Abstract
The outermost proteinaceous layer of bacterial spores, called the coat, is critical for spore survival, germination, and, for pathogenic spores, disease. To identify novel spore coat proteins, we have carried out a preliminary proteomic analysis of Bacillus subtilis and Bacillus anthracis spores, using a combination of standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis separation and improved two-dimensional electrophoretic separations, followed by matrix-assisted laser desorption ionization-time of flight and/or dual mass spectrometry. We identified 38 B. subtilis spore proteins, 12 of which are known coat proteins. We propose that, of the novel proteins, YtaA, YvdP, and YnzH are bona fide coat proteins, and we have renamed them CotI, CotQ, and CotU, respectively. In addition, we initiated a study of coat proteins in B. anthracis and identified 11 spore proteins, 6 of which are candidate coat or exosporium proteins. We also queried the unfinished B. anthracis genome for potential coat proteins. Our analysis suggests that the B. subtilis and B. anthracis coats have roughly similar numbers of proteins and that a core group of coat protein species is shared between these organisms, including the major morphogenetic proteins. Nonetheless, a significant number of coat proteins are probably unique to each species. These results should accelerate efforts to develop B. anthracis detection methods and understand the ecological role of the coat.
Bacteria build a variety of precisely positioned macromolecular structures. These include the divisome, which mediates cell division (43); the flagellum (2); and the type III secretory apparatus, critical for pathogenesis in many organisms (40, 42). To understand how such structures are built and how they function, it is critical to identify their protein components. In this report, we seek to identify the proteins that make up the coat that encases spores of Bacillus subtilis and Bacillus anthracis.
Spores are produced by many species of bacilli and clostridia in response to severe external stress (71, 72). These highly resilient dormant cell types are able to withstand extremes of temperature, radiation, chemical assault, time, and even the vacuum of outer space (58). Upon the return of favorable environmental conditions, spores can readily convert to actively growing vegetative cells through a process known as germination (48, 64). These abilities enable spores not only to survive in extreme conditions but, in some species, to cause significant disease. In the case of B. anthracis, contact between the host and the spore is essential for infection (17). The most prominent structural feature common to all bacterial spores is a multilayered proteinaceous shell called the coat (19, 21, 31). The coat is critical for resistance properties as well as germination. It provides mechanical integrity and excludes large toxic molecules while, at the same time, allowing small nutrient molecules to penetrate and interact with the germination receptors located toward the spore interior (49, 64).
Thin-section electron microscopy of the B. subtilis spore reveals two major layers in the coat: a lightly staining lamellar inner layer and a darkly staining outer layer (4, 80). The morphogenetic coat protein CotE directs the assembly of most, if not all, outer coat proteins and some of the inner coat proteins (23). Additional morphogenetic proteins, such as SafA (YrbA) and SpoVID, further guide coat protein deposition (8, 62, 63, 77). The B. anthracis coat is much thinner than that of B. subtilis, and its layered architecture is less striking, although inner and outer coat layers can still be discerned (20, 28, 30, 47, 66). In many bacilli, including B. anthracis but not B. subtilis, the spore is encased by an additional structure known as the exosporium. The exosporium surrounds the entire spore, including the coat, from which it is structurally and biochemically distinct (45). Notably, the exosporium is heavily glycosylated (26), appears to be composed of relatively few protein species, and harbors at least one major glycoprotein (73). Its function is unknown, although it appears not to have a major role in pathogenesis (73).
Approximately 30 confirmed or putative B. subtilis coat proteins have been identified, but how they participate in spore survival and germination remains, for the most part, obscure (21). A subset of coat proteins has significant, although poorly understood, roles in assembly. Importantly, deletion of any one of the large number of remaining coat proteins has little or no detectable effect on phenotype. As a result, the functions of most of the coat proteins are unknown, and furthermore, most roles of the coat cannot be ascribed to specific proteins. It is very likely that many coat functions are emergent properties of the interactions of multiple coat proteins (79). Therefore, a detailed understanding of the coat's role may require knowledge of most or all of its components. Even though a large number of coat proteins have been found, it is clear that additional coat proteins remain undetected (19). Identification of these proteins will require a comprehensive characterization of the coat at the molecular level, which has not yet been achieved.
Proteomics offers a powerful platform for the analysis of components of many macromolecular assemblies through separation of complex protein mixtures by two-dimensional (2D) electrophoresis and identification by mass spectrometry (MS) (see, for example, references 27 and 33). Several excellent proteomic studies of B. subtilis have been carried out. Notable among these are a recent alkaline 2D map and a comprehensive 2D map (12, 61). These studies focused on the analysis of soluble proteins from vegetative cells. In contrast, analysis of spore proteins has been hindered by the requirement for solubilization of tightly associated coat proteins (4). Recent advances in protein solubilization and separation made in our laboratories (51, 52, 65), coupled with significant increases in MS sensitivity and the availability of genomic sequence data from several organisms, have made the identification of proteins from hard-to-dissect biological samples more realistic. Here, we take advantage of these technological improvements to identify spore proteins in B. subtilis and B. anthracis.
MATERIALS AND METHODS
General methods.
B. subtilis (PY79) (81) or B. anthracis (Sterne strain, from Paul Jackson, Los Alamos National Laboratory) spores were prepared essentially identically by exhaustion in Difco sporulation medium (16). The only difference was that, for the case of B. anthracis, initial growth was on Luria-Bertani agar plates supplemented with 5 g of nutrient broth (Difco) per liter. Spores were washed three times in double-distilled H2O and then resuspended in double-distilled H2O. Resuspended spores were either used immediately or stored at 4°C for no longer than 3 weeks. For 1D gel electrophoresis of B. subtilis and B. anthracis spore extracts, we prepared protein samples as described previously (41).
2D electrophoresis.
2D gel electrophoresis was performed as described previously (65) with the following modifications during the isoelectric focusing (IEF) step. Forty-five microliters of the boiled 2% sodium dodecyl sulfate (SDS) or 2% lithium dodecyl sulfate (LDS) solubilized suspension was mixed by sonication with 405 μl of 1.1× rehydration buffer (7.7 M urea, 2.2 M thiourea, 2.2 mM tributylphosphine, 0.55% [vol/vol] Biolytes 3 to 10 [Bio-Rad, Hercules, Calif.], and 44 mM Tris base) containing 2% benzoylamidopropyldimethylammoniopropanesulfonate (C8φ) and 1% Triton X-100 (TX-100) and allowed to incubate with constant mixing at 30°C. Samples were spun at 13,000 × g to remove insoluble material. Immobilized pH gradients (IPGs) (18 cm; Amersham Pharmacia, Uppsala, Sweden) were rehydrated overnight in 450 μl of solubilized spore suspension in rehydration buffer. IEF was carried out as described previously (65) with the addition of frequent filter paper wick changes during IEF.
MALDI-MS and MS/MS analysis.
Manually excised Coomassie blue protein spots from 1D gels were digested with 150 to 300 ng of modified porcine trypsin (Promega, Madison, Wis.) in 30 μl of freshly prepared 100 mM ammonium bicarbonate at 37°C overnight as described previously (65). Protein spots from 2D gels were manually excised from the gel with a 2-mm dermal punch and transferred to a 96-well plate. The spots were automatically digested with a Micromass MassPREP Station (Micromass Ltd., Manchester, United Kingdom). Briefly, the silver-stained gel pieces were destained with two washes of a 1:1 mixture of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate followed by dehydration of the gel pieces with acetonitrile. Protein spots were reduced and alkylated with 10 mM dithiothreitol and 55 mM iodoacetamide, both buffered with 100 mM ammonium bicarbonate, followed by a wash with 100 mM ammonium bicarbonate and two dehydration cycles with acetonitrile. Protein digestion was performed with the addition of 25 μl of 6-ng/μl sequencing-grade modified trypsin solution in 100 mM ammonium bicarbonate (Promega) and incubation of the solution at 37°C for 4.5 h. The resulting peptides were extracted in an aqueous solution of 1% (vol/vol) formic acid-2% (vol/vol) acetonitrile. A 2.0-μl quantity of extracted peptides and 1.6 μl of α-cyano-4-hydroxycinnamic acid (10 mg/ml in 50% acetonitrile and 0.1% trifluoroacetic acid) were mixed in the dispensing tip and spotted onto a Micromass 96-well matrix-assisted laser desorption ionization (MALDI) target plate.
