Abstract
We generated random transposon insertion mutants to identify genes involved in light input pathways to the circadian clock of the cyanobacterium Synechococcus elongatus PCC 7942. Two mutants, AMC408-M1 and AMC408-M2, were isolated that responded to a 5-h dark pulse differently from the wild-type strain. The two mutants carried independent transposon insertions in an open reading frame here named ldpA (for light-dependent period). Although the mutants were isolated by a phase shift screening protocol, the actual defect is a conditional alteration in the circadian period. The mutants retain the wild-type ability to phase shift the circadian gene expression (bioluminescent reporter) rhythm if the timing of administration of the dark pulse is corrected for a 1-h shortening of the circadian period in the mutant. Further analysis indicated that the conditional short-period mutant phenotype results from insensitivity to light gradients that normally modulate the circadian period in S. elongatus, lengthening the period at low light intensities. The ldpA gene encodes a polypeptide that predicts a 7Fe-8S cluster-binding motif expected to be involved in redox reactions. We suggest that the LdpA protein modulates the circadian clock as an indirect function of light intensity by sensing changes in cellular physiology.
Light is a major environmental signal that synchronizes circadian rhythms in various organisms to coordinate the daily oscillations of biological phenomena with the sidereal day. The input pathways that receive and transmit environmental signals to the central circadian oscillator of circadian systems vary among organisms, particularly in the mechanisms of perception of light signals. In mammals, light that sets the circadian clock is received by membrane-associated opsin proteins in the eyes that carry retinal chromophores (15, 39). In Drosophila, both opsin-based photopigments and the blue light photoreceptor cryptochrome function as photoreceptors for input pathways (39). In Neurospora, the putative transcription factors WC-1 and WC-2, which interact with FRQ, a critical element of the circadian system, are necessary for light perception (10, 25). In higher plants, phytochromes and cryptochrome transduce light signals to the clock (38).
Cyanobacteria are the only prokaryotes demonstrated thus far to have bona fide circadian systems that have the properties of those in eukaryotic organisms (13). In various cyanobacterial genera, the circadian clock has been shown to regulate such diverse biological activities as nitrogen fixation (14), amino acid uptake (8), and gene expression (22, 24). Several genes related to circadian clock function have been identified in the unicellular cyanobacterium Synechococcus elongatus PCC 7942, a strain for which many genetic tools are available. These include the kaiA, kaiB, and kaiC genes, essential components for circadian rhythms (16).
One basic aspect of a circadian rhythm is the free-running period (FRP), which is the duration of the oscillation, peak to peak or trough to trough, under constant environmental conditions (4). Several loci in S. elongatus are known to affect the FRP under conditions of constant light and temperature. Many alleles of the kaiA, kaiB, and kaiC genes affect the FRP (16). An extra copy of the pex gene, which encodes a protein of unknown function, extends the circadian period by about 2 h (23). The sasA gene, which encodes a histidine protein kinase, amplifies circadian oscillation and may be a major conduit for temporal information out of the clock to the genes it controls (17). The residual rhythmicity in an sasA mutant has a short FRP. The only component known to be involved in an input pathway for resetting the clock is the histidine protein kinase CikA, which belongs to the bacteriophytochrome family of proteins (32). Mutants defective in cikA have a short FRP.
In this study, we identified the ldpA (light-dependent period) gene of S. elongatus PCC 7942 as encoding a new component of an input pathway to the cyanobacterial clock. Its predicted product is a previously undescribed protein that carries iron-sulfur cluster-binding motifs. Disruption of ldpA significantly attenuated the ability of S. elongatus to modulate the FRP under different light intensities. Our results suggest that the circadian clock is modulated by the redox state in cyanobacteria, acting at least in part through the iron-sulfur protein LdpA.
MATERIALS AND METHODS
Strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All Synechococcus strains were grown in modified BG-11 medium (BG-11 M) (6) or on BG-11 M agar plates under continuous light (100 microeinsteins m−2 s−1) at 30°C. Spectinomycin (5 μg ml−1), chloramphenicol (7.5 μg ml−1), kanamycin (5 μg ml−1), or gentamicin (1 μg ml−1) was used as needed for selection of S. elongatus transformants.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) or genotype |
|---|---|
| E. coli strains | |
| AM1452a | HB101 with helper plasmid pRL528 and transposon plasmid pAM1037 |
| AM1460 | HB101 with plasmid pRK2013b to provide conjugal transfer functions |
| DH10B | Host for plasmids |
| S. elongatus strains | |
| PCC 7942 | Wild type |
| C22ab | Mutant of AMC149; point mutation in kaiC gene (C3535T) causes short circadian period (formerly called SP22) |
| AMC149 | PpsbAI::luxAB reporter inserted into NSI (Spr) |
| AMC403 | PpsbAI::luxAB reporter inserted into NSII (Kmr) |
