Abstract
Penicillin-binding proteins (PBPs) catalyze the final, essential reactions of peptidoglycan synthesis. Three classes of PBPs catalyze either trans-, endo-, or carboxypeptidase activities on the peptidoglycan peptide side chains. Only the class A high-molecular-weight PBPs have clearly demonstrated glycosyltransferase activities that polymerize the glycan strands, and in some species these proteins have been shown to be essential. The Bacillus subtilis genome sequence contains four genes encoding class A PBPs and no other genes with similarity to their glycosyltransferase domain. A strain lacking all four class A PBPs has been constructed and produces a peptidoglycan wall with only small structural differences from that of the wild type. The growth rate of the quadruple mutant is much lower than those of strains lacking only three of the class A PBPs, and increases in cell length and frequencies of wall abnormalities were noticeable. The viability and wall production of the quadruple-mutant strain indicate that a novel enzyme can perform the glycosyltransferase activity required for peptidoglycan synthesis. This activity was demonstrated in vitro and shown to be sensitive to the glycosyltransferase inhibitor moenomycin. In contrast, the quadruple-mutant strain was resistant to moenomycin in vivo. Exposure of the wild-type strain to moenomycin resulted in production of a phenotype similar to that of the quadruple mutant.
The structural element of the bacterial cell wall is peptidoglycan (PG), and the importance of this structure is apparent in the number of antimicrobial agents targeting its components and the enzymatic reactions leading to its synthesis. The glycosyltransferase (GT) activity carrying out one of the final enzymatic reactions in PG synthesis is an attractive target, because the enzymes performing that activity are highly conserved and believed to be essential (9, 10). PG is composed of glycan strands cross-linked by peptide side chains. The glycan strands are synthesized by a GT that adds lipid-linked disaccharide pentapeptide subunits to nascent glycan strands. The pentapeptides are then utilized by a transpeptidase to cross-link adjacent glycan strands (reviewed in reference 4). Most of the proteins involved in these final enzymatic reactions are penicillin-binding proteins (PBPs), which are divided into three classes based on the presence of conserved functional domains (9, 10). Only the class A PBPs have an N-terminal domain that contains the conserved amino acid sequences found in all GTs clearly demonstrated to polymerize PG. Class B PBPs have a different N-terminal domain; and although some researchers have associated GT activity with class B PBPs (15), others have been unable to reproduce those results (2). Both classes have C-terminal penicillin-binding domains containing the transpeptidase activity that cross-links peptide side chains. Some species also have monofunctional glycosyltransferases (MGTs) that contain the conserved amino acid sequences found in the class A PBP GT domain but which lack a penicillin-binding domain (7, 39; B. G. Spratt, J. Zhou, M. Taylor, and M. J. Merrick, Letter, Mol. Microbiol. 19:639-640, 1996).
The fact that many species contain multiple class A PBPs (10) and that removal of one or more of those proteins results in little or no effect on PG polymerization or cell viability demonstrates their functional redundancies (6, 24, 33). However, a function of class A PBPs has been demonstrated to be essential in both Escherichia coli and Streptococcus pneumoniae. Although loss of either PBP 1a or PBP 1b from E. coli was tolerated, loss of both was lethal, even in the presence of PBP 1c and an MGT (6, 40). A similar result was seen with S. pneumoniae, for which removal of PBPs 1a and 2a was lethal despite the presence of PBP 1b and an MGT (14, 25). Bacillus subtilis has four class A PBPs and no MGTs, as determined from sequence analysis of the B. subtilis genome (19). The genes ponA, pbpD, pbpF, and pbpG encode the class A PBPs 1, 4, 2c, and 2d, respectively (24, 30-32). Construction of null mutations in each class A PBP-encoding gene revealed that only loss of PBP 1 resulted in phenotypic changes: a decrease in growth rate along with changes in cell morphology (24, 27, 33) and PG structure (5). The growth rate and morphological changes were more pronounced in strains lacking PBPs 1 and 4 (33), and a strain lacking PBPs 2c and 2d was unable to properly construct spore PG (24). However, strains lacking any three of the four class A PBPs were viable (24).
In this communication, we demonstrate that a B. subtilis strain lacking all four class A PBPs is viable and produces PG of relatively normal structure. Phenotypic and morphological changes in the quadruple mutant are greater than those noted in the single PBP 1 and triple mutants. The glycosyltransferase activity present in the quadruple mutant is demonstrated in vitro and shown to be sensitive to moenomycin. Finally, we show that the quadruple mutant is resistant to high concentrations of moenomycin in vivo while exposure of the wild-type strain causes phenotypic changes similar to those of the quadruple mutant.
MATERIALS AND METHODS
Bacterial growth and transformation.
All strains of B. subtilis listed in Table 1 were derivatives of strain 168. Natural transformation was performed as previously described (3). Transformants were selected and maintained with appropriate antibiotics as follows: spectinomycin (100 μg/ml), kanamycin (10 μg/ml), and erythromycin (0.5 μg/ml) plus lincomycin (12.5 μg/ml, macrolide-lincosamide-streptogramin B resistance). Cultures were grown with shaking at 37°C in 2× SG medium (21) without antibiotics except where noted. Membranes were prepared from vegetative B. subtilis cultures at an optical density at 600 nm (OD600) of 0.5 as previously described (30). PBPs were labeled by using 125I-penicillin X and detected as previously described (24).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotypea | Transformation
|
Source or reference | |
|---|---|---|---|---|
| Donor | Recipient | |||
| DPVB42 | ΔpbpD | DPVB30 | DPVB40 | 24 |
| DPVB45 | ΔpbpG::Kn | pDPV35 | PS832 | 24 |
| DPVB46 | ΔpbpD ΔpbpF::Ermr | PS1869 | DPVB42 | 24 |
| DPVB49 | ΔpbpD ΔpbpG::Kn ΔpbpF::Ermr | DPVB45 | DPVB46 | 24 |
| DPVB56 | ΔpbpG::Kn ΔpbpF::Ermr | DPVB45 | PS1869 | 24 |
| DPVB57 | ΔpbpD ΔpbpG::Kn | DPVB45 | DPVB42 | 24 |
| DPVB61 | ΔponA::Sp ΔpbpG::Kn | PS2062 | DPVB45 | 24 |
| DPVB62 | ΔponA::Sp ΔpbpD ΔpbpG::Kn | PS2062 | DPVB57 | 24 |
| DPVB63 | ΔponA::Sp ΔpbpG::Kn ΔpbpF::Ermr | PS2062 | DPVB56 | 24 |
| DPVB66 | Sp downstream of and opposing ponA | pDPV43 | PS832 | This work |
| DPVB67 | Sp downstream of ponA and opposing ypoC | pDPV44 | PS832 | This work |
| DPVB68 | ΔponA::Sp ΔpbpD | PS2062 | DPVB42 | 24 |
| DPVB69 | ΔponA::Sp ΔpbpD ΔpbpF::Ermr | PS2062 | DPVB46 | 24 |
| DPVB87 | ΔponA::Sp ΔpbpD ΔpbpF::Ermr ΔpbpG::Kn | PS2062 | DPVB49 | This work |
| DPVB88 | ΔponA::Sp ΔpbpD ΔpbpF::Ermr ΔpbpG::Kn | PS2062 | DPVB49 | This work |
| PS832 | Prototrophic revertant of strain 168 | Laboratory stock | ||
| PS1869 | ΔpbpF::Ermr | pDPC89 | PS832 | 31 |
| PS2062 | ΔponA::Sp | pDPC197 | PS832 | 30 |
| PS2061 | ΔprfA::Sp | pDPC195 | PS832 | 30 |
| PS2251 | ΔponA::Sp ΔpbpF::Ermr | PS2062 | PS1869 | 33 |
Abbreviations: Ermr, resistance to erythromycin and lincomycin; Sp, resistance to spectinomycin; Kn, resistance to kanamycin.
