Abstract
yfiK was discovered as a gene augmenting cysteine production when it was overexpressed in an industrial Escherichia coli production strain. The gene product is an integral membrane protein with about six predicted transmembrane helices; it belongs to the RhtB family of export proteins. YfiK overproduction from a plasmid leads to drastic and parallel secretion of O-acetylserine and cysteine into the medium but only when the organism possesses a serine transacetylase that is feedback insensitive to cysteine. Externally provided O-acetylserine obviated this requirement for cysteine secretion both in the yfiK-carrying transformant and in the wild type. A ΔyfiK mutant did not show any phenotype, and it exported O-acetylserine and cysteine when transformed with a plasmid carrying ydeD, a previously characterized, alternate O-acetylserine/cysteine exporter. Since a ydeD-yfiK double mutant showed the same pattern, it appears that YfiK and YdeD act independently. The necessity for the cell to regulate the size of the internal pool of O-acetylserine via synthesis of exporter proteins could be connected to the fact that this compound (when supplied externally) inhibits growth. Overexpression of either ydeD or yfiK leads to alleviation of this inhibition paralled by increased resistance to azaserine, which is an analog of O-acetylserine.
The capacity of microorganisms to export compounds into the extracellular space is well known and has been connected mainly with the protection of cells against toxic compounds. For instance, resistance to antibiotics, heavy metal ions, or organic acids may reside in the ability to specifically or nonspecifically expel such compounds. The mechanisms involved range from the change of membrane permeability via coupling to osmotic pressure or the inversion of uptake systems to the existence of specific exporters (4, 5, 19).
Information obtained in the last few years has revealed that bacteria also may possess specific export systems for substrates, intermediates, or end products of their normal metabolism. Examples are sugar exporters of the Set family (for “Sugar Efflux Transporter”), with three homologs in Escherichia coli and related proteins in Yersinia pestis and Deinococcus radiodurans (18). Two of the sugar exporters belong to systems catalyzing proton-coupled lactose export. Another example is YdeA from E. coli, which was identified as an efflux system for l-arabinose (3).
A very dynamic field of research has also developed in the characterization of amino acid efflux systems since industrial production of amino acids significantly benefits from this research (6). This work has resulted in many examples of exporters of amino acids, like the members of the RhtB family, which can be differentiated into the RhtB and LysE subfamilies. In E. coli, RhtB confers insensitivity to homoserine and homoserine lactone, which are both metabolic regulators, whereas RhtC is involved in threonine export (32). A database search of the bacterial and archaeal genome sequences available disclosed the existence of 225 genes with obvious sequence similarity to the gene encoding RhtB. They resemble each other in the molecular masses of their products, the topology of the membrane-spanning helices in their products, and the existence of at least three conserved sequence signatures (1). The physiological functions of most of them are still unknown.
The LysE subfamily shares the signature sequences with the RhtB subfamily, and their members also possess characteristic sizes and membrane topology. LysE from Corynebacterium glutamicum is the paradigmatic example and one of the biochemically best characterized systems. LysE exports lysine and arginine at identical rates. Its physiological function involves the export of lysine and/or arginine after bacterial growth on oligopeptides rich in these amino acids. The export is energized by proton antiport (2, 4, 5, 27, 28). The expression of lysE is regulated by the LysR-type regulator LysG in C. glutamicum, with lysine, arginine, citrulline, or histidine as coregulators. Genes for members of the LysE-type export system are present in the genomes of many bacteria.
Recently, export systems for threonine in C. glutamicum (the thrE gene) and for cysteine and O-acetylserine in E. coli (the ydeD gene) were identified that do not fit into the RhtB superfamily. ThrE differs from members of the RhtB superfamily by its larger size and a larger number of membrane-spanning helices, and it exports both threonine and serine (25).