| AMC408 | PpurF::luxAB inserted into NSII (Cmr), PpsbAI::luxCDE inserted into NSI (Spr) |
| AMC408-M1 | Mutant of AMC408 with transposon insertion at nucleotide 729 of ldpA ORF |
| AMC408-M2 | Mutant of AMC408 with transposon insertion at nucleotide 303 of ldpA ORF |
| AMC548 | Derivative of C22a with PpsbAI::luxCDE in NSII (Cmr) |
| AMC664 | Derivative of AMC408 with ldpA inactivated by recombination with pAM2188 |
| AMC665 | Derivative of AMC548 with ldpA inactivated by recombination with pAM2188 |
| AMC693 | PpurF::luxAB reporter gene fusion inserted into NSI (Smr) |
| AMC694 | Derivative of AMC693 with ldpA inactivated by recombination with pAM2180 and psbAI::luxCDE gene fusion introduced into NSII (Cmr) by recombination with pAM1706 |
| AMC695 | Derivative of AMC693 with ldpA inactivated by recombination with pAM2181 and psbAI::luxCDE gene fusion introduced into NSII (Cmr) by recombination with pAM1706 |
| AMC696 | Derivative of AMC693 with ldpA inactivated by recombination with pAM2180 and ectopic copy of ldpA and PpsbAI::luxCDE gene fusion introduced into NSII (Cmr) by recombination with pAM2186 |
| AMC697 | Derivative of AMC693 with ldpA inactivated by recombination with pAM2181 and ectopic copy of ldpA and PpsbAI::luxCDE gene fusion introduced into NSII (Cmr) by recombination with pAM2186 |
| Plasmids | |
| pAM1037c | Tn5 plasmid derivative pRL1058 further modified by insertion of 0.7-kb fragment into XbaI site to add outward-reading promoters from Anabaena sp. strain PCC 7120 glnA and rbcL genes |
| pAM1706 | Vector that transfers PpsbAI::luxCDE fusion to NSII (subsite NS2.2, BglII) of S. elongatus chromosome |
| pAM2180 | Tn5-containing plasmid derivative recovered from mutant AMC408-M1 by KpnI digestion and circularization |
| pAM2181 | Tn5-containing plasmid derivative recovered from mutant AMC408-M2 by KpnI digestion and circularization |
| pAM2182 | pBluescript II SK+ containing 3.7-kb EcoRI-HpaI fragment of pAM2180 at EcoRI-SmaI sites |
| pAM2183 | PBluescript II SK+ containing 2.3-kb HpaI-BglII fragment of pAM2181 at SmaI-BamHI sites |
| pAM2184 | Entire ORF and 5′- and 3′-flanking regions of ldpA inserted into pBluescript II SK+ at EcoRI-BamHI sites |
| pAM2185 | pBluescript II KS+ containing 1.3-kb PvuII fragment including ldpA ORF at EcoRV site in same orientation as lacZ promoter |
| pAM2186 | Derivative of pAM1706 with PlacZ::ldpA fusion upstream of psbAI::luxCDE |
| pAM2188 | Gentamicin resistance cassette inserted into AlwNI site of pAM2188 |
Transposon mutagenesis.
A modified Tn5 transposon was introduced into wild-type AMC408 by conjugal transfer from Escherichia coli of pAM1037, a derivative of pRL1058, as described previously (20). AMC408 carries a PpurF::luxAB reporter that exhibits class 2 circadian bioluminescence rhythms (24). Restriction enzymes and modifying enzymes were purchased from Promega and used as directed by the manufacturer. DNA flanking the transposon was recovered by extracting genomic DNAs from exconjugants, digesting them with KpnI, circularizing them with T4 DNA ligase, and introducing them as plasmids into E. coli DH10B by electroporation (2, 20). Kmr plasmids were recovered, and nucleotide sequences of the genomic regions flanking the transposons were determined by cycle sequencing (dye terminator cycle sequencing ready reaction; ABI PRISM; PE Applied Biosystems, Foster City, Calif.) with primers AMO134 and AMO285, which are complementary to each end of the transposon.
Sequence analysis and Fe-S motif determination.
Sequence motif determination was performed by using multiple approaches. The full-length LdpA sequence was first used to search against the nonredundant protein database and the protein structural database (PDB) at the National Center for Biotechnology Information by using BLAST (1) (http://www.ncbi.nlm.nih.gov/BLAST/). In this search, we used the BLOSUM45 amino acid substitution matrix, with a gap opening penalty of 11, an extension penalty of 1, an Expect value of 10, and a word size of 3. A more sensitive database search using a Bayesian algorithm (37) was carried out with the BALSA program (http://bayesweb.wadsworth.org/balsa/balsa.html). Sequence motifs were further searched for the LdpA aminoacid sequence by using the FingerPRINTScan (http://www.bioinf.man.ac.uk/fingerPRINTScan/), eMOTIF Search (http://motif.stanford.edu/emotif/emotif-search.html), Block (http://blocks.fhcrc.org/blocks/blocks_search.html), and InterPro (http://www.ebi.ac.uk/interpro/) servers. Structural fold prediction was done by using a threading server, BIOINBGU (http://www.cs.bgu.ac.il/∼bioinbgu/form.html). Multiple-sequence alignment of LdpA with three ferredoxins with known 7Fe-8S structures was done by using the T-COFFEE server (http://www.ch.embnet.org/software/TCoffee.html), followed by manual refinement. To assess the statistical significance of sequence similarities between LdpA and 7Fe-8S ferredoxins, we used an alignment-independent program, PRSS, that calculates the probability of similarities of randomly shuffled and unshuffled sequences by using a distance matrix Monte Carlo procedure (29). The analysis was done in the Biology Workbench web server (http://workbench.sdsc.edu/). The gap-opening penalty was set at 12, and the gap-extending penalty was set at 2, and 1,000 global shuffles were performed by using the BLOSUM50 scoring matrix.
Construction of plasmids for complementation and disruption of ldpA.