Plasmid construction.
A 1,189-bp HincII-EcoRV fragment containing the downstream end of ponA and an inversely oriented gene, ypoC, was cloned into the HincII site of pUC19 to create pDPC254. This plasmid was then digested with ClaI, treated with the Klenow fragment of DNA polymerase to blunt the ends, and ligated with a 1,193-bp EcoRV-HincII fragment from pDG1726 (12) containing a spectinomycin resistance gene. This ligation resulted in two plasmids, pDPV43 and pDPV44, in which the spectinomycin gene was inserted in opposite directions. pDPV43 and pDPV44 were used to transform PS832 with selection for spectinomycin resistance to construct DPVB66 and DPVB67, respectively, in which the plasmids were inserted into the chromosome via a single crossover. This placed the spectinomycin resistance marker downstream of ponA, which had no effect on growth (data not shown).
PCR and Southern blot analysis.
PCR was used to demonstrate the presence of wild-type and mutant alleles of class A PBP-encoding genes. All nucleotide positions correspond to the coding sequences reported in the B. subtilis genome sequence (19) and on the SubtiList website (http://genolist.pasteur.fr/SubtiList/). Primers located from 645 to 621 bp upstream of ponA and from 2,725 bp within ponA to 6 bp downstream produced PCR products of 3,396 bp from wild-type ponA and 2,970 bp from ΔponA::Sp. Primers located from 53 to 75 bp within pbpD and from 1,855 bp within pbpD to 7 bp downstream produced PCR products of 1,824 bp from wild-type pbpD and 342 bp from ΔpbpD. Primers located from 11 bp upstream to 9 bp within pbpF and 2,136 bp within pbpF to 10 bp downstream produced PCR products that were subsequently digested with PvuII. Digestion resulted in fragment sizes of 1,068 and 1,098 bp from wild-type pbpF and an uncut fragment size of 2,169 bp from ΔpbpF::Erm. Primers located from 416 to 396 bp upstream of pbpG and from 191 to 210 bp downstream of pbpG produced PCR products that were subsequently digested with PvuII. Digestion resulted in fragment sizes of 463, 498, and 1,600 bp from wild-type pbpG and an uncut fragment size of 2,461 bp from ΔpbpG::Kn.
Two probes were used to verify the presence of each wild-type and mutant allele by Southern blotting. For each gene, one probe was complementary to a sequence within the deleted region and the other probe was complementary to a sequence at the 3′ end of the gene. For ponA, the interior probe was complementary to bases 535 to 1449 and the exterior probe was complementary to bases 1769 to 2745 of the coding sequence. For pbpD, the interior probe was complementary to bases 270 to 1621 and the exterior probe was complementary to bases 1591 within pbpD to 6 bp downstream of the coding sequence. For pbpF, the interior probe was complementary to bases 629 to 1237 and the exterior probe was complementary to bases 1447 within pbpF to 10 bp downstream of the coding sequence. For pbpG, the interior probe was complementary to bases 164 to 1105 and the exterior probe was complementary to bases 1251 within pbpG to 210 bp downstream of the coding sequence. Probes were prepared by restriction digestion of PCR products obtained from either wild-type chromosomal DNA (ponA, pbpF, and pbpG) or plasmid DNA (pDPC142, pbpD [32]) by using the primers described above followed by isolation of the desired fragments from agarose gels. Probes were labeled and detected by using the ECF random prime labeling and signal amplification system (Amersham Pharmacia Biotech) and a STORM 860 PhosphorImager and ImageQuant software (Molecular Dynamics).
Microscopy.
For electron microscopy, 10-ml samples from B. subtilis cultures grown to an OD600 of 0.5 were centrifuged at 5,000 × g for 5 min at 4°C. Cell pellets were suspended in 840 μl of 0.5 M NaPO4 (pH 7.0) plus 50 μl of 25% electron microscopy grade glutaraldehyde (Sigma) and held at 4°C overnight. Cells were washed four times in cold 0.1 M NaPO4 (pH 6.7), suspended in 1% osmium in phosphate buffer, and held at 4°C overnight. Cells were washed in 0.5 M NH4Cl and then suspended and pelleted in 2% agar. Dehydration was performed at 30, 50, 70, 95, and 100% ethanol and then in a 1:1 solution of ethanol and Spurr's resin overnight. Samples were then suspended in 100% Spurr's resin overnight before being cured. Samples were sectioned, placed on 200-mesh copper grids, and stained with 1% uranyl acetate for 12 min and Reynold's lead for 5 min. Samples were viewed by using a JEOL 100 CX-II transmission electron microscope at an accelerating voltage of 80 kV.
Samples for fluorescence and phase-contrast microscopy were prepared by growing cultures to an OD600 of 0.5 for vegetative cultures or for 2.5 and 24 h following exposure to 80 μg of moenomycin/ml. For fluorescence microscopy, cells were pelleted from 0.5 ml of culture, resuspended in 200 μl of Tris-EDTA (TE) buffer (10 mM Tris HCl [pH 7.5] and 1 mM EDTA), and applied to a poly-l-lysine-coated coverslip. Staining was performed as described previously (29). The coverslip was placed on a slide spotted with 10 μg of FM4-64 [N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide]/ml and 2 μg of DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride)/ml in either TE buffer or Slow Fade (all stains and the Slow Fade Antifade kit were obtained from Molecular Probes). Cells were visualized with a Nikon Microphot-SA microscope equipped with a UV-1B cube (DAPI) and a G-ZA cube (FM4-64) with a Nikon Fluor 100×/1.30 objective. Images were collected with a DAGE-MTI CCD100 video camera and DAGE InstaGater and pseudocolored by using ISEE software (Inovision Inc.) on a Silicon Graphics O2 computer. For phase-contrast microscopy, cells were pelleted from 0.5 ml of culture and fixed in 0.5 ml of 4.4% paraformaldehyde and 0.01% glutaraldehyde in 28 mM NaPO4 (pH 7.0) for 15 min at room temperature and 30 min on ice. Cells were washed twice in 0.5 ml of TE buffer and resuspended in 50 μl of TE buffer. Cells were visualized with an Olympus Provis AX70 microscope equipped with an Olympus UPlanF1 100×/1.30 Oil Ph3 objective. Images were collected with a Colorview 12 video camera and Olympus MicroSuite-B3 software. Fluorescence and phase-contrast images were transferred to a Macintosh G4 computer and processed for publication by Adobe Photoshop version 4.0.1.
Determination of in vivo moenomycin sensitivity.
