Abstract
The gram-negative, oral bacterium Actinobacillus actinomycetemcomitans has been implicated as the causative agent of several forms of periodontal disease in humans. When cultured in broth, fresh clinical isolates of A. actinomycetemcomitans form tenacious biofilms on surfaces such as glass, plastic, and saliva-coated hydroxyapatite, a property that probably plays an important role in the ability of this bacterium to colonize the oral cavity and cause disease. We examined the morphology of A. actinomycetemcomitans biofilm colonies grown on glass slides and in polystyrene petri dishes by using light microscopy and scanning and transmission electron microscopy. We found that A. actinomycetemcomitans developed asymmetric, lobed biofilm colonies that displayed complex architectural features, including a layer of densely packed cells on the outside of the colony and nonaggregated cells and large, transparent cavities on the inside of the colony. Mature biofilm colonies released single cells or small clusters of cells into the medium. These released cells adhered to the surface of the culture vessel and formed new colonies, enabling the biofilm to spread. We isolated three transposon insertion mutants which produced biofilm colonies that lacked internal, nonaggregated cells and were unable to release cells into the medium. All three transposon insertions mapped to genes required for the synthesis of the O polysaccharide (O-PS) component of lipopolysaccharide. Plasmids carrying the complementary wild-type genes restored the ability of mutant strains to synthesize O-PS and release cells into the medium. Our findings suggest that A. actinomycetemcomitans biofilm growth and detachment are discrete processes and that biofilm cell detachment evidently involves the formation of nonaggregated cells inside the biofilm colony that are destined for release from the colony.
Actinobacillus actinomycetemcomitans is a gram-negative, nonmotile coccobacillus that colonizes the human oral cavity (20). A. actinomycetemcomitans has been implicated as the causative agent of several forms of severe periodontal disease, including localized juvenile periodontitis, early-onset periodontitis, and rapidly progressive periodontitis (37). Infrequently, A. actinomycetemcomitans can enter the submucosa and cause nonoral infections, including bacteremias, infective endocarditis, and localized abscesses (17). When cultured in broth, fresh clinical isolates of A. actinomycetemcomitans form extremely tenacious biofilms on surfaces such as glass, plastic, and saliva-coated hydroxyapatite (7, 8, 11, 13-16, 20, 29, 30). Nearly all of the cells grow attached to the surface, while the broth remains clear and is often sterile (7). The dense biofilm that forms on the surface is resistant to removal by agents such as detergents, proteases, heat, sonication, and vortex agitation (7) and exhibits increased resistance to antimicrobial agents compared with that exhibited by cells grown in planktonic form (6). Tight adherence has been shown to play an important role in the ability of A. actinomycetemcomitans to colonize the mouths of rats (9) and probably plays an equally important role in its ability to colonize humans. Tenacious surface attachment is dependent on the presence of long, bundled adhesive pili (fimbriae) that form on the surface of the cell (29). Mutations in flp-1, which encodes the major fimbrial protein subunit, result in cells that fail to produce fimbriae or attach to surfaces (15).
Kaplan and Fine have recently shown that A. actinomycetemcomitans biofilm colonies are capable of releasing single cells or small clusters of cells into liquid medium and that these released cells can attach to the surface of the culture vessel and form new biofilm colonies, enabling the biofilm to spread (18). In the present report, we describe the morphology of A. actinomycetemcomitans biofilm colonies that were grown attached to glass and plastic surfaces and the dynamics of biofilm cell detachment. Our findings suggest that A. actinomycetemcomitans utilizes a novel mechanism of biofilm cell detachment that may involve the release of cells from inside the biofilm colony.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The A. actinomycetemcomitans strains used in this study are listed in Table 1. Mutagenesis of strain CU1000N with transposon IS903φkan was carried out as previously described (19). Bacteria were grown in Trypticase soy broth (BD Biosystems) supplemented with 6 g of yeast extract and 8 g of glucose/liter at 37°C in 10% CO2. Single-cell suspensions were prepared as previously described (18). Culture vessels were 35- or 100-mm-diameter tissue culture-treated polystyrene petri dishes (Corning no. 340165 and Falcon no. 353003, respectively). Bacteria for scanning electron microscopy were grown on 20- by 25-mm pieces of acid-washed borosilicate glass slides (Fisher) placed in 35-mm-diameter petri dishes. Four milliliters of medium was used in 35-mm-diameter dishes, and 20 ml was used in 100-mm-diameter dishes.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference |
|---|---|---|
| A. actinomycetemcomitans strains | ||
| CU1000 | Wild-type clinical isolate | 8 |
| CU1000N | Spontaneous nalidixic acid-resistant mutant of CU1000 | 14 |
| JK1002 | CU1000N wzt::IS903φkan; Kmr | 19 |
| JK1017 | CU1000N rmlA::IS903φkan; Kmr | This study |
| JK1022 | CU1000N dspA::IS903φkan; Kmr | 19 |
| Plasmids | ||
| pJAK16 | Broad-host-range, mobilizable IncQ vector; Cmr; tac promoter, lacIq | 32 |
| pJK595 | pJAK16 with wzt of CU1000 expressed from tac promoter | This study |
| pJK596 | pJAK16 with dspA of CU1000 expressed from tac promoter | This study |
| pJK597 | pJAK16 with rmlA of CU1000 expressed from tac promoter | This study |
Kmr, kanamycin resistant; Cmr, chloramphenicol resistant.