An intact copy of ldpA was reconstructed from the transposon insertion-containing alleles recovered from AMC408-M1 and AMC408-M2. We excised a 3.7-kb EcoRI-HpaI fragment that includes the N terminus-encoding and upstream flanking regions of ldpA from pAM2180 (Kmr plasmid recovered from AMC408-M2; Fig. 2) and inserted it into EcoRI- and SmaI-digested pBluescriptII SK+ to create pAM2182. The 2.3-kb HpaI-BglII fragment, which encodes the C-terminal portion and downstream flanking region of ldpA, was excised from pAM2181 (Kmr plasmid recovered from AMC408-M1) and inserted into SmaI- and BamHI-digested pBluescriptII SK+ to create pAM2183. After complete digestion of pAM2182 with EcoRI and its partial digestion with PstI, a 2.9-kb EcoRI-PstI fragment was cloned into EcoRI- and PstI-digested pAM2183 to create pAM2184. pAM2184 contains the entire open reading frame (ORF) and the upstream and downstream flanking regions of ldpA. The 1.3-kb PvuII fragment that contains ldpA was excised from pAM2184 and cloned into EcoRV-digested pBluescriptII SK+ to create pAM2185. A 1.7-kb PvuII fragment in which ldpA is downstream of the lacZ promoter was excised from pAM2185 and cloned into SmaI-digested pAM1706 to create pAM2186, a vector that targets inserted genes to an S. elongatus locus called neutral site II (NSII; GenBank accession no. U44761). In this plasmid, ldpA is inserted upstream of the luxCDE genes (present to direct synthesis of the aldehyde substrate of the luciferase reporter) but transcribed divergently. The recreated transposon mutants AMC694 and AMC695 were transformed with pAM2186 to create complemented strains AMC696 and AMC697, respectively.
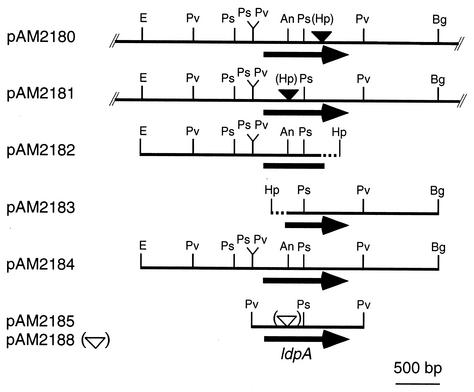
FIG. 2.
Physical map of the ldpA region of the S. elongatus chromosome and plasmids used to reconstruct intact and disrupted ldpA alleles. Restriction sites are indicated by the following abbreviations: E, EcoRI; Pv, PvuII; Ps, PstI; An, AlwNI; Bg, BglII; Hp, HpaI. Insertion sites of the transposon and Gmr cassette are indicated by filled and open triangles, respectively. Broken lines indicate regions derived from the transposon. Plasmids pAM2180 and pAM2181 were recovered by restriction digestion and circularization from genomic DNA of mutants AMC408-M1 and AMC408-M2, respectively.
For directed disruption of ldpA, pAM2185 was partially digested with AlwNI and made blunt ended by using T4 DNA polymerase. Into the linearized plasmid, we inserted a 2.0-kb PvuII-SmaI fragment from pAM2055 that includes a gentamicin resistance (Gmr)-encoding gene to create pAM2188. We transformed wild-type PpurF::luxAB reporter strain AMC408 and short-period (22 h) mutant AM548 with pAM2188 to create AMC664 and AMC665, respectively. Complete segregation of the mutant allele was confirmed by PCR with primers 5′-AGAACTTCGGGATGGGC-3′ and 5′-GAACGTCTAACAGGACG-3′, which cross the insertion site of the gene encoding Gmr.
Assay of bioluminescence rhythms.
The original Tn5 insertion mutants were identified by bioluminescence monitoring with a Packard TopCount luminometer (2). Briefly, mutant exconjugants grown on BG-11 M agar in continuous light (LL) were inoculated into BG-11 M and incubated on a rotary shaker for 2 weeks. Cyanobacterial suspensions (10 μl) at an optical density at 750 nm (OD750) of 0.7 were used to inoculate BG-11 M agar pads in 96-well sample plates. These plates were incubated under standard LL conditions for 24 h and then subjected to a 12-h dark interval to synchronize the clocks of all of the cells in the population prior to bioluminescence monitoring on the TopCount luminometer. Illumination around the TopCount stackers results in a light gradient over the 96-well measuring plates, such that outer wells (columns 1 and 12) receive a fluence of approximately 225 microeinsteins m−2 s−1 and the inner wells (columns 6 and 7) receive approximately 50 microeinsteins m−2 s−1.
For measurement of the bioluminescence rhythm under turbidostatic cultivation conditions, 150-ml cyanobacterial suspensions with an OD730 of 0.7 to 1.0 were used to inoculate 1.4 liters of BG-11 M in flat bottles. The cultures were bubbled with air under standard LL conditions and incubated until the OD730 reached 0.25. The cultures were then subjected to a 12-h dark interval to synchronize their circadian rhythms. Cell density was monitored by measurement of infrared light penetration into the cell suspension. An increase in OD730 triggered a switch that activated a pump to add fresh medium and keep an OD730 of 0.25. An aliquot (24 ml) was withdrawn every hour, and bioluminescence was measured by a photomultiplier tube apparatus. The output of the photomultiplier tube was amplified electronically and read by a computer analog-to-digital converter. The reading was averaged over the measurement period and plotted on the ordinate in millivolts as bioluminescence. Phase shifts and FRPs were calculated by regression procedures as described previously (32).
RESULTS
Screening for light input mutants.
We generated random transposon mutations in a PpurF::luxAB bioluminescent reporter strain of S. elongatus PCC 7942 (AMC408) and screened for mutants affected in light input pathways to the circadian system. In all screenings, a 12-h dark interval or a temperature cycle was used for synchronization of the circadian rhythm first, and then photic signals (5- or 12-h dark interval) were applied at specific times of the day that induce a circadian phase shift in the wild type. We chose exconjugants that showed phases different from that of the wild-type strain after the second dark pulse as candidate input mutants.