Samples from cultures grown to an OD600 of 0.5 were diluted 100-fold in 2× SG medium and plated in the presence of various concentrations of moenomycin (Intervet, Inc.). Colony number and morphology were examined after incubation at 37°C for 24 h. The effects of moenomycin exposure to cells grown in liquid culture were examined by growing each strain to an OD600 of 0.5 prior to adding moenomycin. Incubation was continued and, at various time points, samples were diluted in 2× SG and plated on Luria-Bertani agar lacking moenomycin.
Peptidoglycan analysis.
Cultures (100 ml) were grown to an OD600 of 0.5, chilled by swirling in ice water for 5 min, and centrifuged at 15,000 × g for 10 min at 4°C. Pellets were suspended in 2 ml of 4°C water, added dropwise to 50 ml of boiling 4% sodium dodecyl sulfate (SDS), and boiled for 30 min. Suspensions were allowed to cool, centrifuged at 12,000 × g for 10 min at room temperature, and washed with water until free of SDS. Pellets were suspended in 1 ml of 100 mM Tris HCl (pH 7.5) and incubated with 100 μg of α-amylase (Sigma) at 37°C for 2 h. DNase I (10 μg), RNase A (50 μg), and MgSO4 (20 mM) were added, and incubation was continued at 37°C for 2 h. Trypsin (100 μg) and CaCl2 (10 mM) were added, and incubation was at 37°C overnight. SDS was added to 1%, and the solution was boiled for 15 min, diluted into 7 ml of water, and then centrifuged at 12,000 × g for 10 min at 20°C. The pellet was suspended and washed twice in 8 ml of water, once in 8 M LiCl, and twice more in water. The pellet was then lyophilized, suspended in 1 ml of 49% hydrofluoric acid, and rocked for 48 h at 4°C to remove teichoic acids. The peptidoglycan was collected by centrifugation at 13,000 × g for 5 min and washed three times with water. The pellet was then suspended in 1 ml of 100 mM NH4HCO3 and incubated with 5 units of calf intestinal alkaline phosphatase (Promega) at 37°C overnight. The sample was boiled for 5 min, centrifuged at 13,000 × g for 15 min, and washed three times with water. Muramidase digestion of the peptidoglycan, reduction of the soluble muropeptides, and high-pressure liquid chromatography separation of muropeptides was performed as previously described (5, 11). Muropeptide identification were performed by amino acid analysis (11) and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry with a Kratos Analytical Kompact SEQ instrument and software.
Glycosyltransferase activity.
Cultures were grown to an OD600 of 0.5 in 1% tryptone, 0.5% yeast extract, 0.25% K2HPO4 (pH 7.3) with glucose added to a final concentration of 0.1% just before use (34). Cultures were centrifuged at 9,000 × g for 10 min at room temperature. Protoplasts were produced essentially as described by Taku and Fan (36). Briefly, the pellet was suspended in 20 ml of buffer A (50 mM Tris HCl [pH 7.5], 20 mM MgCl2, 15% sucrose) plus 1.25 mg of lysozyme/ml and incubated at 37°C for 1 h. Protoplasts were pelleted at 9,000 × g for 10 min at 4°C and then washed twice more with 10 ml of buffer A. The final protoplast pellet was then suspended in 2.5 ml of buffer B (50 mM Tris HCl [pH 7.5], 20 mM MgCl2, 13% sucrose), and the protein content was determined by using the Lowry assay (22).
Protoplasts containing 18 μg of protein were incubated for 2 h at 37°C with 3 nmol of UDP-N-acetyl-muramic acid-pentapeptide and 5.64 nmol of UDP-N-acetyl-d-[U-14C]glucosamine (UDP-NAG) (266 mCi/mmol; Amersham Pharmacia Biotech) in a total volume of 40 μl of 50 mM Tris HCl (pH 7.5)-10 mM MgCl2-6.5% sucrose. UDP-N-acetyl-muramic acid-pentapeptide was prepared from B. subtilis cultures as previously described (23, 26). Reactions were stopped by heating at 95°C for 5 min. When used, lysozyme (2 mg/ml) or moenomycin (16 μg/ml) was added at the start of the reaction. Reactions were spotted on thick chromatography paper (Fisher Scientific) and separated by using ascending paper chromatography with isobutyric acid and 1 M NH4OH (5:3) for 12 h (35). During solvent migration, polymerized PG remained at the origin while the free precursors migrated away from the origin. Chromatogram signal intensities were integrated by using a STORM 860 PhosphorImager and ImageQuant software (Molecular Dynamics).
RESULTS
Construction and verification of a ponA pbpD pbpF pbpG mutant.
Each mutation in a class A PBP-encoding gene was a deletion that removed ≥57% of the coding sequence, including the five most highly conserved motifs of the N-terminal GT domains (10). The deleted regions in the ponA, pbpF, and pbpG alleles were replaced by antibiotic resistance cassettes, ensuring that the C-terminal domains were not expressed. The pbpD mutation was an in-frame deletion removing 79% of the coding sequence, which also included the penicillin-binding active-site serine (24). Construction of the quadruple mutant was performed by transforming a pbpD pbpF pbpG triple mutant (DPVB49) (24) with limiting chromosomal DNA from a ponA mutant (PS2062). As controls, DPVB49 was transformed separately with limiting chromosomal DNA from PS2061 (ΔprfA::Spr), DPVB66 (Spr downstream of ponA), and DPVB67 (Spr downstream of ponA) and DPVB56 (pbpF pbpG) was transformed with chromosomal DNA from PS2062. All the chromosomal DNA preparations used for the transformations were equal in concentration, and quadruple-mutant transformants were obtained at approximately the same frequency as the strains produced in the control transformations. This indicates that the viability of the quadruple mutant was not dependent on the presence of any type of suppressor mutation. Two independent isolates (DPVB87 and DPVB88) of the quadruple mutant were selected for further analysis.
Due to the unexpected result that the quadruple mutant was viable, the presence of each mutation was verified by both PCR and Southern blot analysis (data not shown). For the Southern blot analysis, two probes were utilized for each gene. One probe was complementary to a sequence interior to the deletion, whereas the second probe was complementary to a sequence exterior to the deleted region. The probes complementary to a sequence exterior to a deleted region verified the presence of each wild-type or mutant allele. The probe interior to the deleted region of each gene verified the presence or absence of that complementary sequence, indicating that the wild-type sequence had not appeared elsewhere in the chromosome via some nonhomologous recombination event.
We demonstrated the absence of class A PBPs 1, 2c, and 4 from the respective strains using 125I-labeled penicillin X (data not shown). PBP 2d is not highly expressed during vegetative growth (28) and is therefore not visualized in the PBP profile of vegetative cultures. We found that the band intensities of PBPs 2c, 3, and 4 did not change relative to each other in any strain; however, the band intensities of two class B PBPs, 2a and 2b, did change in relation to each other and to PBP 3. Quantitative analysis indicated that, relative to PBP 3, expression of PBPs 2a and 2b increased 51% ± 17% (mean ± standard deviation) (n = 3) and 22% ± 12% (n = 3), respectively, in strains lacking PBP 1, and more so in strains lacking PBPs 1 and 4, 87% ± 31% (n = 9) and 52% ± 19% (n = 9), respectively. We cannot formally exclude the possibility that PBPs 2c, 3, and 4 all decrease under these conditions. However, we feel that their relative stability and the differential changes in PBPs 2a and 2b are suggestive of increases in the last two. This is consistent with a previous observation that PBP 2a expression appeared to be elevated in strains lacking PBP 1 and either PBP 2c or PBP 4 (33).