Analysis with the PerSeptive Biosystems Voyager-DE-STR was performed as described previously (65). The resulting peptide mass fingerprints were searched by using a local copy of the program MS-Fit (a component of the Protein Prospector package) against an internal database as described below. For peptide mass fingerprinting searches, MALDI spectra were analyzed with the Micromass MassLynx 3.5 software package. Spectra were manually recalibrated with the 2,211.104-Da and 842.509-Da trypsin autodigestion peptides, and the data were exported to text files. Monoisotopic peptide masses were obtained manually or by using an in-house virtual instrument created in the LabView graphical programming language (G. Rymar and P. Andrews, unpublished data). The resulting peptide mass fingerprints were searched by using a local copy of the program MS-Fit against an internal database. A mass accuracy of 75 ppm was used. A maximum of one missed enzymatic cleavage and modification of cysteines by carbamidomethylation plus possible modification by acrylamide were considered during the searches.
For MS/MS, 0.5 μl of gel digest was spotted onto the target plate followed by 0.5 μl of α-cyano-4-hydroxycinnamic acid (5 mg/ml in 50% acetonitrile-1% trifluoroacetic acid), and the spot was allowed to air dry at room temperature. MS/MS was performed on a Micromass MALDI-quadrupole time-of-flight (TOF) Ultima mass spectrometer. This instrument features an unattenuated nitrogen laser, operating at 337 nm and firing at 10 Hz, which is rastered over the sample spot (2.5-mm diameter) at 1 Hz. Ions are introduced into the instrument by a nitrogen flow (5 lb/in2) that also serves to cool the ions. Ions are selected for MS/MS in the quadrupole, with the mass tolerance set to ±5 Da. This window was closed to 2 Da if another peptide occurred within the 5-Da window of the peptide of interest. The selected ion was fragmented in the hexapole collision cell, with the argon gas pressure set to 25 lb/in2, and the collision energy varied from 50 to 150 V according to the mass of the parent ion. The parent and product ions are resolved in the TOF region, equipped with a Reflectron which produces monoisotopic resolution for ions from 100 to 4,000 Da, and detected by a microchannel plate detector set to 2,250 V. Parent ion masses were taken from the peptide mass fingerprint spectrum after internal calibration with trypsin autolysis peaks. Product ion masses were calibrated by instrument calibration only. Parent and product ion masses were submitted to Mascot Ions Search, searching the National Center for Biotechnology Information (NCBI) nonredundant database for all species, for protein identification. The parent ion tolerance was set to 100 ppm, and the product ion tolerance was set to 0.5 Da. Alternative modes of searching (no enzyme, specifying posttranslational modifications (PTMs), and searching additional databases) were performed as needed. An identification based on MS/MS on one peptide was considered adequate if the Mascot score was above the significance level, although in many cases we obtained additional confirmation by MS/MS on a second peptide or from a peptide mass fingerprint database search (Table 1).
TABLE 1.
B. subtilis proteins identified from a 1D gel
| Band | Protein | MALDI MSa | MS/MSb | Predicted molecular mass (kDa)/pI | Observed migration (kDa) |
|---|---|---|---|---|---|
| 1 | CotA | Yes | 1 (1,514) | 58.5/5.91 | 71 |
| 2 | CotB | 2 (1,305, 1,530) | 43.0/9.47 | 62 | |
| 3 | YaaH | Yes | 2 (1,414, 1,366) | 48.6/5.72 | 53 |
| 3 | YpeP | 2 (1,542, 1,635) | 51.2/6.31 | 53 | |
| 3 | PhoA | 1 (2,555) | 50.2/9.50 | 53 | |
| 4 | YdhD | 2 (1,317, 1,784) | 49.0/9.01 | 48 | |
| 5 | YtaA | 1 (1,401) | 41.2/5.16 | 46 | |
| 6 | CotS | 1 (1,430) | 41.1/6.61 | 44 | |
| 7 | CotG | Yes | 2 (1,249, 1,676) | 24.0/10.26 | 40 |
| 8 | CotR | 2 (1,562, 2,123) | 35.4/9.87 | 37 | |
| 9 | YusA | 1 (1,998) | 30.4/8.26 | 35 | |
| 10 | SleB | Yes | 1 (1,800) | 34.0/9.27 | 33 |
| 11 | YckK | 1 (1,893) | 29.4/7.64 | 31 | |
| 12 | CotE | Yes | 2 (1,994, 1,549) | 21.0/4.36 | 26 |
| 12 | YrbB | 2 (2,498, 1,650) | 19.5/8.89 | 26 | |
| 13 | CotJC | Yes | 1 (1,909) | 21.7/5.08 | 25 |
| 13 | YhcN | 2 (1,851, 2,249) | 21.0/5.56 | 25 | |
| 15 | CwlJ | 1 (1,589) | 16.4/9.38 | 17 | |
| 16 | YpzA | 1 (1,551) | 10.1/4.3 | 18 | |
| 16 | SspE | Yes | 9.3/8.19 | 18 | |
| 17 | SspE | Yes | 4 (1,481, 1,812, 1,871, 2,968) | 9.3/8.19 | 15 |
| 18 | YxeE | 2 (1,585, 1,821) | 14.7/8.03 | 12 | |
| 18 | YjdH | 1 (1,303) | 15.3/5.25 | 12 | |
| 18 | CspD | 1 (1,922) | 7.3/4.51 | 12 | |
| 18 | Hbs | 1 (1,393) | 9.9/8.96 | 12 | |
| 19 | CotF | Yes | 1 (1,634) | 18.7/6.98 | 8 |
| 20 | CotF | 2 (1,452, 1,744) | 18.7/6.98 | 5 | |
| 20 | YodI | 1 (1,070) | 9.2/10.28 | 5 | |
| 20 | SspA | 2 (1,656, 1,879) | 7.1/4.94 | 5 |
Top MALDI MS hit.
Number of peptides providing fragment ions. Numbers in parentheses are sizes.
Creating a peptide mass fingerprint searchable B. anthracis database.
The latest releases of the B. anthracis genome sequence were made available prior to publication by The Institute for Genomic Research (Rockville, Md.). Sequence data were in the form of several large contigs in FASTA format. Contigs were assembled into a single contiguous DNA sequence in FASTA format with a script written in the Perl programming language. Putative open reading frame (ORF) predictions were carried out with the interpolated Markov model gene prediction package GLIMMER (68). Briefly, a trained model was created, with the program Build-icm, on a data set of sequences from closely related species available in the NCBI nonredundant database. Complete nucleotide sequences of known and predicted chromosomal ORFs from B. anthracis, Bacillus cereus, Bacillus thuringiensis, Bacillus halodurans, and B. subtilis were used to train the model. The program Glimmer2 was used to predict putative ORFs from the concatenated DNA sequence file based on the trained model. FASTA-formatted nucleotide sequences of predicted ORFs were obtained from the output of Glimmer2 by using an in-house Perl program. The resulting nucleotide sequences were used to retrain the model with Build-icm, and the gene prediction process was repeated with the newly trained model. The predicted ORF nucleotide sequences were translated to amino acid sequences in reading frame 1 with the Transeq program from the EMBOSS suite of molecular biology applications (http://www.emboss.org/). Rapid preliminary annotation was carried out with scripts written in the Python programming language to submit each predicted sequence to BLAST (3), COGNITOR (78), FramePlot (36), SignalP2 (59), ProtParam (http://us.expasy.org/tools/protparam.html), PSORT (57), and SOSUI (32). The translated and annotated sequences were concatenated into a file containing known and predicted protein sequences from B. anthracis, B. cereus, B. thuringiensis, B. halodurans, and B. subtilis. The resulting file was indexed for searching with MS-Fit by using the Faindex program from the Protein Prospector package.