Scanning electron microscopy.
Biofilm colonies grown on glass slides were washed once with phosphate-buffered saline and fixed in 2% (vol/vol) cold (4°C) freshly prepared glutaraldehyde in 0.2 M phosphate buffer (pH 7.2) overnight. Colonies were postfixed in 1% cold osmium tetroxide for 2 h. The glutaraldehyde was removed by vacuum aspiration, and the colonies were washed once with phosphate-buffered saline and then dehydrated through a graded acetone series. Samples were critical point dried, mounted on copper stubs, and vacuum coated in a Polaron sputter-coating unit (E5000) with approximately 10-nm-thick Au-Pd deposited from a circular target mounted 3 cm from the sample. Samples were examined in a JEOL 25S scanning electron microscope operating at 15 kV and photographed on type 665 positive-negative Land film.
Thin sectioning.
Biofilm colonies grown in polystyrene petri dishes were washed and fixed as described above. After dehydration through graded ethanol, the cells were removed from the dishes by using propylene oxide. The floating biofilm layers were washed in several changes of propylene oxide, pelleted by low-speed centrifugation, and embedded in Epon 812. One-micron-thick and ultrathin sections were prepared on an LKB III ultramicrotome.
Light microscopy.
One-micron-thick sections were stained in 1% toluidine blue-borax, coverslipped, and examined with an Olympus IMT inverted microscope. Light photomicrographs were taken with an Olympus DP10 digital camera or with a Polaroid MP4 system using type 667 film.
Transmission electron microscopy.
Ultrathin sections were stained with uranyl acetate and lead acetate and examined in a Philips 300 transmission electron microscope operating at 60 kV. Photographs were made on Kodak electron imaging film with type 4463 photographic emulsion.
Ninety-six-well microtiter plate biofilm detachment assay.
Biofilm colonies were grown on polystyrene rods suspended in broth in the wells of a 96-well microtiter plate. Cells that detached from the biofilms fell to the bottom of the well, where they attached to the surface and formed new biofilm colonies. The amount of biofilm growth on the bottom of the well, which was proportional to the number of cells that detached from biofilm colonies on the rods, was measured by staining with crystal violet. The detachment assay was carried out as follows.
(i) Construction of apparatus.
The lid of a 96-well polystyrene flat-bottomed tissue culture plate (Falcon no. 353072) was modified as follows. First, 96 1.5-mm-diameter holes were drilled in the lid, with the position of each hole corresponding to the center of one of the 96 wells. Then, an 11-mm-long polystyrene rod (1.5-mm-diameter; Plastruct Corp., City of Industry, Calif.) was placed in each hole (with one end of the rod flush against the top of the lid) and secured with trichloromethane plastic solvent. When this modified lid was placed on a 96-well microtiter plate bottom, the rods were suspended in the wells, with the bottom of each rod being approximately 2 mm above the bottom of the well. The modified lid was sterilized by soaking in 70% ethanol for 30 min and air drying in a biological safety cabinet.
(ii) Inoculation and incubation of polystyrene rods.