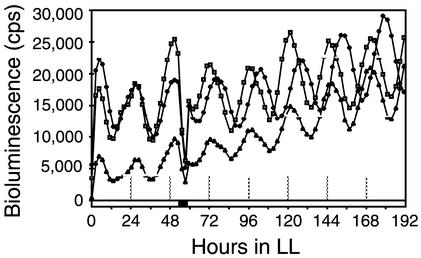
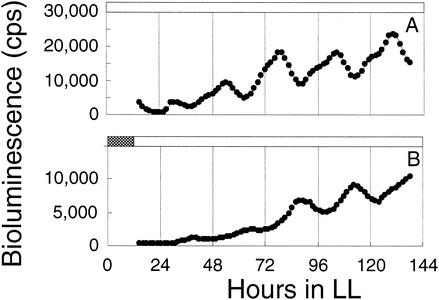
We used three screening protocols to accommodate mutants that might or might not be synchronized initially by photic signals. All of the protocols used a 5-h dark pulse that, for wild-type S. elongatus, would reset the relative phasing of circadian rhythms by as much at 10 h, depending on the stage in the circadian cycle at which it was administered (32). The protocols were as follows: (i) a 12-h dark interval to synchronize rhythms, followed by a transfer to LL and a 5-h dark pulse that started 53 h after the transfer to LL (56 mutant candidates among 700 exconjugants); (ii) a temperature cycle of 12 h at 40°C, 12 h at 30°C, and 12 h at 40°C in LL, followed by a 12-h dark interval that began 36 h after the end of the last 40°C pulse (234 mutant candidates among 2,600 exconjugants); and (iii) a temperature cycle of 12 h at 40°C, 12 h at 30°C, and 12 h at 40°C in LL, followed by a 5-h dark pulse that began 53 h after the end of the last 40°C pulse (93 mutant candidates among 700 exconjugants). From these candidate input mutants, we chose five exconjugants that reproducibly showed phases different from that of the wild-type strain after the phase-resetting dark pulse and recovered the transposons and flanking DNA as Kmr plasmids. The mutants were recreated by transforming AMC408 with the plasmids that had been linearized by cleavage with the restriction enzyme used for their recovery. Two mutants, AMC408-M1, which was originally isolated by protocol 3, and AMC408-M2, isolated by protocol 1, retained the phenotypes expected for input mutants (Fig. 1). These mutants also had a slightly shorter free-running circadian period than the wild-type strain under these screening conditions (Fig. 1).
FIG. 1.
Traces of circadian rhythms of bioluminescence from PpurF::luxAB reporter strains. Symbols: diamonds, wild type; triangles, mutant AMC408-M1; squares, mutant AMC408-M2. Hour zero in LL indicates the time point of the end of the 12-h dark interval used for initial synchronization. The black bar indicates the time of application of a 5-h dark pulse for phase resetting.
Identification of transposon insertion sites.
To identify the sites of mutations in AMC408-M1 and AMC408-M2, we determined the nucleotide sequences of the genomic regions flanking the transposons. Nucleotide sequencing revealed that the two strains carry independent insertions in the same ORF, as shown in Fig. 2. We designated this ORF the ldpA gene on the basis of phenotypic characteristics described below.
Translation of the ldpA ORF and comparison to similar sequences in public databases suggested that the start codon is a GTG and that LdpA is a soluble protein of 352 amino acid residues. Putative ldpA homologs (percent amino acid identity) are encoded in the genomes of Synechocystis sp. strain PCC 6803 (sll0031, 49%), Anabaena sp. strain PCC 7120 (alr2308, 44%), Nostoc punctiforme ATCC 29133 (44.0%), marine Synechococcus sp. strain WH8102 (43%), and Prochlorococcus marinus strain MED4 (35%), indicating conservation among diverse cyanobacteria.
Determination of an LdpA Fe-S motif.
The deduced amino acid sequence of LdpA was first searched against the National Center for Biotechnology Information nonredundant database by using a gapped BLAST (1). Seven significant database hits (E < 10−6) were recovered that were all annotated as hypothetical proteins from recently sequenced cyanobacterial genomes. A similar BLAST search against a more restricted database for protein structures (PDB) was done that generated no significant hits. However, the best hit (PDB code 1H98) was a ferredoxin with 3Fe-4S and 4Fe-4S centers from Thermus thermophilus (Table 2). A more sensitive database search was conducted with a Bayesian algorithm (37) that identified significant hits as a ferredoxin from Clostridium acidurici (PDB code 1FCA) with two 4Fe-4S centers and another from Azotobacter vinelandii with 7Fe-8S centers (Table 2).
TABLE 2.
Determination of Fe-S center types in LdpA by using multiple sequence motif search methods
| Center type | Statistical supporta
|
||||||
|---|---|---|---|---|---|---|---|
| BLAST against PDB | BALSA | FingerPRINTScan | eMOTIF | Block | InterProb | BIOINGBUc | |
| 4Fe-4S | 5.10 × 10−6 | 9.90 × 10−6 | + | ||||
| 3Fe-4S | 1.34 × 10−3 | + | |||||
| 7Fe-8S | 0.40 | 3.68 × 10−4 | 4.40 × 10−8 | 1.20 × 10−5 | + | 14.8 | |
| 8Fe-8S | 1.70 × 10−6 | 12.7 | |||||
Only the most significant (or best) hits are shown. Statistical support (P or E value) is indicated whenever available.
For the InterPro search, + means a positive search result with no quantitative value.
In the BIOINBGU threading search, the statistical confidence level is 12.