RESULTS AND DISCUSSION
Analysis of B. subtilis spore proteins by 1D and 2D electrophoresis.
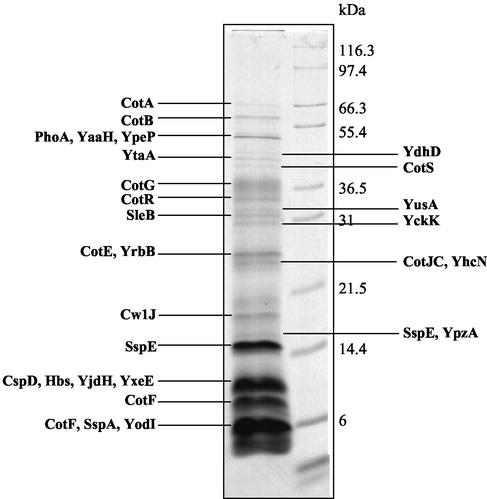
While 1D analysis may not possess the separation power of 2D electrophoresis, it has several advantages over 2D analysis, including the ability to separate hydrophobic proteins such as those found in membranes, the ability to tolerate significantly higher protein loads, and the ability to resolve proteins linearly across the pH scale. Thus, in order to identify proteins difficult to resolve on 2D gels, we first identified proteins in the major bands visualized on Coomassie blue-stained 1D gels (Fig. 1). B. subtilis spore samples were prepared for SDS-polyacrylamide gel electrophoresis (PAGE) analysis as described in Materials and Methods. From these studies, we identified 19 bands comprising 27 unique proteins (Table 1). Not surprisingly, we identified a number of bona fide coat proteins such as CotA (18), CotB (18), CotE (83), CotF (15), CotG (67), CotJC (70), CotR (41), CotS (1), CwlJ (7), and YaaH (38). We also identified YxeE, identified in spores previously and known to be a good coat protein candidate (76). We identified eight spore proteins that are very unlikely to be in the coat (CoxA [YrbB] [74], CspD [29, 69], Hbs [46], PhoA [34], SleB [10, 53-55], SspA [21], SspE [21], and YhcN [6]). Most importantly, we identified the unknown hypothetical proteins YckK, YdhD, YjdH, YodI, YpeP, YpzA, YtaA, and YusA, which we regard as candidate coat proteins.
FIG. 1.
B. subtilis coat proteins resolved by 1D standard SDS-15% PAGE. The gel was stained with Coomassie blue G-250. The indicated bands were prepared for MALDI MS and MS/MS mapping as described in Materials and Methods. Protein identifications correspond to those described in Table 1.
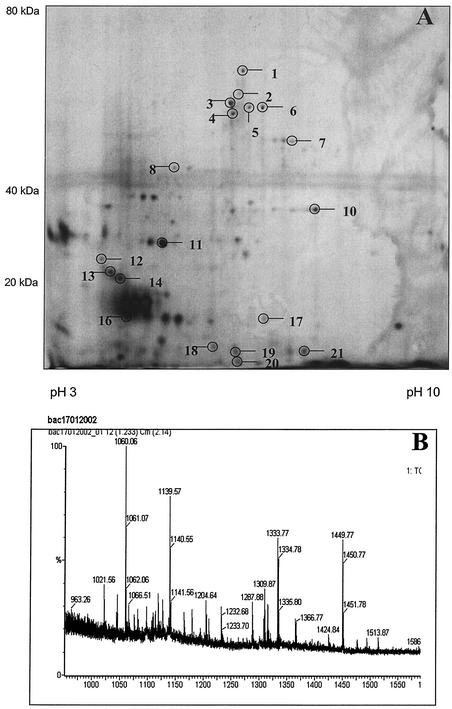
As anticipated, several Coomassie blue-staining bands on our 1D gels contained more than one protein (bands 3, 12, 13, 16, 18, and 20 [Table 1]). Since one long-term goal is to examine how the proteomic profile changes in various coat gene mutant backgrounds and during the course of sporulation, it was critical to develop 2D separation of coat proteins such that the level of individual proteins could be monitored. We found that conventional solubilization techniques (60) and even aggressive solubilization techniques designed for separation of membrane proteins by 2D electrophoresis, involving strong chaotropes, amidosulfobetaine detergents, and strong reducing agents (14, 50, 65), proved ineffective for solubilization of B. subtilis coat-enriched fractions. Large insoluble pellets remained after centrifugation of coat samples solubilized under these conditions (data not shown). Use of the ionic detergents SDS and LDS or chaotropes like guanidium-HCl resulted in much better solubilization. However, these reagents are incompatible with IEF at the concentrations used for solubilization. Several groups have suggested solubilization by these strong agents followed by dilution or detergent exchange with IEF-compatible reagents (25, 50). We investigated the use of strong ionic detergent solubilization followed by dilution with large volumes of IEF-compatible reagents as a method for preparing first-dimension samples. Coat-enriched B. subtilis fractions were solubilized in a modified 1D loading buffer containing SDS or LDS. These samples were subsequently resolubilized in an excess of IEF-compatible reagents and subjected to 2D electrophoresis. Excellent separation was obtained when IEF was carried out for extended periods up to 120 kV · h, with frequent wick changes to remove unfocused material. The best results were seen when initial solubilization was carried out on very fresh samples with small volumes of SDS buffer, followed by dilution with standard rehydration buffer containing 2% C8φ plus 1% TX-100 (Fig. 2 and 3). We identified 19 spots consisting of 14 unique proteins from pH 3 to 10 gels (Fig. 2 and Table 2). Not surprisingly, we found a number of proteins also found in the 1D gels such as CotA, CotE, CotJC, and YaaH as well as coat proteins not found on the 1D gel (CotC and CotY). A protein unlikely to be a coat protein, CggR (24), was also identified. Finally, we identified YhbA, YirY, YisY, YnzH, YopQ, YvdP, and YwqH, which we regard as candidate coat proteins.
FIG. 2.
Preliminary master gel of B. subtilis coat preparations and MALDI analysis. (A) pH 3 to 10 gel of B. subtilis coat preparations. Sample was solubilized in 10% LDS buffer. The solution was diluted in an excess of standard solubilization buffer containing 2% C8φ and 1% TX-100 and resolved by IEF. The second dimension was carried out on an SDS-11% polyacrylamide gel and silver stained (56). Spots identified by peptide mass fingerprinting are circled and numbered, and the identifications are presented in Table 2. (B) MALDI-MS spectrum for spot 1.
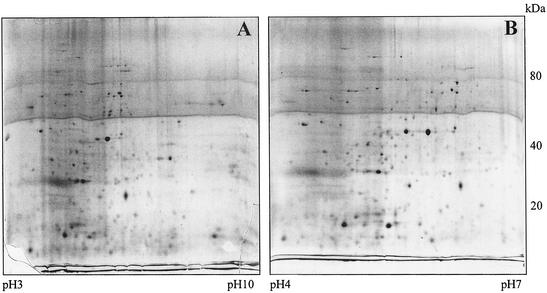
FIG. 3.
Improved 2D separation. One hundred eighty microliters of a spore pellet was solubilized in 100 μl of 1× SDS buffer after being boiled for 10 min. Ninety microliters of this solution was added to 810 μl of 1.1× 2% C8φ-1% TX-100 in standard rehydration buffer and incubated at 30°C for 1 h before rehydration. Four hundred fifty microliters each of this solution was run on a pH 3 to 10 (A) and a pH 4 to 7 (B) IEF strip. Frequent changes of filter paper wicks during IEF and focusing up to 120 kV · h until the dye ran out completely at the acidic end greatly improved separation. The second dimension is as in Fig. 2.
