Abstract
Ribonucleotide reductase (RNR) catalyzes the essential production of deoxyribonucleotides in all living cells. In this study we have established a sensitive in vivo assay to study the activity of RNR in aerobic Escherichia coli cells. The method is based on the complementation of a chromosomally encoded nonfunctional RNR with plasmid-encoded RNR. This assay can be used to determine in vivo activity of RNR mutants with activities beyond the detection limits of traditional in vitro assays. E. coli RNR is composed of two homodimeric proteins, R1 and R2. The R2 protein contains a stable tyrosyl radical essential for the catalysis that takes place at the R1 active site. The three-dimensional structures of both proteins, phylogenetic studies, and site-directed mutagenesis experiments show that the radical is transferred from the R2 protein to the active site in the R1 protein via a radical transfer pathway composed of at least nine conserved amino acid residues. Using the new assay we determined the in vivo activity of mutants affecting the radical transfer pathway in RNR and identified some residual radical transfer activity in two mutant R2 constructs (D237N and W48Y) that had previously been classified as negative for enzyme activity. In addition, we show that the R2 mutant Y356W is completely inactive, in sharp contrast to what has previously been observed for the corresponding mutation in the mouse R2 enzyme.
By catalyzing the reduction of all four ribonucleotides to the corresponding deoxyribonucleotides, the enzyme ribonucleotide reductase (RNR) is essential for all living organisms containing DNA. The genome of Escherichia coli codes for three different RNR enzymes: the nrdAB locus encodes a class Ia enzyme, the nrdEF locus encodes a class Ib enzyme, and the nrdDG locus encodes a class III enzyme and its accompanying activase (2, 10). Only the nrdAB genes are expressed during aerobic growth, and only the nrdDG genes are expressed under anaerobic conditions. The nrdEF locus is only known to be active during oxidative stress (14), and conditional mutations in nrdA or nrdB are lethal under restrictive conditions during aerobic growth (8, 9).
At present, two established methods are used for measuring RNR activity: one utilizes radioactively labeled substrate and monitors product formation, and the other monitors NADPH consumption (24, 27). Monitoring of NADPH consumption is an indirect method for measuring RNR activity and detects a high degree of background activity. The use of radioactively labeled substrate is a direct method, and the detected background is about 0.5 to 3% depending on protein preparation and substrate purity. A factor that influences the results of these assays is the relative amount of contamination by chromosomally encoded wild-type protein, since the mutant E. coli RNR components are expressed and purified from E. coli cells that also have a chromosomal nrdAB operon encoding wild-type RNR. Traditional assays require that the mutant constructs be expressed and purified prior to the activity measurement. Some mutant proteins are unstable in vitro and are degraded during this time-consuming procedure.
In this study, we have used an E. coli strain with conditional lethal mutations in the nrdA and nrdB genes to develop an in vivo assay with selection for active class Ia RNR at the bacterial colony stage. This assay can be used to determine in vivo RNR activity of mutant forms of the enzyme. In general, in vivo assays are very sensitive methods and could in theory be as sensitive as the reversion or mutation frequency of the gene. Our method is based on extrachromosomal complementation of cells with conditional lethal mutations in both chromosomal genes encoding the major aerobic RNR. The assay enables plasmid-encoded RNR subunits to complement the chromosomally encoded defective RNR under specific selective conditions. With a detection limit of ≤10−7 in plating efficiency, it is several orders of magnitude more sensitive than traditional activity assays, and cell survival rates as low as 10−6 can be distinguished.
We have used the in vivo system to determine the plating efficiency of a set of E. coli class Ia RNR mutants earlier shown to have low or no activity by classical in vitro methods. The R1 protein encoded by the nrdA gene and the R2 protein encoded by the nrdB gene are both needed for enzyme activity (for a recent review, see reference 6). The R1 protein contains the active site where a radical-based catalytic reaction takes place (12, 18), and the R2 protein contains a stable tyrosyl radical involved in the reaction mechanism (11). Structural data (15, 16, 28), phylogenetic studies (10), and site-directed mutagenesis experiments (1, 3-5, 11, 13, 19, 21, 22) show that the two radical sites are connected via a radical transfer pathway (RTP) that is operational during catalysis and composed of at least nine conserved residues (Fig. 1). Previous site-directed mutagenesis experiments have involved the E. coli R1 and R2 proteins and the mouse R2 protein. By using the in vivo assay on mutants affecting the RTP we have been able to differentiate between mutant R2 proteins with some residual activity (D237N and W48Y) and completely inactive ones.