To determine more accurately the type of Fe-S centers likely to be present in LdpA, we analyzed the sequence by using multiple motif prediction programs that have different sensitivities for certain domains because of intrinsic differences in algorithms and database content (Table 2). Indeed, our searches indicated the presence of 4Fe-4S, 3Fe-4S, and 7Fe-8S types of clusters with significant statistical support. Furthermore, a structural fold recognition analysis carried out for LdpA generated a most significant hit for a 7Fe-8S ferredoxin (PDB code 5FD1 from A. vinelandii) and second most significant hit for an 8Fe-8S ferredoxin (PDB code 1CLF from C. pasteurianum). The result demonstrates a high probability that the LdpA protein adopts a tertiary structure similar to that of the 7Fe-8S ferredoxin in A. vinelandii. In reviewing all of the available evidence, the consensus appears to be that LdpA has two Fe-S centers, one for 3Fe-4S and another for 4Fe-4S. A more refined sequence alignment between LdpA and proteins with known 7Fe-8S centers (3, 7, 26) showed full conservation of all Cys residues at the putative Fe-S center domain (Fig. 3). Statistical support (P values) for each pairwise comparison (29) between LdpA and known 7Fe-8S proteins is shown in Table 2. The entropy-based analysis without influence of the alignment shows that the sequences are indeed significantly similar, with 100% of the comparisons having P values smaller than 0.001, which is significant enough to establish a direct homologous relationship.
FIG. 3.
Comparison of the deduced amino acid sequence encoded by the ldpA gene of S. elongatus PCC 7942 and ferredoxins with known 7Fe-8S centers. LdpA, GenBank accession no. AY136759; T. thermophilus ferredoxin, SwissProt accession no. P03942; Bacillus schlegelii ferredoxin, SwissProt accession no. Q45560; A. vinelandii FdI, SwissProt accession no. P00214. Only the Fe-S center domain is shown. Conserved Cys residues are highlighted, and corresponding Fe-S centers are indicated by brackets. The PDB codes for the ferredoxins from A. vinelandii, T. thermophilus, and B. schlegelii are 1F5B, 1H98, and 1BC6, respectively. Pairwise sequence similarities between LdpA and other ferredoxins were assessed by an alignment-independent PRSS test. Statistical significance (P values) for the pairwise sequence comparison is indicated on the right of the sequence alignment.
Insertional inactivation of ldpA and complementation of the mutant phenotype by a wild-type allele.
We created a Gmr interruption allele of ldpA by introducing the drug resistance cartridge into an AlwNI site that is near the insertion site of the transposon in mutant AMC408-M2. This reconstructed mutant had the short-period phenotype observed in the original transposon mutants (Table 3). To confirm that ldpA alone is responsible for the phenotype, we introduced a wild-type ldpA gene into ldpA Tn5 mutants. For this experiment, we used strains AMC694 and AMC695, which carry the M1 and M2 Tn5 mutations in a background with a reporter configuration different from that of AMC408. A heterologous promoter, PlacZ, was provided in the vector because sequence data suggested that ldpA is downstream of a thioredoxin reductase gene with which it might be cotranscribed; thus, the ldpA fragment might not have its own promoter (data not shown). As shown in Table 3, the FRPs of both AMC694 and AMC695 were restored to the wild-type FRP by the introduction of ldpA (AMC696 and AMC697, respectively). We concluded that loss of function of ldpA is responsible for the short-period phenotypes in the collection of ldpA mutants.
TABLE 3.
FRPs of ldpA mutants and complemented derivatives
| Strain | Avg FRP a (h) ± SD (n = 6) |
|---|---|
| Wild type | 25.6 ± 0.06 |
| AMC694 (ldpA) | 24.6 ± 0.18 |
| AMC665 (ldpA) | 24.5 ± 0.31 |
| AMC695 (ldpA) | 24.7 ± 0.18 |
| AMC696 (ldpA+) | 25.8 ± 0.28 |
| AMC697 (ldpA+) | 25.5 ± 0.49 |
FRPs were measured under white light (25 microeinsteins m−2 s−1).
Characterization of the light input phenotype of ldpA mutants.
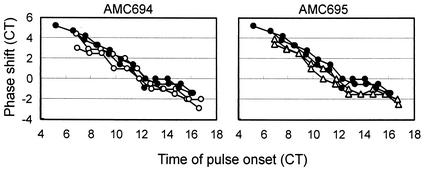
To elucidate the presumed input pathway defect of the mutants, we performed a series of phase-resetting experiments. A dark pulse administered at different times in the circadian cycle reproducibly causes phase shifts that differ with respect to magnitude and direction in S. elongatus (32). We compared the relationships between the timing of the 5-h dark pulse relative to the circadian cycle and the resultant phase shift in the wild type and in ldpA mutants. The results are plotted as phase response curves (PRCs) in Fig. 4. The shapes of the PRCs of AMC694 and AMC695 are almost identical to that of the PRC of the wild-type strain. This indicates that both mutants retain the wild-type ability to shift the phase of the circadian rhythm.
FIG. 4.
PRCs for 5-h dark pulses in the wild-type strain (solid circles) and ldpA mutants AMC694 (○) and AMC695 (▵). PRCs for duplicate samples of each strain are shown. Circadian time (CT) is a normalization for the difference between the circadian periods of the strains; 1 circadian h = 1/24 of a full circadian cycle. Positive values represent phase advances, and negative values represent phase delays.