The microtiter plate bottom was filled with medium (100 μl per well), and each well was inoculated with a single 2- to 3-day-old colony from an agar plate by using a sterile toothpick. The modified lid was then placed on the inoculated plate to submerge the polystyrene rods in the inoculated medium, and the plate was incubated for 24 h to allow bacteria to adhere to the rods. The lid was then transferred to a fresh microtiter plate containing prewarmed medium and incubated for an additional 24 h to allow biofilm cells to detach from the rods.
(iii) Measuring detached cells.
The lid was removed, and the plate was washed extensively under running tap water to remove loosely adherent cells. The wells were filled with 100 μl of Gram-staining reagent (2 g of crystal violet, 0.8 g of ammonium oxalate, 20 ml of ethanol per 100 ml), and the plate was incubated at room temperature for 10 min. The plate was rewashed extensively under running tap water to remove unbound dye. The wells were then filled with 100 μl of ethanol, and the plate was incubated at room temperature for 10 min to solubilize the dye. The optical density (at 590 nm) of the ethanol-dye solution in each well was measured by using a Bio-Rad Benchmark microplate reader.
Plasmids and DNA techniques.
The plasmids utilized in this study are listed in Table 1. PCRs, plasmid DNA manipulations, and DNA sequence analyses were carried out as previously described (19). Plasmids containing wild-type wzt, dspA (dispersal; formerly ORFf1), and rmlA genes (pJK595, pJK596 and pJK597, respectively) for use in genetic complementation experiments were constructed as follows. First, the wzt, dspA, and rmlA genes from A. actinomycetemcomitans strain CU1000 (19) were amplified by PCR with primers that introduced a KpnI restriction site 14 bp upstream from the ATG initiation codon of each gene and a BamHI restriction site 1 bp downstream from the stop codon of each gene. PCR products were digested with KpnI and BamHI and ligated into the KpnI and BamHI restriction sites of the broad-host-range plasmid vector pJAK16 (14), which placed each gene under control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter. The resulting plasmids were subjected to DNA sequence analysis to confirm the sequence of the insert. Complementing plasmids were mobilized into mutant strains by using the RK2 oriT-defective mutant plasmid pRK21761 as previously described (32). Plasmid-harboring strains were grown in broth supplemented with 3 μg of chloramphenicol/ml and 1 mM IPTG.
RESULTS
Microscopic analysis of A. actinomycetemcomitans biofilm colonies.
Fig. 1 shows light and electron microscopic analyses of A. actinomycetemcomitans biofilm colonies grown from single-cell suspensions inoculated onto borosilicate glass slides and into tissue culture-treated polystyrene petri dishes. The A. actinomycetemcomitans biofilm colony morphologies (as determined by light microscopy at a magnification of ×40) and biofilm detachment phenotypes (see below) on these two surfaces, as well as those of colonies grown on untreated polystyrene and polycarbonate surfaces, were indistinguishable (data not shown). The photographs shown in Fig. 1 are representative of approximately 5 to 25 biofilm colonies examined for each sample. All of the colonies examined displayed morphologies that were indistinguishable from those shown in Fig. 1.
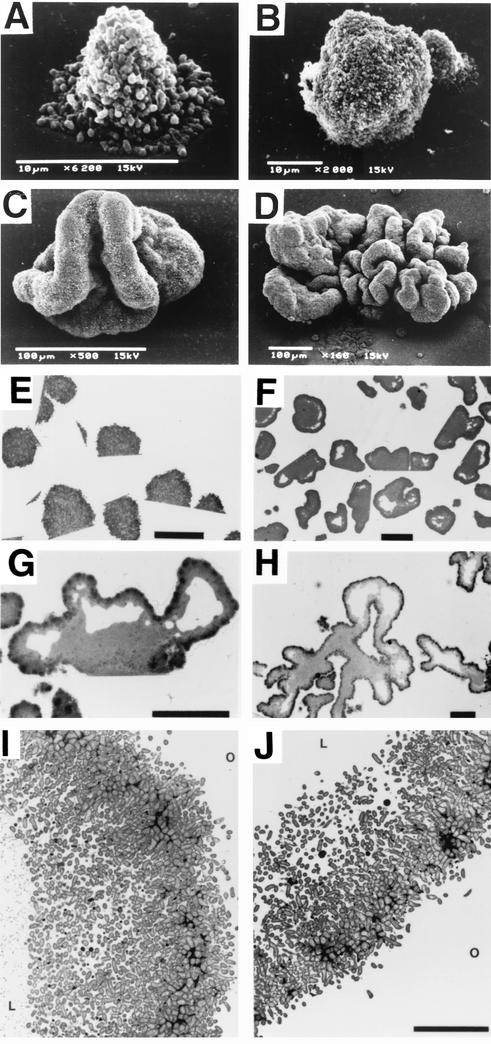
FIG. 1.
