Abstract
We have cloned a two-component regulatory system (phoR2-phoP2) of Myxococcus xanthus while searching for genes that encode proteins with phosphatase activity, where phoR2 encodes the histidine kinase and phoP2 encodes the response regulator. A second system, phoR3-phoP3, was identified and isolated by using phoP2 as a probe. These two systems are quite similar, sharing identities along the full-length proteins of 52% on the histidine kinases and 64% on the response regulators. The predicted structures of both kinases suggest that they are anchored to the membrane, with the sensor domains being located in the periplasmic space and the kinase domains in the cytoplasm. The response regulators (PhoP2 and PhoP3) exhibit a helix-loop-helix motif typical of DNA-binding proteins in the effector domains located in the C-terminal region. Studies on two single-deletion mutants and one double-deletion mutant have revealed that these systems are involved in development. Mutant fruiting bodies are not well packed, originating loose and flat aggregates where some myxospores do not reshape properly, and they remain as elongated cells. These systems are also involved in the expression of Mg-independent acid and neutral phosphatases, which are expressed during development. The neutral phosphatase gene is especially dependent on PhoP3. Neither PhoP2 nor PhoP3 regulates the expression of alkaline phosphatases and the pph1 gene.
Myxococcus xanthus is a gram-negative bacterium that undergoes a spectacular developmental cycle upon starvation that is unique among the prokaryotes. When nutrients are depleted, cells aggregate in certain points and originate colored macroscopic structures known as fruiting bodies. Inside the fruiting bodies, cells differentiate, and the long and thin vegetative rods become ovoid-resistant cells known as myxospores, which are covered with a thick coat. Myxospores are able to germinate when nutritional conditions are restored (for reviews, see references 5 and 6). In order to culminate the developmental cycle, five signals have to be exchanged by cells (35), and a large number of genes must be expressed in a sequential fashion (14, 17, 18).
During the last decade, it was found that M. xanthus possesses a large family of eukaryotic-style protein kinases (12, 46). Although their exact roles have not been elucidated yet, they most likely participate in signal transduction pathways, as the eukaryotic kinases do. Pkn1, the first M. xanthus kinase that was characterized, is expressed exclusively during the developmental cycle, at the onset of sporulation. This kinase is required for normal development, and deletion of its gene leads to immature fruiting bodies and a low yield of myxospores (28). Other kinases, such as Pkn5 and Pkn6, are expressed at very low levels during vegetative growth, and their expressions peak during development (45). In contrast, Pkn2 is expressed only during vegetative growth, reaching a maximum at the beginning of the stationary phase (39). The levels of Pkn2 decrease upon starvation. Pkn9 is expressed at very high levels during both vegetative growth and the developmental cycle (10). All the kinases so far characterized phosphorylate at either serine or serine and threonine, but none of them phosphorylate at tyrosine, although phosphotyrosine has been identified in M. xanthus (7). The existence of a large number of kinases and the fact that they can be expressed during vegetative growth, development, or both indicate that phosphorylation at serine and threonine must be one of the important events that regulate the peculiar life cycle of this bacterium.
As a counterpart, other proteins must exist with ability to dephosphorylate the substrates phosphorylated by the kinases. Five different phosphatase activities have been reported in M. xanthus, two of which (Mg-dependent acid and alkaline phosphatases) are detected during vegetative growth, while the other three (Mg-independent acid, neutral, and alkaline phosphatases) are detected during development (43). No relationship between these five activities and the serine/threonine kinases has been established, nor is it known what type of proteins is responsible for these activities. It is even possible that each activity is originated by more than one protein. More recently, a protein phosphatase, Pph1, has been identified, and its gene has been cloned (38). This gene is expressed during both vegetative growth and development, but its expression peaks during early aggregation. Pph1 seems to be involved in the frizzy signal transduction system, since the gene was cloned during a search for proteins that interact with FrzZ by using the yeast two-hybrid system. This same method has also revealed that Pph1 interacts with Pkn5 (38). So far, pph1 is the only phosphatase-encoding gene that has been cloned in M. xanthus.
In our laboratory, we have searched for genes that encode proteins with phosphatase activity. An M. xanthus genomic library was used to transform Escherichia coli, and selection of positive colonies was carried out on Luria-Bertani (LB) medium containing BCIP (5-bromo-4-chloro-3-indolylphosphate). BCIP is a substrate that turns blue when cleaved by phosphatases so that positive colonies should exhibit this color (26). By this strategy, we have cloned four different genes. One of them encodes a lipoprotein, MlpB (23), and another encodes a permease for glycerol 3-phosphate, GlpT (26). In the present study, we report the characterization of a third fragment, cloned by this method, which contains an operon carrying two genes designated phoR2 and phoP2. PhoR2 and PhoP2 show similarities to histidine kinases and response regulators, respectively. Furthermore, by Southern blot hybridization with phoP2 as a probe, we have cloned a second two-component system designated PhoR3-PhoP3. These two systems are involved in aggregation, sporulation, and the expression of phosphatases in M. xanthus.
MATERIALS AND METHODS
Materials.
A digoxigenin DNA labeling and detection kit and all molecular biology enzymes were purchased from Roche Molecular Biochemicals. Oligonucleotides were purchased from Gibco BRL, and kits for plasmid preparation were purchased from Roche Molecular Biochemicals and Promega.
