Abstract
Expression of the pspABCDE operon of Escherichia coli is induced upon infection by filamentous phage and by many other stress conditions, including defects in protein export. Expression of the operon requires the alternative sigma factor σ54 and the transcriptional activator PspF. In addition, PspA plays a negative regulatory role, and the integral-membrane proteins PspB and PspC play a positive one. In this study, we investigated whether the suggested protein-protein interactions implicated in this complex regulatory network can indeed be demonstrated. Antisera were raised against PspB, PspC, and PspD, which revealed, in Western blotting experiments, that PspC forms stable sodium dodecyl sulfate-resistant dimers and that the hypothetical pspD gene is indeed expressed in vivo. Fractionation experiments showed that PspD localizes as a peripherally bound inner membrane protein. Cross-linking studies with intact cells revealed specific interactions of PspA with PspB and PspC, but not with PspD. Furthermore, affinity-chromatography suggested that PspB could bind PspA only in the presence of PspC. These data indicate that regulation of the psp operon is mediated via protein-protein interactions.
Infection of Escherichia coli with filamentous bacteriophage f1 induces strongly the expression of the pspABCDE (phage-shock protein) operon (7; reviewed in reference 26). The operon encodes at least four proteins. PspA, encoded by the first gene of the operon, is a 26-kDa protein localized both peripherally bound to the inner membrane and in the cytosol (7, 21). PspB is anchored in the inner membrane by a hydrophobic N-terminal segment and has a C-terminal cytoplasmic domain (8, 20). The product of the third gene, PspC, consists of an N-terminal cytoplasmic domain, a single transmembrane segment, and a periplasmic C-terminal domain containing a leucine zipper motif (20). The putative product of the fourth open reading frame, PspD, has not been detected yet, possibly because it would comigrate with PspB in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8). The sequence of the putative PspD protein does neither reveal a signal sequence nor a transmembrane segment, suggesting that the protein would be cytoplasmic if the gene were expressed. PspE contains a signal sequence and localizes in the periplasm (8, 20, 25). We recently demonstrated that PspE is a rhodanese-related enzyme (1).
It has been demonstrated that expression of the gene IV product of filamentous phage leads to the induction of the psp operon (7, 19). The gene IV product is a member of the secretin family and forms large multimeric channels in the outer membrane, through which the phage is extruded. Expression of other members of the secretin family, involved in type II or type III protein secretion, also led to the induction of the psp operon in E. coli (3, 15, 23). Furthermore, various other stress conditions, including extreme heat shock, osmotic shock (7), exposure to organic solvents (22) or ionophores, and prolonged incubation under alkaline conditions (34), have been reported to be inducing stimuli. In our laboratory, it has been observed that the psp operon is induced under conditions that are known to block the protein export apparatus in the inner membrane. For example, expression of a mutant form of outer membrane protein PhoE, containing a stretch of eight consecutive hydrophobic amino acids in its mature domain, led to a strong induction of the operon (21). Quantitative in vivo determination of the membrane potential across the inner membrane demonstrated that the proton motive force (PMF) specifically decreased in a pspA mutant strain upon expression of this mutant PhoE protein (20). This result suggested that PspA is involved in the maintenance of the PMF, which may dissipate by proton leakage when the Sec channel is obstructed. It is not clear whether all the different stimuli for induction of psp expression interfere with protein translocation, but all of them probably injure the cell membrane integrity. Therefore, PspA may play a general role in maintaining the integrity of the inner membrane, rather than being directly involved in protein translocation.
Transcription of the pspABCDE operon is initiated from a promoter located upstream of the pspA gene by σ54-containing RNA polymerase (8). Expression of σ54-dependent genes generally requires an activator of the enhancer-binding protein (EBP) family (28, 35). Such a protein binds to upstream activating sequences and is usually inactive until it is modified in its N-terminal regulatory domain either by specific phosphorylation or by the binding of an effector molecule. The pspF gene, which is located upstream of the pspABCDE operon and transcribed in the opposite direction, encodes the activator of the psp operon (18). Although PspF belongs to the EBP family of activators, the protein lacks an N-terminal regulatory domain, and it is constitutively active both in vivo as in vitro (17). PspF binds to sites overlapping its promoter, thereby regulating its own expression, and its production is not affected by stimuli that induce the pspABCDE operon (16). Thus, regulation of the expression of the pspABCDE operon appears to be different from the EBP modification pathway used by other σ54-dependent systems.
In addition to PspF, other proteins appear to affect the transcription of the psp operon. It has been shown that psp expression is negatively regulated by PspA (32), but this protein does not bind DNA (10). Recently, it has been demonstrated that PspA directly interacts with PspF to inhibit transcription (11). The PspB and PspC proteins act cooperatively as positive regulators of the operon, possibly by relieving the PspA-mediated repression (8, 32, 33). Interestingly, whereas PspB and PspC are not necessary for psp expression during heat shock, they are absolutely required for induction of the operon upon phage infection and only partially required for induction by osmotic shock and ethanol treatment (32). Taken together, the regulatory circuit of the psp operon appears to be very complex and probably involves several protein-protein interactions. In this study, we sought evidence for this hypothesis by identifying such interactions.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The E. coli K-12 strain CE1224 carries a chromosomal deletion including the phoE gene (31), and its derivatives CE1343 (21), CE1417, and CE1418 (20) carry a kanamycin resistance cassette in the pspA, pspB, and pspC genes, respectively. Strains DH5α (14) and SG13009, which carries plasmid pREP4 (Qiagen) encoding the lac repressor and a kanamycin resistance marker, were used for cloning. Cells were grown at 37°C in L broth (31) supplemented with 0.4% glucose or in a synthetic phosphate-limited medium (30). When appropriate, ampicillin (100 μg/ml) or kanamycin (25 μg/ml) was added to the media for plasmid maintenance. To evaluate their sensitivity to bile salts, cells were streaked on BBL MacConkey (Becton Dickinson) plates, followed by incubation at 30°C for 24 h.