Microscopic analyses of A. actinomycetemcomitans CU1000 biofilm colonies. (A to D) Scanning electron micrographs of day 0.5 (A), day 1 (B), day 2 (C), and day 4 (D) colonies grown on glass slides. (E to H) Thin sections of day 1 (E), day 2 (F), day 3 (G), and day 4 (H) colonies grown on polystyrene. Panels E and F show multiple biofilm colonies. (I and J) Transmission electron micrographs of the outer surface of day 3 (I) and day 4 (J) colonies grown on polystyrene. O, outside of the colony; L, the internal lacuna. Bars = 25 μm (E), 100 μm (F to H), and 10 μm (J).
Figure 1A to D shows scanning electron micrographs of A. actinomycetemcomitans strain CU1000 biofilm colonies that were grown attached to glass slides. Very young, umbonate colonies (Fig. 1A) became domed or ovoid by day 1 (Fig. 1B) and distended and lobate by day 2 (Fig. 1C). By days 3 and 4, the surface of the colony was covered with a complex series of lobes and invaginations (Fig. 1D). Thin sections showed that day 1 colonies contained densely packed cells (Fig. 1E). By day 2, colonies had developed a dark-staining outer layer consisting of densely packed cells, a light-staining interior consisting of loosely packed, nonaggregated cells, and large, transparent internal cavities (lacunae) that formed near the outer surface of the colony (Fig. 1F). By days 3 and 4, the colonies were highly invaginated and contained lacunae both in the interior of the colony and in the exterior lobes, and these lacunae occupied a large volume of the colony (Fig. 1G and H). Transmission electron micrographs confirmed that cells on the outer surface of the colony were densely packed, whereas cells located in the interior of the colony were loosely packed and nonaggregated (Fig. 1I and J). The cellular ultrastructure of strain CU1000 revealed by transmission electron microscopy was similar to that of other strains of A. actinomycetemcomitans (1, 12, 21).
Dispersal of A. actinomycetemcomitans biofilm colonies grown in polystyrene petri dishes.
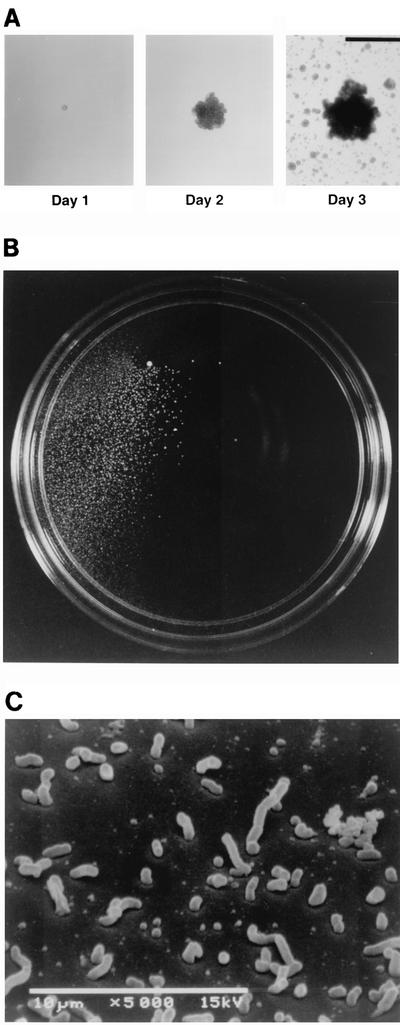
Figure 2A shows the growth over 3 days of a single A. actinomycetemcomitans biofilm colony in a polystyrene petri dish containing broth. By day 3, numerous small satellite colonies were growing on the surface of the dish. Satellite colonies covered the entire surface of the dish and were not visibly attached to mature biofilm colonies or to each other. The number of mature biofilm colonies growing on the surface of each dish was the same as the number of colonies that grew on an agar plate inoculated with the same inoculum, indicating that satellite colonies arose from cells released by mature biofilm colonies into the medium and not from slow settlers from the planktonic phase. Figure 2B shows a 100-mm-diameter petri dish inoculated with 1 CFU of strain CU1000 and incubated for 3 days. Approximately 105 satellite colonies grew in a localized area adjacent to the mature colony. Figure 2C shows an electron micrograph of the surface of a glass slide in an area approximately 5 mm away from a mature colony after 2 days of growth. The surface was covered with numerous single cells and small clusters of cells, suggesting that A. actinomycetemcomitans biofilm colonies released small units of adherent cells into the medium.
