Abstract
The group A Streptococcus (GAS) sof gene encodes the serum opacity factor protein, which is capable of opacifying mammalian sera and binding at least two host proteins, fibronectin and fibrinogen. The sof gene exists in approximately 50% of clinical isolates, and there is a classical association of so-called nephritogenic strains with the opacity factor-positive phenotype. In both a type emm49 strain and a type emm12 strain, the sequences upstream of the 5′ end of sof and downstream of the putative terminator were determined to be nearly identical to a region in the M type 1 genome approximately 10 kb upstream of the emm1 gene. This close genetic linkage is likely reflected in the strict correlation of opacity factor phenotype with specific emm genotypes. A new fibronectin-binding protein gene, sfbX, was discovered immediately downstream of sof in emm12 and emm49 strains and in several other sof-positive strains. The sof and sfbX genes were found to be expressed on the same transcription unit, which was correlated with the putative promoter and rho-independant terminator sequences that flank these two genes. The sfbX genes from different emm types are predicted to encode ∼650-residue surface-bound proteins sharing 89 to 92% sequence identity. SfbX residues approximately 1 to 480 are not highly similar to those of other known proteins, with the closest match being the Staphylococcus aureus coagulase protein. The remaining portions of these proteins (residues 481 to 650) contain four putative fibronectin-binding repeats highly similar to those of other streptococcal fibronectin-binding proteins and a potential LP(X)SG cell wall anchor motif. Targeted in-frame allelic-exchange mutagenesis, complementation, and heterologous-expression studies found that serum opacification is encoded by sof alone and that sfbX encodes a fibronectin-binding function. A recombinant SfbX protein was found to bind immobilized fibronectin and to partially inhibit GAS adherence to fibronectin. The sfbX gene was found to be present only in sof-positive strains, and together these genes could influence the spectrum of tissues colonized by sof-positive GAS.
Group A Streptococcus (GAS) is a major human pathogen associated with a wide spectrum of mucosal and invasive infections. A characteristic of 40 to 50% of GAS strains is the ability to opacify mammalian serum due to the presence of serum opacity factor (Sof, OF), encoded by the sof gene (2, 25). GAS Sof is an ∼110-kDa extracellular and cell surface-bound protein with lipoproteinase and host protein-binding activities that possesses a C-terminal LP(X)SG cell wall anchor motif (10, 22, 38). Serum opacification has been reported to be the result of Sof-mediated cleavage of apoprotein A1, causing aggregation and insolubility of high-density lipoprotein (42). The Sof reaction can be inhibited in a strain-specific manner by postimmune antisera against Sof-positive GAS strains (anti-OF) and is generally (but not always) predictive of M protein serotype or emm type (2, 31). Sof lipoproteinase activity and anti-OF specificity both map to a hypervariable “enzymatic domain” of ∼700 amino acids located toward the N terminus of the protein (24), while fibronectin- and fibrinogen-binding activity maps to a region of highly conserved tandem repeats located just before the C-terminal membrane anchor (9, 10, 22, 38). Sof appears to function as a virulence factor, since insertional activation of the sof gene in a serotype 2 GAS isolate led to notable reductions in mortality in a mouse intraperitoneal infection model (10).
Expression of Sof is under positive control of the GAS multiple-gene regulator Mga (formerly VirR) (33, 36), but restriction mapping indicated that the sof gene was situated at least 15 kb apart from emm (encoding M protein) and scpA (encoding the C5a peptidase) (38), which are coregulated by the closely situated mga gene product. Very recently, the complete genome sequences of three strains of GAS have been elucidated (3, 11, 47), accomplishments that promise to accelerate the pace of research discoveries in the field. However, the strains SF370, MGAS315, and MGAS8232, used for genome sequencing, like others belonging to the M1, M3, and M18 serotypes, do not opacify serum and do not contain the sof gene. In the present study, we report a series of experiments that allowed us to map the location of sof on the GAS genome. Concurrently, we discovered that sof is invariably transcriptionally linked to a second and novel open reading frame (ORF) whose product has sequence features of a bifunctional GAS fibronectin-binding surface protein. Precise, in-frame allelic-exchange mutagenesis and heterologous expression are used to analyze the two-gene sof operon.
MATERIALS AND METHODS
Bacterial strains and culture and transformation conditions.
Bacterial strains used in this study are listed in Table 1. GAS was grown in Todd-Hewitt broth (THB) or on Todd-Hewitt agar plates; for antibiotic selection 2 μg of erythromycin (EM) or 1 μg of chloramphenicol (CM) per ml was added to the media. Escherichia coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates; antibiotic selection utilized 100 μg of ampicillin per ml, 500 μg of EM per ml, or 5 μg of CM per ml. Lactococcus lactis was grown in M17 broth (Difco) supplemented with 1% glucose (GM17) or on GM17 agar plates with selection by EM at 5 μg/ml. GAS strains were rendered transformable by electroporation through growth in THB plus 0.3% glycine as described for Streptococcus agalactiae (12). Similarly, L. lactis was made transformable by growth in GM17 plus 2.5% glycine (17). After electroporation (Eppendorf 2510; 1,500 V), cells were incubated in THB plus 0.25 M sucrose (GAS) or GM17 media-20 mM MgCl2-2 mM CaCl2 (L. lactis) for 1 to 2 h prior to antibiotic selection on agar media.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| GAS clinical isolates | ||
| NZ131 | T14 OF+, emm49 glomerulonephritis isolate | 45 |
| 5448 | T1 OF−, emm1.0 necrotizing fasciitis-STSSa isolate | 20 |
| SS635 | T12 OF−, emm12 Lancefield M type 12 reference strain, sof12 sfbX+ | 27 |
| 2108-98 | T12 OF−, emm12 blood isolate, sof12 sfbX+ | This study |
| 3833-98 | T12 OF−, emm12 blood isolate, sof12 sfbX+ | This study |
| 27-01 | T25 OF+, emm75 blood isolate, sof75 sfbX+ | This study |
| 140-01 | T28, OF+, emm87.3 blood isolate, sof87 sfbX+ | This study |
| 149-01 | T8/25/Imp19 OF+, emm92 isolate, sof92 sfbX+ | This study |
| 142-01 | T-NT OF+, emml 14 isolate from joint | This study |
| GAS mutants | ||
| NZ131:sofΔcat | NZ131 with in-frame allelic replacement of sof by cat | This study |
| NZ131:sfbxΔcat | NZ131 with in-frame allelic replacement of sfbX by cat | This study |
| NZ131:sof+sfbxΔcat | NZ131 with in-frame allelic replacement of sof-sfbX by cat | This study |
| E. coli | ||
| DH5α | endA1 hsdR17(rK−mK+) sup E44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-arg F)U169 [φ80dlac Δ(lacZ)M15] | 50 |
| MC1061 | F−araD139 Δ(araABC-leu)7696 Δ(lac)X74galU galK hsdR2 (rK−mK+) mcrB1 rpsL (Strr) | 49 |
| Top 10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 Δ(lac) X74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | 14 |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| L. lactis | ||
| NZ9000 | MG1363 (lacking nisin operon); pepN::nisRK | 26 |
| Plasmids | ||