Plasmids pJP379, pJP380, and pJP381 (20), derived from plasmid pJF119HE (13), harbor pspB, pspABCDE, and pspA, respectively, under control of the tac promoter. Plasmid pMR05H2 encodes a mutant PhoE protein with an insertion of eight consecutive hydrophobic amino acids (2). Its expression leads to a strong induction of the phage-shock response (21).
For the construction of a glutathione S-transferase (GST) fusion with PspA, primers HA-005 (5′-GAGGATCCTATGGGTATTTTTTCTCGCTT-3′) and HA-006 (5′-TGGCCCGGGTTATTGATTGTCTTGCTTCA-3′) were used for amplification of the pspA gene. The PCR was performed with Pwo polymerase (Roche) and pJP380 as the template, with an annealing temperature of 60°C for 60 s and extension at 68°C for 60 s in 30 cycles. The resulting PCR product was agarose gel purified; digested with BamHI and SmaI, for which sites were contained in the primers (underlined); and cloned into the corresponding sites of the GST fusion vector pRP269 (29). After sequencing, it appeared that the pspA part of the construct was truncated, resulting in the expression of a GST-PspA polypeptide in which the last 32 amino acid residues of PspA were replaced by the amino acids GSTSMHKLEFIVTD, encoded by the expression vector pRP269. Since no other clones were obtained, expression of a full-length GST-PspA hybrid may be lethal. The construct obtained was designated pGST-PspAΔC.
Plasmids pGST-PspBcyt, pGST-PspCpp, and pHis-PspC were constructed as follows. Primers HA-011 (5′-CTGGATCCTAGCAATCGTTCTGGTCGCAG-3′) and HA-002 (5′-TGCCCCGGGTTAGCGATCCCTCCAGTTCG-3′) were used for the amplification of a pspB fragment encoding the cytoplasmic part of PspB (amino acid residues 25 to 74), primers HA-036 (5′-TCATTTGCGCTGGATCCAATGCC-3′) and HA-004 (5′-TAACCCGGGTCACAGTTGACGGAAACGGC-3′) were used for the amplification of a pspC fragment encoding the periplasmic part of PspC (amino acid residues 65 to 119), and primers HA-003 (5′-GGGGATCCAATGGCGGGCATTAATCTCAA-3′) and HA-004 were used to amplify the full-length pspC gene. These primers contain BamHI or SmaI sites (underlined). The PCRs with Taq polymerase and pJP380 as the template were performed with an annealing temperature of 50°C for 60 s and extension at 68°C for 60 s in 30 cycles. The resulting PCR products were cloned in PCR II TOPO (Invitrogen) according to the manufacturer's instructions, resulting in TOPO-PspBcyt, TOPO-PspCpp, and TOPO-PspC, respectively. From these plasmids, the relevant fragments were excised with BamHI and SmaI and ligated into the corresponding sites either of pRP269, yielding pGST-PspBcyt and pGST-PspCpp, or of plasmid pQE31 (Qiagen), resulting in pHis-PspC.
The nucleotide sequences of all constructs resulting from PCR amplifications were confirmed by sequence analysis on an ABI 377 sequencer with the Dye-Terminator kit (Perkin-Elmer).
Purification of GST fusion proteins.
Overnight cultures of strain CE1343 carrying pGST-PspAΔC, pGST-PspBcyt or pGST-PspCpp, were diluted 1:20 in 800 ml of L broth supplemented with ampicillin and glucose and incubated at 37°C until an optical density at 660 nm (OD660) of 0.6 was reached. Then, isopropyl-β-d-thiogalactopyranoside (IPTG) (100 μM final concentration) was added and incubation was continued for 2.5 h at 37°C. The cells were chilled on ice and collected by centrifugation at 5,000 × g for 10 min at 4°C. The supernatant was removed, and the pellet was either resuspended as described below or stored at −20°C. All subsequent steps for purification of the GST fusion proteins were done at 0 or 4°C. The pellet was resuspended in 20 ml of a solution containing 30 mM Tris-HCl (pH 8.0), 25% sucrose, and 1 mM dithiothreitol (DTT) supplemented with Complete protease inhibitor (Roche) according to the manufacturer's instructions. The cell suspension was disintegrated in a French press twice at 8,000 lb/in2. The intact cells and cell envelopes were removed by centrifugation at 9,600 × g for 20 min and at 113,600 × g for 90 min, respectively. The supernatant was loaded, at a flow rate of 1 ml/min, onto a 10-ml GST-affinity column (Pharmacia) equilibrated with mouse tonicity phosphate-buffered saline (MTPBS) (150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4 [pH 7.3]) (29) supplemented with 1% Triton X-100. The column material was washed with 50 ml of MTPBS buffer without Triton X-100, at a flow rate of 2 ml/min. The GST fusion proteins were eluted with MTPBS containing 125 mM glutathione. After dialysis overnight against MTPBS, fractions containing GST fusion protein were stored in aliquots at −20°C at a protein concentration of 2 mg/ml. Protein concentrations were estimated from the absorbance at 280 nm in a Unicam UV1 spectrophotometer (Spectronic), assuming an extinction coefficient of 1.0 for a solution with a protein concentration of 1.0 mg/ml.