FIG. 2.
Dispersal of A. actinomycetemcomitans CU1000 biofilm colonies in broth cultures. (A) Pictures, taken 1, 2, and 3 days after inoculation, of a single colony from a 35-mm-diameter petri dish inoculated with ≈25 CFU. Bar = 1 mm. (B) A 100-mm-diameter petri dish inoculated with 1 CFU of strain CU1000 and incubated for 3 days. Colonies appear as white spots on a dark background. (C) Scanning electron micrograph of the surface of a glass slide approximately 5 mm away from a mature biofilm colony after 2 days of growth. The glass slide contained five mature colonies.
Isolation of A. actinomycetemcomitans biofilm dispersal mutants.
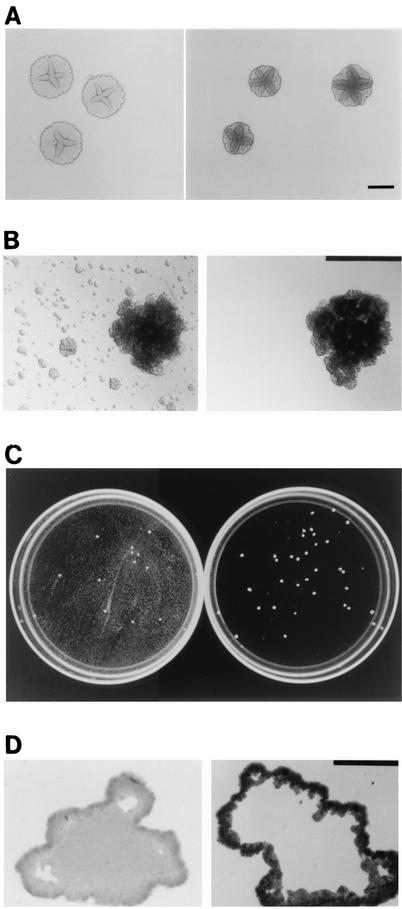
Colonies of A. actinomycetemcomitans grown on nutrient agar plates exhibit a complex morphology characterized by a rough surface texture, an irregular edge, a star-shaped internal structure, and invasive growth and pitting of the agar surface (2, 8, 11, 13, 20) (Fig. 3A, left panel). We reasoned that some components of the A. actinomycetemcomitans colony morphology on agar might reflect components of the biofilm colony morphology observed in broth and that mutants that exhibit gross alterations in their colony morphology on agar may exhibit corresponding alterations in their biofilm colony morphology and may therefore be good candidates for biofilm developmental mutants. We used transposon IS903φkan (32), which carries a cryptic kanamycin resistance (Kmr) gene that is expressed only after successful transposition into an actively transcribed gene, to mutagenize A. actinomycetemcomitans strain CU1000N (14), a spontaneous nalidixic acid-resistant variant of strain CU1000 that exhibits the same surface attachment, biofilm formation, and biofilm dispersal phenotypes as the parent strain (15) (see below). We selected three Kmr mutants (out of ≈1,000) that exhibited a colony morphology on agar that was rougher than the wild-type A. actinomycetemcomitans rough-colony phenotype (Fig. 3A, right panel). Figure 3B shows the biofilm colony morphology and dispersal phenotype in broth for strain CU1000N (left panel) and for one of these rough-colony mutants (JK1017; right panel). Strain JK1017 formed tightly adherent biofilm colonies that were similar in appearance to those of strain CU1000N but which failed to disperse in broth. Figure 3C shows 100-mm-diameter petri dishes inoculated with a small inoculum of strain CU1000N (left panel) and of mutant strain JK1017 (right panel). In the petri dish inoculated with strain CU1000N, satellite colonies appeared as a haze that covers the surface of the dish. This haze was absent from the dish inoculated with strain JK1017. Figure 3D shows thin sections of 2-day-old biofilm colonies of strains CU1000N (left panel) and JK1017 (right panel). Colonies of strain JK1017 were similar in size and shape to those of strain CU1000N but lacked internal, nonaggregated cells. The thin section shown in Fig. 3D (right panel) is representative of approximately 25 other JK1017 colonies examined, all of which displayed nearly identical morphologies. The agar and biofilm colony morphologies and biofilm dispersal phenotypes of the two other rough-colony mutants that we isolated (JK1002 and JK1022) were identical to those of strain JK1017 (data not shown).