Bacterial strains, growth conditions, and plasmids.
M. xanthus DZF1 was routinely grown at 30°C in liquid CTT (11) with shaking. CTT agar plates contained 1.5% Bacto agar (Difco). M. xanthus grows vegetatively on these two media. When necessary, kanamycin (40 μg/ml) or galactose (10 mg/ml) was added. In addition, other media with less nutrients, such as 1/2CTT, 1/5CTT, and 1/10CTT, which contain only 1/2, 1/5, and 1/10 as much Bacto Casitone as CTT agar, respectively, have been used to analyze the phenotypes of mutants.
E. coli DH5 (9) was used for transformation with the M. xanthus genomic library. E. coli DH5α (9) and CL83 (20) were used for routine transformations. E. coli strains were grown at 37°C in LB medium (24), which was supplemented with ampicillin (50 μg/ml), kanamycin (25 μg/ml), and/or BCIP (40 μg/ml), when needed.
pUC19 (41) and pBluescript SK+ (Stratagene) were used for routine cloning. The kanamycin resistance gene was obtained from plasmid pUC7Skm(Pst−), which was kindly provided by S. Inouye (University of Medicine and Dentistry of New Jersey). For construction of a double-deletion mutant, plasmid pKG-2 was used (40). This plasmid contains a positive-negative KG (kanamycin-galactokinase) cassette that allows the construction of multigene deletions by using only the kanamycin resistance gene as a marker.
DNA manipulations and sequencing.
M. xanthus chromosomal DNA was prepared by the method described by Avery and Kaiser (2). Plasmids were prepared by using the kits mentioned above in accordance with the instructions of the manufacturers. Routine techniques for restriction digestion, ligation, and transformation were used (32). DNA probes were labeled with digoxigenin, and hybridization was carried out at 42°C in 50% formamide under the conditions previously reported (13). A kit for color detection with Nitro Blue Tetrazolium and BCIP was used.
Plasmids were introduced into E. coli by transformation and into M. xanthus by electroporation by using the conditions reported by Kashefi and Hartzell (16).
DNA sequencing was performed by using the dideoxy terminator method (33) in an Applied Biosystem 373 DNA sequencer. Appropriate oligonucleotides were designed as primers. Comparison of the deduced protein sequences with others in the databases was performed with the FASTA program, version 3.3t06 (30).
Construction of deletion strains.
Three different deletion mutants have been constructed, namely, ΔphoR2-phoP2, ΔphoR3-phoP3, and ΔphoR2-phoP2-phoR3-phoP3. For deletion of system 2, fragment SmaI(a)-SmaI(b) (Fig. 1A) was cloned into pUC19 digested with SmaI, the proper orientation was selected, and the plasmid was named pAM2S2. On the other hand, fragment ApaI(b)-ApaI(c) (Fig. 1A) was cloned into pBluescript digested with ApaI. The plasmid with the proper orientation was selected and designated pAM2A1. A 2-kb fragment obtained after digestion of plasmid pAM2S2 with EcoRI and HindIII was ligated to pAM2A1 digested with the same enzymes. The resulting construct, designated pAM2AEH, was then digested with SalI and ligated to the kanamycin resistance gene obtained from pUC7SKm(Pst−). This plasmid (pAM2AEH1) contains fragments upstream and downstream of the operon phoR2-phoP2, with 75% of the operon replaced by the kanamycin resistance gene. This final plasmid was linearized with ScaI for electroporation to M. xanthus. Several kanamycin-resistant colonies were selected to confirm the double crossover event by Southern blot hybridization. The ΔphoR2-phoP2 strain was designated JM2.
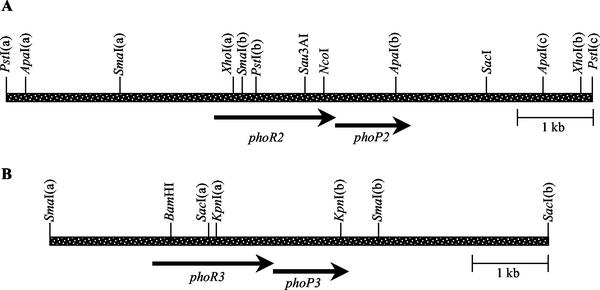
FIG. 1.
Restriction map of DNA fragments containing phoR2-phoP2 (A) and phoR3-phoP3 (B).
For deletion of the system 3, fragments SmaI(a)-BamHI and KpnI(b)-SacI(b) (Fig. 1B) were cloned into pUC19 to originate plasmids pAM3B2 and pAM3K, respectively. pAM3K was digested with HindIII and blunt-ended with T4 DNA polymerase. This linearized plasmid was digested with PstI and ligated to a 1.6-kb PstI-SmaI fragment isolated from pAM3B2 to give plasmid pΔR3P3. This construct was linearized by digestion with PstI and ligated to a kanamycin resistance gene obtained from pUC7SKm(Pst−). In the resulting plasmid, pΔR3P31, a kanamycin resistance gene has replaced 90% of the operon phoR3-phoP3. Electroporation of M. xanthus was carried out with ScaI-digested pΔR3P31. Kanamycin-resistant colonies were confirmed for replacement of the operon by the kanamycin resistance gene as mentioned above. The ΔphoR3-phoP3 strain was designated JM3.