Antisera.
Polyclonal rabbit antisera were raised against the purified GST-PspBcyt and GST-PspCpp proteins at Eurogentec, Liège, Belgium. Antibodies directed against PspD were obtained by immunizing rabbits with synthetic peptides corresponding to amino acids 1 to 14 (MNTRWQQAGQKVKP) and 60 to 73 (LSRAANKLAQRYKR) of PspD using the Double X system for peptide immunization (Eurogentec).
In vivo cross-linking of Psp proteins.
To induce expression of the chromosomal pspABCDE operon, cells carrying plasmid pMR05H2 were grown under phosphate limitation for 5 h at 30°C as previously described (6) to induce expression of the plasmid-encoded mutant PhoE protein. Alternatively, the Psp proteins were produced from plasmid pJP380 by growing cells in L-broth to the logarithmic phase and then adding 100 μM IPTG to the medium and continuing incubation for 1 h at 37°C. Cells were harvested by centrifugation, washed once with 0.9% NaCl, and resuspended in 125 mM HEPES (pH 7.3) at an OD660 of 1.0. The cell suspension (1.0 ml) was treated with 100 μM dithiobis(succinimydylpropionate) (DSP) (Pierce) at 25°C for 15 min, and the cross-link reaction was quenched with 50 μl of 1 M Tris-HCl (pH 8.0). After incubation for 5 min at 0°C, cells were collected in a microcentrifuge (14,000 × g, 1 min), and proteins were solubilized in sample buffer for 10 min at 100°C. The proteins were separated by SDS-PAGE on 11 or 15% polyacrylamide gels and transferred to nitrocellulose filters (pore size, 0.45 μm; Schleicher and Schuell) using a Mini Trans-Blot cell (Bio-Rad Laboratories). Immunoincubations were performed essentially as described previously (27), using polyclonal antisera directed against PspA (21), PspB, PspC, or PspD. After incubation with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antiserum (Biosource International), blots were developed with 4-chloro-1-naphthol-H2O2 as the substrate or with SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's recommendations.
Extraction of inner membrane proteins.
Cells of CE1224, carrying pJP379, pJP380, or pJP381, were grown aerobically at 37°C in L broth, containing ampicillin, to an OD660 of 0.1. After addition of 100 μM IPTG, growth was continued for 3 h. Inner membrane vesicles (IMVs) were prepared as previously described (9). Subsequently, 10 μl of IMVs (OD280= 25) was extracted with 80 μl of buffer B (50 mM triethanolamine-acetate [pH 7.5], 250 mM sucrose, 3 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride), to which was added 10 μl of 10% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). The solution was tumbled on a rotating wheel for 1 h at room temperature and then centrifuged at 20,000 × g for 45 min at 4°C. Incubation of IMVs of PspABCDE- or PspA-overproducing strains with CHAPS for 15 min at 0°C resulted in hardly any release of PspA, whereas incubation at room temperature for 1 h released PspA more efficiently. The CHAPS extracts were kept at 0°C.
In vitro cross-linking of Psp proteins.
Aliquots (5 μl) of IMVs (OD280 = 1.25) were mixed with 90 μl of 125 mM HEPES. Subsequently, the IMVs were treated either with 5 μl of 4 mM DSP or with 5 μl of 4% formaldehyde for 20 min at 0°C. The cross-linking reaction was quenched with 150 μl of 1 M Tris-HCl (pH 8.0), followed by addition of 250 μl of 20% trichloroacetic acid. After incubation at 0°C for 1 h, proteins were collected by centrifugation in a microcentrifuge for 15 min at maximal speed and washed once with cold acetone. The pellet fraction was air dried and solubilized in sample buffer for 20 min at 37°C. Proteins were separated by SDS-PAGE, followed by Western blotting and immunoincubation with PspA antiserum as described above.
GST-PspA and GST-PspB pull-down assays.
A 50-μl aliquot of 50% (vol/vol) GST-affinity agarose beads, equilibrated with MTPBS, was mixed with 400 μl of MTPBS and 50 μl of purified GST-PspAΔC or GST-PspBcyt. The mixture was tumbled for 1 h at room temperature to allow for binding of the GST fusion proteins to the beads, followed by centrifugation at 500 × g for 1 min, and the supernatant was discarded. Subsequently, the agarose beads were incubated with 40 μl of buffer A and 40 μl of CHAPS extracts from IMVs of cells overproducing PspA, PspB, or PspABCDE. After tumbling for 1 h at room temperature, the agarose beads were washed twice with 100 μl of MTPBS. Proteins were eluted with MTPBS containing 125 mM glutathione and analyzed by SDS-PAGE followed by silver staining (4) or Western blotting with antisera directed against PspA and PspB. N-terminal sequencing by Edman degradation was performed as described previously (12).
Subcellular localization of PspD.
Cells of strain CE1224 carrying pJP380 were grown aerobically at 37°C in L-broth, and expression of Psp proteins was induced as described above. Outer and inner membranes were separated by selective ultracentrifugation as previously described (9). Peripherally bound membrane proteins were extracted from IMVs with a buffer containing 50 mM Tris-HCl-4 M urea (pH 7.8). Protein patterns were analyzed by SDS-PAGE and Western blotting using polyclonal antiserum against PspD.
RESULTS
In vivo cross-linking of PspA and associated proteins.