FIG. 3.
Phenotypes of A. actinomycetemcomitans wild-type strain CU1000N (left side of panels) and biofilm dispersal mutant JK1017 (right side of panels). (A to C) Shown are colony morphology on agar (bar = 2 mm) (A), biofilm colony morphology in broth (bar = 1 mm) (B), and growth in 100-mm-diameter petri dishes (C), all after 3 days of growth. (D) Thin sections of 2-day-old biofilm colonies grown on polystyrene (bar = 100 μm).
Characterization of A. actinomycetemcomitans biofilm dispersal mutants.
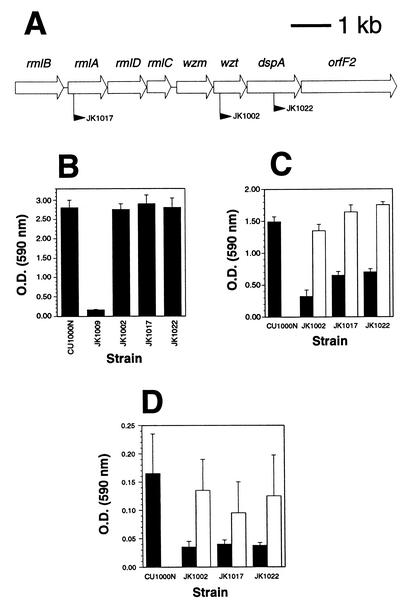
We mapped the transposon insertion sites in the three biofilm dispersal mutants by using inverse PCR (19, 33). The mapping of the transposon insertions in two of these mutants (JK1002 and JK1022) has been described in a previous study (19). All three insertions were located in genes necessary for the synthesis of the serotype f-specific O polysaccharide (O-PS) component of lipopolysaccharide in strain CU1000 (Fig. 4A) (19). These genes included rmlA, encoding glucose-1-phosphate thymidylyl transferase, which catalyzes the first step in the biosynthesis of dTDP-l-rhamnose, the activated nucleotide sugar precursor of A. actinomycetemcomitans O-PS carbohydrates (24, 36); wzt, encoding the hydrophilic ATPase component of an ATP-binding cassette (ABC) membrane transporter involved in the export and assembly of lipopolysaccharide components (5); and dspA, encoding a putative glycosyltransferase (19). All three biofilm dispersal mutants exhibited wild-type surface adherence properties in broth (Fig. 4B), reduced or absent O-PS antigenic side chains (Fig. 4C, filled bars) (19), and reduced biofilm cell detachment as measured by a 96-well biofilm detachment assay (Fig. 4D, filled bars).
FIG. 4.
Characterization of A. actinomycetemcomitans biofilm dispersal mutants. (A) Genetic map of an 8.7-kb region of the gene cluster responsible for the synthesis of serotype f-specific O-PS in strain CU1000 (19). Open arrows indicate open reading frames and direction of transcription. Gene names are shown above the arrows, and the filled arrowheads indicate the location and direction of transcription of IS903φkan insertions in three different mutant strains. (B) Adherence to polystyrene was measured using a 96-well microtiter plate binding assay (15). Adherence is proportional to the optical density (O.D.) values; mean values plus standard errors for triplicate samples are shown. Strain JK1009 contains a transposon insertion in flp-1 which results in complete loss of surface attachment (15). (C) Synthesis of O-PS in wild-type and mutant strains. O-PS was detected with an enzyme-linked immunosorbent assay using anti-CU1000 rabbit antiserum as previously described (19). The amount of O-PS is proportional to the O.D. values; mean values plus standard errors for triplicate samples are shown. Filled bars indicate the levels of O-PS from strains carrying vector plasmid pJAK16, and open bars indicate the levels from mutants JK1002, JK1017, and JK1022 carrying complementing plasmids pJK595, pJK597, and pJK596, respectively. (D) Detachment of cells from biofilm colonies grown on polystyrene rods as measured by a 96-well biofilm detachment assay. Cell detachment is proportional to the O.D. values; mean results plus standard errors for 8 to 15 wells for each strain are shown. Bars are as described for panel C.