ydeD (eamA) has been detected as a gene augmenting the yield in a cysteine-overproducing strain (9). YdeD belongs to the PecM family of transporters, with PecM itself being a transporter for indigoidine of Erwinia chrysanthemi (22). When YdeD is overproduced, export of O-acetylserine (OAS) or cysteine is so efficient that the cysteine regulon is not induced during growth on a poor sulfur source like sulfate. This deficiency is suppressed when a rich sulfur source like thiosulfate is supplied (9). In the course of this work, a second gene was identified during the gene bank screening for cysteine overproducers. When sequenced, it was identified as yfiK, which had been recognized as an RthB family member in the work of Aleshin et al. (1). The present investigation demonstrates that YfiK represents a putative cysteine and OAS exporter alternative to YdeD.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains and plasmids used in this study are listed in Table 1. The media used were Luria-Bertani (LB) (21) or SM1 minimal medium with 1.5% glucose as the carbon source (9). Where indicated, SM1 was supplemented with LB medium, thiosulfate, or OAS. When required, antibiotics were added as follows: ampicillin, 100 μg/ml; kanamycin sulfate, 50 μg/ml; tetracycline, 10 μg/ml; and chloramphenicol, 30 μg/ml. In growth experiments, the cultures were incubated aerobically with vigorous shaking at 37°C. Growth was monitored by measurement of the optical density at 600 nm (OD600).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant featurea | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | F− (φ80d lacIqZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ (lacZYA-argF)U169 | 30 |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rpsR | 7 |
| MC4100Δ299 | MC4100 ydeD::Kan Kmr | 9 |
| W3110 | F− λ− IN (rrnD-rrnE) 1 rph-1 | 11 |
| W3110Δ299 | W3110 ydeD::Kan Kmr | This work |
| W3110 yfiK | W3110 with a deletion in yfiK (Δ14-161) | This work |
| W3110-DK | W3110 ΔyfiK ydeD::kan Kmr | This work |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| K38 | HfrC phoA4 pit-10 tonA22 ompF627 relA1 λ+ | 20 |
| JM15 | F−cysE50 tfr-8 | CGSC no. 5042 |
| JM16 | F−cysE50 tfr-8 lac::serA-cysEX | —b |
| Plasmids | ||
| pACYC184-LH | Tetr | 29 |
| pET3a | Apr, T7φ10 | Novagen |
| pET3a-yfiK | pET3a, yfiK under the control of the T7φ10 promoter | This work |
| pG2 | pACYC184-LH with yfiK under the control of the GAPDH promoter P1, Tetr | —b |
| pG7 | pG2 cysEX Tetr | —b |
| pGP1-2 | T7 gene 1, cI857, Kmr | 26 |
| pHCEX | pACYC184-LH, cysEX Tetr | —b |
| pKO3 | cat repA(Ts) sacB M13 ori Kmr Cmr | 17 |
| pKO3-yfiK | pKO3, ΔyfiK (codons 14-161) | This work |
| pKP290 | pACYC184-LH containing the constitutive E. coli GAPDH promoter P1 | 9 |
| pKP291 | pKP290 with ydeD under the control of the GAPDH promoter P1 | 9 |
| pUC19 | AprlacZ′ ′IPOZ' | 31 |
| pUC19-ΔyfiK | pUC19, ΔyfiK (codons 14-161) | This work |
Apr, Kmr, Tetr, Cmr, resistance to ampicillin, kanamycin, tetracycline, and chloramphenicol, respectively. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Patent strains or plasmids from the Consortium für Elektrochemische Industrie GmbH, Munich, Germany.
To determine growth inhibition by OAS, we used SM4 minimal medium [12 g of Na(NH4)HPO4 · 4H2O per liter, 3 g of KH2PO4 per liter, 0.312 g of Mg(NO3)2 · 6H2O per liter, 14.7 mg of CaCl2 · 2H2O per liter, 0.1 g of NaCl per liter, 1.4 mg of FeCl2 · 4H2O per liter, 1 g of trisodium citrate · 2H2O, per liter, 4.6 g of Na2S2O3 · 5H2O per liter, 5 mg of thiamine per liter, trace elements as in SM1 medium [9]) buffered at pH 6.0 with 100 mM morpholineethanesulfonic acid (MES), containing 0.5% glucose as the carbon source, and supplemented with different concentrations of OAS.
To determine the MIC of azaserine, serial dilutions of azaserine in LB or SM1 medium were performed and inoculated to obtain an OD600 of 0.001, and growth of the respective strain was monitored after 24 h of incubation.
Construction of plasmids.
All DNA manipulations were performed by standard procedures (23) and as recommended by the enzyme manufacturers. All plasmid constructions were made in E. coli DH5α (30) or XL-1 Blue (Stratagene).
A plasmid designated pG1, which contained the genes yfiK-yfiD-ung, was isolated from a gene bank containing chromosomal Sau3AI DNA fragments from E. coli JM15 cloned in pKP290 (9). By removal of an AspI-BfrI fragment, yfiD and ung were deleted from the plasmid to yield plasmid pG2. Plasmid pG7 was further derived by insertion of the cysEX allele as a SacI-NsiI fragment from pHCEX into the SacI-NsiI sites of pG2.