Growth and morphology of the ponA pbpD pbpF pbpG mutant strains.
Doubling times of the quadruple-mutant strains were 62 and 63 min (Table 2). This is threefold slower than the doubling times of the wild-type strain and the triple mutant lacking PBPs 2c, 2d, and 4 and twofold slower than the triple mutants lacking PBP 1 (Table 2) (24, 33). The fact that the quadruple mutant grows slower than the triple mutant lacking PBPs 1, 2c, and 4 suggests that, unlike the situation in the wild type (28), in this triple mutant PBP 2d may be expressed during vegetative growth. When samples of quadruple-mutant cultures were plated, colonies of various sizes arose. However, subsequent streaking of those various-sized colonies resulted in uniform colony sizes from all, and growth in liquid medium revealed uniform growth rates (data not shown). These results suggest that suppressor mutations were not appearing at a high frequency in these quadruple-mutant strains. We believe that the variations in colony size resulted from a wide variation in cell size (see below) in the liquid cultures. A filamentous cell with more than five times the mass of a cell of wild-type dimensions could rapidly divide under the slower-growth conditions on a plate. A colony resulting from a large cell could therefore be significantly larger than that derived from a smaller cell. Size variation was less pronounced among the slower-growing cells within a colony, resulting in more uniform colony size upon restreaking.
TABLE 2.
Growth rates of class A PBP mutant strainsa
| Strain | PBP phenotype | Doubling timeb (min) |
|---|---|---|
| PS832 | Wild type | 20 |
| PS1869 | PBP2c− | 20 |
| DPVB42 | PBP4− | 20 |
| DPVB45 | PBP2d− | 20 |
| PS2062 | PBP1− | 25 |
| DPVB49 | PDP2c− PBP2d− PBP4− | 21 |
| DPVB63 | PBP1− PBP2c− PBP2d− | 28 |
| DPVB69 | PBP1− PBP2c− PBP4− | 28 |
| DPVB62 | PBP1− PBP2d− PBP4− | 31 |
| DPVB87 | PBP1− PBP2c− PBP2d− PBP4− | 62 |
| DPVB88 | PBP1− PBP2c− PBP2d− PBP4− | 63 |
Growth was in liquid 2× SG medium at 37°C.
Doubling times are averages from at least three separate experiments.
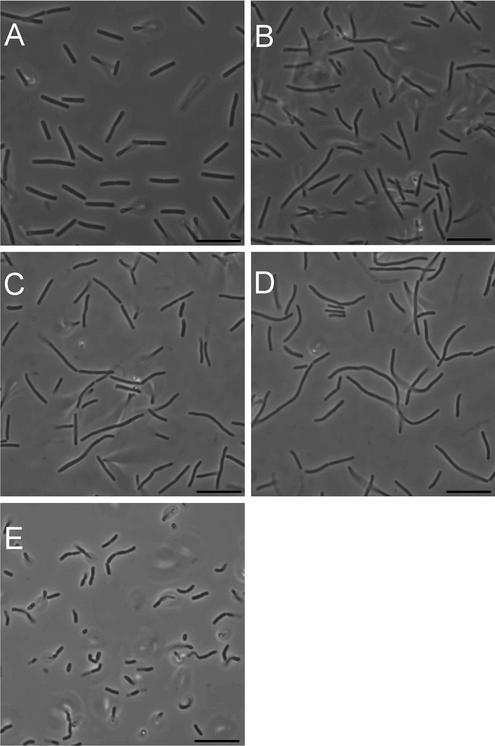
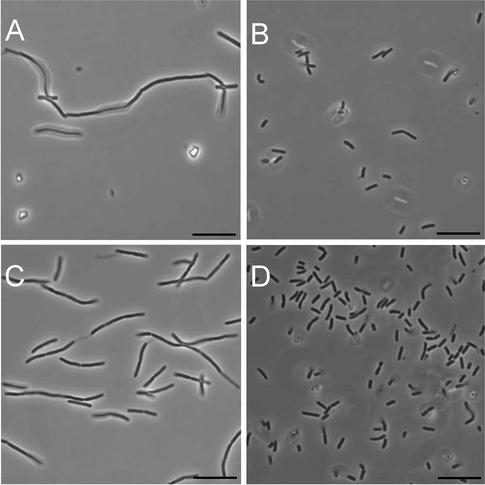
As previously described (18, 27, 33), B. subtilis strains lacking PBP 1 grew slowly and had a smaller cell radius and their cultures contained a population of cells that were slightly longer, bent, and multinucleoid. The cell length and bending were more pronounced in a larger number of PBP 1− PBP 4− cells and were even more pronounced in the PBP 1− PBP 2c− PBP 4− strain (33). When using phase-contrast microscopy, we noted a large variation in cell length among the populations of the quadruple-mutant strains and many individual cells appeared even longer and more bent than any produced by the triple-mutant strains (Fig. 1). Quantitative analysis of the cell lengths revealed that while many of the quadruple-mutant cells were of normal length, 30% of the cells were significantly longer than the entire wild-type population (Fig. 2). Upon entry into stationary phase, the filamentous cells underwent division to produce a population of relatively homogenous length (Fig. 1E).
FIG. 1.
Phase-contrast microscopy of class A PBP mutants. Cells were photographed from exponentially growing (A to D) and 24-h (E) cultures of PS832 (wild type [A]), PS2062 (PBP 1− [B]), DPVB69 (PBP 1− PBP 2c− PBP 4− [C]) and DPVB87 (PBP 1− PBP 2c− PBP 2d− PBP 4− [D and E]). Bars, 10 μm.
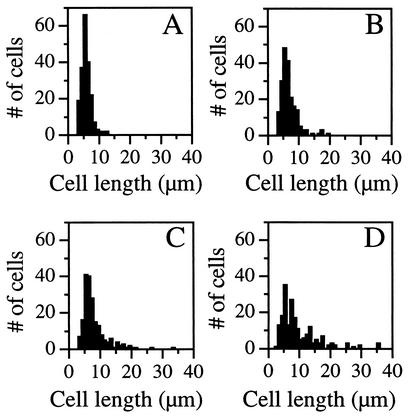
FIG. 2.
Cell length distributions of class A PBP mutants. Cell were harvested from exponentially growing cultures and examined under phase-contrast microscopy. Cell lengths (n = 200 for each strain) were determined from digital images. (A) PS832 (wild type); (B) PS2062 (PBP 1−); (C) DPVB69 (PBP 1 PBP 2c PBP 4−); (D) DPVB87 (PBP 1 PBP 2c PBP 2d PBP 4−). In the wild-type culture 3% of the cells were ≥10 μm. This increased to 9% in the PBP 1− culture, 20% in the PBP 1 PBP 2c PBP 4− culture, and 33% in PBP 1 PBP 2c PBP 2d PBP 4− cultures.