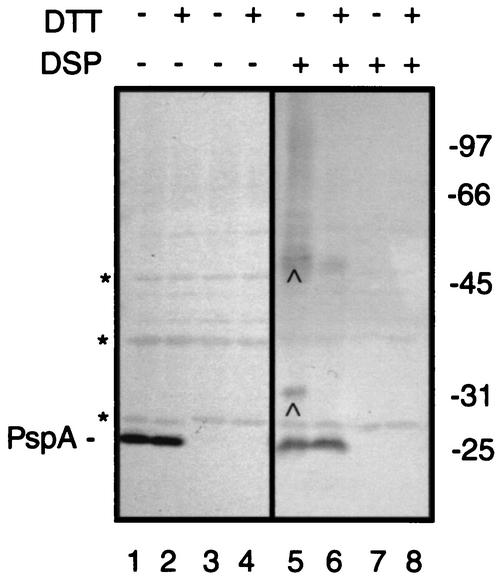
A pilot experiment was conducted to investigate whether PspA interacts with other proteins. Cells of strain CE1224 and of its pspA::kan derivative CE1343, both carrying pMR05H2, were grown under phosphate limitation to induce expression of the plasmid-encoded mutant PhoE protein. Expression of this mutant PhoE protein strongly induces expression of the psp operon (21). After treatment of the cells with the membrane-permeable, thiol-cleavable, homobifunctional cross-linker DSP, cross-linked complexes were analyzed by SDS-PAGE and immunoblotting with a polyclonal antiserum directed against PspA. In addition to monomeric PspA, which migrates with an apparent molecular weight (Mr) of 26,000, several cross-linked protein complexes were observed (Fig. 1, lane 5). One of them migrated with an Mr of approximately 32,000, whereas a broader band migrated with an Mr of about 52,000. In addition, a smear of cross-linked complexes with an Mr larger than 52,000 up to the top of the gel was observed. Neither the monomeric PspA, nor the cross-linked complexes were detected in the case of the pspA mutant strain (Fig. 1, lane 7), demonstrating the specificity of these bands. Furthermore, DSP-dependent cross-linking of proteins is reversible, and all complexes were found to dissociate upon addition of DTT to the sample buffer (Fig. 1, lane 6). However, a small fraction of the 52-kDa adduct in the protein sample remained intact upon boiling in the presence of DTT. This incomplete dissociation of the DSP-induced cross-link was always observed when the adduct was present in large quantities in the protein sample. It was recently reported that PspA can form dimers or higher-ordered oligomers in vivo (10), and the 52-kDa adduct could represent a dimeric form. However, since the band is broad, it may contain several different cross-linked complexes. It is not clear whether the smear with an Mr of >52,000 contains specific cross-linked adducts to PspA or protein aggregates. Since the calculated molecular mass of PspB is ∼7 kDa, the 32-kDa complex could be an adduct of PspA with PspB.
FIG. 1.
Detection of PspA-associated proteins by in vivo cross-linking. Cells of strain CE1224 (lanes 1, 2, 5, and 6) or its pspA::kan derivative CE1343 (lanes 3, 4, 7, and 8), both carrying plasmid pMR05H2, were grown under phosphate limitation to induce the expression of mutant PhoE protein and, consequently, of the psp operon. After incubation of the cells with DSP (lanes 5 to 8), proteins were boiled in sample buffer, with (+) or without (−) DTT as indicated, and separated by SDS-11% PAGE, and this was followed by Western blotting using antibodies directed against PspA. The position of monomeric PspA is indicated. Asterisks depict proteins that nonspecifically react with anti-PspA antiserum. Relevant cross-linked adducts are indicated by carets. Blots were developed with 4-chloro-1-naphthol-H2O2 as the substrate. The positions of molecular mass marker proteins are indicated at the right (in kilodaltons).
Antisera directed against PspB, PspC, and PspD.
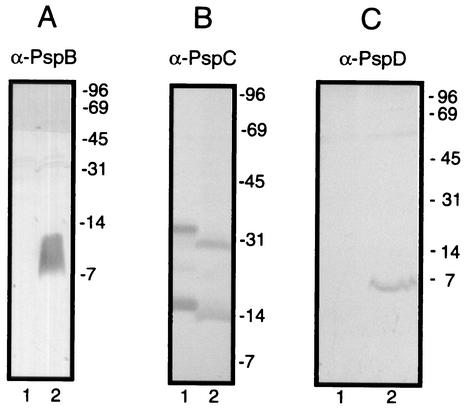
To be able to assess whether the cross-linked complexes contain, in addition to PspA, any of the other proteins encoded by the psp operon, antisera were raised against GST-PspB and GST-PspC fusions and against synthetic PspD peptides. The antiserum directed against PspB reacted specifically with a diffuse band of approximately 8 to 10 kDa in total protein samples of wild-type cells expressing PspB from plasmid (Fig. 2A, lane 2), whereas this band was absent in the pspB::kan strain CE1417 (Fig. 2A, lane 1). The antiserum raised against the GST-PspC fusion recognized two major bands of 14 and 28 kDa, respectively, in inner membranes of cells overexpressing PspABCDE from pJP380 (Fig. 2B, lane 2). Both bands were absent in IMVs of the pspC::kan strain CE1418 (data not shown). Moreover, also two bands, but both with a slightly higher Mr, were detected when inner membranes from cells producing His-tagged PspC protein were analyzed (Fig. 2B, lane 1). Therefore, both bands represent products of the pspC gene. We assume that the 28-kDa band represents a dimeric form of PspC, although it is remarkable that this putative dimer did not dissociate upon boiling in 2% SDS-containing sample buffer supplemented with DTT. The antiserum directed against two synthetic PspD peptides reacted specifically with a band of approximately 7 kDa in total protein samples of wild-type cells expressing the PspABCDE proteins from plasmid, whereas this band was not detectable in total protein samples of cells carrying the vector pJF119HE (Fig. 2C, lanes 1 and 2).