For deletion of both systems, plasmid pΔR3P3 was digested with PstI and ligated to a 4.3-kb fragment obtained by digestion of pKG-2 (40). The construct thus obtained was named pΔR3P3KG2. SacI-linearized pΔR3P3KG2 was introduced into M. xanthus by electroporation, and several kanamycin-resistant colonies were analyzed by Southern blot hybridization, as described above, to confirm the replacement of the phoR3-phoP3 operon by the KG2 system. Removal of the kanamycin resistance and galK genes was carried out in a medium with galactose, as described by Ueki et al. (40). The resulting ΔphoR3-phoP3 kanamycin-sensitive strain, designated JM32, was used to electroporate linearized plasmid pAM2AEH1 (see above) in order to delete system 2. Replacement of system 2 by the kanamycin resistance gene was also confirmed by Southern blot hybridization. The final ΔphoR2-phoP2-phoR3-phoP3 strain was designated JM322.
Developmental conditions.
Fruiting bodies of M. xanthus DZF1, JM2, JM3, and JM322 were obtained on CF, TPM, or MCM medium. CF medium contains 10 mM Tris-HCl (pH 7.6), 1 mM K2NaPO4 (pH 6.8), 8 mM MgSO4, 0.02% (NH4)2SO4, 0.015% Bacto Casitone, 0.2% sodium citrate, 0.1% sodium pyruvate, and 15 g of Bacto agar/liter (8). TPM medium contains 10 mM Tris-HCl (pH 7.6), 1 mM KH2PO4-K2HPO4 (pH 7.5), 5 mM MgSO4 and 15 g of Bacto agar/liter (18). MCM medium contains 10 mM MOPS [3-(N-morpholino)propanesulfonic acid] (pH 7.6), 4 mM MgCl2, 2 mM CaCl2, and 8 g of agarose/liter (31). The main differences between these three media are that CF contains a small amount of nutrients, which allows a few divisions, and MCM contains no phosphate. Cells were grown in liquid CTT at approximately 3 × 108 cells/ml and concentrated to 1.5 × 109 cells/ml in TM buffer (10 mM Tris-HCl, pH 7.6, 1 mM MgSO4). For analysis of aggregation, one unique drop of 10 μl was spotted in the center of a 5.5-cm petri dish containing the appropriate medium. For counting spores, 10 drops of 20 μl each were spotted on a 9-cm petri dish containing MCM medium. At different times, the fruiting bodies of one plate were harvested and resuspended in 200 μl of TM buffer. Fruiting bodies were dispersed, and rod-shaped cells were disrupted by sonication. Myxospores were counted in a Petroff-Hausser chamber.
Studies on germination were carried out on CTT agar plates as previously reported (44).
Transmission and scanning electron microscopy.
For transmission electron microscopy, myxospores were harvested from 72-h fruiting bodies on MCM medium and treated as described by Mueller and Dworkin (27), with slight modifications. Photographs were taken in a Zeiss TEM902 transmission electron microscope at 80 kV. For scanning electron microscopy, 72-h fruiting bodies obtained on MCM medium were fixed with glutaraldehyde vapors for 24 h at room temperature. They were then postfixed with osmium tetroxide under the same conditions described above. Dehydration was accomplished by a graded series of ethanol. Samples were then critical-point dried and sputter coated with gold. Photographs were taken in a Zeiss DSM950 scanning electron microscope.
Determination of enzymatic activities.
For the preparation of cell extracts during development, several 20-μl drops of concentrated cultures containing 4.5 × 109 cell/ml were spotted on CF or MCM medium. After incubation, fruiting bodies were harvested by scraping the surface of the plates, resuspended in 200 μl of glass beads, equilibrated in TM buffer, and sonicated as described previously (28). Cell debris was removed by centrifugation. For preparation of cell extracts during vegetative growth, we followed the same procedure described above, but the medium was CTT agar, and sonication was carried out in the absence of glass beads. The amount of protein in the supernatants was determined by using the Bio-Rad protein assay. Phosphatase activities were determined as described by Weinberg and Zusman (43) by using p-nitrophenyl phosphate as substrate, and β-galactosidase activity was determined as described by Kroos et al. (18).
Nucleotide sequence accession numbers.
The nucleotide sequences of the DNA fragments containing the operons phoR2-phoP2 and phoR3-phoP3 have been deposited in the GenBank database with the accession numbers AF157830 and AF157829, respectively.
RESULTS
Cloning, sequencing, and analysis of the sequences of phoR2 and phoP2 genes.