FIG. 1.
Scheme of the RTP of E. coli RNR.
MATERIALS AND METHODS
Materials.
Primers used for sequencing were obtained and purified by Scandinavian Gene Synthesis AB. All plasmids were prepared with Qiagen plasmid midi/mini kit obtained from Qiagen. Hydroxyurea (HU) and thymine were obtained from Sigma. Stock solutions of HU (0.3 M) were prepared in sterile water immediately before use, and thymine (2 mg/ml) was dissolved in 0.05 mM NaOH. Restriction enzymes used for subcloning were SacI obtained from Boehringer and BsrGI from New England Biolabs. Ligations were performed with T4 DNA ligase obtained from U.S. Biochemicals.
Plasmids and vectors.
Plasmid pAL7 was originally constructed for simultaneous expression of both subunits of E. coli RNR (11). DNA (5.7 kb) containing the nrdA gene with its upstream promoter region and the downstream nrdB gene was cloned in 2.3 kb of pBR322 between the HindIII and PvuII sites. A repeated region (25) of 207 bp between the two nrd genes was deleted from pAL7 to obtain the pAL21 plasmid. MPpAL21 and MPpTB2 were constructed by adding two cleavage sites, SacI before and BsrGI after the nrdB gene.
Plasmid pTB1 and pTB2, containing the nrdA (pTB1) and nrdB (pTB2) genes (1, 3), are derivatives of pTZ18R obtained from Amersham Pharmacia Biotech.
Site-directed mutagenesis.
The Y356W and W48Y mutations and the cleavage sites, SacI and BsrGI, were inserted into MPpAL21, pAL21, and pTB2 by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit from Stratagene. This method is PCR based and can be performed on any double-stranded DNA. The oligonucleotides used for mutagenesis were (underlining denotes mismatched nucleotides and boldface type denotes recognition sequence for restriction enzymes), for BsrGI in pAL21 and pTB2, d(5′-GCTGTGCCAGGATGTACACCCTTCCCTT-3′) and d(5′-GAAGGGAAGGGTGTACATCCTGGCACAGC-3′); for SacI in pAL21, d(5′-GATCTGATATTGAGCTCAACAGGACACACTC-3′) and d(5′-GAGTGTGTCCTGTTGAGCTCAATATCAGATC-3′); for SacI in pTB2, d(5′-GTGTCCTGTTGGGAGCTCAATGCCGGATAAGGC-3′) and d(5′-GCCTTATCCGGCATTGAGCTCCCAACAGGACAC-3′); and for W48Y in MPpAL21, d(5′-GCAGCTCTCTTTCTTCTATCTGCCGGAAGAAGTTGACG-3′) and d(5′-CGTCAACTTCTTCCGGACGATAGAAGAAAGAGAGCTG-3′) (the prefix “d” indicates deoxy). Verifications of mutations and sequencing of the entire genes were done by the Sanger chain termination DNA sequencing method using a DYEnamic ET terminator cycle sequencing kit. The MPpAL21 plasmid used as the starting construct contained, in addition to the desired cleavage sites, a mutation in the R1 gene, resulting in a valine 361-to-alanine substitution. Sequence analysis showed that this mutation was also present in pAL21 but not in pAL7. All three plasmid constructs were phenotypically indistinguishable in all assays.
Subcloning of point mutations.
The mutations were subcloned from pTB1 and pTB2 into MPpAL21. To be able to subclone the different mutations, cleavage sites SacI or BsrGI were moved from MPpTB2 to the different mutant pTB2 plasmids; BsrGI sites were added to D237A and D237N, and SacI sites were added to D84A, H118A, and Y122F. These cleavage sites were then used to move the mutations from the pTB2 derivatives to MPpAL21. Mutations in pTB1—C439A, Y7301F, Y731F, and E441D—were subcloned by using already-existing cleavage sites.
Bacterial strains.
E. coli MV1190 [Δ(lac-proAB) thi supE Δ(srl-recA)306::Tn10/F′ traD36 proAB lacIq ZΔM15], obtained from Bio-Rad, was used for cloning. E. coli KK535 (thr leu thi deo tonA lacY supE44 recA nalA nrdA nrdB), obtained from O. Karlström, Department of Microbiology, University of Copenhagen, carries conditional lethal mutations in both structural genes of RNR, nrdA-1 and nrdB-1, resulting in a temperature- and HU-sensitive phenotype.