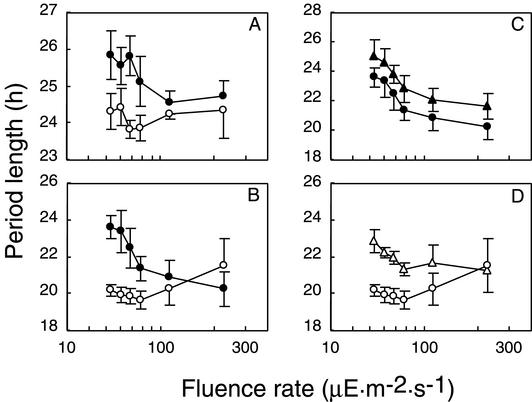
The FRPs of ldpA mutants were shorter than that of the wild-type strain by approximately 1 h under light conditions of 25 microeinsteins m−2 s−1 (Table 3). Light is known to affect circadian systems not only with respect to phase resetting but also by influencing the FRP (4). Generally, increasing light intensity shortens the FRP in diurnal organisms and phototrophic organisms, including unicellular algae (9) and higher plants (34). As shown in Fig. 5A, the FRP of wild-type S. elongatus decreases with increasing light intensity by about 1 h. However, ldpA mutants show no substantive change in period over this range of light intensities (Fig. 5A).
FIG. 5.
Effect of white light fluence on the FRP. Panels A and B show the FRPs of the wild type (A) and C22a (B) (•) and those genetic backgrounds with ldpA disrupted (○). Panels C and D show the FRPs of C22a (C) and C22a with ldpA disrupted (D) with (▵, ▴) and without DCMU addition (○, •). DCMU was added to media at a final concentration of 0.2 μM at the beginning of a 12-h dark interval. Error bars indicate standard deviations (n = 8).
To examine this phenotype in more detail, we inactivated ldpA in the short-period mutant C22a (22 h), which has a point mutation in the kaiC gene (16). The FRP of C22a is more sensitive to the change in light intensity than is the wild-type strain, varying by about 3 h over the light gradient tested (Fig. 5B). Inactivation of ldpA decreased the sensitivity of the period to light intensity in C22a as well (Fig. 5A). These results suggest that the primary effect of inactivation of ldpA is attenuation of the ability to increase the period length with decreasing light intensity, not a defect in the ability to phase shift in response to a dark pulse. This specific phenotype led us to name the locus light-dependent period gene A.
Effect of a photosynthesis inhibitor on the circadian period.
The expectation that LdpA carries Fe-S clusters suggests involvement in electron transfer and redox reactions (5). One possibility is that LdpA mediates a redox signal from the photosynthetic apparatus to the circadian oscillator. To test this, we examined the effect of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), an inhibitor of the photosystem II reaction center, on the free-running circadian period. If the putative LdpA protein transmits photosynthetic information, addition of DCMU would be expected to have a smaller effect on the FRP of an ldpA mutant than on that of a wild-type strain. As shown in Fig. 5C, addition of a sublethal concentration of DCMU increased the FRP in C22a but did not block sensitivity to different light intensities. DCMU also increased the FRP of the C22a-ldpA mutant. The period of the C22a-ldpA mutant was still shorter than that of C22a in the presence of DCMU (Fig. 5D). These results indicate that photosynthetic activity contributes to the light input pathways that modulate FRP, but the putative LdpA protein is not the sole component that transmits light intensity signals to the circadian clock.
Disruption of ldpA had pleiotropic effects. The ldpA disruptants grew slightly more slowly than the wild type, with a doubling time of 7.3 h instead of the 6.1 h of the wild-type strain under a light fluence of 34 microeinsteins m−2 s−1. ldpA mutants were also yellowish green in color because of a phycocyanin content that was only 50 to 70% of that of the wild-type strain. Because a slower growth rate would lead to a lower cell density and greater light penetration into the culture, we wanted to exclude the possibility that the short-period phenotype of ldpA mutants is an indirect effect of the slow growth rate. We compared the FRPs of an ldpA mutant and the wild-type strain under turbidostatic culture conditions in which the OD730 was maintained at 0.25. The mutant showed a consistently shorter period (24.2 h) than the wild-type strain (25.2 h) under 16 microeinsteins of white light m−2 s−1.
DISCUSSION
Mutants identified by a screen for circadian-resetting defects revealed the involvement of a previously undescribed Fe-S protein in the modulation of the circadian period of S. elongatus. The primary defect in the mutants is not an inability to reset per se, as is seen in mutants that lack the CikA histidine protein kinase (32). Rather, ldpA mutants are insensitive to the period lengthening under low light fluence that occurs in the wild-type strain. As a result, even under a gradient of light intensities, ldpA mutants always exhibit an FRP that is at the short end of the spectrum observed for the wild type. Thus, at the highest light intensities we use routinely in the laboratory, the mutant and wild-type strains have equivalent FRPs (Fig. 5). The screen for phase-resetting mutants used a monitoring setup in which a light gradient was present across the 96-well plates; we do not know the fluence received by the controls with which these mutants were compared when they were originally identified. Our current understanding of the phenotype suggests that they were running with a 1-h shorter FRP than the controls, such that the timing of the resetting dark pulse fell at a slightly different point during the circadian cycle for the wild type and the ldpA mutants. With these data in mind, we now pair mutants and controls in the same columns of sample plates for screening (27). Despite the serendipitous identification of these mutants in the resetting screen, ldpA does seem to be involved in an input pathway to the clock but affecting a parametric (recurrent period-modulating) rather than nonparametric (single phase-resetting) aspect. Identification of LdpA provides a means by which to gain insight into the biological mechanisms behind Aschoff's rule, the observation that circadian periods tend to decrease with an increase in light intensity (4).