In our laboratory, we have tried to clone genes that encode proteins with phosphatase activity by using a strategy that can be summarized as follows. An M. xanthus genomic library was constructed by partial digestion of chromosomal DNA with Sau3AI and purification of 3- to 5-kb fragments. These fragments were ligated to pUC19, digested with BamHI, and dephosphorylated. The library was used to transform E. coli, and selection of positive colonies was carried out on LB medium containing BCIP, a substrate of phosphatases that turns blue when it is hydrolyzed (26). One of the positive colonies harbored a plasmid that contained a fragment of approximately 7 kb. When a portion of this fragment was sequenced, we could identify two open reading frames (ORFs), one of them with 609 codons, 93% of which showed either G or C in the third position, a codon preference typical of M. xanthus genes. The other ORF encoded a protein of 238 amino acids and had a G+C content of the wobble position of 95%. The restriction map of the fragment encompassing these genes is shown on Fig. 1A. These genes overlap in the sequence ATGA, where ATG is the initiation codon for the downstream gene and TGA the termination codon for the upstream gene. When the deduced amino acid sequences for these genes were compared with others deposited in the databases, we found that the upstream gene encoded a protein whose C-terminal domain (about 232 amino acids) exhibited significant similarities to histidine kinases, especially with those involved in the expression of the phosphate regulon. It shared 28.81% identity with PhoR from Pseudomonas aeruginosa (1), 27.46% with PhoR from Bacillus subtilis (34), and 25.92% with PhoR from E. coli (22). The amino-terminal region did not resemble that of any other protein in the databases. The M. xanthus gene was then designated phoR2.
The downstream gene encoded a protein that exhibited high homologies along the full length of the protein to response regulators, sharing 42.23% identity with PhoB from Sinorhizobium meliloti (4), 40.97% with PhoB from P. aeruginosa (1), and 38.69% with PhoP from B. subtilis (19). These response regulators are also involved in the expression of phosphatases and the phosphate regulon in these bacteria. The M. xanthus gene has been designated phoP2. A 2.1-kb Sau3AI-SacI fragment containing the phoP2 gene (Fig. 1A) cloned in pUC19 was responsible for originating E. coli blue colonies on LB medium supplemented with BCIP.
The sequence of PhoR2 predicted a protein with a molecular weight of 67,047, with a putative signal peptide sequence in the amino terminal portion and a transmembrane domain from residues 357 to 376 (Fig. 2A). As a consequence, the sensor domain of PhoR2 must be located in the periplasmic space, and the histidine kinase domain must be located in the cytoplasm. PhoP2 is a cytoplasmic protein with a molecular weight of 26,718. This protein also consists of two domains, a response regulator domain located in the N-terminal region of the protein, and an effector domain in the C-terminal region, where a helix-loop-helix structure typical of DNA-binding proteins of the OmpR family (25) is present (Fig. 2B). The domain organization of PhoR2 and PhoP2, the similarities with other proteins, and the fact that a fragment containing the genes for these proteins originates E. coli blue colonies on LB medium with BCIP clearly indicate that system 2 must function by sensing external stimuli to activate the transcription of phosphatase genes.
FIG. 2.
Alignment of PhoR2 with PhoR3 (A), and PhoP2 with PhoP3 (B). Identical amino acids are boxed in black, and functionally similar amino acids are boxed in gray. ▾, putative cleavage site of the signal peptide in both kinases; ♦, amino acids of the transmembrane domain; *, amino acids that are conserved in response regulators; , amino acids that constitute the two helixes of the helix-loop-helix motif.
Cloning, sequencing, and analysis of the sequences of phoR3 and phoP3 genes.
When several DNA fragments containing part of the operon phoR2-phoP2 were used as probes in Southern blot hybridization experiments, we could observe that the phoP2 gene highlighted two different fragments in each M. xanthus chromosomal DNA digestion (Fig. 3). Several of these fragments were cloned, and the restriction map of that region is shown on Fig. 1B. When part of this fragment was sequenced, we detected two ORFs with 607 and 233 codons that also overlap in the sequence ATGA. The upstream gene encoded a protein that was also similar in the C-terminal domain to histidine kinases; this gene has been designated phoR3. PhoR3 exhibited 31.83% identity with KpdD from E. coli (42), 31.64% with IrlS from Burkholderia pseudomallei (15), and 31.62% with SenX from Mycobacterium bovis (36). It also showed homologies with kinases of the phosphate regulon. PhoR3 is a protein with a deduced molecular weight slightly smaller than that of PhoR2 (66,885), but it also shows the existence of a signal peptide sequence and a transmembrane domain between residues 358 and 377 (Fig. 2A). Therefore, the topology of this protein in the membrane must be identical to that of PhoR2.
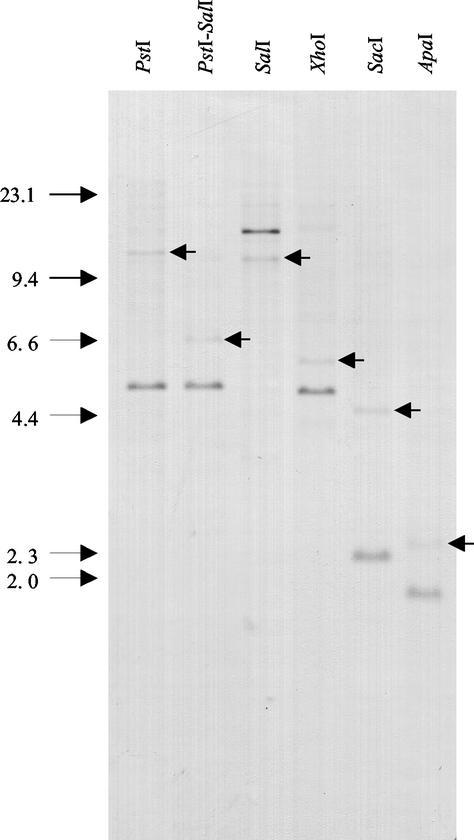
FIG. 3.