| Chromosomal walking | ||
| pBluescript II | ColE1 ori Amp lacZα | Stratagene |
| Allelic-exchange mutagenesis | ||
| pCR2.1-TOPO | ColE ori Ampr KnrlacZα; T-A cloning vector | Invitrogen |
| pHY304 | Temperature-sensitive pVE6007Δ derivative, EmrlacZα/MCSb of pBluescript | 37 |
| pACYC184 | rep(p15A) Cmr Tetr | 40 |
| pSof-TV | pCR2.1 with 3,786-bp NZ131 DNA fragment containing sof | This study |
| pSfbX-TV | pCR2.1 with 2,589-bp NZ131 DNA fragment containing sfbX | This study |
| pSof/SfbX-TV | pCR2.1 with 5,947-bp NZ131 DNA fragment containing sof-sfbX | This study |
| pSofΔcat-TV | In vivo PCR recombination derivative of pSof-TV with cat replacing sof gene precisely in frame, Ampr Cmr | This study |
| pSfbXΔcat-TV | In vivo PCR recombination derivative of pSfbX-TV with cat replacing sfbX gene precisely in frame, Ampr Cmr | This study |
| pSof/SfbXΔcat-TV | In vivo PCR recombination derivative of pSof/SfbX-TV with cat replacing sof-sfbX genes precisely in frame, Ampr Cmr | This study |
| pSofΔcat-KO | pHY304 containing Δsof cat allele and flanking DNA, Emr Cmr | This study |
| pSfbXΔcat-KO | pHY304 containing ΔsfbX cat allele and flanking DNA, Emr Cmr | This study |
| pSof/SfbXΔcat-KO | pHY304 containing Δsof-sfbX cat allele and flanking DNA, Emr Cmr | This study |
| Complementation and heterologous expression | ||
| pDC123 | E. coli-streptococcus shuttle vector, JS-3 replicon, phoS Cmr (cat) | 8 |
| pDCerm | pDCerm derivative with Ermr (erm of Tn916ΔE) replacing cat | This study |
| pAJI-sof | pDCerm plus NZ131 sof gene behind vector and native promoters | This study |
| pAJI-sfbX | pDCerm plus NZ131 sfbX gene behind vector promoters | This study |
| Expression of recombinant SfbX | ||
| pCRT7/CT-TOPO | ColE ori, Ampr Zeor, T7 promoter, T-A cloning, V5 epitope, His6 | Invitrogen |
| pStbX-V5-His | Complete sfbX ORF cloned in pCRT7/CT-TOPO for C-terminal tag | This study |
STSS, staphylococcal toxic shock syndrome.
MCS, multiple cloning site.
Sequence analysis and genome mapping.
Single-primer PCR, with three successive cycling parameters, was used as described previously (21) to amplify the genomic sequence adjacent to the sof genes from type emm49 and type emm12 GAS strains. For single-primer PCR, 20 cycles employing a 30-s denaturation at 94°C, 50°C annealing (30 s), and 72°C elongation (3 min) were followed by 30 cycles of 94°C denaturation (30 s), 30°C annealing (30 s), and 72°C elongation (2 min). This second set of parameters was immediately followed by 30 cycles of 94°C (30 s), 50°C (30 s), and 72°C (2 min), before a final 7-min extension at 72°C. Primer R2 (5′-TTTCTAAGTTTATACTTACAATTTGTCA-3′) was used with a primer R-generated template to sequence the region immediately upstream of sof. Primer F2 was used with a primer F-generated template to sequence the region immediately downstream of sof. Subsequent rounds of single primer PCR using new primers from the generated sequence allowed further extension in the 5′ and 3′ directions. All primary sequencing data generated from the single primer template were reconfirmed by using a template amplified conventionally from genomic DNA with the use of two primers. For the PCRs described below for amplifying ∼9-kb regions, 10 cycles of denaturation (94°C, 15 s), annealing (55°C, 30 s), and elongation (72°C, 8.75 min) were followed by 20 cycles with the same parameters except for the addition of 20-s increments to each successive elongation. To amplify the ∼9-kb region between the sfbX and mga genes in a type emm12 strain, primers 12 mga2 (accession no. AE006624, U02342, AB016537, and M58461 are the 5′ mga sequences from emm1, emm12, emm23, and emm6, respectively) (5′-TACATACATTACCTTGTATACCCTTCT-3′) and sfbF4 (5′-GTGCCTGAGGCTGGTGGTCAGAGTGG-3′) were used. To amplify the ∼9-kb region between sfbX and mga in a type emm49 strain, primers chosen were 49 mga5R (derived from the GenBank accession no. U31919 sequence upstream of mga49) (5′-GGGTTTTATCAGCTGATTTTTCTGG-3′) and sfbF5 (5′-GACACTTGCTAGCATTTCGGTGTAAAA-3′). Primers sfbF1 (5′-GCAGTGATTTCTAGGCTTAGCAAGCATA-3′) and sfbR1 (5′-GTTTTGTCGGTGTTGCGACGTTTTT-3′) were used to amplify the sfbX region, predicted to encode the mature SfbX proteins from various GAS strains, including type emm12, emm49, emm75, emm87, emm92, and emm114 strains.
RT-PCR analysis.
Cells of strain NZ131 were disrupted with the Bio 101 Fast Prep for 40 s at speed 6, and total RNA was isolated with the FastRNA isolation kit as described by the manufacturer. The RNA was treated with DNase I, extracted with phenol-chloroform, and precipitated with ethanol prior to resuspension in diethyl pyrocarbonate-treated water. The Epicentre MasterAmp reverse transcription-PCR (RT-PCR) kit was used as described by the manufacturers with 0.1 μg of the RNA preparation and primers sofF1-3p (5′-CTGGAGACAAACGTGAAGCATCCTTTAC-3′) and either sfb5p-R1 (5′-GGGAACGTTCTCCTAATTTTGGACT-3′) or sfb5p-R2 (5′-GGGTTGCGTCATGCTGATGGTTATG-3′). Absence of contaminating DNA was confirmed by the lack of PCR amplification of RNA preparations prior to reverse transcription.
Targeted mutagenesis of sof, sfbX, and sof-sfbX.
PCR was used to amplify NZ131 chromosomal DNA fragments containing sof, sfbX, and sof-sfbX with several hundred base pairs of upstream and downstream sequence in each case. Specific primers used were sofF (5′-GGAGAAAAAGATTAAAAATCAAGGTC-3′; 360 bp upstream of the sof start ATG), sofR (5′-GGACTTGTTGCTTTTAATGC-3′; 340 bp downstream of the last sof codon), sfbXF (5′-TGGAGACAAACGTGAAGCAT-3′; 280 bp upstream of the sfbX start ATG), and sfbXR (5′-TCTTCAGATAGGACACCGCA-3′; 358 bp downstream of the last sfbX codon). The PCR products were T-A cloned in pCR2.1 (Invitrogen). Vectors pSof-TV and pSfbX-TV served as templates for inverse PCRs using (i) a reverse primer immediately upstream of the start codon of the cloned ORF(s) and (ii) a forward primer immediately after the stop codon of the cloned ORF(s); these primers were designed with 25-bp 5′ extensions corresponding to the start and end of the chloramphenicol acetyltransferase gene (cat), respectively. The resultant linearized PCR products, alternatively containing an in-frame deletion of sof, and sof plus sfbX, were used to transform E. coli Top 10 together with an ∼650-bp PCR amplicon of the complete cat gene from pACYC184. In vivo recombination events were identified by screening for E. coli tarnsformants exhibiting Ampr and Cmr; recombinant plasmids were verified by PCR and restriction analysis to contain an in-frame replacement of the target gene(s) with cat. The mutated sequences in which cat replaced the sof (sofΔcat), sfbX (sfbXΔcat), and sof-sfbX (sof+sfbXΔcat) genes and flanking DNA were subcloned as BamHI/XbaI fragments to the temperature-sensitive Emr vector pHY304 to produce knockout vectors pSofΔcat-KO, pSfbXΔcat-KO, and pSof/SfbXΔcat-KO, respectively. NZ131 was transformed with each knockout plasmid, and single recombination events were identified at 37°C under EM selection. Selection was relaxed by serial passage at 30°C without antibiotics, and double-crossover events were identified by detecting NZ131 mutants exhibiting Cmr but Ems. Allelic exchanges of sof, sfbX, and sof-sfbX in the NZ131 chromosome were confirmed by (i) PCR using cat primers with upstream and downstream primers and (ii) absence of amplification of the wild-type gene(s).