FIG. 2.
Specificity of anti-PspB, anti-PspC and anti-PspD antisera. (A) Total protein samples of pspB::kan strain CE1417 (lane 1) and strain CE1224 expressing PspB from pJP379 (lane 2) were separated by SDS-20% PAGE, and this was followed by immunoblotting with antiserum directed against PspB (α-PspB) (dilution, 1:1,000). (B) IMVs of strain SG13009[pREP4] expressing His-tagged PspC from pHis-PspC (lane 1) and of strain CE1224 expressing PspABCDE from pJP380 (lane 2) were separated by SDS-15% PAGE, and this was followed by immunoblotting with antiserum directed against PspC (α-PspC) (dilution, 1:1,000). (C) Total protein samples of strain CE1224 carrying pJP119HE (lane 1) or expressing PspABCDE from pJP380 (lane 2) were separated by SDS-15% PAGE, and this was followed by immunoblotting with antiserum directed against PspD (α-PspD) (dilution, 1:1,000). Blots were developed with 4-chloro-1-naphthol-H2O2 as the substrate. The positions of molecular mass marker proteins are indicated at the right of each panel (in kilodaltons).
Subcellular localization of PspD.
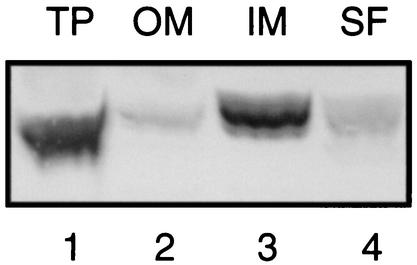
Since the experiments described in the previous paragraph provided the first evidence that the hypothetical pspD gene, which was so far only recognized as an open reading frame, indeed encodes a protein, it was of interest to determine the subcellular localization of this protein. The soluble fraction, inner membranes, and outer membranes of pJP380-containing cells expressing all proteins of the pspABCDE operon were isolated and analyzed by SDS-PAGE and Western blotting. Although the PspD protein sequence does not reveal any stretch of hydrophobic amino acid residues long enough to span the membrane, the protein appeared to be present mostly in the inner membrane fraction (Fig. 3, lane 3). When these membranes were extracted with 4 M urea, the PspD protein was partly recovered in the urea-soluble fraction (results not shown). Together, these data indicate that PspD is a peripherally bound inner membrane protein.
FIG. 3.
Subcellular localization of PspD. Cells of strain CE1224 carrying pJP380 were grown in L-broth, and expression of the plasmid-encoded psp genes was induced by the addition of 100 μM IPTG. After 3 h of incubation, cells were harvested and either directly analyzed (total proteins [TP]) or fractionated. After subjecting the cells to a French press, the outer membrane (OM) and inner membrane (IM) fractions and the soluble fraction (SF) (i.e., the combined periplasmic and cytoplasmic fraction) were obtained by differential ultracentrifugation. The fractions obtained were analyzed by SDS-PAGE followed by Western blotting with anti-PspD antiserum. The blot was developed with 4-chloro-1-naphthol-H2O2 as the substrate.
Complex formation in vivo of overproduced Psp proteins.
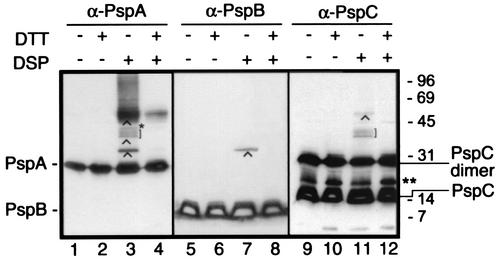
To obtain a high cross-linking efficiency, cross-linking experiments were conducted with cells that overproduced the PspABCDE proteins from plasmid. CE1224 cells containing pJP380 were induced with IPTG and treated with DSP. Analysis by Western blotting with antibodies directed against PspA revealed again the 32-kDa complex, the broader band of ∼52 kDa, and the smear of >52 kDa (Fig. 4, lane 3). In addition, three specific cross-linked protein complexes with Mrs of about 40,000 to 45,000 were detected (Fig. 4, lane 3). Blots were also incubated with the antisera directed against PspB and PspC. The PspB antiserum reacted with the 32-kDa band in the cross-linked sample (Fig. 4, lane 7), while the PspC antiserum reacted with two of the three cross-linked complexes with Mrs of about 40,000 to 45,000 and (faintly) with a cross-linked complex of ∼60 kDa (Fig. 4, lane 11). We assume that the ∼60-kDa complex represents a PspC dimer-PspA complex. Incubation of the blots with PspD antiserum demonstrated the presence of monomeric PspD but did not reveal PspD-containing cross-linked adducts (data not shown). Since the spacer arm of the cross-linker DSP is fairly long (12 Å), the identification of cross-linked complexes reflects a close proximity, but not necessarily a direct interaction between the cross-linked partners. Therefore, similar experiments were performed with formaldehyde as the cross-linking agent. Formaldehyde induces a one-atom bridge between neighboring proteins. Comparison of DSP- and formaldehyde-induced cross-linking revealed very similar cross-linking patterns (results not shown). Taken together, these data demonstrate that PspA interacts with PspB and PspC in vivo, but not with PspD.