Southern blot hybridization of M. xanthus chromosomal DNA digested with different restriction enzymes. The fragment NcoI-ApaI(b) encompassing phoP2 (Fig. 1A) was used as a probe. The arrows within the lanes point to the second band that hybridizes with this probe in each digestion. The arrows to the left of the gel indicate molecular sizes in kilobases.
The downstream gene also encoded a protein which belongs to the family of the response regulators and which has been designated PhoP3. Unlike PhoR3, PhoP3 exhibited the highest similarities with proteins described as positive regulators of the phosphate regulon, such as PhoB from S. meliloti (40.30% identity) (4), PhoP from B. subtilis (39.39%) (19), and PhoB from P. aeruginosa (38.56%) (1). PhoP3 has a deduced molecular weight of 26,172, and it will most likely be found to be a DNA-binding protein, too (Fig. 2B).
Fruiting-body formation, sporulation, and germination in deletion mutants.
As described in Materials and Methods, three different strains have been constructed. JM2 harbors a deletion of the PhoR2-PhoP2 system, JM3 has a deletion of the PhoR3-PhoP3 system, and JM322 has a deletion of both. In order to analyze the roles of these systems during development, the three deletion mutants were plated on several low-nutrient media, such as CF, TPM, and MCM. On CF medium, we observed that JM2 and JM3 strains could make normal fruiting bodies and could do so with the same timing as the wild-type strain. However, the fruiting bodies of the single-deletion strains were visibly smaller (Fig. 4A). The double-deletion mutant, by contrast, did not develop well, and even at 72 h of starvation, aggregation had not been completed yet (Fig. 4A). This means a delay in aggregation of over 48 h, since the wild-type strain is quite well aggregated at 24 h of development.
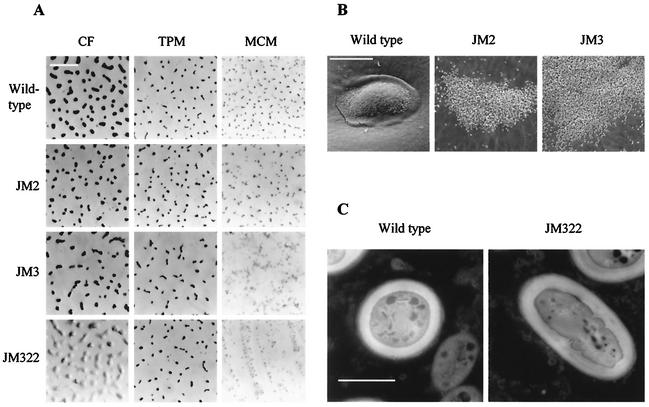
FIG. 4.
Morphologies of fruiting bodies and myxospores of M. xanthus DZF1 and deletion mutants. (A) The shapes and sizes of fruiting bodies on different starvation media are shown; the pictures were taken on a light microscope. Bar, 1 mm. (B) Fruiting bodies of M. xanthus DZF1, JM2, and JM3 on MCM medium are shown; the pictures were taken in a scanning electron microscope. Bar, 50 μm. (C) Wild-type and JM322 myxospores are shown; the pictures were taken on a transmission electron microscope. Bar, 1 μm.
On TPM medium, we could not observe any significant difference in either timing or shape between the fruiting bodies of the wild-type strain and those of the mutants (Fig. 4A).
The major differences were found on MCM, a medium that lacks phosphate. On this medium, wild-type fruiting bodies were much smaller than on CF, but this might be due to the total absence of nutrients, which does not allow bacterial growth. However, fruiting bodies appeared as isolated and raised mounds (Fig. 4A). The morphology of JM2 fruiting bodies was similar to that of the wild-type strain, but spores were not as perfectly packed (Fig. 4B). By contrast, JM3 and JM322 did not make mounds. Cells tended to aggregate on certain points, but they failed to pile up and build discrete mounds, remaining as loose aggregates, where they differentiated into myxospores (Fig. 4A and B). In the three mutants, spores did not seem to be embedded in an extracellular matrix as did those of the wild-type strain, and they appeared very independent from one another (Fig. 4B). The number of myxospores produced by the mutants was very similar to that of the wild-type strain (data not shown). However, approximately 5 to 10% of the myxospores did not reshape completely, and they did not become ovoid (Fig. 4C). The length of these defective myxospores was shorter than that of vegetative rods. The thicknesses of the coats were very similar in both types of myxospores (Fig. 4C). Germination of mutant myxospores seemed to be normal, since the percentages of colonies originated by myxospores on rich medium were very similar in the wild type and all the mutant strains (data not shown). All these results indicate that both PhoR2-PhoP2 and PhoR3-PhoP3 play an important role in aggregation and also in sporulation, contributing to the achievement of the proper shape of fruiting bodies and myxospores.
