10.6 | 5.3 |
| Probable peptidase | PA3461 | 4.9 | 10.3 | 4.0 |
| Probable bacterioferritin | PA4880 | 5.2 | 11.0 | 1.6 |
| Unknown | ||||
| Hypothetical protein | PA0988 | 5.0 | 0.1 | 0.3 |
| Hypothetical protein | PA1323 | 4.5 | 9.0 | 3.0 |
| Hypothetical protein | PA1324 | 3.8 | 9.5 | 2.7 |
| Conserved hypothetical protein | PA2146 | 14.4 | 15.0 | 13.5 |
| Hypothetical protein | PA2747 | 3.0 | 7.4 | 0.0 |
| Hypothetical protein | PA3691 | 3.7 | 8.5 | 2.5 |
| Hypothetical protein | PA3843 | 7.5 | −0.8 | −1.2 |
| Hypothetical protein | PA4139 | −5.9 | −2.4 | −1.1 |
| Hypothetical protein | PA5482 | 3.2 | 7.8 | −0.8 |
| Secreted factors (cytotoxins) | ||||
| ADP-ribosyltransferase | exoT | 5.3 | −3.2 | −3.0 |
| Adenylate cyclase | exoY | 6.1 | −2.7 | −1.3 |
| Protein secretion (type III secretion system) | ||||
| Type III secretion proteins | 5.8d | −2.6 | 0.4 | |
| Chaperone | spcU | 5.0 | −2.2 | −0.5 |
| Probable chaperone | PA3842 | 6.0 | −4.3 | −2.4 |
From the Pseudomonas Genome Project website (www.pseudomonas.com). Proposed gene names for the 2-ketogluconate utilization operon are in parentheses (20).
Steady-state values (from Table 1) are for log-phase cells grown at 37°C in a glucose minimal medium containing 0.3 M NaCl.
Gene expression 15 or 60 min after addition of NaCl (0.3 M final) to a log-phase culture grown in a glucose minimal medium at 37°C. A positive number indicates an up-regulation of the gene after NaCl addition. Values are the averages of two trials.
Average expression of 36 genes encoding the type III secretion apparatus (see Table 1).
The most striking differences between gene expression in cells grown under steady-state osmotic stress conditions and in cells subjected to osmotic up-shock are the total number of genes affected and the number of regulatory genes represented. For instance, the expression of 431 genes was changed at least threefold during osmotic up-shock with 0.3 M NaCl compared to that of 116 and 81 genes for cells grown under steady-state osmotic stress conditions with 0.3 M NaCl and 0.7 M sucrose, respectively. Moreover, only TTSS regulatory genes are represented in the steady-state osmotic stress regulon (Table 1), whereas the expression of 26 genes encoding proteins of known or probable regulatory function was changed at least threefold within 15 min of osmotic up-shock with 0.3 M NaCl (Table 3).
TABLE 3.
Effect of osmotic up-shock on expression of genes associated with regulation or adaptation and protection in P. aeruginosa PA14
| Functional classa | Genea | Fold regulation (min after up-shock)b
|
|||
|---|---|---|---|---|---|
| 15 | 30 | 45 | 60 | ||
| Regulation | |||||
| Anti-sigma factor | mucA | 3.4 | 3.2 | 0.8 | 0.6 |
| Negative regulator | mucB | 3.8 | 3.4 | 1.0 | 0.8 |
| Sigma factor | algU | 4.0 | 2.9 | 0.1 | 0.5 |
| Response regulator | algR | 4.2 | 3.7 | 2.5 | 1.8 |
| Sensor kinase | kinB | 5.4 | 3.1 | −0.5 | −0.6 |
| Response regulator | algB | 4.6 | 3.4 | −0.7 | −1.4 |
| Sensor kinase | phoQ | 5.1 | 4.5 | 5.8 | 5.1 |
| Response regulator | phoP | 4.3 | 4.0 | 5.3 | 4.5 |
| Transcriptional regulator | pchR | 3.5 | 5.1 | 4.3 | 5.0 |
| Transcriptional regulator | rtcR | 4.4 | 3.4 | 0.1 | −0.8 |
| Transcriptional regulator | rhlR | −3.4 | −1.1 | 0.5 | 0.3 |
| Probable transcriptional regulator | PA0236 | −3.5 | −4.4 | −3.0 | −1.2 |
| Probable transcriptional regulator | PA0448 | 4.5 | 3.2 | 1.7 | 0.9 |
| Probable transcriptional regulator | PA1226 | 3.4 | 3.1 | 1.7 | 1.6 |
| Probable transcriptional regulator | PA2417 | 4.7 | 3.3 | 1.9 | 1.6 |
| Probable transcriptional regulator | PA2588 | −3.6 | −2.8 | −2.9 | −2.6 |
| Probable transcriptional regulator | PA2665 | 3.6 | 2.2 | 0.7 | 0.3 |
| Probable transcriptional regulator | PA3689 | 3.6 | 3.2 | 0.8 | 0.8 |
| Probable transcriptional regulator | PA4135 | −3.8 | −4.0 | −1.9 | −0.7 |
| Probable transmembrane sensor | PA0471 | 3.2 | 3.4 | 2.7 | 3.6 |
| Probable response regulator | PA4296 | −4.0 | 0.4 | 3.4 | 2.1 |
| Probable sensor/regulator hybrid | PA1243 | 8.2 | 6.9 | 1.9 | 0.9 |
| Probable sensor/regulator hybrid | PA2177 | 5.3 | 6.4 | 2.7 | 1.8 |
| Probable sensor/regulator hybrid | PA3462 | 5.3 | 4.5 | 1.5 | 0.7 |
| Probable sigma-70 factor, ECFc | PA2896 | 7.4 | 6.5 | 2.7 | 2.2 |
| Probable sigma-70 factor, ECF | PA0472 | 3.8 | 3.4 | 2.6 | 3.7 |
| Adaptation/protection | |||||
| Catalase HPII | katE | 11.4 | 13.9 | 11.0 | 4.7 |
| Osmotically inducible lipoprotein | osmE | 8.2 | 7.5 | 3.1 | 0.5 |
| Probable DNA-binding stress protein | PA0962 | 5.3 | 4.8 | 2.2 | 1.4 |
| Probable cold shock protein | PA1159 | −4.4 | −4.1 | −3.0 | −1.5 |
From the Pseudomonas Genome Project website (www.pseudomonas.com).