Transformation and screening.
CFU were determined as the number of colonies on plates at 42°C containing HU after transformation of 1 ng of MPpAL (wild-type or mutant) construct by electroporation into 40 μl of KK535. Luria broth (40 μl) was added, and the transformation mixture was incubated at 30°C for 90 min for phenotypic expression and plated on Luria agar containing thymine (50 μg/ml), carbenicillin (50 μg/ml), and 0 to 12 mM HU. The plates were then incubated for 2 days at 42°C, and the colonies were counted.
To determine the plating efficiency, KK535/MPpAL strains were grown in L broth containing thymine (50 μg/ml) and carbenicillin (50 μg/ml) at 30°C. At an A640 of 0.5, a dilution series was made, and 100-μl aliquots from representative dilutions were plated on L agar containing thymine (50 μg/ml), carbenicillin (50 μg/ml), and 0 to 12 mM HU and incubated at 42°C for 2 days.
Determination of growth rate and cell morphology.
Transformants of KK535/pAL7, /pAL21, /MPpAL21, /MPpAL(D237E), and /MPpAL(E441D) were plated on L agar containing thymine (50 μg/ml), carbenicillin (50 μg/ml), and HU (3 mM) and incubated at 42°C. A colony from a freshly prepared plate was grown at 42°C in liquid medium containing thymine (50 μg/ml), carbenicillin (50 μg/ml), and HU (3 mM), and to start the experiments log-phase cultures were diluted in the same medium to an A640 of 0.03. The diluted cultures were grown at 42°C, and their absorbances were recorded every hour until lag phase was reached.
In another set of experiments transformants of KK535/MPpAL21, /MPpAL(W48Y), /MPpAL(Y122F), /MPpAL(D237E), /MPpAL(D237N), and /MPpAL(E441D) were grown logarithmically in nonselective medium at 30°C and then diluted 10 times in selective medium (3 mM HU) at 42°C. Growth rates were measured at 1-h intervals for approximately 8 h, and the cell morphology in the different cultures was observed in a Leica DM IRB light microscope.
RESULTS
Development and evaluation of an in vivo-complementation assay for RNR activity.
Our in vivo assay is based on complementation, where defective chromosomally encoded RNR is complemented by plasmid encoded RNR. The recipient strain KK535 contains conditional lethal mutations (nrdA-1 and nrdB-1) in both genes encoding class Ia RNR, which results in a temperature- and HU-sensitive phenotype (8, 9). It has been shown that plasmids expressing both RNR subunits can complement the defective chromosomally encoded RNR of KK535 when these transformants are grown under restrictive conditions (20). The plasmid construct MPpAL was derived from pAL21 that in turn is derived from pAL7 (11). MPpAL21 differs from pAL21 by two restriction cleavage sites on either side of the R2 gene, SacI before and BsrGI after the R2 gene. Cleavage sites SacI and BsrGI were used to subclone the specific point mutations from MPpTB2 derivatives into the MPpAL21 vector, whereas mutations from pTB1 derivatives could be transferred by using restriction sites already existing in the pAL7 derivative.
Using the KK535/MPpAL system we varied the HU concentration to identify suitable selection conditions to distinguish a plasmid encoding an active RNR from one encoding an inactive RNR component. MPpAL21 wild type and four mutant constructs—MPpAL(Y122F), MPpAL(C439A), MPpAL(D237E), and MPpAL(E441D)—were used. The mutant R2 Y122F protein is completely inactive due to the loss of the essential radical that normally is located on Tyr122 (11). Cys439 in R1 is proposed to take part in the reaction mechanism as a transient radical during catalysis (26), and the mutant R1 protein C439A is inactive in traditional enzymatic activity assays (1). Asp-237 in R2 is proposed to be a member of the RTP. The mutant D237E can still perform RNR catalysis, but at a considerably lower rate; in traditional assays its activity is about 7% of the wild-type activity (4). The E441D mutant in the R1 protein has about 8% of the wild-type activity; this residue is located at the R1 active site and participates as a general base in the reduction of substrate (17).