During vegetative growth, we have not observed any difference between the mutants and the wild-type strain, either in the generation time, the cell density reached by the cultures at the stationary phase, or the lysis phase when liquid CTT medium was used. In liquid cultures with less nutrient, this experiment could not be performed properly because the strain JM2 originated aggregates. On solid media, by contrast, we observed that mutant JM2 formed fruiting bodies and myxospores in a rich medium such as 1/5CTT in only 24 h (Table 1). Conversely, the wild-type strain and the rest of the mutants originated small amounts of fruiting bodies only after a 48-h incubation on 1/10CTT medium (Table 1). JM2 fruiting bodies on 1/5CTT are identical to those observed on CF medium (see Fig. 4A). It is interesting to note that the double-deletion mutation restored the wild-type phenotype on these media (Table 1).
TABLE 1.
Fruiting-body and myxospore formation of several strains at 24 and 48 h on culture media with different concentrations of nutrientsa
| Medium | DZF1 |
JM2 |
JM3 |
JM322 |
||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| CTT | − | − | − | − | − | − | − | − |
| 1/2CTT | − | − | − | − | − | − | − | − |
| 1/5CTT | − | − | + | ++ | − | − | − | − |
| 1/10CTT | − | + | + | ++ | − | + | − | + |
−, no presence of fruiting bodies and myxospores; +, presence of some aggregates with a small amount of myxospores; ++, number of fruiting bodies and spores similar to that observed on CF agar.
Roles of the two-component systems in the expression of phosphatases.
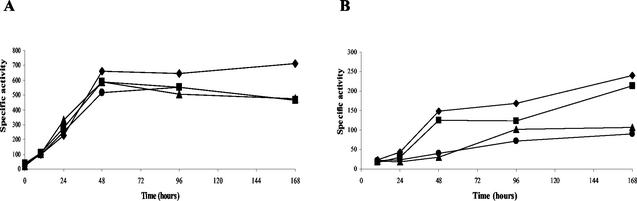
We have analyzed the activities of the five phosphatases reported by Weinberg and Zusman (43) in the three mutants. No significant differences in either phosphatase activities (Mg-dependent acid and alkaline phosphatases) during vegetative growth or Mg-independent alkaline phosphatase activities during development were found in any of the mutants (data not shown). However, compared to the wild-type strain, the three mutants showed a diminished Mg-independent acid phosphatase activity, which amounted to approximately 70% of that of the wild type at 168 h of development (Fig. 5A). When neutral phosphatase was determined, we could observe that strain JM2 showed only a slight decrease in activity compared to the wild-type strain (to 90% of the wild-type activity at 168 h of development). The deletion strain lacking system 3 exhibited only 40% of the activity of the wild-type strain at 168 h of development, and the double-deletion mutant exhibited 33% at that time. These results indicated that the expression of the gene(s) for Mg-independent acid phosphatase is dependent on both PhoP2 and PhoP3. The expression of the gene(s) for neutral phosphatase strongly depends on PhoP3, and it is also dependent on PhoP2, but to a lesser extent. However, the dependency on these two systems is not absolute, since 33% of neutral and 70% of acid phosphatase activities still remain in the JM322 strain. The rest of the phosphatases, including the two alkaline phosphatases, are completely independent on PhoR2-PhoP2 and PhoR3-PhoP3.
FIG. 5.
Determination of phosphatase activities in M. xanthus DZF1 (♦), JM2 (▪), JM3 (▴), and JM322 (•). Results for Mg-independent acid phosphatase (A) and Mg-independent neutral phosphatase (B) are shown. Specific activity is expressed as nmol of p-nitrophenol per mg of protein per min. The results shown are the averages of results of three experiments.
Since M. xanthus neutral phosphatase has not been characterized and Pph1 hydrolyzes p-nitrophenyl phosphate at neutral pH (38), we analyzed whether pph1 is under control of PhoP3. For this purpose, we introduced by electroporation a plasmid with a pph1-lacZ fusion into DZF1 and JM32 strains. The latter strain harbors a deletion in system 3 and is kanamycin sensitive (see Materials and Methods). Positive kanamycin-resistant colonies were then analyzed for β-galactosidase activity. The results obtained revealed that the deletion strain showed the same levels of β-galactosidase activity as the wild type (data not shown). Therefore, we can conclude that pph1 is independent of PhoP3.
DISCUSSION
While searching for genes that encode proteins with phosphatase activity in M. xanthus, we have cloned two fragments that encode two different two-component regulatory systems, designated PhoR2-PhoP2 and PhoR3-PhoP3. The fragment containing the phoR2-phoP2 operon is able to originate E. coli colonies with the ability to degrade BCIP. Since none of these proteins are able to hydrolyze BCIP by themselves, they must induce the expression of E. coli phosphatases.