To demonstrate that O-PS is directly involved in the detachment process, we cloned the wild-type rmlA, wzt, and dspA genes downstream from an inducible promoter on a broad-host-range plasmid and then introduced these recombinant plasmids into the mutant strains by conjugation with an Escherichia coli donor strain. Plasmids carrying complementing wild-type rmlA, wzt, and dspA genes (pJK597, pJK595, and pJK596, respectively) restored the ability of mutant strains to synthesize O-PS (Fig. 4C, open bars) and release cells into broth (Fig. 4D, open bars). These findings indicate that synthesis of serotype f-specific O-PS is necessary for detachment of cells from A. actinomycetemcomitans CU1000N biofilm colonies.
DISCUSSION
The morphology of A. actinomycetemcomitans biofilm colonies revealed in the present study was similar to that of the tower- and mushroom-shaped biofilm colonies commonly observed in other bacteria, including Pseudomonas aeruginosa, P. fluorescens, Vibrio parahaemolyticus (22), P. putida (4), V. cholerae (34), and Salmonella spp. (28). In addition, our results indicate that A. actinomycetemcomitans biofilm colonies exhibit a distinct, phenotypic life cycle characterized by adherence of planktonic cells to a surface, the growth of asymmetric, lobed microcolonies that display complex morphological features, and the subsequent release of cells from the biofilm colony.
Our findings constitute the first report of defined bacterial mutants that are deficient in biofilm cell detachment. We showed that genes required for biofilm detachment in A. actinomycetemcomitans were not required for surface attachment or biofilm colony formation, indicating that biofilm detachment is a distinct process. Our findings suggest that this process involves the formation of nonaggregated cells inside the biofilm colony that are destined for release from the colony. This proposed detachment mechanism differs from the two previously proposed mechanisms of biofilm cell detachment, namely, erosion (the continuous release of single cells or small clusters of cells) and sloughing (the rapid detachment of large portions of the biofilm) (31).
Our results demonstrate that the synthesis of O-PS is required for A. actinomycetemcomitans biofilm cell detachment but not for surface attachment or biofilm colony formation. O-PS has been shown to play a role in biofilm development in other bacteria. Surface attachment and biofilm formation are decreased in O-PS mutants of E. coli (10), V. cholerae (25), and Serratia marcescens (27) but are increased in O-PS mutants of P. aeruginosa (23) and P. fluorescens (35). These latter findings suggest that biofilm cells of A. actinomycetemcomitans O-PS mutants may fail to detach because they are hyperattached to the inside of the colony. O-PS has also been shown to be necessary for multicellular development in Myxococcus xanthus, suggesting that O-PS could mediate cell-to-cell and cell-to-substratum interactions that are critical for cellular differentiation (3). These findings raise the possibility that biofilm growth and detachment in A. actinomycetemcomitans may be controlled by a genetically regulated developmental pathway (26). It is also possible that defects in O-PS may have pleiotropic effects on other extracellular structures, such as pili or fimbriae (10), which are required for biofilm development and biofilm cell detachment in A. actinomycetemcomitans.
Acknowledgments
We thank David Figurski for helpful comments and Rupal Shah and Aseel Toni for technical assistance.