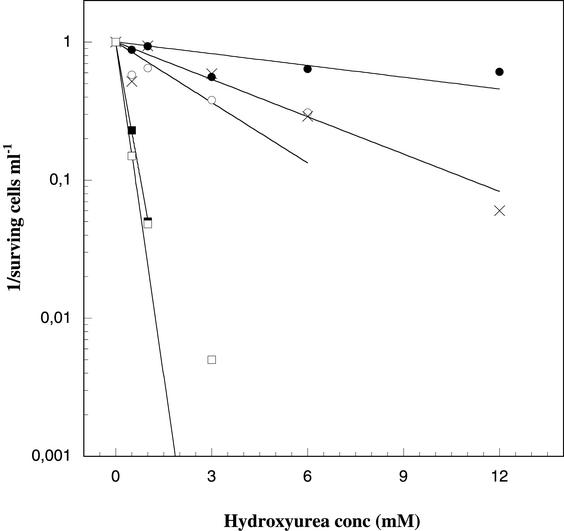
Figure 2 shows that the plating efficiencies at 3 mM HU and 42°C were less than 10−7 for cells containing the Y122F and C439A constructs, whereas cells containing MPpAL21(wt) plated to about 80%, and the two low-activity constructs D237E and E441D plated to about 40 to 60%. At higher concentrations of HU (6 and 12 mM) the plating efficiency for the wild-type construct was still better than 60% but was more notably reduced in the two low activity constructs. Further studies were restricted to 3 mM HU, where the chromosomally encoded RNR of the KK535 cells is clearly insufficient to sustain growth since no colonies could be detected in the Y122F and C439A constructs, and the plating efficiency of the low-activity constructs E441D and D237E is only moderately lower than that of the wild-type construct.
FIG. 2.
HU-dependent plating efficiency for different constructs are plotted as 1/surving cells ml−1 versus HU concentration. Shown are data for KK535/MPpAL21 (•), KK535/MPpAL(D237E) (○), KK535/MPpAL(E441D) (×), KK535/MPpAL(Y122F) (▪), and KK535/MPpAL(C439A) (□).
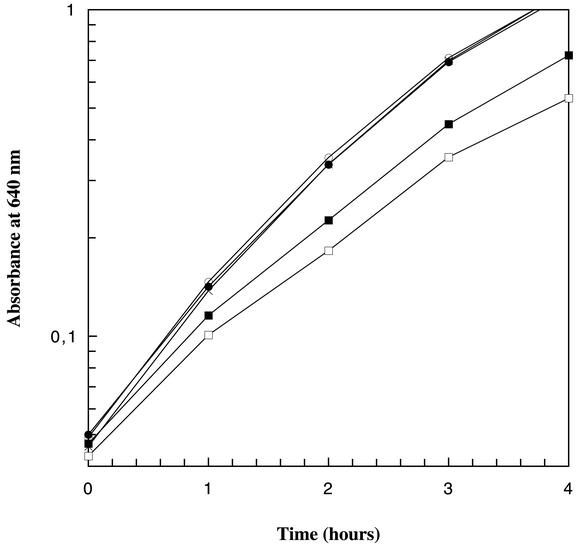
The growth rates of the low-activity constructs at the restrictive condition were compared to that of the wild-type construct (Fig. 3). Cells containing MPpAL21(wt) have a doubling time of 46 min, those containing MPpAL(D237E) have a doubling time of 59 min, and those containing MPpAL(E441D) have a doubling time of 55 min. When comparing the growth rates for the pAL7, pAL21, and MPpAL21 constructs we found the same in all three (Fig. 3). These data confirm that the mutant constructs can complement the defective chromosomally encoded RNR under restrictive growth conditions and also show that differences in growth rates of wild-type and mutant constructs can be distinguished.
FIG. 3.
Growth curves measured as absorbance at 640 nm detected for different constructs in KK535 cells grown under selective conditions (at 42°C with 3 mM HU). Shown are data for KK535/pAL7 (•), KK535/pAL21 (○), KK535/MPpAL21 (×), KK535/MPpAL(E441D) (▪), and KK535/MPpAL(D237E) (□).