TABLE 2.
Proteins identified by MALDI-TOF MS from 2D separation of B. subtilis spore coat fractions
| Spot no. | MOWSE score | No. of peptides matched | MWb/pI | Name | Function |
|---|---|---|---|---|---|
| 1 | 2.15E+06 | 11 | 58,499.3/5.91 | CotA | Multicopper oxidase |
| 2 | 5.50E+05 | 13 | 48,637.0/5.72 | YaaH | Unknown protein |
| 3 | NAa | 9 | 48,534.4/7.5 | YhbA | Unknown protein |
| 4 | 1.14E+06 | 10 | 58,499.3/5.91 | CotA | Multicopper oxidase |
| 5 | 8.49E+03 | 6 | 50,085.0/6.12 | YvdP (CotQ) | Similar to coat protein CotQ |
| 6 | 8.16E+02 | 5 | 53,503.9/5.41 | YopQ | Unknown protein |
| 7 | NA | 8 | 129,334/5.44 | YirY | Unknown protein |
| 8 | NA | 8 | 37,382.4/5.8 | CggR | Transcriptional regulator |
| 10 | 3.44E+04 | 6 | 30,559/6.8 | YisY | Unknown protein |
| 11 | NA | 7 | 21,695.8/5.1 | CotJC | Coat protein |
| 12 | 1,453 | 4 | 20,977/4.4 | CotE | Morphogenetic coat protein E |
| 13 | 7,589 | 5 | 20,977/4.4 | CotE | Morphogenetic coat protein E |
| 14 | 6.26E+05 | 8 | 20,977/4.4 | CotE | Morphogenetic coat protein E |
| 16 | NA | 5 | 18,728/5.02 | CotY | Coat protein Y |
| 17 | NA | 5 | 15,857/5.01 | YwqH | Unknown protein |
| 18 | NA | 5 | 14,785/9.4 | CotC | Coat protein C |
| 19 | NA | 5 | 14,785/9.4 | CotC | Coat protein C |
| 20 | NA | 5 | 14,785/9.4 | CotC | Coat protein C |
| 21 | 5.62E+03 | 6 | 11,562.5/6.28 | YnzH (CotU) | Coat protein |
NA, identifications were made with the MASCOT algorithm or with the Micromass Protein Lynx software package.
MW, molecular weight.
B. subtilis coat proteins.
About 30 coat proteins or coat protein candidates have been described elsewhere (21). We found 12 of these known proteins and 17 additional coat proteins or coat protein candidates. Of these, we regard YtaA, YvdP, and YnzH as highly likely to be coat proteins, and we have renamed them CotI, CotQ, and CotU, respectively. We regard YtaA as a coat protein based on (i) significant similarity to CotS (BLAST score of 3E-25), as already noted in the B. subtilis genome sequence annotation, and (ii) its gene being adjacent to, and oriented away from, the operon harboring cotS. We had already identified YvdP in a separate study and showed that it is synthesized at a late time in sporulation and associated with the spore in a manner dependent on the coat morphogenetic protein CotE (41, 83; M. Otte and A. Driks, unpublished observations). Furthermore, its gene is adjacent to cotR and transcribed divergently from it. cotR (41) and cotQ could share upstream regulatory sequences. We regard YnzH as a coat protein because it has also been shown to be synthesized late in sporulation and assembled in a CotE-dependent manner (Otte and Driks, unpublished). As was previously noted in the B. subtilis genome sequence annotation (http://genolist.pasteur.fr/SubtiList/), the YnzH sequence is strikingly similar to that of CotC.
Of the 14 additional coat protein candidates, we predict that many will also be bona fide coat proteins. Of particular interest are YisY, very likely to be a chloride peroxidase; CotQ, highly similar to reticuline oxidase from plants; and YdhD, predicted to be a cortex-lytic enzyme (39). Precedence for a cortex-lytic enzyme in the coat is provided by CwlJ (7). Several other uncharacterized proteins identified in our study are also good coat protein candidates. For example, YhbA is an iron-sulfur cluster-binding protein and likely the first gene in a six-gene operon. Most of the other genes in the operon (yhbB, yhbD, yhbE, and yhbF) are found only among the bacilli and clostridia, consistent with a role in sporulation. YpeP is a novel protein that appears to be cotranscribed with ypeQ and transcribed divergently from the operon containing ypdP and ypdQ. YpdQ is predicted to be a cell wall synthesis protein. Interestingly, the gene downstream of the ypeP operon encodes YpzA, another protein identified in our study.
The possibility that CotQ and YisY are oxidases is notable in light of the recent demonstration that CotA is a multicopper oxidase of the laccase class (35, 44) and the similarity of CotJC to peroxidase (70). Oxidases can participate in a wide variety of biosynthetic activities. Potentially, they could play a role in coat protein cross-linking, as has been suggested previously (4, 19, 31), in detoxification of environmental contaminants, or in symbiosis with other soil organisms (22). The identification of these enzymes in the coat is consistent with the view that the coat plays active roles in spore protection and germination, rather than acting solely as a passive barrier (21, 23). Whatever their roles, CotA (18), CotJC (70), and CotQ (Otte and Driks, unpublished) are dispensable for coat resistance and germination, as usually measured in the laboratory.
We found 12 previously identified B. subtilis coat proteins (Tables 1 and 2). Several coat proteins are known to be present in a form smaller than the presumed translation product, including CotF, CotT, and SafA (5, 11, 15, 75). Consistent with this and extending the list of processed coat proteins, we found that CotA, CotC, CotE, and CotF were present in multiple forms. The two forms of CotF are the known mature forms (15). CotC was present in several spots of different mobilities and with a charge very different from its predicted pI of 8.6 (Fig. 2), possibly the result of proteolysis or modification during coat assembly. Likewise, we found multiple forms of CotE. YjdH, YodI, and YirY are three additional proteins that may also be processed based on their migration (Fig. 1 and 2). YirY is a particularly striking example. YirY is predicted to be a 129-kDa protein that migrates at ∼50 kDa (Fig. 2). We predict either that YirY is processed or that the original annotation that separated YirY into two distinct proteins (YirY and YirZ) is, in fact, correct. Finally, some proteins migrate slower than predicted, including CotA, CotB, CotG, and YpzA (Fig. 1 and 2). These proteins may be modified or cross-linked to additional protein.
Deletions of most of the known coat proteins have minimal or undetectable phenotypes in standard laboratory assays, as already pointed out (21). Given the complexity of coat composition and the likely enzymatic roles of some of the components, it seems reasonable to suppose that the coat has functions beyond those measured in the laboratory. These may include roles for coat-associated enzymes in coat protein cross-linking and degradation of environmental toxins, symbiosis with plants, and protection against competitor organisms. An additional important function of the coat is in germination. While this role has been appreciated for some time (4), it is only recently that its molecular basis has begun to become understood. For example, in addition to the discovery that the cortex-lytic enzyme CwlJ is in the coat (7), the Moir laboratory has shown that the gerP operon, likely encoding coat protein(s), participates in germination (9). Specifically, that study indicates that the coat does more than merely act as a passive sieve through which germinants flow.