REFERENCES
- 1.Berthold, P., D. Forti, I. R. Kieba, J. Rosenbloom, N. S. Taichman, and E. T. Lally. 1992. Electron immunocytochemical localization of Actinobacillus actinomycetemcomitans leukotoxin. Oral Microbiol. Immunol. 7:24-27. [DOI] [PubMed] [Google Scholar]
- 2.Blix, I. J. S., H. R. Preus, and I. Olsen. 1990. Invasive growth of Actinobacillus actinomycetemcomitans on solid medium (TSBV). Acta Odontol. Scand. 48:313-318. [DOI] [PubMed] [Google Scholar]
- 3.Bowden, M. G., and H. B. Kaplan. 1998. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 30:275-284. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, B. B., C. Sternberg, J. B. Andersen, L. Eberl, S. Moller, M. Givskov, and S. Molin. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fine, D. H., D. Furgang, and M. L. Barnett. 2001. Comparative antimicrobial activities of antiseptic mouthrinses against isogenic planktonic and biofilm forms of Actinobacillus actinomycetemcomitans. J. Clin. Periodontol. 28:697-700. [DOI] [PubMed] [Google Scholar]
- 7.Fine, D. H., D. Furgang, J. B. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063-1076. [DOI] [PubMed] [Google Scholar]
- 8.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 9.Fine, D. H., P. Goncharoff, H. Schreiner, K. M. Chang, D. Furgang, and D. H. Figurski. 2001. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch. Oral Biol. 46:1065-1078. [DOI] [PubMed] [Google Scholar]
- 10.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the dsbA gene affecting Escherichia coli biofilm formation. FEMS Microbiol. Lett. 173:403-409. [DOI] [PubMed] [Google Scholar]
- 11.Haase, E. M., J. L. Zmuda, and F. A. Scannapieco. 1999. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect. Immun. 67:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt, S. C., A. C. R. Tanner, and S. S. Socransky. 1980. Morphology and ultrastructure of oral strains of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. Infect. Immun. 30:588-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inouye, T., H. Ohta, S. Kokeguchi, K. Fukui, and K. Kato. 1990. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 69:13-18. [DOI] [PubMed] [Google Scholar]
- 14.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, first representative of a new pilin gene subfamily, is required for nonspecific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 16.Kagermeier, A. S., and J. London. 1985. Actinobacillus actinomycetemcomitans strains Y4 and N27 adhere to hydroxyapatite by distinctive mechanisms. Infect. Immun. 47:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan, A. H., D. J. Weber, E. Z. Oddone, and J. R. Perfect. 1989. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev. Infect. Dis. 11:46-63. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, J. B., and D. H. Fine. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl. Environ. Microbiol. 68:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, J. B., M. B. Perry, L. L. MacLean, D. Furgang, M. E. Wilson, and D. H. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 69:5375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, E. O., and H. W. Tatum. 1962. Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. J. Infect. Dis. 111:85-94. [DOI] [PubMed] [Google Scholar]
- 21.Lai, C.-H., M. A. Listgarten, and B. F. Hammond. 1981. Comparative ultrastructure of leukotoxic and non-leukotoxic strains of Actinobacillus actinomycetemcomitans. J. Periodontal Res. 16:379-389. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, J. R., D. R. Korber, B. D. Hoyle, J. W. Costerton, and D. E. Caldwell. 1991. Optical sectioning of microbial biofilms. J. Bacteriol. 173:6558-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makin, S. A., and T. J. Beveridge. 1996. The influence of A-band and B-band lipopolysaccharides on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology 142:299-307. [DOI] [PubMed] [Google Scholar]
- 24.Nakano, Y., N. Suzuki, Y. Yoshida, T. Nezu, Y. Yamashita, and T. Koga. 2000. Thymidine diphosphate-6-deoxy-l-lyxo-4-hexulose reductase synthesizing dTDP-6-deoxy-l-talose from Actinobacillus actinomycetemcomitans. J. Biol. Chem. 275:6806-6812. [DOI] [PubMed] [Google Scholar]
- 25.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiß, and J. Reidl. 2001. Characterization of Vibrio cholerae El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 27.Palomar, J., A. M. Leranoz, and M. Viñas. 1995. Serratia marcescens adherence: the effect of O-antigen presence. Microbios 81:107-113. [PubMed] [Google Scholar]
- 28.Prouty, A. M., W. H. Schwesinger, and J. S. Gunn. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70:2640-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosan, B., J. Slots, R. J. Lamont, M. A. Listgarten, and G. M. Nelson. 1988. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol. Immunol. 3:58-63. [DOI] [PubMed] [Google Scholar]
- 30.Slots, J., H. S. Reynolds, and R. J. Genco. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect. Immun. 29:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoodley, P., S. Wilson, L. Hall-Stoodley, J. D. Boyle, H. M. Lappin-Scott, and J. W. Costerton. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triglia, T., M. Peterson, and D. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, V., and M. Fletcher. 1996. Pseudomonas fluorescens adhesion and transport through porous media are affected by lipopolysaccharide composition. Appl. Environ. Microbiol. 62:100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida, Y., Y. Nakano, T. Nezu, Y. Yamashita, and T. Koga. 1999. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-d-fucose. J. Biol. Chem. 274:16933-16939. [DOI] [PubMed] [Google Scholar]
- 37.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]