The 60% plating efficiency of D237E and E441D constructs compared to wild type (42°C with 3 mM HU) was at first surprising. We had expected an equal plating efficiency for both wild-type and the viable mutant constructs together with a smaller colony size of the mutants due to their lower intrinsic growth rate. Colonies of the low-activity constructs were indeed too small to be observed on plates after 1 day of incubation, but they were readily detected on plates analyzed after 2 days. The same number of colonies was observed after 5 and 7 days of incubation as after 2 days. The cell morphologies of the mutant constructs in liquid restrictive medium were elongated compared to the wild-type construct, plausibly due to disturbed cell division caused by suboptimal deoxynucleotide synthesis. The reduced plating efficiency may thus be explained by their changed cell morphology.
Assessing the activity of mutations affecting the RTP in RNR with the in vivo complementation assay.
Eight biochemically well-characterized RTP mutations in the nrdA or nrdB genes were cloned into the MPpAL vector and transformed into KK535 (1 ng of DNA per 2 × 1011 cells). The number of CFU at 42°C in presence of 3 mM HU was determined as well as the plating efficiency of each transformed strain under restrictive conditions. In addition to the biochemically well-characterized constructs, we also included three RTP mutations in nrdB (W48Y, D237A, and Y356W) that have been less well characterized in vitro and the E441D nrdA mutation, which affects the active-site mechanism.
The CFU for the different MPpAL constructs were in agreement with previous in vitro activity measurements for purified protein. The mutants that have been characterized as inactive in vitro were unable to form colonies under restrictive conditions directly after transformation, whereas mutants with 7 to 8% activity in vitro (D237E and E441D) formed about 60 and 40% of colonies compared to the wild-type construct directly after transformation (Table 1).
TABLE 1.
CFU and plating efficiencies for MPpAL constructs with a disturbed reaction mechanism for ribonucleotide reductionj
| MPpAL construct | Mean no. of CFU ± SDa | Mean plating efficiency ± SDb | Enzyme activity (nmol/min · mg)k |
|---|---|---|---|
| Wild type | 836 ± 101 | 0.51 ± 0.12 | 1.0 |
| C439A | 0 | ≤10−7 | ND (≤0.05g) |
| E441D | 322 ± 76 | 0.37 ± 0.07 | 0.08h |
| Y730F | 0 | ≤10−7 | ND (≤0.015i) |
| Y731F | 0 | ≤10−7 | ND (≤0.015i) |
| W48Y | 0 | 2.3 × 10−6 ± 0.9 × 10−6 | NDc |
| D84A | 0 | ≤10−7 | ND (≤0.01d) |
| H118A | 0 | ≤10−7 | ND (≤0.02d) |
| Y122F | 0 | ≤10−7 | ND (≤0.01e) |
| D237A | 0 | ≤10−7 | NAf |
| D237N | 1 ± 1 | 1.7 × 10−5 ± 0.6 × 10−5 | ND (≤0.002f) |
| D237E | 423 ± 85 | 0.33 ± 0.09 | 0.07f |
| Y356W | 0 | ≤2 × 10−7 | NDc |
The number of CFU was determined as the amount of colonies plated under restrictive conditions directly after transformation of 1 ng of DNA into approximately 2 × 1011 electrocompetent KK535 cells. Values are based on results of three to nine experiments.
The plating efficiencies were determined as the fraction of colonies under restrictive conditions compared to the number of colonies under nonrestrictive conditions when different amounts of bacteria were plated. Values are based on results of two to six experiments.
Huque and Slaby, unpublished results.
Per reference 19.
Per reference 11.
Per reference 4.
Per reference 1.
Per reference 17.
Per reference 5.
CFU and plating efficiencies were determined in KK535 cells with 3 mM HU at 42°C.
Abbreviations: ND, not detected; NA, not available.
The plating efficiencies for KK535 cells containing the different MPpAL constructs are also shown in Table 1. The low-activity constructs D237E and E441D had plating efficiencies of about 0.4 compared to those grown under nonrestrictive conditions. Cells containing the Y122F, C439A, H118A, D84A, Y730F, Y731F, D237A, and Y356W constructs had plating efficiencies less than or equal to 10−7. Interestingly, we were able to detect a low but significant level of surviving cells in other mutant constructs, 2 × 10−5 for D237N and 2 × 10−6 for W48Y, suggesting that these mutations confer some residual enzyme activity. To establish that surviving colonies did not result from reversion of the inserted mutation, 10 colonies each of W48Y and D237N were sequenced. All of them were found to contain the desired mutation. We also compared the cell morphologies in liquid cultures after transfer from nonselective to selective conditions and found that W48Y and D237N cultures consisted of severely elongated cells, a phenotype that plausibly explains their impaired plating efficiencies. Our data show that the in vivo assay is sensitive and that very small differences in cell survival can be distinguished among these constructs.