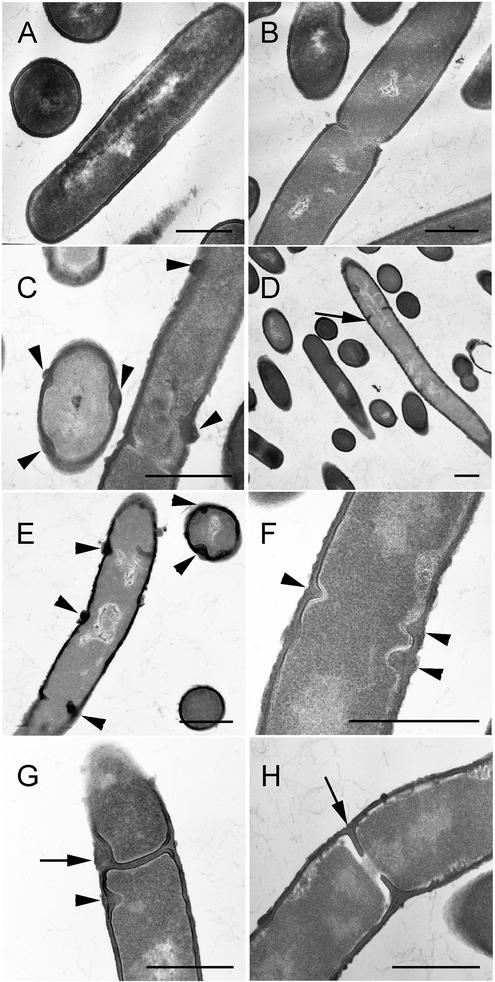
Electron micrographs of the quadruple mutants also show long, bent, multinucleoid cells (Fig. 3). Measurements of laterally cross-sectioned cells indicate that these cells had reduced diameters, similar to those previously observed in a PBP 1− strain (33). Relative to the wild type, the cell diameter decreased by 25% ± 15% (n = 384) in the PBP 1− strain, by 21% ± 11% (n = 685) in the PBP 1− PBP 4− strain, and by 20% ± 10% (n = 527) in the quadruple-mutant strains. This is consistent with earlier observations of narrow cell diameters in strains lacking PBP 1. It was previously noted that the PBP 1− strain had wall material synthesized in aberrant masses along the cylindrical wall (27). We also noticed these formations (Fig. 3C and E to H), which appeared to occur more often in the PBP 1− PBP 2c− PBP 2d− PBP 4− strains, and some of these masses protruded into the cytoplasm (Fig. 3F through G). In some cases the positioning of these protrusions suggested that they are incomplete septa. Previously, strains lacking PBP 1 were shown to have a defect in septation (27). Although the PBP 1 and quadruple mutants contained some defects in septum formation (Fig. 3G), there were also many apparently normal septa (FIG. 3H). However, we observed a number of septa that appeared to impinge upon a chromosome (Fig. 3D and H).
FIG. 3.
Electron microscopy of class A PBP mutants. Cells obtained from exponentially growing cultures of PS832 (wild type [A and B]), PS2062 (PBP 1− [C and D]), and DPVB87 (PBP 1− PBP 2c− PBP 2d− PBP 4− [E to H]) were fixed, sectioned, and stained for transmission electron microscopy. Arrowheads in C, E, F, and G indicate abnormal cell wall material. Arrows in G indicate irregularly formed septa, and in D and H they indicate septa impinging upon the chromosome. Bars, 0.5 μm.
Strains were prepared for fluorescence microscopy to further examine potential irregularities in division septa upon loss of multiple class A PBPs. Staining with FM4-64, a membrane stain, showed irregular placement of what appeared to be division septa in all strains lacking at least PBP 1 and this was exacerbated in the quadruple-mutant strains (data not shown). In most cases, the chromosomes were segregated throughout the cell. However, many areas in which division septa appeared to be synthesized into an area of the cytoplasm containing DNA were visualized. Due to resolution restrictions, we were unable to differentiate division septa and those protrusions of cell wall material seen in the electron micrographs.
Growth and morphology in the presence of moenomycin.
Moenomycin has previously been shown to specifically inhibit the GT activity performed by class A PBPs (37) due to its structural similarity to the lipid II precursor (20). We tried to determine the minimal concentration needed to inhibit growth of B. subtilis strains by plating 105 bacteria in the presence of 1 to 16 μg of moenomycin/ml (data not shown). For all strains containing PBP 1, the number of colonies decreased approximately 100-fold as the concentration of moenomycin increased from 0 to 16 μg/ml. As moenomycin increased from 0 to 8 μg/ml, the colony sizes decreased but on each plate the colonies were of uniform size. At a moenomycin concentration of 16 μg/ml, approximately 1% of the colonies grew significantly faster than the others. Strains lacking PBP 1 displayed a different phenotype. While colony number decreased with increasing moenomycin concentration, approximately 1,000-fold from 0 to 16 μg of moenomycin/ml, colony size did not change as much. At a moenomycin concentration of 16 μg/ml, all the colonies were of uniform size and were as large as those on the plates with 4- and 8-μg/ml concentrations. Comparison of the PBP 1− strain to multiple mutants lacking additional class A PBPs revealed that the multiple mutants displayed progressively fewer effects from exposure to moenomycin. The quadruple mutant had only a 10-fold decrease in colony number between concentrations of moenomycin of 0 and 16 μg/ml, and all the colonies on all the plates were of uniform size.
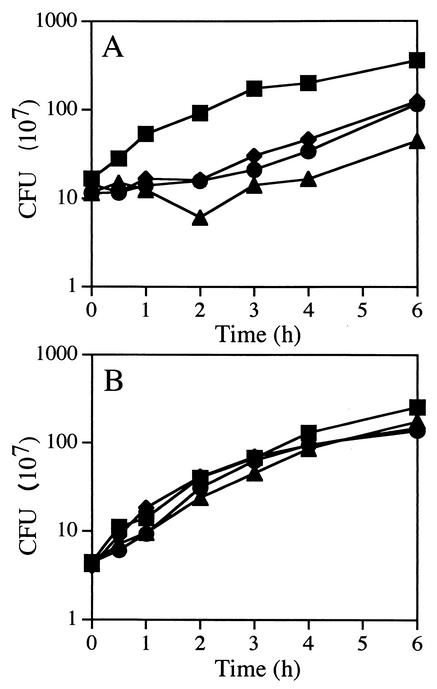
To determine the viability of cells grown in liquid medium containing moenomycin, cultures were grown to an OD600 of 0.5, at which time moenomycin was added at concentrations of 16, 40, and 80 μg/ml. At various time points, samples were plated in the absence of moenomycin to determine viable counts. Wild-type cultures entered a lag phase during which the number of CFU remained constant for up to 4 h depending on the concentration of moenomycin (Fig. 4A). The OD of the culture exposed to 80 μg of moenomycin/ml doubled during the first 30 min but increased only 25% during the next 2 h and another 20% during the next 3 h, suggesting that not only cell division but also growth of the cells was impeded. Similar results were seen for class A PBP mutant strains that contained PBP 1 (data not shown). Growth of a quadruple-mutant culture exposed to the same moenomycin concentrations was not affected (Fig. 4B). This also correlated with the OD of the culture, which continued to increase at the same rate as that of the culture not exposed to the antibiotic. Again in this experiment, class A PBP mutant strains lacking PBP 1 exhibited a phenotype similar to that of the quadruple mutant (data not shown).