To obtain plasmid pET3a-yfiK, plasmid pG2 was treated with XbaI and BclI and the resulting 867-bp fragment was ligated into pET3a (Novagen) treated with XbaI and BamHI, resulting in yfiK under the control of the phage T7 promoter.
Construction of a chromosomal yfiK deletion mutant.
To generate an in-frame deletion in yfiK (codons 14 to 161), an inverse PCR with oligonucleotides 5 (5′-TTTGAATTCGGCGGGGCATCTG-3′) and 6 (5′-TTTGAATTCAGGGTGTAAGTCC-3′) was performed with pG2 as the template. The resulting plasmid was used as template for PCR with oligonucleotides 7 (5′-AAAGGATCCATAACCCCAAACC-3′) and 8 (5′-TTTAAGCTTACTAAGCGGAAGAGGG-3′), generating BamHI and HindIII restriction sites. The PCR fragment was treated with these enzymes and ligated into BamHI- and HindIII-linearized vector pUC19, resulting in plasmid pUC19-ΔyfiK. To generate a chromosomal deletion in yfiK, plasmid pKO3 (17) was treated with NotI and the protruding ends were refilled with Klenow enzyme before the plasmid was ligated with a BamHI-HindIII fragment from pUC19-ΔyfiK treated with Klenow enzyme, resulting in plasmid pKO3-yfiK. E. coli W3110 was transformed with this plasmid, and the wild-type yfiK was replaced by its truncated allele by homologous recombination by the procedure of Link et al. (17), to give W3110ΔyfiK. The presence of the correct deletion was verified by PCR analysis.
Construction of chromosomal ydeD insertion mutants.
The wild-type ydeD gene from strain W3110 or W3110ΔyfiK was replaced by its disrupted allele from strain MC4100Δ299 (9) via P1 transduction (21) to give W3110Δ299 and W3110-DK, respectively.
Radioactive labeling of YfiK.
pET3a-yfiK was transferred into strain K38/pGP1-2, and radioactive labeling of YfiK with l-[35S]methionine was performed essentially by the method of Tabor and Richardson (26) with modifications described by Daßler et al. (9).
Cellular localization of YfiK.
Six times as many [35S]methionine-labeled cells (strain K38/pGP1-2/pET3a-yfiK) as used normally (see above) were mixed with 12 times as many nonlabeled but otherwise equally treated cells as used normally. They were resuspended in 600 μl of breaking buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 2 mM dithiothreitol, 20 μg of phenylmethylsulfonyl fluoride per ml) and broken by sonication. Unbroken cells and cellular debris were removed by centrifugation at 30,000 × g for 15 min followed by ultracentrifugation of the S30 extract at 100,000 × g for 2 h. The resulting pellet (P100) and supernatant (S100) fractions were adjusted to identical sample buffer volumes (15) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Cysteine determination.
The concentration of cysteine in culture supernatants was determined by the method of Gaitonde as previously described (10). Before ninhydrin was added, putative cystine in the samples was reduced by incubation with 3 mM dithioerythritol in 100 mM Tris-HCl (pH 8.5) for 10 min. This assay method also determines the cysteine occluded in the 2-methyl-2,4-thiazolidinedicarboxylic acid, which is a reaction product of cysteine with pyruvate.
Amino acid determination.
For detection of amino acids other than cysteine in culture supernatants, samples were reacted with o-phthaldialdehyde-mercaptopropionic acid and analyzed by high-performance liquid chromatography on a Hypersil AA-ODS column (200 mm by 2.1 mm) under the conditions recommended by the manufacturer (Agilent Technologies, Waldbronn, Germany).
RESULTS
YfiK is a membrane-integral protein.
yfiK was originally detected in a search for genes which, when overexpressed in a cysteine-overproducing mutant of E. coli, greatly augmented the yield of the amino acid in the medium. In plasmid pG2, yfiK overexpression is mediated by the constitutively active promoter of the E. coli gapA gene. Plasmid pG7 carries, in addition to yfiK, the gene (cysEX) for a serine O-acetyltransferase (SAT) with a mutation rendering it insensitive to feedback inhibition by l-cysteine (16).