Complementation studies and heterologous expression of sof and sfbX.
An Emr derivative of streptococcus-E. coli shuttle vector pDC123 (8) was constructed as follows. Primers designed with 5′ extensions containing HaeII recognition sites were used to amplify the Emr marker and its constitutive promoter from transposon Tn916ΔE (41). The amplicon was cloned into the HaeII site of pDC123 upstream of the cat gene but behind the vector's constitutive tet and cat promoters. The intermediate vector pDC123erm was identified by Cmr and Emr and confirmed by restriction and PCR analysis. The cat gene was then deleted from pDC123erm by inverse PCR and blunt end religation to give pDCerm. This expression vector confers Emr, retains blue-white selection based on multiple cloning site interruption of the enterococcal phoS gene, and places cloned genes behind the tet, cat, and erm promoters on the plasmid backbone. The NZ131 sof and sfbX genes were cut from pSof-TV and pSfbX-TV with KpnI and BamHI plus XbaI, respectively, and subcloned to expression vector pDCerm prepared with the compatible restriction enzymes. The resultant constructs, pAJ1sof and pAJ1sfbX, placed the respective genes behind the constitutive promoters on the plasmid backbone. These two vectors were used to transform electrocompetent emm1 GAS strain 5448 and L. lactis strain NZ9000. Transformants were identified by Emr and confirmed by restriction enzyme and PCR analysis of purified plasmid preps. Vectors pAJ1sof and pAJ1sfbX were also used for complementation of the allelic-exchange mutants NZ131:sofΔcat and NZ131:sfbXΔcat, respectively; pDCerm was likewise introduced into the parent strain and allelic-exchange mutants as a vector-only control.
Serum opacification assays.
A modified version of the microplate method of Johnson and Kaplan was used (19). Bacteria were grown to stationary phase overnight and then pelleted by centrifugation. One hundred microliters of supernatant was removed and added to 1 ml of horse serum (Gibco-BRL) in individual wells of a 24-well cell culture plate. The plates were incubated overnight at 37oC in 5% CO2. Two milliliters of normal saline was then added to each well, and opacification was observed.
Fibronectin-binding ELISA for GAS.
Fibronectin (Sigma; F0895) was diluted in phosphate-buffered saline (PBS) to 20 μg/ml, and 100 μl was used to coat the bottoms of 96-well tissue culture plates (Costar) for 2 h at 37°C. The wells were then blocked with Dulbecco's PBS-5% bovine serum albumin (BSA) for 2 h at room temperature and subsequently washed three times with PBS-1% BSA. GAS was grown to an optical density at 600 nm (OD600) of 0.4 (∼108 CFU/ml) and diluted in PBS, and 106 CFU in 100 μl was inoculated into each well. PBS alone was used in control wells. The plate was centrifuged at 800 × g for 10 min then incubated at 37°C for 30 min. After three washes with PBS-1% BSA, 85 μl of diluted M49 (1:5,000) or M1 (1:500) immune rabbit typing serum (provided by R. Facklam, Centers for Disease Control and Prevention [CDC], Atlanta, Ga) was added to each well, and the plates were incubated at room temperature for 1 h. The plates were again washed with PBS-1% BSA, followed by addition of 85 μl of the secondary antibody (anti-rabbit immunoglobulin [Ig]-horseradish peroxidase [HRP]; NA934; Amersham Life Sciences; diluted 1:7,000) to each well and incubation at room temperature for 30 min. After another wash with PBS-1% BSA, 200 μl of developing solution (tetramethylbenzidine diluted in 0.1 M sodium acetate-hydrogen peroxide) was added, and 50 μl of 1.2 M sulfuric acid was used to stop the reaction after 90 min. The OD450 of each well was measured in an enzyme-linked immunosorbent assay (ELISA) plate reader. Fibronectin-binding measurements were performed in triplicate and repeated three (emm49 GAS NZ131 and mutants) or two (emm1 GAS 5448 and transformants) times. Data were compared to standard curves constructed from serial dilutions of each of the wild-type GAS strains. For fibronectin-binding ELISAs involving the pAJ1sof- and pAJ1sfbX-complemented mutants, all bacterial cultures were grown under EM selection. Wild-type and mutant strains transformed with the vector-only control pDCerm were used for comparison. The complemented-mutant studies were performed in triplicate and repeated four times. For inhibition experiments with the recombinant SfbX protein (see below), the ELISA was performed as described above with an inoculum of 6 × 105 wild-type GAS NZ131 cells and preincubation of the fibronectin-coated wells with serial twofold dilutions of the recombinant SfbX protein ranging from 728 to 1.4 ng.
Fibronectin-binding assay for L. lactis.
Fibronectin-coated 96-well plates were blocked with PBS-5% BSA for 2 h and subsequently washed with PBS-Tween (0.05%). L. lactis strain NZ9000 and the same strain harboring either plasmid pAJ1sof or pAJ1sfbX were grown to OD600 of 0.4 and diluted in PBS to allow 106 colonies/100 μl to be inoculated into each well. PBS alone was used in control wells. The plate was spun at 2,000 rpm for 10 min and then allowed to incubate at 37°C for 30 min. After a wash with PBS-Tween, 50 μl of THB, 50 μl of PBS, and 20 μl of MTS-PES reagent (Promega; Celltiter 90AQ One solution) were added to each well and the plate was placed in a 37°C, 5% CO2 incubator for 4 h. The OD490 was measured in a plate reader, and data were compared to a standard curve constructed from serial dilutions of L. lactis NZ9000. Assays were performed in triplicate and repeated three times.
Expression and purification of recombinant SfbX.