FIG. 4.
Effects of moenomycin on growth of B. subtilis. Cultures of PS832 (wild type [A]) and DPVB87 (PBP 1− PBP 2c− PBP 2d− PBP 4− [B]) were exposed to either 0 (▪), 16 (•), 40 (♦), or 80 (▴) μg of moenomycin/ml. CFU values are averages from three separate experiments. Moenomycin was added when the OD reached 0.5. At this culture density, growth of the wild-type strain had begun to slow, reducing the difference between the growth rates of the wild-type and quadruple-mutant strains.
Cells exposed to 80 μg of moenomycin/ml for 2.5 h were observed by using both phase-contrast and fluorescence microscopy. Phase-contrast microscopy showed that wild-type cells became more filamentous and bent (Fig. 5A), a phenotype similar to what was seen in the quadruple mutant under normal growth conditions (Fig. 1B). These cells elongated two- to eightfold, indicating that the increase in OD corresponded to the increase in cell mass, but not cell division, which is consistent with the small change in CFU observed over that time period. Furthermore, irregularly spaced septa were visualized by fluorescence microscopy (data not shown). The quadruple mutant was largely unaffected by the presence of moenomycin. Figure 5C shows that the general population of the quadruple-mutant cells may be slightly more filamentous than when grown in the absence of moenomycin (Fig. 1D). After exposure to moenomycin for 4 to 6 h, cells from both the wild-type and the quadruple-mutant cultures began to shorten (data not shown). At 24 h following exposure to moenomycin, the cell populations from both wild-type (Fig. 5B) and quadruple-mutant (Fig. 5D) strains had shortened, similar to what was seen in a 24-h culture of the quadruple mutant in the absence of moenomycin (Fig. 1E).
FIG. 5.
Effects of moenomycin on the morphology of B. subtilis. PS832 (wild type [A and B]) and DPVB87 (PBP 1− PBP 2c− PBP 2d− PBP 4− [C and D]) cells were exposed to 80 μg of moenomycin/ml for 2.5 h (A and C) and 24 h (B and D) and examined by phase-contrast microscopy. Bars, 10 μm.
Structural analysis of vegetative PG from wild-type and class A PBP mutant strains.
Atrih et al. (5) found that PG from exponentially growing wild-type B. subtilis cells was cross-linked by 29% of the peptide side chains and that cross-linking decreased in strains lacking PBP 1. Our analysis of PG from our exponentially growing wild-type strain demonstrated that 26% of the muramic acid residues had peptides involved in cross-linking (Table 3). This percentage decreased to 24% in strains lacking at least PBP 1 and 22% in all strains lacking at least PBPs 1 and 4, including both quadruple-mutant strains (Table 3).
TABLE 3.
Structural parameters of B. subtilis PGa
| Strain | PBP(s) missing | % Muramic acid with cross-linked peptide | % Muropeptideb
|
||||
|---|---|---|---|---|---|---|---|
| Monomer | Dimer | Trimer | Tetramer | 1,6-Anhydro | |||
| PS832 | Wild type | 26.4 ± 0.5 | 48.2 ± 0.9 | 49.0 ± 0.7 | 2.5 ± 0.3 | 0.3 | 0.2 |
| PS1869 | 2c | 25.6 ± 0.6 | 49.7 ± 1.0 | 47.5 ± 0.6 | 2.5 ± 0.3 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| DPVB45 | 2d | 26.6 ± 1.1 | 47.9 ± 2.0 | 49.0 ± 1.6 | 2.7 ± 0.4 | 0.4 | 0.3 ± 0.1 |
| DPVB56 | 2c, 2d | 25.9 ± 0.9 | 49.3 ± 1.6 | 47.7 ± 1.3 | 2.7 ± 0.3 | 0.3 | 0.3 ± 0.1 |
| DPVB42 | 4 | 25.4 ± 0.4 | 50.1 ± 0.9 | 47.5 ± 0.9 | 2.2 ± 0.02 | 0.2 | 0.3 |
| DPVB46 | 2c, 4 | 25.3 ± 0.5 | 50.4 ± 1.0 | 47.1 ± 0.7 | 2.3 ± 0.3 | 0.3 | 0.3 |
| DPVB57 | 2d, 4 | 25.3 ± 0.2 | 50.2 ± 0.3 | 47.1 ± 0.3 | 2.3 ± 0.1 | 0.3 | 0.3 |
| DPVB49 | 2c, 2d, 4 | 25.5 ± 0.2 | 50.0 ± 0.3 | 47.4 ± 0.2 | 2.4 ± 0.1 | 0.3 | 0.2 |
| PS2062 | 1 | 24.2 ± 0.5 | 52.4 ± 1.1 | 45.7 ± 1.1 | 1.6 ± 0.1 | 0.3 ± 0.1 | 0.4 ± 0.1 |
| PS2251 | 1, 2c | 23.4 ± 0.5 | 53.9 ± 1.0 | 44.3 ± 1.1 | 1.5 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| DPVB61 | 1, 2d | 22.5 ± 2.6 | 55.5 ± 5.0 | 42.8 ± 4.7 | 1.3 ± 0.4 | 0.3 | 0.4 ± 0.1 |
| DPVB63 | 1, 2c, 2d | 24.1 ± 0.1 | 52.6 ± 0.2 | 45.3 ± 0.2 | 1.8 | 0.3 | 0.3 ± 0.1 |
| DPVB68 | 1, 4 | 21.7 ± 1.5 | 56.9 ± 3.0 | 42.3 ± 2.8 | 0.6 ± 0.1 | 0.1 | 0.2 |
| DPVB69 | 1, 2c, 4 | 23.2 ± 1.1 | 54.1 ± 2.3 | 44.8 ± 2.4 | 0.9 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| DPVB62 | 1, 2d, 4 | 22.3 ± 0.8 | 55.7 ± 1.6 | 43.4 ± 1.5 | 0.8 ± 0.1 | 0.1 | 0.2 |
| DPVB87 | 1, 2c, 2d, 4 | 21.7 ± 0.7 | 56.8 ± 1.5 | 42.2 ± 1.6 | 0.8 | 0.2 ± 0.1 | 0.2 |
| DPVB88 | 1, 2c, 2d, 4 | 21.9 ± 0.1 | 56.5 ± 0.1 | 42.6 ± 0.1 | 0.8 ± 0.1 | 0.2 | 0.2 |
Values are averages of at least two independent analyses with errors of 1 standard deviation. In cases where no error is indicated, the values of the multiple analyses were identical.
Percentage of total muropeptides identified as either un-cross-linked monomers or cross-linked dimers, trimers, and tetramers and those muropeptides containing 1,6-anhydrodisaccharide muropeptides.
In vitro glycosyltransferase activity of the PBP 1− PBP 2c− PBP 2d− PBP 4− strain.