DISCUSSION
A logical question concerns whether a purified mutant protein, assayed in vitro, really behaves as the in vivo enzyme does. It has been difficult to evaluate RNR mutants with very low activity in vitro, especially in the E. coli enzyme since the protein is expressed and purified from E. coli cells (23) and therefore always contains a low amount of chromosomally encoded wild-type RNR protein. The background activity in traditional E. coli RNR activity assays ranges from 0.5 to 3% of wild-type activity (7) depending on the degree of expression and also the enzyme activity of the wild-type control. In this work we present a sensitive and fast in vivo assay for RNR activity based on complementation of chromosomally encoded defective RNR components by plasmid encoded enzyme.
Platz et al. have shown that it is possible to complement the temperature- and HU-sensitive conditional lethal phenotype of KK535 cells with plasmid-encoded wild-type RNR (20). By using the MPpAL constructs, containing the two wild-type RNR genes, nrdA and nrdB expressed from the nrd promoter, we were able to reverse the conditional lethal phenotype of KK535 cells very efficiently. As a first step we determined the level of HU resistance where we would knock out the chromosomally encoded RNR and affect the plasmid-encoded enzyme as little as possible. At 3 mM HU we knocked out the chromosomally encoded enzyme completely (cell survival ≤ 10−7), as seen from the data on the negative controls Y122F, which lacks the essential R2 tyrosyl radical (11), and C439A, which affects a cysteine residue in R1 active site that has been suggested to be a key residue in the reaction mechanism (1, 13). At the same concentration the wild type and the two low-activity mutants D237E in the R2 protein (4) and E441D in the R1 protein (17) still plated to a reasonably high efficiency. At higher HU concentrations the differences in plating efficiencies between the wild type and the low-activity mutants became more pronounced. It has been shown with traditional enzyme assays that the two low-activity mutants, D237E and E441D, have a lower kcat than the wild-type enzyme in vitro and plausibly also produce fewer deoxyribonucleotides for DNA synthesis in vivo. Our observations that the low-activity mutants under selective conditions (42°C and 3 mM HU) have lower growth rates, smaller colony sizes, slightly elongated cell morphologies and lower plating efficiencies compared to the wild-type plasmid construct support this assumption.
We also determined the CFU of transformation mixtures and the plating efficiency of transformed strains for a collection of mutants that affect the reaction mechanism in different ways (Table 1). The D237E and E441D constructs (the only mutants in this collection that show activity in the traditional activity assay) were the only ones that formed colonies when transformation mixtures were plated directly under selective conditions. This makes the system perfect for a direct screening of low activity mutants such as in e.g., random mutagenesis experiments.
The plating efficiencies of eight mutants (D84A, Y122F, H118A, D237A, Y356W, Y730F, Y731F, and C439A) affecting side chains involved in the RTP of RNR (Fig. 1) were ≤10−7, suggesting that these mutants are completely inactive. In a related in vitro study the purified E. coli Y356W enzyme had no activity in the NADPH assay (Y. Huque and A. Slaby, unpublished results). Our Y356W results in the E. coli system differ from the results obtained with the corresponding purified Y370W protein from the mouse enzyme. Mouse Trp370 had 1.7% activity compared to the wild-type protein, whereas the mutant protein Phe370 was completely inactive (21). The observed activity in mouse Y370W and the loss of activity in the Y370F mutant mouse enzyme were explained by the capacity of a tryptophan residue to form hydrogen bonds, which is lacking in a phenylalanine residue (21). The difference observed in the mouse Y370W and the E. coli Y356W enzymes might be a result of subtle structural differences between the two enzymes.
In theory chromosomally encoded RNR proteins can interfere with the plasmid-encoded proteins and therefore affect the result. The activity of a plasmid-encoded subunit might be masked when in complex with an inactive chromosomally encoded subunit. However the RNR encoded by the multicopy plasmid is in excess of the chromosomally encoded RNR, which therefore should have little consequence on the enzymatic activity. The close agreement between the data reported here and earlier results using traditional activity assays demonstrates the reliability of our new method.