FIG. 4.
Complex formation of Psp proteins. Cells of strain CE1224, expressing PspABCDE from plasmid pJP380, were treated with DSP followed by SDS-PAGE and Western blotting with anti-PspA (α-PspA), anti-PspB (α-PspB), and anti-PspC (α-PspC) antisera. Where indicated, samples were boiled in the presence of DTT prior to SDS-PAGE to reverse the cross-linking. The positions of monomeric PspA, PspB, and PspC and the dimeric form of PspC are indicated. Relevant cross-linked complexes are indicated by carets, square brackets, and a single asterisk. The double asterisk indicates a band that reacts specifically with anti-PspC antiserum, but the exact composition is unknown. The positions of the molecular mass marker proteins are indicated at the right (in kilodaltons). Blots were developed with SuperSignal West Pico chemiluminescent substrate.
In vitro binding of proteins to GST-PspA and GST-PspB.
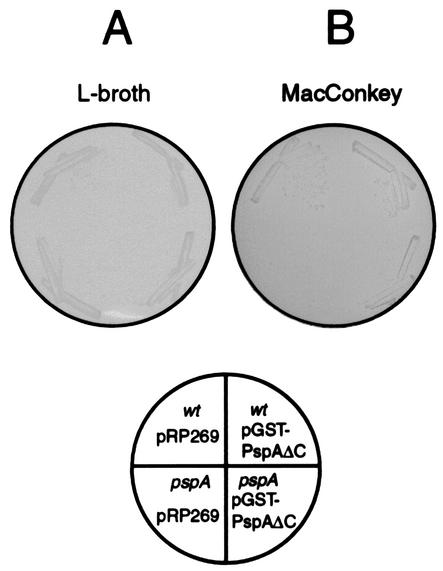
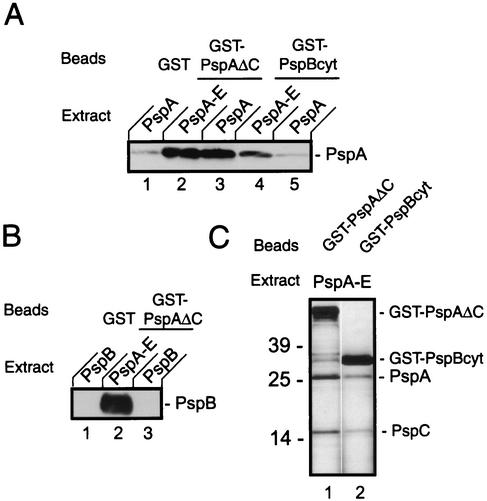
Since the detection of complexes by cross-linking is limited by the availability and the proximity of reactive groups, we also employed a GST pull-down assay to analyze interactions between Psp proteins. A GST-PspA fusion was genetically constructed which, after sequencing, appeared to lack the last 32 amino acids of PspA. To test whether the truncated GST-PspA fusion is functional in vivo, we made use of the observation that the pspA::kan strain CE1343, in contrast to its parental strain CE1224, is unable to grow on MacConkey plates, probably due to the presence of bile salts in this medium, which could cause membrane damage (Fig. 5). The growth defect of strain CE1343 in the MacConkey medium was largely suppressed by introduction of plasmid pGST-PspAΔC. Since the GST-PspAΔC construct was apparently functional and also soluble, in contrast to, for example, a full-length His-tagged PspA, which fractionated with the inner membrane (unpublished observation), we decided to use this construct in the GST pull down assays. Furthermore, the cytoplasmic domain of PspB was fused to GST, yielding the chimeric protein GST-PspBcyt. The purified proteins GST-PspAΔC and GST-PspBcyt were bound to GST-affinity beads and incubated with CHAPS-solubilized inner membranes of PspABCDE-, PspA-, or PspB-overproducing strains. After extensive washing, the beads were eluted with glutathione, and proteins coeluting with GST-PspAΔC and GST-PspBcyt from the beads were identified by SDS-PAGE and Western blotting. Some nonspecific binding of PspA to beads charged with GST was observed (Fig. 6A, lane 1). However, when CHAPS-solubilized inner membranes from a PspABCDE-overproducing strain were incubated with beads charged either with GST-PspAΔC or with GST-PspBcyt, much more efficient binding of PspA was found (Fig. 6A, lanes 2 and 4, respectively). Similarly, PspA from cells overproducing only PspA bound to GST-PspAΔC (Fig. 6A, lane 3), confirming that PspA can form oligomers as reported previously (10). Remarkably, whereas PspA from PspABCDE-overproducing cells did bind to GST-PspBcyt (Fig. 6A, lane 4), PspA of cells overproducing only PspA did not (Fig. 6A, lane 5), suggesting that another Psp protein is required to mediate binding of PspA to PspB. In agreement with this observation, PspB did bind to GST-PspAΔC, when it was extracted from the IMVs of a PspABCDE-overproducing strain (Fig. 6B, lane 2) but not when it was extracted from cells overproducing PspB only (Fig. 6B, lane 3). Considering the localization of PspE in the periplasm, we suspect that PspC is the additional Psp protein required for PspA-PspB binding, but we cannot exclude the possibility that PspD is required for this binding. In support of this hypothesis, silver staining of the gels revealed a band with an Mr of 16,000 when extracts of IMVs of PspABCDE-overproducing cells were bound to GST-PspAΔC and GST-PspBcyt-charged beads (Fig. 6C, lanes 1 and 2, respectively). N-terminal sequencing of the protein identified the band as PspC. In conclusion, these data confirm interactions between PspA and PspB and between PspA and PspC and suggest that PspA binds to PspB only in the presence of another Psp protein, probably PspC.