Several Arabidopsis thaliana mutants have conditional FRP changes depending on the light intensity. Deficiency in phyA (defect in one of the phytochrome photoreceptors) causes an exaggerated increase in FRP relative to that of the wild type in response to decreased red light fluence (34). A cry1 (cryptochrome blue light receptor) mutation similarly affects the response to decreasing blue light (34). The gi-1 weak allele of GI, which encodes a membrane-spanning protein, shortens the FRP and reduces the rate of period lengthening with decreasing fluence (28). Conversely, mutation of ZTL lengthens the FRP and increases the rate of period lengthening with decreasing light intensity (35). Among these, the phyA mutant also has a resetting defect, taking a longer time that the wild type for re-entrainment under 10-h light-10-h dark cycles (34).
In contrast to phyA or cry1 Arabidopsis mutants, disruption of ldpA decreased the rate of change in FRP over a range of light intensities. This is most similar to Arabidopsis GI mutants (28). It is likely that the putative LdpA protein has a function that represses the light sensitivity of the circadian clock such that it tends to run slower at decreased light intensity. Another mutation that causes a short-period phenotype in S. elongatus PCC 7942 is in pex, a gene that encodes a protein of unknown function (23). We expect that cyanobacteria have several input pathways that modulate the FRP, as do plants.
Addition of a sublethal concentration of the photosystem II inhibitor DCMU also increased the FRP. This suggests that photosynthesis can be a part of the light input pathway of the circadian system in cyanobacteria. This is consistent with the finding that pulsed addition of a lethal concentration of DCMU at various times during the circadian cycle resets the rhythm similarly to a dark pulse (K. Okamoto and T. Kondo, unpublished results), as shown here for one time point (Fig. 6). In the eukaryotic alga Lingulodinium polyedrum (formerly Gonyaulax polyedra), a sublethal concentration of DCMU shortens the FRP (30). DCMU also inhibits phase shifting in response to a light pulse in L. polyedrum (19) and in Chlamydomonas reinhardtii (18). Thus, a connection between photosynthetic electron transport and light input pathways of the circadian system is a common characteristic among some phototropic organisms.
FIG. 6.
Resetting of circadian rhythms by DCMU. Cyanobacterial suspensions (3 ml) with an OD730 of 0.3 were exposed a synchronizing 12-h dark interval, and then 1 μM DCMU (A) or 0.05% ethanol (solvent used for DCMU; B) was added. After incubation for 12 h under standard light conditions, cells were collected by centrifugation, washed twice with fresh BG-11 M, and suspended in 3 ml of BG-11 M. Samples (10 μl) were inoculated onto sample plates, and circadian rhythms were measured. The interval of DCMU treatment is indicated by the gray bar in panel B.
The deduced sequence of LdpA predicts that it is a 7Fe-8S protein similar to some ferredoxins (3, 7, 26). This suggests that LdpA is involved in reduction, oxidation, or electron transfer (5). This biochemical function is consistent with a role that represses the sensitivity of the circadian clock to light. We propose that photosynthetic activity (favors reduction) negatively regulates the activity of LdpA, repressing the sensitivity of the circadian clock to light. This model can explain the observation that the short-period phenotype of ldpA mutants is prominent only at low light intensity (Fig. 5).
Alternatively, the putative LdpA protein may function as a redox sensor unrelated to light per se (5). In the SoxR protein of E. coli, oxidative stress changes the redox state in the [2Fe-2S] cluster and leads to a change in its activity as a transcription factor (11, 12). In the FNR protein, exposure to oxygen causes the conversion of [4Fe-4S] to [2Fe-2S] and this conversion causes a change in protein conformation that represses its transcription factor activity (21). The cellular redox state was recently suggested to modulate the mammalian circadian system (31). The reduced phycocyanin content in an ldpA mutant, like the short period, is typical of the phenotype of wild-type S. elongatus cells incubated at a high light intensity. However, in the mutant, the antenna pigment does not increase when cells are incubated under low-light conditions (data not shown). Recent reports indicate that responses to a high light intensity and nutrient limitation in this organism share pathway components, indicating an integration of photoreception and metabolic signals (33, 36). LdpA may lie within such a pathway. Whether the connection is direct or indirect, LdpA seems to adjust the cyanobacterial clock to changes in the light environment.
Acknowledgments
We thank S. Canales, J. L. Ditty, N. Ivleva, H. Min, and D. Bell-Pedersen for helpful comments regarding the manuscript.
This work was supported by National Science Foundation grant MCB-9982852 to S.S.G.; grants 11233203 and COE 13CE2005 to T.K. from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; and a JSPS fellowship to M.K.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, C. R., N. F. Tsinoremas, J. Shelton, N. V. Lebedeva, J. Yarrow, H. Min, S. S. Golden, H. Iwasaki, A. Tanabe, C. H. Johnson, and T. Kondo. 2000. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305:527-542. [DOI] [PubMed] [Google Scholar]
- 3.Aono, S., S. Nakamura, R. Aono, and I. Okura. 1994. Cloning and expression of the gene encoding the 7Fe type ferredoxin from a thermophilic hydrogen oxidizing bacterium, Bacillus schlegelii. Biochem. Biophys. Res. Commun. 201:938-942. [DOI] [PubMed] [Google Scholar]
- 4.Aschoff, J. 1981. Freerunning and entrained circadian rhythms, p. 547-548. In J. Aschoff (ed.), Handbook of behavioral neurobiology, vol. 4. Biological rhythms. Plenum Press, New York, N.Y.