These two systems exhibit high identity in both sequence and domain organization with each other. Both histidine kinases seem to be anchored to the membrane, with the sensor domain located in the periplasmic space and the kinase domain in the cytoplasm. The sensor domains of PhoR2 and PhoR3 share 46% identity, and the kinase domains share 61% identity (Fig. 2A). These proteins belong to class I of histidine kinases (3). Similarities between PhoP2 and PhoP3 are even more pronounced, since they have response regulator domains with 57% identity and effector domains that resemble those of DNA-binding proteins with 70% identity (Fig. 2B). If we consider functionally similar amino acids, these homologies rise by about 10%. Furthermore, the identity in the helix 2 motif, which is predicted to be the DNA recognition domain (25), is 100% (Fig. 2B). The fact that the sensor domains of the kinases share approximately 55% similarity and that the effector domains of the response regulators reach 80% identity, with 100% identity in helix 2, suggests that the histidine kinases respond to the input of different signals to activate the expression of the same genes by their cognate response regulator. In other words, different signals may be transduced through these two systems to elicit similar cell responses.
In most bacteria, alkaline phosphatases are expressed only during phosphate starvation as a result of the activation by two-component systems (37). In this study, we have confirmed that, in spite of the similarities between PhoP2 and PhoP3 and other response regulators involved in the activation of the phosphate regulon, neither of the two alkaline phosphatases of M. xanthus is under control of any of these systems. This is not surprising if we take into consideration that expression of phosphatases in M. xanthus seems to follow a different scheme from that used in other bacteria, since it is independent from starvation of phosphate. In fact, M. xanthus always exhibits a high phosphatase activity and blue color on media with BCIP during both vegetative growth and development and on media with or without phosphate (43; this study).
The gene for neutral phosphatase is indeed dependent on PhoP3 and PhoP2, but this activity is not completely abolished in a double-deletion mutant. The 30% remaining activity can be attributed to several reasons. (i) Phosphatase activity might be due to different proteins, and only some of the genes that encode them could be PhoP2-PhoP3 dependent. (ii) The expression of the gene(s) that encodes the phosphatase(s) could be carried out by the RNA polymerase without need of activation by a two-component system. (iii) A third two-component system could be involved in the expression of the phosphatase(s). In our laboratory, the fourth DNA fragment cloned during the search for M. xanthus phosphatases also encodes a two-component system, designated PhoR1-PhoP1 (unpublished results), which could be responsible for the expression of the remaining acid, neutral, and alkaline phosphatase activities. Characterization of system 1 undoubtedly will shed some light on the complex regulation of phosphatase expression in M. xanthus.
Regarding the phenotype of the mutants on aggregation and sporulation, we do not know whether the defects observed can be explained exclusively on the basis of the decrease in the levels of Mg-independent acid and neutral phosphatases. Phosphate is important for the developmental cycle, since wild-type fruiting bodies are considerably smaller on MCM medium than on CF or TPM medium (see Fig. 4A). However, the fact that the double-deletion mutant exhibits a clear delay in development in a medium with phosphate, such as CF, leads us to think that probably some other genes, different from those encoding phosphatases, could be under control of these two-component systems. In B. subtilis, the system PhoR-PhoP is responsible for the activation of the expression of 23 genes (29). Moreover, PhoP has been reported to inhibit the expression of the operons tagAB and tagDEF, which encode proteins involved in teichoic acid synthesis (21). Without doubt, the expected number of genes under control of the three M. xanthus systems must be high, and it is even tempting to speculate that they control the expression of developmental genes. In order to clarify the remaining questions, we have focused our research on the characterization of the system PhoR1-PhoP1, the signals that activate the three kinases, and the genes that are under control of the response regulators.
Acknowledgments
We thank Sumiko Inouye (University of Medicine and Dentistry of New Jersey) for providing plasmids containing kanamycin resistance genes and KG systems, and Anke Treuner-Lange (Forschungszentrum der Universität Giessen, Germany) for providing the pph1-lacZ plasmid.
This work has been supported by the Ministerio de Educación y Cultura, Dirección General de Enseñanza Superior, Spain (grant number PB98-1359). A.M.-M. and J.C.-L. are predoctoral fellows from the Ministerio de Educación y Cultura, Spain.
REFERENCES
- 1.Anba, J., M. Bidaud, M. L. Vasil, and A. Lazdunski. 1990. Nucleotide sequence of the Pseudomonas aeruginosa phoB gene, the regulatory gene for the phosphate regulon. J. Bacteriol. 172:4685-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery, L., and D. Kaiser. 1983. In situ transposon replacement and isolation of a spontaneous tandem duplication. Mol. Gen. Genet. 19:99-109. [DOI] [PubMed] [Google Scholar]
- 3.Bilwes, A. M., L. A. Alex, B. R. Crane, and M. I. Simon. 1999. Structure of CheA, a signal-transducing histidine kinase. Cell 96:131-141. [DOI] [PubMed] [Google Scholar]
- 4.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puehler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dworkin, M. 1996. Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60:70-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dworkin, M., and D. Kaiser. 1993. Myxobacteria II. ASM Press, Washington, D.C.