We note that less than half of the known coat proteins were found in our experiments. This is likely to be for several reasons. First, we have not exhausted our analysis of faint bands present on 1D gels (Fig. 1) or of focused spots on the 2D gels (Fig. 2). As MS/MS technologies become more sensitive, we are certain that we will also identify these proteins. Second, the identification of any protein by MS relies on the ionization efficiency of the trypsin fragments and the sensitivity of the mass spectrometers. MALDI-MS mapping is sensitive but requires multiple peptides for identification. MS/MS identification with the quadrupole TOF requires fewer peptides but often a stronger ionization signal. Thus, low-abundance proteins or those with only a few detected peptides may be missed. Third, many of the coat proteins are basic and would not be resolved on the 2D gels used in this study. Although the theoretical separation on 3 to 10 IPGs should allow for separation of proteins up to pI 10, we rarely focus proteins with pIs greater than 8 with these IPGs (51, 65). The development of separation procedures for alkaline coat proteins, similar to what has been achieved for membrane proteins (51), will be required to visualize these proteins. Fourth, small proteins (such as the 29-amino-acid SpoVM) will be absent from the 2D gels and are not amenable to MALDI-MS mapping, as the required number of peptides to identify the protein would not be available. Finally, some of the covalently cross-linked coat proteins may not have been solubilized in this study and would, therefore, be missed altogether. Despite these limitations, we have identified many of the known coat proteins and have uncovered 14 new coat candidates (Table 3). Our survey is clearly less than saturated, and it is likely that a significant number of novel coat protein species remain to be discovered. Future analysis by MS/MS coupled with the use of alkaline 2D gels (51) should readily lead to a proteome map of most, if not all, spore proteins. Such a proteomic map will be instrumental for biochemical characterization of coat assembly.
TABLE 3.
Known and predicted spore coat proteins in B. subtilis and B. anthracis
| Proteina | Comments on B. subtilis protein | B. anthracis protein identification no.b | No. of amino acids identical/total no. of amino acids in length (%) | Comments on B. anthracis protein |
|---|---|---|---|---|
| B. subtilis previously known spore coat proteins | ||||
| CotA | Multi copper oxidase | 655766 | 39/133 (29) | Two regions of similarity to CotA |
| 51/213 (24) | ||||
| CotB | Encoded downstream of cotH | 654287 | 47/156 (30) | Adjacent genes |
| 654288 | 55/153 (33) | |||
| CotC | None | |||
| CotD | 655458 | 32/70 (45) | Likely homolog | |
| CotE | Controls assembly of most outer coat proteins and some inner coat proteins | 657736 | 106/181 (58) | Homolog |
| CotF | Proteolytically Processed from a 23-kDa precursor | 656983 | 97/154 (62) | Homolog |
| CotG | Gene is divergent from cotH | None | Lack of homology could be due to lack of complexity | |
| CotH | Gene is divergent from cotG, upstream of cotB | 655906 | 198/356 (55) | Homolog |
| CotJA | Encoded in cotJABC operon | 654751 | 46/66 (69) | Homolog |
| CotJC | Encoded in cotJABC operon | 1326c | 170/189 (89) | 654749 is a gene fragment. 1326 is the homolog |
| 654749 | 63/68 (92) | |||
| 656966 | 48/201 (23) | |||
| CotM | Encoded in sspO sspP cotM operon | 657526 | 26/111 (23) | 657526 is CotM-like |
| 656113 | 20/83 (24) | |||
| CotR | Gene is adjacent to and divergent from cotQ (yvdP) | None | ||
| CotSA | Encoded in cotSA cotS operon. Gene is divergent from cotI (ytaA) | 658792 | 57/215 (26) | |
| 655436 | 59/221 (26) | |||
| 653730 | 52/212 (24) | |||
| CotS | Encoded in cotSA cotS operon | 653420 | 48/183 (26) | C terminus only |
| CotT | Proteolytically processed from an 8-kDa precursor | None | ||
| CotV | Encoded in cotVWXYZ operon | None | Slightly similar to several ORFs | |
| CotW | Encoded in cotVWXYZ operon | None | Slightly similar to several ORFs | |
| CotY | Encoded in cotVWXYZ operon | 1889c | 53/154 (34) | Both CotY/Z-like |
| 655129 | 54/153 (35) | |||
| CotZ | Encoded in cotVWXYZ operon | 655129 | 50/141 (35) | Both CotY/Z-like |
| 1889c | 48/139 (34) | |||
| CwlJ | Cortex-lytic enzyme | 653856 | 87/140 (62) | Both 653856 and 656463 are homologs. 656634 and 657724 have a CwlJ-like C-terminal motif |
| 656463 | 82/141 (58) | |||
| 656634 | 34/119 (28) | |||
| 657724 | 40/126 (31) | |||
| SpolVA | Appears to connect the coat to the forespore and is required for cortex formation | 655410 | 434/492 (88) | Homolog |
| SpoVM | Partially required for SpoIVA localization | 657826 | 21/26 (84) | Homolog |
| SpoVID | Partially required for coat localization | 658491 | 100/300 (33) | Gene fragment? |
| GerPd | Some product(s) of the six-gene gerP operon likely controls entry of germinant into the spore interior | |||
| YaaH | Peptidoglycan hydrolase. Affects germination. Similar to YvbX and YkvQ | 657516 | 207/424 (48) | 657516 is a homolog |
| 657340 | 64/247 (25) | |||
| 654321 | 37/163 (22) | |||
| YabG | Affects coat protein composition and germination. Has protease activity | 653990 | 170/288 (59) | Homolog |
| YrbA (SafA) | Affects germination and coat assembly. 31-kDa species likely to be a product of proteolysis of a 43-kDa precursor | 658457 | 56/112 (50) | Gene fragment? |
| B. subtilis candidate spore coat proteins | ||||
| CotQ (YvdP) | Gene is adjacent to and divergent from cotR | None | ||
| CotU (YnzH) | Similar to CotC | None | Small ORF | |
| CotI (YtaA) | Similar (47% over most of its length) to CotS. Some similarity to YutH. Gene is divergent from cotSA | 653420 | 41/190 (21) | C terminus of 653420 similar to C terminus of CotI |
| YckK | Resembles an ABC transporter | 654795 | 138/267 (51) | 654795 homolog. All are clearly related along entire length |
| 654301 | 81/267 (30) | |||
| 654584 | 75/276 (31) | |||
| YdhD | Predicted to be a cortex-lytic enzyme. Similar to YaaH and YvbX | 657516 657340 | 125/430 (30) 55/213 (25) | |
| YidH | Unknown protein | None | ||
| YhbA | Resembles an iron-sulfur cluster-binding protein | 654477 | 263/375 (70) | Homolog |
| YhdE | Resembles an RRF2 family protein. Slightly similar to YwgB. Transcribed divergently from spoVR | 654902 | 37/130 (28) | All YhdE-like along entire length |
| 658427 | 38/137 (27) | |||
| 657309 | 29/135 (21) | |||
| YirY | Potentially an exonuclease | 657817 | 192/1013 (18) | |
| 656214 | 92/349 (26) | |||
| YisY | Resembles a chloride peroxidase, 45% similar to YdjP | 658807 | 115/268 (42) | 14 more ORFs with weak similarity over the entire sequence |
| YodI | 657487 | 20/42 (47) | Likely homolog, gene fragment | |
| YopQ | 657926 | 36/129 (27) | ||
| YpeP/YpeB | ypeB, ypzA, and cspD are within 1.5-kb region | 656633 | 257/448 (57) | Homolog |
| YpzA | Homolog identified only in B. anthracis, ypeB, ypzA, and cspD are within 1.5-kb region | 655504 | 25/73 (34) | Homolog |
| YusA | ABC transporter substrate-binding protein | 653451 | 165/275 (60) | 653451 and 653452 are adjacent genes |
| 653452 | 156/275 (56) | |||
| 654118 | 103/277 (37) | |||
| 654245 | 95/277 (34) | |||
| YwqH | Some similarity to YxiB | 657122 | 34/135 (25) | |
| YxeF | 657441 | 50/109 (45) | Homolog | |
| Additional proteins identified in this study | ||||
| CspD | Cold shock protein. ypeB, ypzA, and cspD are within 1.5-kb region | 658893 | 57/65 (87) | All appear to be homologs |
| 655505 | 54/63 (85) | |||
| 655043 | 52/63 (82) | |||
| 653638 | 50/63 (79) | |||
| 657446 | 45/56 (80) | |||
| Hsb | Histone-like protein. Gene is downstream from spoIVA | 655411 | 80/89 (89) | All appear to be homologs |
| 656232 | 65/89 (73) | |||
| 5344c | 58/89 (65) | |||
| PhoA | Alkaline phosphatase | 658376 | 256/449 (57) | Homolog |
| SleB | Cortex-lytic enzyme | 656634 | 156/277 (56) | 656634 homolog. 657724, 656463, and 653856 similar to C terminus of SleB |
| 657724 | 68/203 (33) | |||
| 656463 | 36/126 (28) | |||
| 653856 | 34/124 (27) | |||
| SspA | Alpha-type small acid-soluble spore protein | 656989 | 52/67 (77) | All appear to be homologs |
| 656992 | 50/66 (75) | |||
| 654798 | 44/66 (66) | |||
| 658678 | 44/61 (72) | |||
| 655848 | 47/67 (70) | |||
| 655208 | 39/58 (67) | |||
| SspE | Gamma-type small acid-soluble spore protein | |||
| YhcN | Forespore protein, some similarity to Y1aJ | 656632 | 47/182 (25) | |
| 658456 | 52/196 (26) | |||
| YrbB (CoxA) | Cortex protein. Gene is downstream of safA (yrbA) | 658456 | 43/143 (30) | Gene fragment? |
| CggR | Transcriptional regulator | 653588 | 206/338 (60) | Homolog |
Proteins in bold-face were identified in this study.