Interestingly, we observed a low plating efficiency in the D237N and W48Y constructs, although they were below the detection limit in the traditional activity assays. The protein containing the D237N mutation has previously been characterized in vitro (4) but failed to crystallize under standard R2 crystallization conditions. The D237N protein can form a tyrosyl radical in vitro that decays over time (half-life, ∼110 min) but has no detectable electron paramagnetic resonance signal in vivo (M. Ekberg and M. Sahlin, unpublished result). Since D237N at least in vitro can harbor a tyrosyl radical, and since an asparagine theoretically may be engaged in hydrogen bonds, the low but significant plating efficiency of the D237N construct suggests that the asparagine in the 237 position can promote some RTP, albeit at a very low efficiency. The W48Y mutation in E. coli R2 and the corresponding W103Y mutation in mouse R2 have no detectable in vitro activities, but both mutant proteins stabilize a radical at Tyr-122 (Huque and Slaby, unpublished result; (22). Our results suggest that the Tyr-48 construct may allow a very inefficient RTP.
In conclusion, we have shown that the in vivo complementation assay is a fast method to determine apparent in vivo activity of mutant RNR components. With its low detection limit it is considerably more sensitive than the traditional RNR activity assays. Based on the results with mutations in the RTP, we suggest that this method would be a suitable selection system for second-site revertants of negative RNR component in random mutagenesis experiments, e.g., in vitro evolution approaches.
Acknowledgments
M.E. and P.B. contributed equally to this work.
This study was supported by grants from the Swedish Cancer Foundation and the Swedish Research Council.
REFERENCES
- 1.Åberg, A., S. Hahne, M. Karlsson, A. Larsson, M. Ormö, A. Åhgren, and B.-M. Sjöberg. 1989. Evidence for two different classes of redox-active cysteines in ribonucleotide reductase of Escherichia coli. J. Biol. Chem. 264:12249-12252. [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 3.Climent, I., B.-M. Sjöberg, and C. Y. Huang. 1992. Site-directed mutagenesis and deletion of the carboxyl terminus of Escherichia coli ribonucleotide reductase protein R2. Effects on catalytic activity and subunit interaction. Biochemistry 31:4801-4807. [DOI] [PubMed] [Google Scholar]
- 4.Ekberg, M., S. Pötsch, E. Sandin, M. Thunnissen, P. Nordlund, M. Sahlin, and B.-M. Sjöberg. 1998. Preserved catalytic activity in an engineered ribonucleotide reductase R2 protein with a nonphysiological radical transfer pathway—the importance of hydrogen bond connections between the participating residues. J. Biol. Chem. 273:21003-21008. [DOI] [PubMed] [Google Scholar]
- 5.Ekberg, M., M. Sahlin, M. Eriksson, and B.-M. Sjöberg. 1996. Two conserved tyrosine residues in protein R1 participate in an intermolecular electron transfer in ribonucleotide reductase. J. Biol. Chem. 271:20655-20659. [DOI] [PubMed] [Google Scholar]
- 6.Eklund, H., U. Uhlin, M. Färnegårdh, D. T. Logan, and P. Nordlund. 2001. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77:177-268. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson, S., B.-M. Sjöberg, and S. Hahne. 1977. Ribonucleoside diphosphate reductase from Escherichia coli. An immunological assay and a novel purification from an overproducing strain lysogenic for phage λdnrd. J. Biol. Chem. 252:6132-6138. [PubMed] [Google Scholar]
- 8.Fuchs, J. A., and H. O. Karlström. 1973. A mutant of Escherichia coli defective in ribonucleosidediphosphate reductase. 2. Characterization of the enzymatic defect. Eur. J. Biochem. 32:457-462. [PubMed] [Google Scholar]
- 9.Fuchs, J. A., H. O. Karlström, H. R. Warner, and P. Reichard. 1972. Defective gene product in dnaF mutant of Escherichia coli. Nat. New Biol. 238:69-71. [DOI] [PubMed] [Google Scholar]