An in vitro glycosyltransferase assay was used to demonstrate this activity in protoplasts of the wild-type, PBP 1−, PBP 1− PBP 4−, and all triple- and quadruple-mutant strains. As observed previously, we were unable to detect GT activity in purified B. subtilis membrane preparations (34). Protoplasts, prepared in an isotonic sucrose solution, were mixed with labeled, nucleotide-linked PG precursors simultaneously with a twofold dilution of the sucrose. Disruption of the protoplasts during this dilution step allowed access of the precursors to the enzymes involved in production of lipid II. Reaction substrates and products were separated by paper chromatography, in which polymerized PG strands remain at the origin. The radioactivity incorporated into insoluble material by protoplasts of each mutant strain was quantified and expressed as a percentage of that of the wild-type protoplasts (Table 4). In general, PG synthetic activity decreased with successive loss of class A PBPs. Addition of lysozyme to reaction mixtures resulted in significant decreases, usually more than 50%, in radioactivity remaining at the origin, demonstrating that this material was PG. Addition of moenomycin, a specific inhibitor of PG-synthetic GTs, decreased radioactivity remaining at the origin by 70 to 97%. Lysozyme-insensitive, 14C-containing polymers could result from several phenomena. First, some 14C could be incorporated into a product other than PG. Second, some lysed protoplasts may produce inside-out vesicles, which would not allow access of lysozyme to PG synthesized within the vesicle. Finally, synthesis of multiply cross-linked PG with disaccharides would produce trimers and tetramers of cross-linked disaccharide muropeptides upon lysozyme digestion. These multimers may remain at the origin during chromatography. The fact that in all cases the majority of incorporation of radioactivity was sensitive to moenomycin suggests that we are observing mostly PG synthesis and that the last two possibilities are more likely. The fact that the fraction of lysozyme-resistant material increased in strains lacking multiple class A PBPs suggests that PG synthetic activity is decreasing relative to synthesis of other compounds. However, even in the absence of class A PBPs, >70% of incorporation of radioactivity was moenomycin sensitive, suggesting that the majority of incorporation was into PG.
TABLE 4.
Incorporation of 14C-UDP-NAG into glycan by B. subtilis protoplasts
| PBP phenotype | % of radioactivity remaining at origin:
|
||
|---|---|---|---|
| Relative to that of the wild type | That is lost upon addition of:
|
||
| Lysozyme | Moenomycin | ||
| Wild type | 100 | 56 | 97 |
| PBP 1− | 59 | 74 | 96 |
| PBP 1− PBP 4− | 20 | 65 | 91 |
| PBP 2c− PBP 2d− PBP 4− | 52 | 44 | 90 |
| PBP 1− PBP 2c− PBP 2d− | 51 | 77 | 92 |
| PBP 1− PBP 2c− PBP 4− | 19 | 39 | 68 |
| PBP 1− PBP 2d− PBP 4− | 40 | 68 | 79 |
| PBP 1− PBP 2c− PBP 2d− PBP 4− | 31 | 51 | 79 |
| PBP 1− PBP 2c− PBP 2d− PBP 4− | 17 | 38 | 72 |
DISCUSSION
Studies with several species have shown that strains lacking one or more class A high-molecular-weight PBPs were viable, suggesting that the multiple PBPs within a cell have somewhat redundant functions. However, removal of two specific class A PBPs of the three present in either E. coli or S. pneumoniae is lethal (14, 25, 40), indicating that some class A PBP function is essential. In the present study, we have demonstrated that a B. subtilis strain lacking all four class A PBPs is viable, although its growth rate is diminished threefold. The viability of the quadruple-mutant strain, as well as structural analysis of its PG, demonstrates that it can synthesize PG, indicating that an unidentified protein, or protein complex, is performing the required GT activity. We were able to demonstrate that this GT could synthesize PG in vitro and that this activity was sensitive to the GT inhibitor moenomycin.
In contrast to what was observed in vitro, the novel GT activity may be moenomycin insensitive in vivo. When strains containing PBP 1 were exposed to moenomycin, the growth of the culture entered a lag phase and the cells became filamentous and bent, a morphology very similar to what was seen in the quadruple mutant during exponential growth. These cells eventually resumed growth with a morphology similar to that of the quadruple mutant. Cells lacking at least PBP 1 were largely unaffected by exposure to moenomycin. These results suggest that in a wild-type strain PBP 1 is the major GT and that the novel GT is either not expressed or not in a position to take over the major role. In the absence of PBP 1, in the absence of all class A PBPs, or in the presence of moenomycin, the novel GT polymerizes PG in an environment that is inaccessible to moenomycin in vivo. Alternatively, we may have observed the activities of two different GTs: one, observed in vivo, which is moenomycin resistant, and the other, observed in vitro, which is moenomycin sensitive. The abilities of different strains and of individual cells to survive moenomycin exposure and the rate at which they can adapt and reinitiate growth may be a function of two factors, the percentage of their PG synthetic activity that is being carried out by class A PBPs and the percentage of cells at a particular stage of the cell cycle that is inherently sensitive or resistant to moenomycin.
Structural analysis of PG from all class A PBP mutant strains showed that the percentage of peptide side chains involved in cross-linking was ∼24% in all strains that lacked PBP 1 and ∼22% in strains that lacked both PBPs 1 and 4, including the quadruple mutant. These are 9 and 15% decreases, respectively, from the 26% cross-linking found in the wild type. Atrih et al. (5) found a larger decrease in cross-linked muropeptides in the PBP 1− strain (22% decrease) than in the wild type. However, the PG they used for analyzing strain differences was prepared from late stationary phase cultures, whereas we compared PG from exponentially growing cultures. Decreases in cross-linking may be directly due to the loss of the transpeptidase activity of PBP 1 or to the loss of PBP 1-containing complexes that bring other PBPs or wall-modifying proteins to sites of PG incorporation.
In the absence of PBP 1, the PG synthetic machinery produces variability in cell morphology, wall thickness, and septation (27, 33). The role that PBP 1 plays in the control of wall formation is apparently important, but how it interacts with other proteins or cell structures to determine cell morphology is currently unknown. Recent data showed the presence of helical filaments, made up of MreB and Mbl monomers, lying just underneath the cytoplasmic membrane of B. subtilis (17). These proteins play roles in determining cell morphology (1, 17), and the helical filaments have been suggested to potentially exert spatial control on the PG synthetic machinery (17). If PBP 1 is part of the pathway through which these proteins exert their effect on maintenance of a straight rod shape with consistent diameter, then the other class A PBPs and a novel GT must be able to at least partially fill this role. Furthermore, Pedersen et al. found that in 48% of cells lacking PBP 1, FtsZ localization was disrupted (27), suggesting that there is interplay between the wall synthetic machinery containing PBP 1 and the cell division apparatus involving FtsZ. The question of what protein-protein interactions might be occurring between PG synthesizing machinery (PBP 1?), proteins that regulate placement of cell division septa (Fts proteins), and proteins required for maintenance of cell morphology (helical filaments and class B PBPs) is an intriguing area for future studies. Presumably, all these systems can also interact to some degree with the novel GT in order to produce rod-shaped cells and septa, suggesting that even in the presence of PBP 1 this novel GT may be part of the PG synthetic apparatus.