NP _ number accessible through NCBI.
Our working ORF identification; gene not in public database.
It is unknown which proteins encoded in the gerP operon are coat proteins. Tallies of coat proteins in the text do not include those potentially encoded by the gerP operon.
Analysis of B. anthracis spore proteins by 1D electrophoresis.
In spite of its importance to spore survival, germination, and pathogenesis, the protein composition of the B. anthracis coat has received very little study. None of the coat proteins in this organism have been identified, nor has coat assembly been characterized, although one exosporium protein has been characterized (73) and some spore-associated proteins have been identified in the close relative B. cereus (13). Comparison of the B. subtilis and B. anthracis genomes shows that coat proteins with key roles in morphogenesis are present in both organisms (Table 3), and therefore, it is plausible that coat assembly follows largely the same program in the two species (20). Interestingly, this same analysis suggested that there are important differences in the protein compositions of the coats of these organisms, possibly among the outer layers. Identification of novel coat proteins in B. anthracis not only will help in understanding coat assembly and function but could also be of significant value in development of vaccines and methods for decontamination, detection, and inhibition of germination.
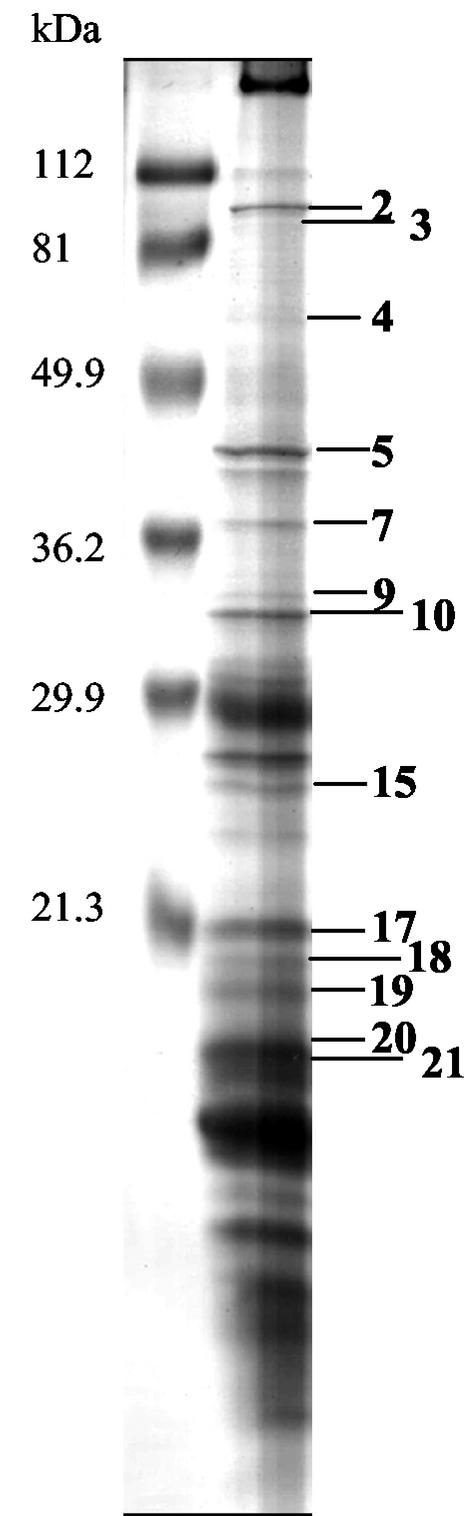
Using standard 1D SDS-PAGE with 15% polyacrylamide gels, we identified 11 B. anthracis spore proteins (Fig. 4 and Table 4). We consider ORF1326 (similar to CotJC), NP_654287 (more distantly related to CotB and YwrJ), and NP_655129 (related to CotY and CotZ) to be coat proteins. NP_657011 encodes a protein that is 49% similar to amino acids 23 to 173 of the predicted 404-amino-acid B. subtilis protein YndF. NP_657011 also resembles GerBC and GerBA (27 and 31% identical, respectively). NP_654944 encodes an unknown protein closely related to a Clostridium protein and therefore is a good candidate for a coat or exosporium protein. Finally, we consider NP_655132 to be a coat protein as the gene is flanked by genes whose products we predict will be coat proteins (NP_655129 and ORF1889, Table 3). In this preliminary study, we did not detect several expected B. anthracis coat proteins (based on analysis of the B. subtilis genome [Table 3]), and therefore, we anticipate that a significant number of coat proteins have yet to be identified. Taken together, the SDS-PAGE analysis (Fig. 4 and Table 4) and the genomic comparisons (Table 3) suggest that the numbers of coat proteins in B. subtilis and B. anthracis are similar, although we predict that spores from each organism will also contain species-specific proteins.
FIG. 4.
B. anthracis coat proteins resolved by 1D standard SDS-15% PAGE. The samples were solubilized with SDS, and the resulting gel was stained with Coomassie blue G-250. Proteins were identified by MALDI-TOF MS, and the identifications are presented in Table 4.
TABLE 4.
Proteins identified by MALDI-TOF MS from 1D of B. anthracis spore coat fractions
| Band | Protein IDa | MOWSE score | No. of peptides matched | Coverage (%) | MWc/pI | Name |
|---|---|---|---|---|---|---|
| 2 | 654830 | 1.49E+11 | 18 | 28 | 91,362.5/5.70 | Cell surface antigen, contains SLH domain |
| 3 | 654030 | 663 | 6 | 7 | 90,530.8/6.06 | 81% identical to B. subtilis ClpC, class III stress response ATPase |
| 4 | 654198 | 1.56E+08 | 10 | 21 | 57,432.2/4.79 | 78% identical to B. subtilis GroEL |
| 5 | 657011 | 1.21E+03 | 4 | 19 | 24,508.4/8.17 | 49% identical to B. subtilis YndF (amino acids 23-173); putative spore germination protein |
| 7 | 656768 | 1.01E+07 | 12 | 36 | 36,244.2/5.06 | Nucleoside hydrolase |
| 9 | 657458 | 8.93E+03 | 6 | 21 | 41,951.3/8.82 | Nucleoside hydrolase |
| 10 | 654944 | 9.59E+08 | 12 | 39 | 38,373.2/6.17 | Unknown protein, also in Clostridium perfringens |
| 15 | 1326b | 3.17E+03 | 5 | 51 | 21,650.7/5.15 | 89% identical to B. subtilis CotJC |
| 17 | 654287 | 17.7 | 3 | 15 | 19,389.8/5.31 | 37% identical to B. subtilis CotB (amino acids 18-146) and 25% identical to YwrJ |
| 18 | 655132 | 3.29E+03 | 3 | 35 | 17,672.4/4.59 | Unknown protein, adjacent to genes encoding CotY/Z-like proteins |
| 19 | 655132 | 168 | 2 | 19 | 17,672.4/4.59 | Unknown protein, adjacent to genes encoding CotY/Z-like proteins |
| 20 | 655129 | 8.36E+03 | 4 | 20 | 16,459.9/4.95 | 35% identical to B. subtilis CotZ and CotY |
| 21 | 655129 | 5.09E+04 | 6 | 47 | 16,459.9/4.95 | 35% identical to B. subtilis CotZ and CotY |
NP _ number accessible through NCBI.