To generate a C-terminal His6-tagged version of the full-length SfbX protein, a PCR amplicon of the entire sfbX ORF (lacking the stop codon) from the NZ131 chromosome was made with primers sfbX-start (5′-ATGACTAAAAAATTTATTAAA-3′) and sfbX-end (5′-GTTTTTGTCGGTGTTGCGACGTTTTT-3′) and T-A cloned into pCR T7/CT-TOPO (Invitrogen). This construct was confirmed by restriction and PCR analysis and named pSfbX-V5-His, as the coding sequences for a C-terminal V5 epitope and polyhistidine were added to the SfbX coding region. Vector pSfbX-V5-His was then transformed into E. coli BL21(DE3) possessing an inducible T7 RNA polymerase. For induction, BL21(DE3)(pSfbX-V5-His) was grown in 500 ml of LB broth plus ampicillin at 37°C to an OD600 of 0.5, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and the mixture was incubated an additional 2.5 h. The bacterial culture was pelleted at 4,200 × g for 20 min, resuspended in 15 ml of lysis buffer (20 mM NaPO4 [pH 7.8], 5 mg of lysozyme/ml, 100 mg of RNase A/ml), vortexed, and incubated at room temperature for 45 min. The lysis mixture was then placed on dry ice alternating with a 37°C water bath (three times for 15 min each), sonicated at midpower three times for 15 s each, and then centrifuged at 14,500 × g for 10 min. The protein-containing supernatant was decanted, filtered (0.2 μm-pore-size filter; Whatman), and passed over a nickel affinity column, and the His-tagged recombinant SfbX was eluted with increasing concentrations (50, 200, 350, and 500 mM) of imidazole buffer. Eluted fractions were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and electroblotted to a nitrocellulose membrane, and recombinant SfbX was detected by immunoblotting with a primary mouse anti-V5 antibody (Invitrogen; 1:5,000) and a secondary anti-mouse IgG-HRP antibody (Sigma; A4416; 1:3,000) and with the ECL chemiluminescence detection system (Amersham Biosciences) used in accordance with the manufacturer's instructions.
Fibronectin-binding ELISA for recombinant SfbX.
Fibronectin-coated 96-well plates were blocked and washed as described above. Serial twofold dilutions of recombinant SfbX protein were made so that amounts of protein ranging from 679 ng to 0.66 ng were added to individual wells in a volume of 100 μl. PBS alone and a control protein (the GAS cysteine protease SpeB with identical C-terminal V5 and His tags) served as controls. The plate was centrifuged at 800 × g for 10 min then incubated at 37°C for 1 h. After three washes with PBS-0.05% Tween, 100 μl of diluted anti-V5 mouse IgG (1:5,000) was added to each well at room temperature for 1 h. Subsequent steps including washes, secondary-antibody detection (anti-mouse IgG-HRP; 1:3,000), tetramethylbenzidine developing reactions, and OD450 measurement were performed as described above. Assays of control wells for each recombinant SfbX protein concentration were performed with omission of the primary anti-V5 antibody. Experiments were repeated three times.
Nucleotide sequence accession numbers.
The new sequences reported in this work are listed in the GenBank under accession no. AY120870 (1,326 bases, including the 1,050-base region upstream of sof49 and 5′ end of sof49), AF335322 (2,606 bases, including the sof49 3′ end, sfbX49, terminator, and short downstream ORF), AF387738 (6,386 bases, including the sof12 upstream sequence, defective sof12 gene, and the complete sfbX12 gene), AY120871 (1,890 bases; encodes SfbX75 plus partial signal sequence), AY120873 (1,881 bases; encodes the complete SfbX92 −10 signal sequence residues), AY120874 (1,881 bases; encodes the SfbX114 −10 signal sequence residues), and AY120872 (1,881 bases; encodes the SfbX87 −10 signal sequence residues).
RESULTS
Chromosomal location of the sof gene and discovery of sfbX.
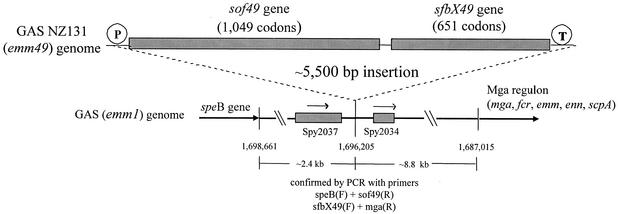
Using the single-primer chromosome walking approach, 1,050- to 1,185-bp regions upstream of sof12 and sof49 were sequenced. An emm12 strain was selected because these strains are unique among strains with classic emm genotypes in possessing the sof gene while lacking a serum opacification phenotype. Bases 1 to 330, immediately upstream of the sof12 and sof49 structural genes, had 92.4% sequence identity; however no significant matching sequences were identified in the type M1 genome. A region of near identity to the M1 genome was found at bases 331 to 1185 upstream of the sof12 structural gene (97.2% identity). This region is unusual in that the largest ORF in either orientation within the 925-bp regions upstream of sof12, sof49, and the corresponding region in the M1 genome (GenBank accession no. AE006625, bases 1119 to 1975) is only 93 codons and in that none of the ORF products have homology to known protein sequences. Bases 930 to 1181 upstream of sof12 and bases 927 to 1050 upstream of sof49 revealed nearly identical ORFs in the same orientation as the sof genes, which had 98 to 99% sequence identity at the 3′ ends with the probable prtM (protein export) genes from the type M1 and M18 genomes (designated ORFs spy2037 [accession no. AE006625] and spym118-2097 [accession no. AE010112], respectively). Similar single-primer chromosome walking analysis downstream of sof12 and sof49 identified the novel 1,953-bp ORF sfbX (see further analysis below). Situated 142 bp downstream of sfbX49 was a 95-codon ORF with complete sequence identity to corresponding ORFs from the M1 and M18 genomes (ORFs spy2034 and spyM18-2092, respectively). In both the type M1 and M18 genomes these ORF pairs (spy2037 and spy2034 and spym18-2097 and spym18-2092) lie approximately 800 bp apart with no significant intervening ORFs. It is within this 800-bp segment that the sof and sfbX genes are situated within the type M12 and type M49 genomes. Approximately 8.5 kb downstream of spy2034 lies the mga gene, and immediately downstream of mga lies the emm1 gene (11). The chromosomal location of the M49 GAS sof and sfbX genes with respect to the M1 chromosomal architecture was further confirmed by PCR analysis as shown in Fig. 1. The two genes are situated approximately 2.4 kb downstream of the M49 speB gene and 8.8 kb upstream of the mga regulon, including the emm49 gene.
FIG. 1.
Chromosomal location of the GAS emm49 serum opacity factor locus (sof49-sfbX49) with respect to the published emm1 genome sequence.
The sof and sfbX genes are cotranscribed.
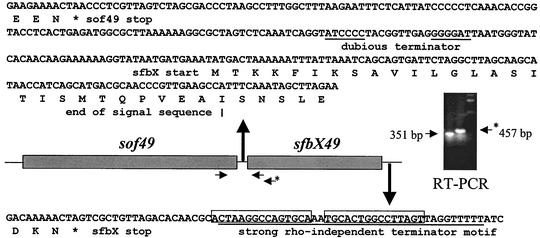
Fifty base pairs upstream of the sof49 gene lies a sequence (5′-TTTAGA-17 bp-TATAAT-3′) identical to the corresponding putative promoter sequence upstream of a sof gene from another strain type (38) with strong homology to the consensus promoter motif (5′-TTGACA-17 bp-TATAAT-3′). A corresponding sequence matching at 25 of 29 positions to the sof49 putative promoter sequence was observed upstream of sof12 (5′-TTTGGG-17 bp-TATAAT-3′). Analysis of the sequence between sof49 and sfbX49 (and sof12 and sfbX12) did not reveal any strong candidate terminator sequences (only a single 7-bp inverted repeat with a 10-bp gap) or the presence of any obvious candidate promoter sequences for independent transcription of sfbX (Fig. 2). In contrast, immediately downstream of the sfbX stop codon a classic rho-independent terminator motif (15-bp hairpin inverted repeat with a 2-bp gap followed by a 9-bp tail containing six Ts) was identified. To determine whether sof and sfbX belong to the same transcriptional unit, RT-PCR analysis was performed with a forward primer from the end of sof and two different reverse primers from the sfbX sequence (Fig. 2). Amplification of products of the expected sizes proved that sof and sfbX represent a two-gene operon.