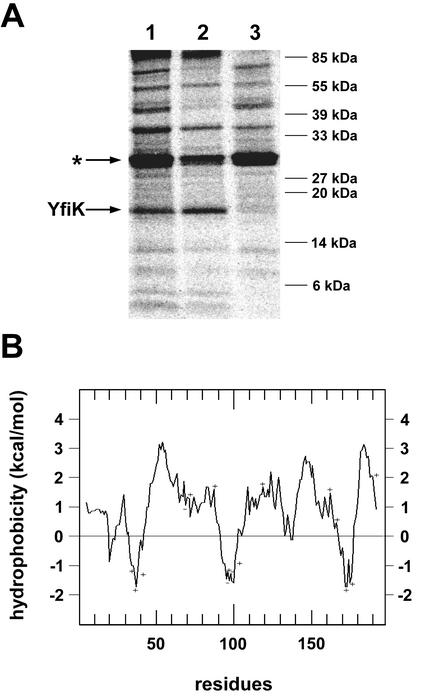
To analyze whether yfiK is transcribed and translated into a protein and, if it is, to localize the gene product in the major cellular compartments, the gene was excised from plasmid pG2 and inserted into BamHI- and XbaI-restricted vector pET3a, yielding plasmid pET3a-yfiK. On this plasmid, yfiK expression is under the control of the phage T7 promoter. pET3a-yfiK was transferred into strain K38 which had been pretransformed with plasmid pGP1-2, and the transformants were labeled with [35S]methionine (26). The cells were lysed, and the lysates were subjected to SDS-PAGE (24) followed by autoradiography (Fig. 1A). In the lane containing the lysate from K38/pGP1-2/pET3a-yfiK, there was one labeled protein band which was not seen in the autoradiogram from lysates of cells containing the empty vector (result not shown). Its apparent size of 17 kDa differed significantly from the 21.2-kDa protein predicted from the gene sequence, which may be a consequence of its extreme hydrophobicity (YfiK contains 69% nonpolar amino acids).
FIG. 1.
Cellular localization of YfiK. (A) Autoradiograph of an SDS-12.5% polyacrylamide gel in which cellular fractions of strain K38/pET3a-yfiK which had been labeled with [35S]methionine in the T7 promoter/polymerase system were separated. The asterisk indicates the migration position of β-lactamase. Lanes: 1, 30,000 × g supernatant; 2, sediment; 3, 100,000 × g supernatant. (B) Hydropathy plot of YfiK obtained by the method of Kyte and Doolittle (14). Charged amino acid residues are marked.
To determine the cellular location of YfiK, labeled cells were broken by sonication, the crude extract was fractionated by centrifugation, and the fractions were analyzed by SDS-PAGE. Figure 1A shows that YfiK was present exclusively in the sediment of the 100,000 × g centrifugation (P100). Treatment of the P100 fraction with 1 M potassium chloride (data not shown) did not detach the protein from the P100 fraction, which is the behavior of membrane-integral proteins. Figure 1B supports this contention since the hydropathy plot (14) predicts the existence of about six membrane-spanning helices.
yfiK overexpression causes cysteine and OAS accumulation in the medium.
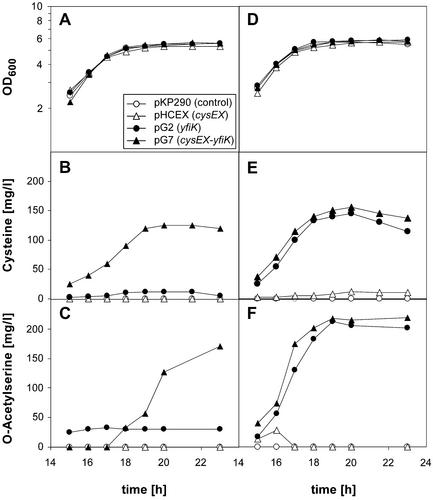
To investigate the initial observation that YfiK augments cysteine production, plasmids pG2 (yfiK) and pG7 (yfiK-cysEX) were introduced into the wild-type strain W3110 and cysteine formation was assessed (Fig. 2B). Plasmid pG7 caused a dramatic accumulation of the amino acid in the medium, whereas transformants carrying plasmids with one gene (yfiK or cysEX) stimulated cysteine formation only slightly or not at all. Thus, yfiK overexpression and a feedback-resistant cysE gene product are both required for the export of the amino acid into the medium. This conclusion is fully supported by the fact that in the transformants carrying pG2 and pG7, cysteine secretion was stimulated to the same extent when the host strain carried the gene for the feedback-insensitive SAT on the chromosome (strain JM16) (Fig. 2E). The onset of cysteine accumulation was in the middle of the exponential growth phase (15 h after inoculation). In these experiments, all transformants of W3110 and JM16 showed identical growth behavior (Fig. 2A and D).