Our working ORF identification; gene not in public database.
MW, molecular weight.
From our initial MALDI mapping of a 1D gel, we identified a homolog of CotJC and proteins related to CotB and CotZ. The CotZ-like protein has two electrophoretic variants (Fig. 2), which may indicate processing or protein modification. In B. subtilis, CotB is on the spore surface (37), CotJC is likely to be in the inner coat (70), and CotZ is part of a relatively insoluble portion of the coat (82). Therefore, if these features hold true in B. anthracis, our approach succeeded in extracting proteins at different positions in the coat and included a protein that we predict will be relatively highly cross-linked. Interestingly, the B. anthracis CotB-like protein is predicted to be much smaller than the B. subtilis homolog (19 and 43 kDa, respectively), and therefore, these proteins may function differently in the two species. The smaller size is not likely due to misannotation, as the B. anthracis protein migrates at 21 kDa (Fig. 4) whereas, in B. subtilis, CotB migrates at 62 kDa (Fig. 1). Among the novel proteins identified, we predict that some will be B. anthracis-specific coat proteins. Although we cannot yet confirm that any of these candidate proteins are in the coat, our general success at extracting bona fide coat proteins, coupled with the likelihood that some B. anthracis coat proteins are not present in B. subtilis (20), suggests that at least some of these candidates are coat components. Nine of the novel proteins that we identified in B. subtilis have homologs in B. anthracis (Table 3), and therefore, we regard them as candidate B. anthracis coat proteins as well. As for B. subtilis coat proteins (see above), we anticipate that additional coat proteins remain to be found.
The complexity of the coat suggests that it has important roles in adaptations to the diverse niches in which bacilli flourish and argues that much more sophisticated tests of coat function are needed. Likewise, much deeper investigations of the ecology of sporeformers will be required if we are to understand the evolutionary pressures that resulted in a protective organelle with such diverse and powerful protective features. Given the likelihood that many properties of the coat are emergent phenomena (see introduction), ascribing coat functions to specific proteins will likely require deleting specific, as yet unknown, combinations of coat protein genes.
Acknowledgments
Generous support was provided by Philip Andrews and the Michigan Life Sciences Consortium. Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org.
This research was funded by the American Cancer Society, grant RSG-01-090-01-MCB (J.R.M.), and the NIH, grants AI053360 (J.R.M.) and GM53989 (A.D.).
E.-M. Lai and N. D. Phadke contributed equally to this work.
REFERENCES
- 1.Abe, A., H. Koide, T. Kohno, and K. Watabe. 1995. A Bacillus subtilis spore coat polypeptide gene, cotS. Microbiology 141:1433-1442. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, S.-I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson, A. I., and P. Fitz-James. 1976. Structure and morphogenesis of the bacterial spore coat. Bacteriol. Rev. 40:360-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson, A. I., H. Y. Song, and N. Bourne. 1989. Gene structure and precursor processing of a novel Bacillus subtilis spore coat protein. Mol. Microbiol. 3:437-444. [DOI] [PubMed] [Google Scholar]
- 6.Bagyan, I., M. Noback, S. Bron, M. Paidhungat, and P. Setlow. 1998. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene 212:179-188. [DOI] [PubMed] [Google Scholar]
- 7.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beall, B., A. Driks, R. Losick, and C. P. Moran, Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 175:1705-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behravan, J., H. Chirakkal, A. Masson, and A. Moir. 2000. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J. Bacteriol. 182:1987-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boland, F. M., A. Atrih, H. Chirakkal, S. J. Foster, and A. Moir. 2000. Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology 146:57-64. [DOI] [PubMed] [Google Scholar]
- 11.Bourne, N., P. C. Fitz-James, and A. I. Aronson. 1991. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J. Bacteriol. 173:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Volker, U. Volker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 13.Charlton, S., A. J. Moir, L. Baillie, and A. Moir. 1999. Characterization of the exosporium of Bacillus cereus. J. Appl. Microbiol. 87:241-245. [DOI] [PubMed] [Google Scholar]
- 14.Chevallet, M., V. Santoni, A. Poinas, D. Rouquie, A. Fuchs, S. Kieffer, M. Rossignol, J. Lunardi, J. Garin, and T. Rabilloud. 1998. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis 19:1901-1909. [DOI] [PubMed] [Google Scholar]
- 15.Cutting, S., L. Zheng, and R. Losick. 1991. Gene encoding two alkali-soluble components of the spore coat from Bacillus subtilis. J. Bacteriol. 173:2915-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutting, S. M., and P. B. Vander Horn. 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 17.Dixon, T. C., M. Meselson, J. Guillemin, and P. C. Hanna. 1999. Anthrax. N. Engl. J. Med. 341:815-826. [DOI] [PubMed] [Google Scholar]
- 18.Donovan, W., L. Zheng, K. Sandman, and R. Losick. 1987. Genes encoding spore coat polypeptides from Bacillus subtilis. J. Mol. Biol. 196:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Driks, A. 1999. The Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Driks, A. 2002. Maximum shields: the armor plating of the bacterial spore. Trends Microbiol. 10:251-254. [DOI] [PubMed] [Google Scholar]
- 21.Driks, A. 2002. Proteins of the spore core and coat, p. 527-536. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, D.C.
- 22.Driks, A. 1999. Spatial and temporal control of gene expression in prokaryotes, p. 21-33. In V. E. A. Russo, D. J. Cove, L. G. Edgar, R. Jaenisch, and F. Salamini (ed.), Development: genetics, epigenetics and environmental regulation. Springer-Verlag, Berlin, Germany.