- 10.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 11.Larsson, A., and B.-M. Sjöberg. 1986. Identification of the stable free radical tyrosine residue in ribonucleotide reductase. EMBO J. 5:2037-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence, C. C., M. Bennati, H. V. Obias, G. Bar, R. G. Griffin, and J. Stubbe. 1999. High-field EPR detection of a disulfide radical anion in the reduction of cytidine 5′-diphosphate by the E441Q R1 mutant of Escherichia coli ribonucleotide reductase. Proc. Natl. Acad. Sci. USA 96:8979-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao, S. S., G. X. Yu, D. Chalfoun, and J. Stubbe. 1992. Characterization of C439SR1, a mutant of Escherichia coli ribonucleotide diphosphate reductase: Evidence that C439 is a residue essential for nucleotide reduction and C439SR1 is a protein possessing novel thioredoxin-like activity. Biochemistry 31:9752-9759. [DOI] [PubMed] [Google Scholar]
- 14.Monje-Casas, F., J. Jurado, M. J. Prieto-Alamo, A. Holmgren, and C. Pueyo. 2001. Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. J. Biol. Chem. 276:18031-18037. [DOI] [PubMed] [Google Scholar]
- 15.Nordlund, P., and H. Eklund. 1993. Structure and function of the Escherichia coli ribonucleotide reductase protein R2. J. Mol. Biol. 232:123-164. [DOI] [PubMed] [Google Scholar]
- 16.Nordlund, P., B.-M. Sjöberg, and H. Eklund. 1990. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature 345:593-598. [DOI] [PubMed] [Google Scholar]
- 17.Persson, A. L., M. Eriksson, B. Katterle, S. Pötsch, M. Sahlin, and B.-M. Sjöberg. 1997. A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu(441) in Escherichia coli ribonucleotide reductase. J. Biol. Chem. 272:31533-31541. [DOI] [PubMed] [Google Scholar]
- 18.Persson, A. L., M. Sahlin, and B.-M. Sjöberg. 1998. Cysteinyl and substrate radical formation in active site mutant E441Q of Escherichia coli class I ribonucleotide reductase. J. Biol. Chem. 273:31016-31020. [DOI] [PubMed] [Google Scholar]
- 19.Persson, B. O., M. Karlsson, I. Climent, J. S. Ling, J. Sanders Loehr, M. Sahlin, and B.-M. Sjöberg. 1996. Iron ligand mutants in protein R2 of Escherichia coli ribonucleotide reductase—retention of diiron site, tyrosyl radical and enzymatic activity in mutant proteins lacking an iron-binding side chain. J. Biol. Inorg. Chem. 1:247-256. [Google Scholar]
- 20.Platz, A., and B.-M. Sjöberg. 1980. Construction and characterization of hybrid plasmids containing the Escherichia coli nrd region. J. Bacteriol. 143:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rova, U., A. Adrait, S. Pötsch, A. Gräslund, and L. Thelander. 1999. Evidence by mutagenesis that Tyr(370) of the mouse ribonucleotide reductase R2 protein is the connecting link in the intersubunit radical transfer pathway. J. Biol. Chem. 274:23746-23751. [DOI] [PubMed] [Google Scholar]
- 22.Rova, U., K. Goodtzova, R. Ingemarson, G. Behravan, A. Gräslund, and L. Thelander. 1995. Evidence by site-directed mutagenesis supports long-range electron transfer in mouse ribonucleotide reductase. Biochemistry 34:4267-4275. [DOI] [PubMed] [Google Scholar]
- 23.Sjöberg, B.-M., S. Hahne, M. Karlsson, H. Jörnvall, M. Göransson, and B. E. Uhlin. 1986. Overproduction and purification of the B2 subunit of ribonucleotide reductase from Escherichia coli. J. Biol. Chem. 261:5658-5662. [PubMed] [Google Scholar]
- 24.Sjöberg, B.-M., and M. Sahlin. 2002. Thiols in redox mechanism of ribonucleotide reductase. Methods Enzymol. 348:1-21. [DOI] [PubMed] [Google Scholar]
- 25.Stern, M. J., G. F. Ames, N. H. Smith, E. C. Robinson, and C. F. Higgins. 1984. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37:1015-1026. [DOI] [PubMed] [Google Scholar]
- 26.Stubbe, J. 1990. Ribonucleotide reductases: amazing and confusing. J. Biol. Chem. 265:5329-5332. [PubMed] [Google Scholar]
- 27.Thelander, L., B.-M. Sjöberg, and S. Eriksson. 1978. Ribonucleoside diphosphate reductase (Escherichia coli). Methods Enzymol. 51:227-237. [DOI] [PubMed] [Google Scholar]
- 28.Uhlin, U., and H. Eklund. 1994. Structure of ribonucleotide reductase protein R1. Nature 370:533-539. [DOI] [PubMed] [Google Scholar]