- 7.Frasch, S. C., and M. Dworkin. 1996. Tyrosine phosphorylation in Myxococcus xanthus, a multicellular prokaryote. J. Bacteriol. 178:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants in Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Hanlon, W. A., M. Inouye, and S. Inouye. 1997. Pkn9, a Ser/Thr protein kinase involved in the development of Myxococcus xanthus. Mol. Microbiol. 23:459-471. [DOI] [PubMed] [Google Scholar]
- 11.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inouye, S., R. Jain, T. Ueki, H. Nariya, C.-Y. Xu, M.-Y. Hsu, B. A. Fernández-Luque, J. Muñoz-Dorado, E. Fárez-Vidal, and M. Inouye. 2000. A large family of eukaryotic-like protein Ser/Thr kinases of Myxococcus xanthus, a developmental bacterium. Microb. Comp. Genom. 5:103-119. [DOI] [PubMed] [Google Scholar]
- 13.Inouye, S., and M. Inouye. 1987. Oligonucleotide-directed site-specific mutagenesis using double-stranded plasmid DNA, p. 181-204. In S. Narang (ed.), Synthesis and applications of DNA and RNA. Academic Press, Orlando, Fla.
- 14.Inouye, M., S. Inouye, and D. R. Zusman. 1979. Gene expression during development of Myxococcus xanthus: pattern of protein expression. Dev. Biol. 68:76-86. [DOI] [PubMed] [Google Scholar]
- 15.Jones, A. L., D. Deshazer, and D. E. Woods. 1997. Identification and characterization of a two-component regulatory system involved in invasion of eukaryotic cells and heavy-metal resistance in Burkholderia pseudomallei. Infect. Immun. 65:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 17.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus xanthus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 18.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. W., and F. M. Hulett. 1992. Nucleotide sequence of the phoP gene encoding PhoP, the response regulator of the phosphate regulon of Bacillus subtilis. Nucleic Acids Res. 20:5848.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerner, C. G., and M. Inouye. 1990. Low copy number plasmids for regulated low level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 18:4631.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, W., S. Eder, and F. M. Hulett. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP∼P. J. Bacteriol. 180:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makino, K., H. Shinagawa, M. Amemura, and A. Nakata. 1986. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J. Mol. Biol. 192:549-556. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Cañamero, M., C. Ortiz-Codorniu, A. L. Extremera, J. Muñoz-Dorado, and J. M. Arias. 2002. mlpB, a gene encoding a new lipoprotein in Myxococcus xanthus. J. Appl. Microbiol. 92:134-139. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Mizuno, T., and I. Tanaka. 1997. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol. Microbiol. 24:665-670. [DOI] [PubMed] [Google Scholar]
- 26.Moraleda-Muñoz, A., J. Carrero-Lérida, A. L. Extremera, J. M. Arias, and J. Muñoz-Dorado. 2001. Glycerol 3-phosphate inhibits swarming and aggregation of Myxococcus xanthus. J. Bacteriol. 183:6135-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller, C., and M. Dworkin. 1991. Effects of glucosamine on lysis, glycerol formation, and sporulation in Myxococcus xanthus. J. Bacteriol. 173:7164-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muñoz-Dorado, J., S. Inouye, and M. Inouye. 1991. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a Gram-negative bacterium. Cell 67:995-1006. [DOI] [PubMed] [Google Scholar]
- 29.Ogura, M., H. Yamaguchi, K.-I. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenbluh, A., and E. Rosenberg. 1989. Sporulation of Myxococcus xanthus in liquid shake flask cultures. J. Bacteriol. 171:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sanger, F., S. Nicklen, and A. R. Coulsson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seki, T., H. Yoshikawa, H. Takahashi, and H. Saito. 1988. Nucleotide sequence of the Bacillus subtilis phoR gene. J. Bacteriol. 170:5935-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 36.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 37.Torriani-Gorini, A., E. Yagil, and S. Silver. 1994. Phosphate in microorganisms: cellular and molecular biology. ASM Press, Washington, D.C.
- 38.Treuner-Lange, A., M. J. Ward, and D. R. Zusman. 2001. Pph1 from Myxococcus xanthus is a protein phosphatase involved in vegetative growth and development. Mol. Microbiol. 40:126-140. [DOI] [PubMed] [Google Scholar]
- 39.Udo, H., J. Muñoz-Dorado, M. Inouye, and S. Inouye. 1995. Myxococcus xanthus, a Gram-negative bacterium, contains a transmembrane protein serine/threonine kinase that blocks the secretion of β-lactamase by phosphorylation. Genes Dev. 9:972-983. [DOI] [PubMed] [Google Scholar]
- 40.Ueki, T., S. Inouye, and M. Inouye. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153-157. [DOI] [PubMed] [Google Scholar]
- 41.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 42.Walderhaug, M. O., J. W. Polarek, P. Voelkner, J. M. Daniel, J. E. Hesse, K. Altendorf, and W. Epstein. 1992. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J. Bacteriol. 174:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberg, R. A., and D. R. Zusman. 1990. Alkaline, acid, and neutral phosphatase activities are induced during development in Myxococcus xanthus. J. Bacteriol. 172:2294-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, S. S., J. Wu, Y. L. Cheng, and D. Kaiser. 1998. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol. Microbiol. 29:1249-1261. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, W., M. Inouye, and S. Inouye. 1996. Reciprocal regulation of the differentiation of Myxococcus xanthus by Pkn5 and Pkn6, eukaryotic-like Ser/Thr protein kinases. Mol. Microbiol. 20:435-447. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, W., J. Muñoz-Dorado, M. Inouye, and S. Inouye. 1992. Identification of a putative eukaryotic-like protein kinase family in the developmental bacterium Myxococcus xanthus. J. Bacteriol. 174:5450-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]