FIG. 2.
Growth (A and D) and excretion of cysteine (B and E) and OAS (C and F) for strains W3110 (wild type) (A to C) and JM16 (cysEX mutant) (D to F) transformed with plasmids pKP290, pHCEX, pG2, and pG7. Only results for the part of the growth phase relevant for cysteine and OAS production are illustrated.
OAS is the immediate biosynthetic precursor of cysteine, and it should be present at increased intracellular levels in a strain harboring the feedback-resistant SAT. We therefore determined whether OAS is secreted into the medium in parallel with cysteine (Fig. 2C and F). Indeed, we found that OAS was also exported at dramatic levels side by side with cysteine; both YfiK overproduction and upregulation of cysteine biosynthesis were prerequisites. The analytical methods used also confirmed the existence of a considerable concentration of pyruvate and acetate in the medium. There was, however, no evidence for the excretion of amino acids other than cysteine and OAS.
Externally supplied OAS influences growth and cysteine excretion.
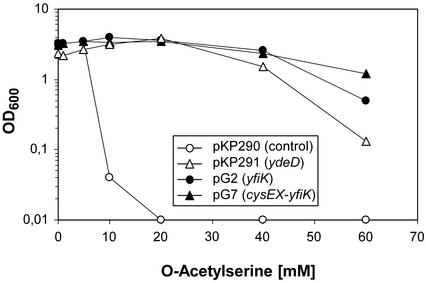
Since the excretion of cysteine and OAS depended, apart from overexpression of yfiK, on the feedback resistance of SAT, we tested whether addition of OAS to the medium could phenotypically mimic the regulatory defect. Since OAS is more stable under slightly acidic conditions, the initial experiments were conducted in SM4 medium whose pH was stabilized at 6.0 with 100 mM MES. Cultures of W3110 and of transformants carrying yfiK, ydeD, or yfiK-cysEX on a plasmid were challenged with different concentrations of OAS, and the OD reached after 24 h of growth was measured. Figure 3 shows that growth of the wild type was already completely blocked by 10 mM OAS and that the presence of either yfiK or ydeD counteracted this inhibition. We concluded that increased levels of OAS are toxic and overexpression of the genes coding for the putative exporter proteins protect against inhibition.
FIG. 3.
Growth inhibition by OAS. E. coli W3110 transformed with plasmid pKP290, pKP291, pG2, or pG7 was inoculated into SM4 minimal medium with 0.5% glucose as the carbon source, buffered at pH 6.0 with 100 mM MES and supplemented with increasing concentrations of OAS. Growth was monitored after 24 h of incubation at 37°C by measurement of the OD600.
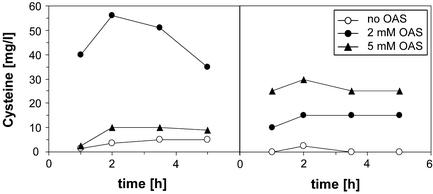
Next, the effect of addition of OAS to the medium on cysteine excretion was analyzed. Transformants of W3110 carrying plasmid pG2 (yfiK) or the vector pKP290 were grown in SM1 medium supplemented with 1.5% glucose and 0.4 g of sodium thiosulfate per liter to the end-exponential phase, and the medium was then supplemented with 2 and 5 mM OAS, respectively. To compare the results with those obtained in previous experiments, the initial pH of the medium was adjusted to 7.0. Samples were taken and analysed for cysteine content (Fig. 4). Figure 4A shows that W3110 carrying pG2 secreted a substantial amount of cysteine in the presence of 2 mM OAS but much less in the presence of 5 mM OAS. Intriguingly, as shown in Fig. 4B, W3110 produced cysteine without overexpression of yfiK (the strain contains the chromosomal copy of the gene) when challenged with OAS. Under these conditions, the level of cysteine formation was lower than in the W3110/pG2 transformant and increased when the concentration of OAS was raised from 2 to 5 mM. The decreased cysteine yield in cultures of W3110/pG2 grown in the presence of 5 mM OAS could be correlated with the export mediated by YfiK, which may lower the internal concentration and compete with the conversion into cysteine.
FIG. 4.