FIG. 5.
Truncated GST-PspA fusion complements pspA mutant CE1343 for growth on MacConkey plates. Cells of wild-type (wt) strain CE1224 and strain CE1343 (pspA), carrying either vector plasmid pRP269 or pGST-PspAΔC, were streaked on L-broth (A) and MacConkey (B) plates containing ampicillin. The plates were incubated at 30°C for 24 h, and the results were photographed.
FIG. 6.
GST-PspA and GST-PspB pull-down assays. Purified GST, GST-PspAΔC, or GST-PspBcyt was bound on a GST-affinity resin, and CHAPS-solubilized IMVs from cells of strain CE1224, overexpressing either PspABCDE, PspA, or PspB from pJP380, pJP381, and pJP379, respectively, were added. After extensive washing, the resin was eluted with glutathione, and eluted proteins were analyzed by SDS-PAGE and either Western blotting with antisera directed against PspA (A) or PspB (B) or silver staining (C). Immunoblots were developed with SuperSignal West Pico chemiluminescent substrate. The positions of GST-PspAΔC, GST-PspBcyt, PspA, PspB, and PspC are indicated. Note that the extract of the PspABCDE overproducer contained equal amounts of PspA and PspB, as did the extracts of the PspA overproducer and of the PspB overproducer, respectively, as determined by Western blotting (data not shown).
DISCUSSION
Induction of the psp operon is regulated in a complex manner, requiring PspF and either PspB or PspC, or both, dependent on the specific inducing conditions. Because the operon is also under negative control of PspA (32), induction can be viewed as overcoming the negative regulatory role of PspA. Consistent with this view, overproduction of PspC (32) or of PspF (18) is sufficient to induce psp expression, indicating titration of PspA by protein-protein interactions. Sequence analysis suggests also that PspA and the cytoplasmic domain of PspB have coiled-coil properties (24), whereas PspC has a leucine zipper motif in its periplasmic domain, all indicative for protein-protein interactions. Recently, it has been demonstrated that PspA is the negative regulator of σ54-dependent transcription of the psp operon, which is probably based on interaction with PspF (10, 11). Taken together, these data suggested that regulation of the psp operon occurs via protein-protein interactions.
In this report, we have used an in vivo cross-linking approach to analyze protein-protein interactions between Psp proteins. The use of the chemical cross-linker DSP allowed for the identification of specific interactions between PspA, PspB, and PspC in intact cells. Western blotting revealed interactions of PspA with PspB and with PspC, but not with PspD. In addition, the cross-linking experiments revealed interactions of PspA with other proteins, resulting in cross-linked adducts with Mrs of ∼45,000 and a smear of >52,000, although the interacting components were not identified. A PspA-PspF adduct, with an expected molecular mass of ∼63 kDa, could be present within the smear, but could not be identified due to unavailability of PspF-specific antiserum. Since evidence for an interaction between PspA and PspF has recently been reported (11), we did not pursue demonstrating such interaction. The interactions between PspA and PspB and between PspA and PspC were confirmed by in vitro affinity experiments. Interestingly, the in vitro interaction between PspA and PspB could only be demonstrated when all Psp proteins were overexpressed, indicating that another Psp protein is required. We speculate that PspC, which also interacted with PspB and PspA, is necessary to induce an efficient complex formation between PspA and PspB. An interaction between PspA and PspF was not revealed in the pull-down assays, probably because PspF, which is expected to be a soluble cytoplasmic protein, was not present in the solubilized inner membranes used in these experiments. Alternatively or in addition, PspF might bind to the C-terminal fragment of PspA, which was missing in the GST-PspA fusion used. Previously, it has been reported that a C-terminally truncated PspA failed to repress the psp operon and, hence, was unable to bind PspF (7).
The Psp proteins protect the cell from dissipation of the PMF under stress conditions (20), probably by maintaining the integrity of the inner membrane. A tempting model for the regulatory pathway is that the periplasmically exposed C-terminal part of PspC, which contains a leucine zipper motif, dimerizes in response to a PMF. Upon dissipation of the PMF, the dimer would dissociate, allowing the cytoplasmic part of PspC to interact with PspB. This would induce binding of PspA to PspB, resulting in PspF release from a PspA-PspF complex. Subsequently, PspF activates the σ54-dependent transcription of the pspABCDE operon. Such a PMF-sensing protein has been described previously in mitochondria. It was demonstrated that dimerization of Tim23, a component of the protein-import machinery of mitochondria, is dependent on a leucine-zipper motif, and it was found that Tim23 dimers were formed in response to the membrane potential (5). The postulated PMF-dependent dimerization of PspC will be a subject for further studies, although such a study will be a difficult task considering the extremely stable PspC dimers that were already detected in the present study.
Acknowledgments
We thank Ria van Boxtel for technical assistance.
This work was supported by grant HPRN-CT-2000-00075 of the European Community.