FIG. 2.
RT-PCR analysis demonstrates that the sof49 and sfbX49 genes are transcribed on the same mRNA and thus represent an operon. Asterisks indicate stop codons. The two underlined sequences between the sof49 and sfbX structural genes indicate an inverted repeat. A putative transcriptional terminator immediately downstream of sfbX is underlined, with the two inverted repeats boxed.
Concordance of sof-sfbX operon with OF phenotype.
A complete sof-sfbX operon was found to be present in several representative OF+ GAS strains including type emm49, emm75, emm87, emm92, and emm114 (formerly st2967) strains. All GAS strains that tested negative for sof by PCR, including type emm1, emm3, emm5, emm6, emm33, emm41, and emm43 strains, were also found to be PCR negative for sfbX. The only classical M protein serotype invariably associated with the opacity-negative phenotype and sof gene sequences is type M12 (2). During this study we noted that the CDC M12 reference strain (isolated more than 50 years ago) and two independent recent clinical isolates each contain a frameshift mutation within sof predicted to result in a truncated Sof precursor protein of 746 amino acids that lacks a portion of the enzymatic domain, all fibronectin-binding repeats, and the C-terminal membrane anchor.
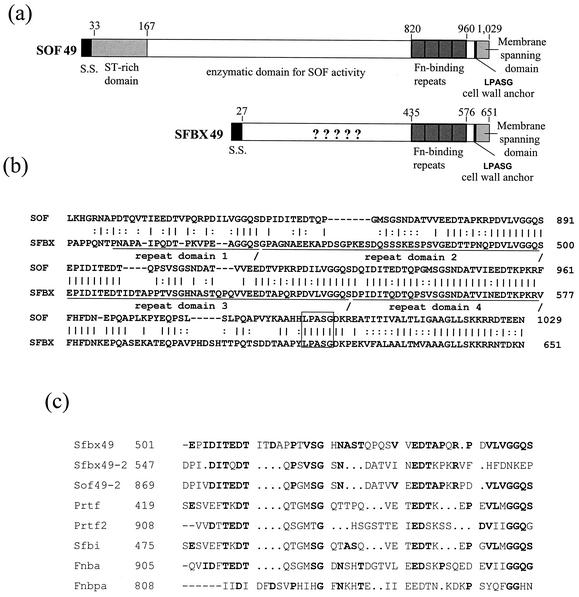
Structural features and conservation of the predicted SfbX protein.
The SfbX49 and SfbX12 proteins had predicted 27-residue leader sequences and wall attachment motifs typical of gram-positive bacterium surface proteins (Fig. 3a). The mature SfbX49 protein (residues 28 to 651) had about 89 to 91% identity with the corresponding sequences of SfbX from type emm12, emm75, emm87, emm92, and emm114 strains. In contrast, the processed forms of Sof proteins are highly divergent (as low as 45% sequence identity). For example, Sof49 and Sof75 have only 60% identity, and Sof114 has only 50 to 56% sequence identity, to either of those two Sof proteins. Thus, the GAS SfbX proteins are much more highly conserved than the hypervariable Sof proteins.
FIG. 3.
(a) Structural comparison of the predicted Sof49 and SfbX49 proteins. S.S., signal sequence; Fn, fibronectin. (b) High degree of homology between Sof and SfbX in the terminal fibronectin-binding repeats (underlined), cell wall anchor (box), and membrane-spanning domain. (c) Alignment of two SfbX49 putative fibronectin-binding repeat regions with similar repeats from other gram-positive bacterium fibronectin-binding proteins. The published sequences for Sof2 (10), PrtF1 and PrtF2 (43), and Sfbi (48) are from Streptococcus pyogenes; FnbpA (44) is from Staphylococcus aureus; and FnbA (28) is from Streptococcus dysgalactiae.
SfbX49 residues 432 to 576 had extensive homology to the fibronectin-binding repeat motifs found in Sof49 (Fig. 3b), other Sof proteins, and other gram-positive bacterium fibronectin-binding proteins. There was a greater degree of homology between the C-terminal 72 SfbX residues and those of various Sof proteins (about 57% identity) than between SfbX and other prototype gram-positive bacterium fibronectin-binding proteins (Fig. 3c). This region (amino acids 577 to 651) lies immediately downstream of the fibronectin-binding repeat motifs and includes the wall attachment motif and N terminus. Other than 37% identity between residues 86 to 140 and a section of the Staphylococcus aureus coagulase residues (residues 59 to 112 of accession no. Q53655), no significant homologies between SfbX and other proteins were observed.
Analysis of SfbX by the Coils method of Lupas (29) revealed alpha-helical regions (residues 31 to 52, 221 to 251, and 299 to 327) with the 7-residue periodicity of hydrophobic residues characteristic of proteins capable of forming a coiled-coil structure. In contrast, the same program run on various full-length Sof proteins revealed no significant periodicity of hydrophobic residues. Proline was the most abundant residue in SfbX proteins (9.2% in SfbX49), followed by aspartate (8.7%), threonine (8%), lysine (8%), and alanine (8%). Two variable-length proline-rich repeat regions were observed within the SfbX proteins, corresponding to SfbX49 residues 348 to 370 and 417 to 448. Such proline-rich regions may be common between functional domains of gram-positive bacterial surface proteins. Examples include the regions linking the choline-binding and extracellular domains of the pneumococcal PspA proteins (52) and linking the Sof fibronectin/fibrinogen-binding domain with its enzymatic domain (38).
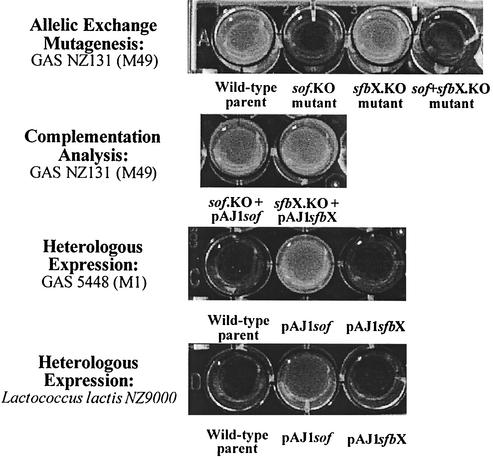
Serum opacification by GAS is solely encoded by the sof gene.
We have shown that the sof gene and the previously unrecognized sfbX gene are cotranscribed. Therefore, earlier experimental results using sof insertion duplication mutants would possibly include polar effects on sfbX transcription. To determine the relative contribution of sof and sfbX genes to the phenotypes of serum opacification and fibronectin binding, we constructed precise, in-frame allelic-exchange sof, sfbX, and sof-sfbX mutants in GAS strain NZ131 (M49). The sof and sfbX genes were also cloned individually in expression vector pDCerm for heterologous-expression analysis. Incubation of GAS culture supernatants with horse serum revealed that deletion of the sof gene resulted in complete loss of opacification, whereas an sfbX deletion mutant maintained wild-type levels of opacification (Fig. 4). The ability to opacify serum could be restored in the sof mutant by reintroduction of the sof gene on a plasmid vector. Heterologous expression of sof conferred the serum opacification phenotype to an M1 GAS strain and to L. lactis; parallel experiments with sfbX did not produce opacification (Fig. 4). Together these results show that GAS serum opacification is uniquely encoded by the sof gene.