The identity of the novel GT polymerizing PG in a strain lacking all class A PBPs is unknown. In the B. subtilis genome (19), we find no protein product containing significant sequence homology to the five motifs that are consistently found in all GT enzymes involved in synthesizing PG (10), indicating the presence of a novel class of GT. It has been proposed (15) and refuted (2, 38) that class B PBPs may possess a GT activity in their N-terminal domains. We found that the relative abundance of two class B PBPs, 2a and 2b, increased in the absence of PBP 1 and more so in cells lacking both PBP 1 and PBP 4. The possibility remains that these two proteins may be involved, alone or in complex with other proteins, in the novel GT activity. Other candidate GT proteins may be those of the SEDS family (mnemonic for “shape, elongation, division, sporulation”) (13) such as RodA, which was enriched (along with a class B PBP) in a membrane preparation that exhibited high levels of GT activity (15, 16). In addition, the B. subtilis genome encodes several uncharacterized proteins with similarity to GTs involved in the polymerization of other polysaccharides (8). Identification of this novel GT in B. subtilis may prove helpful to those researching this activity as a potential antibiotic target site.
Acknowledgments
This work was supported by grant GM56695 from the National Institutes of Health (D.L.P.) and by grants from Sigma Xi (Grants-in-Aid of Research) and the Virginia Polytechnic Institute and State University Graduate Student Associations' Graduate Research Development Program (D.C.M.).
We thank David Nelson and Kevin Young for advice on the use of 125I-penicillin X, Kit Pogliano for advice on fluorescence microscopy, Jill Sible and Rich Walker for advice and use of their microscopes, Bob Grant for the gift of moenomycin, and Marita Seppanen Popham for editing the manuscript.
REFERENCES
- 1.Abhayawardhane, Y., and G. C. Stewart. 1995. Bacillus subtilis possesses a second determinant with extensive sequence similarity to the Escherichia coli mreB morphogene. J. Bacteriol. 177:765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, M., C. Fraipont, N. Rhazi, M. Nguyen-Disteche, B. Lakaye, J. M. Frere, B. Devreese, J. Van Beeumen, Y. van Heijenoort, J. van Heijenoort, and J. M. Ghuysen. 1997. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J. Bacteriol. 179:6005-6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:74-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 5.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Berardino, M., A. Dijkstra, D. Stuber, W. Keck, and M. Gubler. 1996. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesising enzymes. FEBS Lett. 392:184-188. [DOI] [PubMed] [Google Scholar]
- 8.Foster, S. J., and D. L. Popham. 2001. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its close relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 9.Ghuysen, J.-M. 1991. Serine β-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 45:37-67. [DOI] [PubMed] [Google Scholar]
- 10.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Castro, M. J., J. López-Hernández, J. Simal-Lozano, and M. J. Oruña-Concha. 1997. Determination of amino acids in green beans by derivitization with phenylisothiocyanate and high-performance liquid chromatography with ultraviolet detection. J. Chromatogr. Sci. 35:181-185. [Google Scholar]
- 12.Guérout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-337. [DOI] [PubMed] [Google Scholar]
- 13.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 14.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishino, F., and M. Matsuhashi. 1981. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem. Biophys. Res. Commun. 101:905-911. [DOI] [PubMed] [Google Scholar]
- 16.Ishino, F., W. Park, S. Tomioka, S. Tamaki, I. Takase, K. Kunugita, H. Matsuzawa, S. Asoh, T. Ohta, B. G. Spratt, and M. Matsuhashi. 1986. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 261:7024-7031. [PubMed] [Google Scholar]
- 17.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 18.Kleppe, G., W. Yu, and J. L. Strominger. 1982. Penicillin-binding proteins in Bacillus subtilis mutants. Antimicrob. Agents Chemother. 21:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 20.Kurz, M., W. Guba, and L. Vertesy. 1998. Three-dimensional structure of moenomycin A—a potent inhibitor of penicillin-binding protein 1b. Eur. J. Biochem. 252:500-507. [DOI] [PubMed] [Google Scholar]
- 21.Leighton, T. J., and R. H. Doi. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 254:3189-3195. [PubMed] [Google Scholar]
- 22.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 23.Lugtenberg, E. J., A. van Schijndel-van Dam, and T. H. van Bellegem. 1971. In vivo and in vitro action of new antibiotics interfering with the utilization of N-acetyl-glucosamine-N-acetyl-muramyl-pentapeptide. J. Bacteriol. 108:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McPherson, D. C., A. Driks, and D. L. Popham. 2001. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 183:6046-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paik, J., I. Kern, R. Lurz, and R. Hakenbeck. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J. Bacteriol. 181:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, J. T., and A. N. Chatterjee. 1954. Membrane associated reactions involved in bacterial cell wall mucopeptide synthesis. Fed. Proc. 13:466-472. [Google Scholar]
- 27.Pedersen, L. B., E. R. Angert, and P. Setlow. 1999. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J. Bacteriol. 181:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen, L. B., K. Ragkousi, T. J. Cammett, E. Melly, A. Sekowska, E. Schopick, T. Murray, and P. Setlow. 2000. Characterization of ywhE, which encodes a putative high-molecular-weight class A penicillin-binding protein in Bacillus subtilis. Gene 246:187-196. [DOI] [PubMed] [Google Scholar]
- 29.Pogliano, J., N. Osborne, M. D. Sharp, A. Abanes-De Mello, A. Perez, Y. L. Sun, and K. Pogliano. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popham, D. L., and P. Setlow. 1995. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J. Bacteriol. 177:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popham, D. L., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpF gene, which codes for a putative class A high-molecular-weight penicillin-binding protein. J. Bacteriol. 175:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popham, D. L., and P. Setlow. 1994. Cloning, nucleotide sequence, mutagenesis, and mapping of the Bacillus subtilis pbpD gene, which codes for penicillin-binding protein 4. J. Bacteriol. 176:7197-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, P. E. 1971. Peptidoglycan synthesis in bacilli. I. Effect of temperature on the in vitro system from Bacillus megaterium and Bacillus stearothermophilus. Biochim. Biophys. Acta 237:239-254. [PubMed] [Google Scholar]
- 35.Schaller, K., J. V. Holtje, and V. Braun. 1982. Colicin M is an inhibitor of murein biosynthesis. J. Bacteriol. 152:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taku, A., and D. P. Fan. 1979. Dissociation and reconstitution of membranes synthesizing the peptidoglycan of Bacillus megaterium. A protein factor for the polymerization step. J. Biol. Chem. 254:3991-3999. [PubMed] [Google Scholar]
- 37.van Heijenoort, Y., M. Derrien, and J. van Heijenoort. 1978. Polymerization by transglycosylation in the biosynthesis of the peptidoglycan of Escherichia coli K 12 and its inhibition by antibiotics. FEBS Lett. 89:141-144. [DOI] [PubMed] [Google Scholar]
- 38.van Heijenoort, Y., M. Gomez, M. Derrien, J. Ayala, and J. van Heijenoort. 1992. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J. Bacteriol. 174:3549-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Q. M., R. B. Peery, R. B. Johnson, W. E. Alborn, W. K. Yeh, and P. L. Skatrud. 2001. Identification and characterization of a monofunctional glycosyltransferase from Staphylococcus aureus. J. Bacteriol. 183:4779-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yousif, S. Y., J. K. Broome-Smith, and B. G. Spratt. 1985. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 131:2839-2845. [DOI] [PubMed] [Google Scholar]