REFERENCES
- 1.Adams, H., W. Teertstra, M. Koster, and J. Tommassen. 2002. PspE (phage-shock protein E) of Escherichia coli is a rhodanese. FEBS Lett. 518:173-176. [DOI] [PubMed] [Google Scholar]
- 2.Agterberg, M., H. Adriaanse, A. van Bruggen, M. Karperien, and J. Tommassen. 1990. Outer-membrane PhoE protein of Escherichia coli K-12 as an exposure vector: possibilities and limitations. Gene 88:37-45. [DOI] [PubMed] [Google Scholar]
- 3.Akrim, M., M. Bally, G. Ball, J. Tommassen, H. Teerink, A. Filloux, and A. Lazdunski. 1993. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol. Microbiol. 10:431-443. [DOI] [PubMed] [Google Scholar]
- 4.Ansorge, W. 1985. Fast and sensitive detection of protein and DNA bands by treatment with potassium permanganate. J. Biochem. Biophys. Methods 11:13-20. [DOI] [PubMed] [Google Scholar]
- 5.Bauer, M. F., C. Sirrenberg, W. Neupert, and M. Brunner. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87:33-41. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, D., J. Leunissen, J. Verbakel, M. de Jong, H. van Erp, and J. Tommassen. 1986. Periplasmic accumulation of truncated forms of outer-membrane PhoE protein of Escherichia coli K-12. J. Mol. Biol. 189:449-455. [DOI] [PubMed] [Google Scholar]
- 7.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage-shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brissette, J. L., L. Weiner, T. L. Ripmaster, and P. Model. 1991. Characterization and sequence of the Escherichia coli stress-induced psp operon. J. Mol. Biol. 220:35-48. [DOI] [PubMed] [Google Scholar]
- 9.de Cock, H., S. van Blokland, and J. Tommassen. 1996. In vitro insertion and assembly of outer-membrane protein PhoE of Escherichia coli K-12 into the outer membrane. Role of Triton X-100. J. Biol. Chem. 271:12885-12890. [DOI] [PubMed] [Google Scholar]
- 10.Dworkin, J., G. Jovanovic, and P. Model. 2000. The PspA protein of Escherichia coli is a negative regulator of σ54-dependent transcription. J. Bacteriol. 182:311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elderkin, S., S. Jones, J. Schumacher, D. Studholme, and M. Buck. 2002. Mechanism of action of the Escherichia coli Phage-shock protein PspA in repression of the AAA family transcription factor PspF. J. Mol. Biol. 320:23-37. [DOI] [PubMed] [Google Scholar]
- 12.El Khattabi, M., P. Van Gelder, W. Bitter, and J. Tommassen. 2000. Role of the lipase-specific foldase of Burkholderia glumae as a steric chaperone. J. Biol. Chem. 275:26885-26891. [DOI] [PubMed] [Google Scholar]
- 13.Fürste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 14.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer-membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 16.Jovanovic, G., J. Dworkin, and P. Model. 1997. Autogenous control of PspF, a constitutively active enhancer-binding protein of Escherichia coli. J. Bacteriol. 179:5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jovanovic, G., J. Rakonjac, and P. Model. 1999. In vivo and in vitro activities of the Escherichia coli sigma54 transcription activator, PspF, and its DNA-binding mutant, PspFΔHTH. J. Mol. Biol. 285:469-483. [DOI] [PubMed] [Google Scholar]
- 18.Jovanovic, G., L. Weiner, and P. Model. 1996. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J. Bacteriol. 178:1936-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazmierczak, B. I., D. L. Mielke, M. Russel, and P. Model. 1994. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J. Mol. Biol. 238:187-198. [DOI] [PubMed] [Google Scholar]
- 20.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage-shock protein A) of Escherichia coli in maintenance of the proton-motive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 21.Kleerebezem, M., and J. Tommassen. 1993. Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol. Microbiol. 7:947-956. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, H., M. Yamamoto, and R. Aono. 1998. Appearance of a stress-response protein, phage-shock protein A, in Escherichia coli exposed to hydrophobic organic solvents. Microbiology 144:353-359. [DOI] [PubMed] [Google Scholar]
- 23.Koster, M., W. Bitter, H. de Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer-membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789-797. [DOI] [PubMed] [Google Scholar]
- 24.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266:513-525. [DOI] [PubMed] [Google Scholar]
- 25.Mielke, D. L., and M. Russel. 1992. A modified TnphoA useful for single-stranded DNA sequencing. Gene 118:93-95. [DOI] [PubMed] [Google Scholar]
- 26.Model, P., G. Jovanovic, and J. Dworkin. 1997. The Escherichia coli phage-shock protein (psp) operon. Mol. Microbiol. 24:255-261. [DOI] [PubMed] [Google Scholar]
- 27.Pettersson, A., P. van der Ley, J. T. Poolman, and J. Tommassen. 1993. Molecular characterization of the 98-kilodalton iron-regulated outer-membrane protein of Neisseria meningitidis. Infect. Immun. 61:4724-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 30.Tommassen, J., and B. Lugtenberg. 1980. Outer-membrane protein e of Escherichia coli K-12 is coregulated with alkaline phosphatase. J. Bacteriol. 143:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tommassen, J., H. van Tol, and B. Lugtenberg. 1983. The ultimate localization of an outer-membrane protein of Escherichia coli K-12 is not determined by the signal sequence. EMBO J. 2:1275-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiner, L., J. L. Brissette, and P. Model. 1991. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 5:1912-1923. [DOI] [PubMed] [Google Scholar]
- 33.Weiner, L., J. L. Brissette, N. Ramani, and P. Model. 1995. Analysis of the proteins and cis-acting elements regulating the stress-induced phage-shock protein operon. Nucleic Acids Res. 23:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiner, L., and P. Model. 1994. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc. Natl. Acad. Sci. USA 91:2191-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, H., and T. R. Hoover. 2001. Transcriptional regulation at a distance in bacteria. Curr. Opin. Microbiol. 4:138-144. [DOI] [PubMed] [Google Scholar]