FIG. 4.
Contribution of the Sof49 and SfbX49 gene products to opacification of horse serum. Precise in-frame allelic-exchange mutagenesis, complementation analysis, and heterologous-expression studies indicate that sof alone encodes the opacity factor phenotype.
Both sof and sfbX encode fibronectin-binding functions.
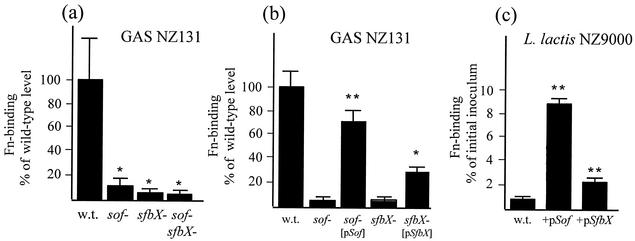
The effect of precise targeted mutagenesis of sof and sfbX on GAS fibronectin binding was assessed by in vitro assays. Marked decreases in fibronectin binding were noted with single-gene deletions of either sof (10.7% ± 5.8% of wild-type level; P = 0.026) or sfbX (7.0% ± 3.8% of the wild-type level; P = 0.016); dual deletion of sof and sfbX produced a further small decrease (6.0% ± 2.0% of the wild-type level), which was not statistically different from that for either single mutant (Fig. 5a). Complementation studies showed that return of the corresponding single gene on an expression plasmid partially restored the fibronectin-binding phenotype of the sof mutant (72% ± 8.6% versus 7% ± 0.7% of wild-type levels; P < 0.00005) and the sfbX mutant (29% ± 5% versus 8.4% ± 0.9% of wild-type levels; P < 0.001) (Fig. 5b). Heterologous expression of sof or sfbX in M1 GAS strain 5448 yielded increases in fibronectin-binding activity (39 and 153%, respectively; data not shown). More-pronounced effects were observed in the low-fibronectin-binding background of the nonpathogenic bacterium L. lactis (0.8% ± 0.1% of input inoculum). Heterologous expression of sof increased L. lactis fibronectin binding by 1,100% (8.8% ± 0.4% of input inoculum; P < 0.0001), while parallel experiments with sfbX increased L. lactis fibronectin binding by 287% (2.3% ± 0.2% of input inoculum; P < 0.0001) (Fig. 5c). Together these data demonstrate that sof and sfbX each possess important fibronectin-binding properties demonstrable in the context of the intact bacterial surface.
FIG. 5.
(a) Contribution of the Sof49 and SfbX49 gene products to the fibronectin (Fn)-binding ability of log-phase GAS assessed by using isogenic allelic-exchange knockout mutants. (b) Complementation of the sof and sfbX knockout mutants by returning the single gene on a plasmid vector. Wild-type (w.t.) and mutant strains transformed with the vector alone (pDCerm) served as controls. (c) Ability of the cloned GAS sof49 and sfbX49 genes to confer fibronectin-binding activity to L. lactis. ∗, P < 0.05; ∗∗, P < 0.0001. The bacterial inoculum was 106 CFU in 100 μl for all experiments.
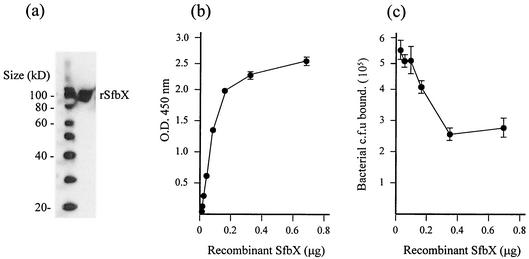
Recombinant SfbX binds fibronectin.
A recombinant SfbX fusion protein bearing C-terminal V5 and His (6) epitopes was expressed in E. coli, purified with an affinity column, and confirmed to migrate at the predicted size of ∼85 kDa by Western blot analysis using an monoclonal anti-V5 antibody (Fig. 6a). In an anti-V5 antibody ELISA, recombinant SfbX exhibited dose-dependent binding to immobilized fibronectin that began to plateau when the concentration reached 2.8 μg/ml (0.2 μg in a 70-μl volume) (Fig. 6b). No ELISA signal was detected in control wells in which either recombinant SfbX or the primary anti-V5 antibody was removed or in parallel experiments using a control recombinant SpeB protein with identical C-terminal fusion epitopes. Finally, addition of SfbX was able to competitively inhibit the ability of wild-type GAS to bind fibronectin in a dose-dependent fashion, the effects of which were seen starting at 1.3 μg of the protein/ml (0.1 μg in 75 ml), culminating in an ∼50% reduction in total GAS adherence (Fig. 6c). The findings obtained with recombinant SfbX corroborate the fibronectin-binding function identified upon targeted knockout of the GAS sfbX gene.
FIG. 6.
(a) Western blot confirmation of recombinant SfbX protein after affinity column purification. (b) Dose-dependent binding of recombinant SfbX protein to immobilized fibronectin. (c) Dose-dependent competitive inhibition of GAS fibronectin-binding by recombinant SfbX.
DISCUSSION
Sof is a unique bifunctional protein anchored to the cell surface of many GAS strains. Our investigations were initiated to map the chromosomal location of the sof gene. In doing so, we made the surprising discovery that sof is linked invariably to a second gene (sfbX) encoding a novel streptococcal fibronectin-binding protein. All GAS strains tested with the capacity to opacify mammalian sera (OF+) were found to possess the two-gene sof-sfbX operon.
Our genomic-mapping studies were performed with GAS type emm49 and emm12 strains, two classical nephritogenic strains (30). GAS strains generally contain one of five different gene patterns at the mga locus based on peptidoglycan-spanning domain-encoding sequences, number, and relative arrangement of emm and emm-like genes. Type emm49 isolates typically correspond to pattern E of emm and emm-like genes and appear to have an equal predisposition to colonize skin and throat (5, 16). Pattern E strains are generally phenotypically OF+ due to the presence of the sof gene and contain emm nested between two additional emm-like genes at the mga locus. In contrast, type emm12 isolates are OF− and have pattern A (only one emm family gene [emm12] at the mga locus). Except for type emm12, emm types that are associated with patterns A to C (for example, type emm1) have invariably been seen to be OF− and to lack sof gene sequences (2). Although type emm12 isolates are invariably OF− and typically correspond to emm and emm-like gene patterns A to C, we show here that these isolates appear to carry a defective sof gene. Based on their pronounced differences in opacity factor phenotype, emm gene homology (emm12 is quite divergent from emm genes from all other sof-positive strains), and mga locus gene arrangement, emm49 and emm12 strains appeared to be ideal candidates for independently verifying the chromosomal location of the sof gene in relation to the completed serotype M1 genome sequence. In addition, these strains are totally divergent at the multilocus sequence type level (see http://www.mlst.net for types emm12 and emm49). In both cases, we found the GAS sof gene together in an operon with sfbX and situated ∼2.4 kb downstream of the gene encoding the cysteine protease SpeB and ∼8.8 kb from the genes of the tandemly linked genes of the mga regulon. This close genetic linkage is likely reflected in the strict correlation of opacify factor phenotype with specific emm genotypes. Despite sof's anomalous chromosomal location upstream of mga, its expression has been shown to be positively regulated by mga (33).