- 5.Beinert, H., R. H. Holm, and E. Munck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. [DOI] [PubMed] [Google Scholar]
- 6.Bustos, S. A., and S. S. Golden. 1991. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J. Bacteriol. 173:7525-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, K., C. A. Bonagura, G. J. Tilley, J. P. McEvoy, Y. S. Jung, F. A. Armstrong, C. D. Stout, and B. K. Burgess. 2002. Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential. Nat. Struct. Biol. 9:188-192. [DOI] [PubMed] [Google Scholar]
- 8.Chen, T. H., T. L. Chen, L. M. Hung, and T. C. Huang. 1991. Circadian rhythm in amino acid uptake by Synechococcus RF-1. Plant Physiol. 97:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, T. S., and T. Roenneberg. 1997. Photobiology of the Gonyaulax circadian system. II. Allopurinol inhibits blue-light effects. Planta 202:502-509. [Google Scholar]
- 10.Froehlich, A. C., Y. Liu, J. J. Loros, and J. C. Dunlap. 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297:815-819. [DOI] [PubMed] [Google Scholar]
- 11.Gaudu, P., N. Moon, and B. Weiss. 1997. Regulation of the soxRS oxidative stress regulon: reversible oxidation of the Fe-S centers of SoxR in vivo. J. Biol. Chem. 272:5082-5086. [DOI] [PubMed] [Google Scholar]
- 12.Gaudu, P., and B. Weiss. 1996. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. USA 93:10094-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golden, S. S., M. Ishiura, C. H. Johnson, and T. Kondo. 1997. Cyanobacterial circadian rhythms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:327-354. [DOI] [PubMed] [Google Scholar]
- 14.Grobbelaar, N., T. C. Huang, H. Y. Lin, and T. J. Chow. 1986. Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol. Lett. 37:173-177.H. T. C. [Google Scholar]
- 15.Hattar, S., H. W. Liao, M. Takao, D. M. Berson, and K. W. Yau. 2002. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishiura, M., S. Kutsuna, S. Aoki, H. Iwasaki, C. R. Andersson, A. Tanabe, S. S. Golden, C. H. Johnson, and T. Kondo. 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281:1519-1523. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki, H., S. B. Williams, Y. Kitayama, M. Ishiura, S. S. Golden, and T. Kondo. 2000. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101:223-233. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, C., T. Kondo, and J. Hastings. 1991. Action spectrum for resetting the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas. 2. Illuminated cells. Plant Physiol. 97:1122-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, C. H., and J. W. Hastings. 1989. Circadian phototransduction: phase resetting and frequency of the circadian clock of Gonyaulax cells in red light. J. Biol. Rhythms 4:417-437. [DOI] [PubMed] [Google Scholar]
- 20.Katayama, M., N. F. Tsinoremas, T. Kondo, and S. S. Golden. 1999. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J. Bacteriol. 181:3516-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 22.Kondo, T., C. A. Strayer, R. D. Kulkarni, W. Taylor, M. Ishiura, S. S. Golden, and C. H. Johnson. 1993. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. USA 90:5672-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutsuna, S., T. Kondo, S. Aoki, and M. Ishiura. 1998. A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 180:2167-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Y., N. F. Tsinoremas, C. H. Johnson, N. V. Lebedeva, S. S. Golden, M. Ishiura, and T. Kondo. 1995. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 9:1469-1478. [DOI] [PubMed] [Google Scholar]
- 25.Loros, J. J., and J. C. Dunlap. 2001. Genetic and molecular analysis of circadian rhythms in Neurospora. Annu. Rev. Physiol. 63:757-794. [DOI] [PubMed] [Google Scholar]
- 26.Macedo-Ribeiro, S., B. M. Martins, P. J. Pereira, G. Buse, R. Huber, and T. Soulimane. 2001. New insights into the thermostability of bacterial ferredoxins: high-resolution crystal structure of the seven-iron ferredoxin from Thermus thermophilus. J. Biol. Inorg. Chem. 6:663-674. [DOI] [PubMed] [Google Scholar]
- 27.Nair, U., J. L. Ditty, H. Min, and S. S. Golden. 2002. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J. Bacteriol. 184:3530-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, D. H., D. E. Somers, Y. S. Kim, Y. H. Choy, H. K. Lim, M. S. Soh, H. J. Kim, S. A. Kay, and H. G. Nam. 1999. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285:1579-1582. [DOI] [PubMed] [Google Scholar]
- 29.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roenneberg, T. 1995. The effects of light on the Gonyaulax circadian system. Ciba Found. Symp. 183:117-133. [DOI] [PubMed] [Google Scholar]
- 31.Rutter, J., M. Reick, L. C. Wu, and S. L. McKnight. 2001. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293:510-514. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz, O., M. Katayama, S. B. Williams, T. Kondo, and S. S. Golden. 2000. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289:765-768. [DOI] [PubMed] [Google Scholar]
- 33.Schwarz, R., and A. R. Grossman. 1998. A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under extreme conditions. Proc. Natl. Acad. Sci. USA 95:11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somers, D. E., P. F. Devlin, and S. A. Kay. 1998. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282:1488-1490. [DOI] [PubMed] [Google Scholar]
- 35.Somers, D. E., T. F. Schultz, M. Milnamow, and S. A. Kay. 2000. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101:319-329. [DOI] [PubMed] [Google Scholar]
- 36.van Waasbergen, L. G., N. Dolganov, and A. R. Grossman. 2002. nblS, a gene involved in controlling photosynthesis-related gene expression during high light and nutrient stress in Synechococcus elongatus PCC 7942. J. Bacteriol. 184:2481-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webb, B. J., J. S. Liu, and C. E. Lawrence. 2002. BALSA: Bayesian algorithm for local sequence alignment. Nucleic Acids Res. 30:1268-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanovsky, M. J., and S. A. Kay. 2001. Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 4:429-435. [DOI] [PubMed] [Google Scholar]
- 39.Zordan, M. A., E. Rosato, A. Piccin, and R. Foster. 2001. Photic entrainment of the circadian clock: from Drosophila to mammals. Semin. Cell Dev. Biol. 12:317-328. [DOI] [PubMed] [Google Scholar]