- 23.Driks, A., and P. Setlow. 2000. Morphogenesis and properties of the bacterial spore, p. 191-218. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 24.Fillinger, S., S. Boschi-Muller, S. Azza, E. Dervyn, G. Branlant, and S. Aymerich. 2000. Two glyceraldehyde-3-phosphate dehydrogenases with opposite physiological roles in a nonphotosynthetic bacterium. J. Biol. Chem. 275:14031-14037. [DOI] [PubMed] [Google Scholar]
- 25.Fountoulakis, M., and B. Takacs. 2001. Effect of strong detergents and chaotropes on the detection of proteins in two-dimensional gels. Electrophoresis 22:1593-1603. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Patrone, M., and J. S. Tandecarz. 1995. A glycoprotein multimer from Bacillus thuringiensis sporangia: dissociation into subunits and sugar composition. Mol. Cell. Biochem. 145:29-37. [DOI] [PubMed] [Google Scholar]
- 27.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 28.Gerhardt, P., and E. Bibi. 1964. Ultrastructure of the exosporium enveloping spores of Bacillus cereus. J. Bacteriol. 88:1774-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graumann, P. L., and M. A. Marahiel. 1999. Cold shock proteins CspB and CspC are major stationary-phase-induced proteins in Bacillus subtilis. Arch. Microbiol. 171:135-138. [DOI] [PubMed] [Google Scholar]
- 30.Hachisuka, Y., K. Kojima, and T. Sato. 1966. Fine filaments on the outside of the exosporium of Bacillus anthracis spores. J. Bacteriol. 91:2382-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 20:95-110. [DOI] [PubMed] [Google Scholar]
- 32.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 33.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 34.Hulett, F. M. 2002. The Pho regulon, p. 193-201. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 35.Hullo, M. F., I. Moszer, A. Danchin, and I. Martin-Verstraete. 2001. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 183:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 37.Isticato, R., G. Cangiano, H. T. Tran, A. Ciabattini, D. Medaglini, M. R. Oggioni, M. De Felice, G. Pozzi, and E. Ricca. 2001. Surface display of recombinant proteins on Bacillus subtilis spores. J. Bacteriol. 183:6294-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kodama, T., H. Takamatsu, K. Asai, K. Kobayashi, N. Ogasawara, and K. Watabe. 1999. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J. Bacteriol. 181:4584-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kodama, T., H. Takamatsu, K. Asai, N. Ogasawara, Y. Sadaie, and K. Watabe. 2000. Synthesis and characterization of the spore proteins of Bacillus subtilis YdhD, YkuD, and YkvP, which carry a motif conserved among cell wall binding proteins. J. Biochem. (Tokyo) 128:655-663. [DOI] [PubMed] [Google Scholar]
- 40.Lee, V. T., and O. Schneewind. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 15:1725-1752. [DOI] [PubMed] [Google Scholar]
- 41.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 42:1107-1120. [DOI] [PubMed] [Google Scholar]
- 42.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 44.Martins, L. O., C. M. Soares, M. M. Pereira, M. Teixeira, T. Costa, G. H. Jones, and A. O. Henriques. 2002. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J. Biol. Chem. 277:18849-18859. [DOI] [PubMed] [Google Scholar]
- 45.Matz, L. L., T. C. Beaman, and P. Gerhardt. 1970. Chemical composition of exosporium from spores of Bacillus cereus. J. Bacteriol. 101:196-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Micka, B., N. Groch, U. Heinemann, and M. A. Marahiel. 1991. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J. Bacteriol. 173:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moberly, B. J., F. Shafa, and P. Gerhardt. 1966. Structural details of anthrax spores during stages of transformation into vegetative cells. J. Bacteriol. 92:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moir, A. 1981. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J. Bacteriol. 146:1106-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moir, A., B. M. Corfe, and J. Behravan. 2002. Spore germination. Cell. Mol. Life Sci. 59:403-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molloy, M. P. 2000. Two-dimensional electrophoresis of membrane proteins using immobilized pH gradients. Anal. Biochem. 280:1-10. [DOI] [PubMed] [Google Scholar]
- 51.Molloy, M. P., N. D. Phadke, H. Chen, R. Tyldesley, D. E. Garfin, J. M. Maddock, and P. C. Andrews. 2002. Profiling the alkaline membrane proteome of Caulobacter crescentus with two-dimensional electrophoresis and mass spectrometry. Proteomics 2:899-910. [DOI] [PubMed] [Google Scholar]
- 52.Molloy, M. P., N. D. Phadke, J. R. Maddock, and P. C. Andrews. 2001. Two-dimensional electrophoresis and peptide mass-fingerprinting of bacterial outer membrane proteins. Electrophoresis 22:1686-1696. [DOI] [PubMed] [Google Scholar]
- 53.Moriyama, R., H. Fukuoka, S. Miyata, S. Kudoh, A. Hattori, S. Kozuka, Y. Yasuda, K. Tochikubo, and S. Makino. 1999. Expression of a germination-specific amidase, SleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J. Bacteriol. 181:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moriyama, R., A. Hattori, S. Miyata, S. Kudoh, and S. Makino. 1996. A gene (sleB) encoding a spore cortex-lytic enzyme from Bacillus subtilis and response of the enzyme to l-alanine-mediated germination. J. Bacteriol. 178:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moriyama, R., S. Kudoh, S. Miyata, S. Nonobe, A. Hattori, and S. Makino. 1996. A germination-specific spore cortex-lytic enzyme from Bacillus cereus spores: cloning and sequencing of the gene and molecular characterization of the enzyme. J. Bacteriol. 178:5330-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mortz, E., T. N. Krogh, H. Vorum, and A. Gorg. 2001. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1:1359-1363. [DOI] [PubMed] [Google Scholar]
- 57.Nakai, K. 2000. Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54:277-344. [DOI] [PubMed] [Google Scholar]
- 58.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nielsen, H., S. Brunak, and G. von Heijne. 1999. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 60.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 61.Ohlmeier, S., C. Scharf, and M. Hecker. 2000. Alkaline proteins of Bacillus subtilis: first steps towards a two-dimensional alkaline master gel. Electrophoresis 21:3701-3709. [DOI] [PubMed] [Google Scholar]
- 62.Ozin, A. J., A. O. Henriques, H. Yi, and C. P. Moran, Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 182:1828-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozin, A. J., C. S. Samford, A. O. Henriques, and C. P. Moran, Jr. 2001. SpoVID guides SafA to the spore coat in Bacillus subtilis. J. Bacteriol. 183:3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paidhungat, M., and P. Setlow. 2002. Spore germination and outgrowth, p. 527-536. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. ASM Press, Washington, D.C.
- 65.Phadke, N. D., M. P. Molloy, S. A. Steinhoff, P. J. Ulintz, P. C. Andrews, and J. R. Maddock. 2001. Analysis of the outer membrane proteome of Caulobacter crescentus by two-dimensional electrophoresis and mass spectrometry. Proteomics 1:705-720. [DOI] [PubMed] [Google Scholar]
- 66.Roth, I. L., and R. P. Williams. 1963. Comparison of the fine structure of virulent and avirulent spores of Bacillus anthracis. Tex. Rep. Biol. Med. 21:394-399. [PubMed] [Google Scholar]
- 67.Sacco, M., E. Ricca, R. Losick, and S. Cutting. 1995. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J. Bacteriol. 177:372-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindler, T., P. L. Graumann, D. Perl, S. Ma, F. X. Schmid, and M. A. Marahiel. 1999. The family of cold shock proteins of Bacillus subtilis. Stability and dynamics in vitro and in vivo. J. Biol. Chem. 274:3407-3413. [DOI] [PubMed] [Google Scholar]
- 70.Seyler, R. W., Jr., A. O. Henriques, A. J. Ozin, and C. P. Moran, Jr. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25:955-966. [DOI] [PubMed] [Google Scholar]
- 71.Sonenshein, A. L. 2000. Endospore-forming bacteria: an overview, p. 133-150. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 72.Sonenshein, A. L., J. A. Hoch, and R. Losick (ed.). 2002. Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 73.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 74.Takamatsu, H., T. Hiraoka, T. Kodama, H. Koide, S. Kozuka, K. Tochikubo, and K. Watabe. 1998. Cloning of a novel gene yrbB, encoding a protein located in the spore integument of Bacillus subtilis. FEMS Microbiol. Lett. 166:361-367. [DOI] [PubMed] [Google Scholar]
- 75.Takamatsu, H., A. Imamura, T. Kodama, K. Asai, N. Ogasawara, and K. Watabe. 2000. The yabG gene of Bacillus subtilis encodes a sporulation specific protease which is involved in the processing of several spore coat proteins. FEMS Microbiol. Lett. 192:33-38. [DOI] [PubMed] [Google Scholar]
- 76.Takamatsu, H., T. Kodama, A. Imamura, K. Asai, K. Kobayashi, T. Nakayama, N. Ogasawara, and K. Watabe. 2000. The Bacillus subtilis yabG gene is transcribed by SigK RNA polymerase during sporulation, and yabG mutant spores have altered coat protein composition. J. Bacteriol. 182:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 181:4986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomas, J. H. 1993. Thinking about genetic redundancy. Trends Genet. 9:395-399. [DOI] [PubMed] [Google Scholar]
- 80.Warth, A. D., D. F. Ohye, and W. G. Murrell. 1963. The composition and structure of bacterial spores. J. Cell Biol. 16:579-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, J., P. C. Fitz-James, and A. I. Aronson. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 175:3757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng, L., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]