It is open to speculation whether GAS strains possessing the sof-sfbX operon are ancestral to sof-sfbX-negative strains or vice versa. The G-C content of the operon (43.4% in the emm49 strain) is not greatly dissimilar to the overall G-C content of the published GAS M1 (38.6%) and M18 (38.5%) genome sequences (11, 47). The presence of the functioning operon is restricted to strains of specific serotypes: those encoding class II M types nearly always opacify serum, while those of class I M types (e.g., M1 and M18) rarely do so (4). Since the DNA sequences immediately flanking the two genes do not possess inverted repeats suggestive of a insertion sequence or homology to bacteriophage or phage-like elements, we hypothesize that OF− GAS strains arose upon deletion of the sof-sfbX operon at an early evolutionary stage coincident with the major delineation of M protein classes I and II.
The ability of several GAS surface-anchored proteins to interact with fibronectin of the extracellular matrix has been shown to play a major role in adherence of the organism to host epithelial cell surfaces (15, 25, 34, 39). It is possible that the presence or absence of the two genes (sof and sfbX) encoding fibronectin-binding functions affects the spectrum of tissue colonization and disease manifestations associated with specific GAS strains. Among other correlations, OF+ strains have been identified with a tropism for skin and the clinical syndrome of impetigo (7). In contrast, the top three GAS genotypes currently associated with invasive (blood-borne) infections including necrotizing fasciitis and toxic shock syndrome in the U.S. are emm1, emm3, and emm12 (CDC, Active Bacterial Core Surveillance, 2000 to 2001). All of these types are OF− due to absence or mutation (emm12) of the sof-sfbX operon. Whereas OF− GAS strains may lack one mechanism for fibronectin-mediated adherence at certain epithelial surfaces, they could perhaps have a competitive advantage in the bloodstream, where circulating fibronectin could theoretically serve as an opsonin for phagocytic clearance by host neutrophils (46, 51). Opacity factor has also been shown to inhibit the beta-hemolytic activity of streptolysin S (35), the potent GAS exotoxin that plays a virulence role in the tissue destruction of necrotizing fasciitis (6, 18).
Earlier mutagenesis studies of the sof gene were performed without knowledge of the presence of the transcriptionally linked sfbX. Our precise in-frame allelic-exchange mutagenesis experiments on a wild-type OF+ emm49 strain confirmed that serum opacification was solely encoded by the sof gene. However, the studies of mutants showed that each gene contributed strongly and independently to the ability of log-phase GAS to bind to immobilized fibronectin. All results were further corroborated by the first-ever heterologous expression of sof and sfbX in a gram-positive species, L. lactis. These experiments strongly suggest that a C-terminal cell wall anchor motif is recognized by the lactococcal sortase to properly attach the proteins to the cell wall and allow their fibronectin-binding and enzymatic domains to interact with the extracellular milieu. The sequence LPASG in Sof and SfbX is properly localized to serve as a cell wall anchor motif and corresponds to the paradigm of LPXTG described by Mazmanian and colleagues, where the X and T residues can vary (32). There exist two known GAS sortases: SrtA appears to be present in all or most M types as well as group C and G strains, while SrtB appears to be present only in a subset of strains. Available data indicate an undefined difference in protein substrate specificities between the two sortases (1). From our BLAST searches, Sof and SfbX are the only proven GAS surface proteins bearing an LPXSG (difference from the LPXTG motif is underlined) candidate anchor motif.
Our successful cloning and expression of a recombinant SfbX facilitated experiments to further confirm the role of this protein in GAS fibronectin binding. The ability of SfbX to mediate GAS fibronectin binding is not wholly unanticipated, since the C-terminal fibronectin-binding repeats are so highly similar to the corresponding fibronectin-binding region of the Sof protein (Fig. 3). The function of the N-terminal domain of SfbX after signal sequence removal is unknown. It is significantly smaller (409 versus 654 amino acids) than the lipoproteinase domain of Sof. The N-terminal regions of Sof proteins are highly varied and immunologically distinct across different GAS emm genotype strains, serving as the basis of alternative serologic or genetic classification schemes (2, 13). In contrast, the SfbX sequence is highly conserved across GAS emm genotypes. Secondary-structure analysis suggests limited hydrophobic residue periodicity characteristic of a protein capable of forming an alpha-helical coiled-coil structure. Such structures are a property of other streptococcal surface proteins such as the M protein, and it has been speculated that they play a role in triggering postinfection autoimmune injury to host tissue antigens bearing similar motifs. However, the M protein is predicted to form a coiled-coil structure along the majority of its entire length while SfbX displays this characteristic only within three restricted areas. No significant homologies with other proteins in the GenBank databases were noted, save for an ∼50-amino-acid stretch bearing 37% identity with the S. aureus coagulase. S. aureus coagulase is an extracellular protein which binds to prothrombin in the host to form a complex called staphylothrombin (23). The protease activity characteristic of thrombin is activated in the complex, resulting in the conversion of fibrinogen to fibrin. As recently reported for Sof (9), the conserved fibronectin-binding repeats of SfbX are anticipated to also bind fibrinogen. Although a specific coagulase activity has never been reported for GAS, it is interesting to speculate that the two domains of SfbX may somehow cooperate to interact with proteins of the host extracellular matrix.
In conclusion, we have mapped the sof gene encoding serum opacity factor to its location on the GAS chromosome. In doing so, we discovered that sof exists in a two-gene operon with sfbX, a novel GAS gene encoding a surface-anchored protein with important fibronectin-binding properties and an N-terminal domain of unknown function. Using the novel GAS mutants, heterologous expression constructs, and the recombinant SfbX protein reagents we have created, future studies will seek to identify the additional function(s) of the SfbX protein and further understand the contributions of the sof-sfbX operon to GAS ecology and human disease pathogenesis.
Acknowledgments
This work was supported by NIH grant AI048694 and the Edward J. Mallinckrodt, Jr., Foundation (V.N.). A.J. is a postdoctoral fellow supported through UCSD institutional NIH training award AI07036.