Gene expression after adding NaCl (0.3 M final) to a log-phase culture growing in a glucose minimal medium at 37°C. Data shown include only those genes not in Table 1 or Table 2 and that showed at least a threefold change in expression 15 min after up-shock. A positive number indicates an up-regulation of the gene after NaCl addition. Values are the averages of two trials.
ECF, extracytoplasmic function.
The up-regulation of algU in osmotically shocked cells (Table 3) was expected given that AlgU has previously been shown to control the expression of genes in Pseudomonas species that confer resistance under a variety of stress conditions, including elevated osmolarity (9, 15). Furthermore, the observation that ORFs PA3459 to PA3461—encoding putative osmoprotectant synthesis proteins—play a significant role in osmoadaptation is consistent with the report by Firoved et al. (6) that expression of PA3459 is AlgU dependent. Transcriptional control of PA3459 by AlgU is likely indirect, however, since the PA3459 promoter region does not possess the consensus sequence typical of AlgU-dependent promoters (6).
Conclusion.
The purpose of this study was to identify genes involved in the osmotic stress response in P. aeruginosa, with a focus on elucidating those determinants that may contribute to survival of the bacterium in diverse and potentially osmotically stressful settings. A critical factor for survival of P. aeruginosa in high-osmolarity environments is the ability to synthesize osmoprotective compounds, i.e., osmoprotectants and hydrophilins. The relative contribution of hydrophilins in conferring osmoprotection in P. aeruginosa has yet to be determined, whereas the importance of osmoprotectants, especially NAGGN, is clear. The identification of osmoprotective genes, and those regulatory factors controlling their expression, is central to understanding how this important opportunistic pathogen survives in osmotically stressful environments.
Acknowledgments
We thank the University of Washington Genome Center for providing mutants, H. Schweizer for providing plasmids, the University of Tulsa Microarray Facility for cDNA sample processing and data analysis, and Adam Switzer for his work on experiments associated with this study.
This publication was made possible by National Institutes of Health grant P20 RR016478 (to A.A.) from the INBRE Program of the National Center for Research Resources.
REFERENCES
- 1.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 2.Corech, R., A. Rao, A. Laxova, J. Moss, M. J. Rock, Z. Li, M. R. Kosorok, M. L. Splaingard, P. M. Farrell, and J. T. Barbieri. 2005. Early immune response to the components of the type III system of Pseudomonas aeruginosa in children with cystic fibrosis. J. Clin. Microbiol. 43:3956-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csonka, L. N. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson, S. H., and R. C. Boucher. 2003. Update on pathogenesis of cystic fibrosis lung disease. Curr. Opin. Pulm. Med. 9:486-491. [DOI] [PubMed] [Google Scholar]
- 5.D'Souza-Ault, M. R., L. T. Smith, and G. M. Smith. 1993. Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl. Environ. Microbiol. 59:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Firoved, A. M., J. C. Boucher, and V. Deretic. 2002. Global genomic analysis of AlgU (σE)-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 184:1057-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 8.Garay-Arroyo, A., J. M. Colmenero-Flores, A. Garciarrubio, and A. A. Covarrubias. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275:5668-5674. [DOI] [PubMed] [Google Scholar]
- 9.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain, M., D. Ramirez, R. Seshadri, J. F. Cullina, C. A. Powers, G. S. Schulert, M. Bar-Meir, C. L. Sullivan, S. A. McColley, and A. R. Hauser. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 42:5229-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pukatzki, S., R. H. Kessin, and J. J. Mekalanos. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 99:3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 15.Schnider-Keel, U., K. B. Lejbolle, E. Baehler, D. Haas, and C. Keel. 2001. The σ-factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHAO. Appl. Environ. Microbiol. 67:5683-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 18.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 20.Swanson, B. L., P. Hager, P. Phibbs, Jr., U. Ochsner, M. L. Vasil, and A. N. Hamood. 2000. Characterization of the 2-ketogluconate utilization operon in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 37:561-573. [DOI] [PubMed] [Google Scholar]
- 21.van Veen, J. A., L. S. van Overbeek, and J. D. van Elsas. 1997. Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61:121-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood, J. M. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63:230-262. [DOI] [PMC free article] [PubMed] [Google Scholar]