REFERENCES
- 1.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 184:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall, B., G. Gherardi, M. Lovgren, R. R. Facklam, B. A. Forwick, and G. J. Tyrrell. 2000. emm and sof gene sequence variation in relation to serological typing of opacity-factor-positive group A streptococci. Microbiology 146:1195-1209. [DOI] [PubMed] [Google Scholar]
- 3.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bessen, D., K. F. Jones, and V. A. Fischetti. 1989. Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. J. Exp. Med. 169:269-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessen, D. E., C. M. Sotir, T. L. Readdy, and S. K. Hollingshead. 1996. Genetic correlates of throat and skin isolates of group A streptococci. J. Infect. Dis. 173:896-900. [DOI] [PubMed] [Google Scholar]
- 6.Betschel, S. D., S. M. Borgia, N. L. Barg, D. E. Low, and J. C. De Azavedo. 1998. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect. Immun. 66:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisno, A. L., and D. L. Stevens. 1996. Streptococcal infections of skin and soft tissues. N. Engl. J. Med. 334:240-245. [DOI] [PubMed] [Google Scholar]
- 8.Chaffin, D. O., and C. E. Rubens. 1998. Blue/white screening of recombinant plasmids in gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91-99. [DOI] [PubMed] [Google Scholar]
- 9.Courtney, H. S., J. B. Dale, and D. L. Hasty. 2002. Mapping the fibrinogen-binding domain of serum opacity factor of group a streptococci. Curr. Microbiol. 44:236-240. [DOI] [PubMed] [Google Scholar]
- 10.Courtney, H. S., D. L. Hasty, Y. Li, H. C. Chiang, J. L. Thacker, and J. B. Dale. 1999. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol. Microbiol. 32:89-98. [DOI] [PubMed] [Google Scholar]
- 11.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillen, C. M., R. J. Towers, D. J. McMillan, A. Delvecchio, K. S. Sriprakash, B. Currie, B. Kreikemeyer, G. S. Chhatwal, and M. J. Walker. 2002. Immunological response mounted by Aboriginal Australians living in the Northern Territory of Australia against Streptococcus pyogenes serum opacity factor. Microbiology 148:169-178. [DOI] [PubMed] [Google Scholar]
- 14.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollingshead, S. K., T. L. Readdy, D. L. Yung, and D. E. Bessen. 1993. Structural heterogeneity of the emm gene cluster in group A streptococci. Mol. Microbiol. 8:707-717. [DOI] [PubMed] [Google Scholar]
- 17.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotic stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humar, D., V. Datta, D. J. Bast, B. Beall, J. C. De Azavedo, and V. Nizet. 2002. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet 359:124-129. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, D. R., and E. L. Kaplan. 1988. Microtechnique for serum opacity factor characterization of group A streptococci adaptable to the use of human sera. J. Clin. Microbiol. 26:2025-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kansal, R. G., A. McGeer, D. E. Low, A. Norrby-Teglund, and M. Kotb. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun. 68:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlyshev, A. V., M. J. Pallen, and B. W. Wren. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. BioTechniques 28:1078-1082. [DOI] [PubMed] [Google Scholar]
- 22.Katerov, V., P. E. Lindgren, A. A. Totolian, and C. Schalen. 2000. Streptococcal opacity factor: a family of bifunctional proteins with lipoproteinase and fibronectin-binding activities. Curr. Microbiol. 40:149-156. [DOI] [PubMed] [Google Scholar]
- 23.Kawabata, S., and S. Iwanaga. 1994. Structure and function of staphylothrombin. Semin. Thromb. Hemost. 20:345-350. [DOI] [PubMed] [Google Scholar]
- 24.Kreikemeyer, B., D. R. Martin, and G. S. Chhatwal. 1999. SfbII protein, a fibronectin binding surface protein of group A streptococci, is a serum opacity factor with high serotype-specific apolipoproteinase activity. FEMS Microbiol. Lett. 178:305-311. [DOI] [PubMed] [Google Scholar]
- 25.Kreikemeyer, B., S. R. Talay, and G. S. Chhatwal. 1995. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol. Microbiol. 17:137-145. [DOI] [PubMed] [Google Scholar]
- 26.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 27.Lancefield, R. C. 1962. Current knowledge of the type specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 28.Lindgren, P. E., M. J. McGavin, C. Signas, B. Guss, S. Gurusiddappa, M. Hook, and M. Lindberg. 1993. Two different genes coding for fibronectin-binding proteins from Streptococcus dysgalactiae. The complete nucleotide sequences and characterization of the binding domains. Eur. J. Biochem. 214:819-827. [DOI] [PubMed] [Google Scholar]
- 29.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266:513-525. [DOI] [PubMed] [Google Scholar]
- 30.Majeed, H. A., A. M. Yousof, J. Rotta, H. Havlickpva, G. Bahar, and K. Bahbahani. 1992. Group A streptococcal strains in Kuwait: a nine-year prospective study of prevalence and associations. Pediatr. Infect. Dis. J. 11:295-303. [PubMed] [Google Scholar]
- 31.Maxted, W. R., J. P. Widdowson, C. A. Fraser, L. C. Ball, and D. C. Bassett. 1973. The use of the serum opacity reaction in the typing of group-A streptococci. J. Med. Microbiol. 6:83-90. [DOI] [PubMed] [Google Scholar]
- 32.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 33.McLandsborough, L. A., and P. P. Cleary. 1995. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of ScpA, FcRA, OF, and M protein. FEMS Microbiol Lett. 128:45-51. [DOI] [PubMed] [Google Scholar]
- 34.Molinari, G., and G. S. Chhatwal. 1999. Role played by the fibronectin-binding protein SfbI (protein F1) of Streptococcus pyogenes in bacterial internalization by epithelial cells. J. Infect. Dis. 179:1049-1050. [DOI] [PubMed] [Google Scholar]
- 35.Pinney, A. M., J. P. Widdowson, and W. R. Maxted. 1977. Inhibition of beta-haemolysis by opacity factor in group A streptococci. J. Hyg. 78:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podbielski, A., A. Kaufhold, and R. Lutticken. 1992. The vir-regulon of Streptococcus pyogenes: coordinate expression of important virulence factors. Immun. Infekt. 20:161-168. [PubMed] [Google Scholar]
- 37.Pritzlaff, C. A., J. C. Chang, S. P. Kuo, G. S. Tamura, C. E. Rubens, and V. Nizet. 2001. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol. Microbiol. 39:236-247. [DOI] [PubMed] [Google Scholar]
- 38.Rakonjac, J. V., J. C. Robbins, and V. A. Fischetti. 1995. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect. Immun. 63:622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha, C. L., and V. A. Fischetti. 1999. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infect. Immun. 67:2720-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubens, C. E., and L. M. Heggen. 1988. Tn916 delta E: a Tn916 transposon derivative expressing erythromycin resistance. Plasmid 20:137-142. [DOI] [PubMed] [Google Scholar]
- 42.Saravani, G. A., and D. R. Martin. 1990. Opacity factor from group A streptococci is an apoproteinase. FEMS Microbiol. Lett. 56:35-39. [DOI] [PubMed] [Google Scholar]
- 43.Sela, S., A. Aviv, A. Tovi, I. Burstein, M. G. Caparon, and E. Hanski. 1993. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10:1049-1055. [DOI] [PubMed] [Google Scholar]
- 44.Signas, C., G. Raucci, K. Jonsson, P. E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon, D., and J. J. Ferretti. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66:219-224. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, W. A., D. L. Hasty, J. M. Mason, and E. H. Beachey. 1982. Fibronectin-mediated binding of group A streptococci to human polymorphonuclear leukocytes. Infect. Immun. 37:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talay, S. R., P. Valentin-Weigand, P. G. Jerlstrom, K. N. Timmis, and G. S. Chhatwal. 1992. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect. Immun. 60:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 50.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, K. D., N. H. Augustine, L. A. Gonzalez, J. F. Bohnsack, and H. R. Hill. 1988. Effects of fibronectin on the interaction of polymorphonuclear leukocytes with unopsonized and antibody-opsonized bacteria. J. Infect. Dis. 158:823-830. [DOI] [PubMed] [Google Scholar]
- 52.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]