Cysteine formation and excretion after OAS supplementation by strain W3110/pG2 (A) and control strain W3110/pKP290 (B). Strains were grown to the late log phase, portions of the cultures were provided with different concentrations of OAS (0 h), and the cysteine content of the medium was measured.
YfiK acts independently of YdeD.
We have previously described a gene (ydeD) from E. coli which, like yfiK, stimulates the excretion of OAS and cysteine into the medium (9). A condition was that the medium be fortified with a source of reduced sulfur, e.g., thiosulfate plus methionine. The existence of two such cysteine and OAS export systems is intriguing and suggested an analysis whether they act independently or cooperatively. To this end, a mutant yfiK gene carrying a deletion which removes amino acids 14 to 161 from the gene product was constructed. It was transferred into the chromosome of W3110, and the W3110 ΔyfiK derivative was transformed with plasmid pKP291 carrying the ydeD gene under control of the gapA promoter (9). Conversely, strain W3110Δ299 (a ydeD mutant) was transformed with plasmid pG2 (yfiK) or pG7 (yfiK-cysEX). The transformants were grown in SM1 medium in the presence of 1.5% glucose and 0.4 g of sodium thiosulfate per liter, and the medium was analyzed for cysteine content. There was no measurable difference in cysteine concentration between cultures of strains containing the wild-type and mutant yfiK alleles or the wild-type and mutant ydeD alleles (data not shown). We concluded that accumulation of cysteine in the medium stimulated by yfiK and in medium stimulated by ydeD overexpression appear to be independently regulated processes.
Next, a ydeD::kan ΔyfiK (strain W3110-DK) double mutant was constructed and analyzed physiologically after transformation with vectors or plasmids carrying either yfiK or ydeD. W3110-DK displays normal growth in minimal medium, indicating that neither of the two genes is essential under laboratory conditions (data not shown). When the transformants carrying both mutant alleles were analyzed, the results were consistent with those obtained with the single-mutant strains (data not shown). The growth of the transformant W3110-DK/pKP291 (overproducing YdeD) was retarded, as described previously for strain W3110/pKP291 (9); cysteine formation by all strains resembled the situation in the respective W3110 wild-type background. It should be emphasized that cysteine formation in W3110-DK/pKP291 initiates in the stationary phase of growth whereas that of W3110-DK/pG7 takes place during the exponential phase (data not shown).
Specificity of YfiK.
It has been shown conclusively that the ydeD gene product specifically exports OAS and cysteine (9) plus two unidentified compounds. These unidentified compounds have since been identified as glutamine and asparagine (8). The specificity of YfiK, on the other hand, extends only to the excretion of OAS and cysteine. Three-dimensional structural models of OAS, glutamine, and, to a lesser extent, asparagine have considerable steric similarities and resemble that of the glutamine antagonist azaserine. Overproduction of YdeD and YfiK might therefore result in increased azaserine resistance. This assumption was tested by determining the MIC. Indeed, when either ydeD or yfiK was present on a plasmid, the MIC of azaserine was increased about fourfold in LB medium and two- to threefold in SM1 minimal medium. The fact that the level of resistance is identical whether or not the transformants overproduce and export OAS or cysteine strongly supports the contention that azaserine resistance is a consequence of increased export.
DISCUSSION
Table 2 presents a comparison of the functional and physiological characteristics of YfiK and YdeD. They differ in their molecular size, they belong to different families of export proteins, and they are not essential for growth under laboratory conditions. Whereas YdeD resembles RhtB (32) and SetA (18) in having broad substrate specificity, that of YfiK appears to be confined to the export of cysteine and OAS. This conclusion is based on the amino acid analysis of the medium after growth under conditions which promote the export of OAS and/or cysteine. One has to bear in mind, however, that other substrates of YfiK might have escaped detection since they are not overproduced. We have tested this possibility by coexpression of yfiK and genes responsible for threonine overproduction but could not detect an increase in the amount of threonine in the medium (our unpublished results).
TABLE 2.
Comparison of structural and functional characteristics of YdeD and YfiK
| Property or function | YdeD | YfiK |
|---|---|---|
| Size (no. of amino acids) | 299 | 195 |
| Start codon | ATG | GTG |
| No. of putative transmembrane helices | 9-10 | 5-7 |
| Protein family | PecM | RhtB |
| Mutant phenotype | None | None |
| Phenotype on overproductiona | NAS deficiency | None |
| Prerequisite for export | None | High intracellular level of OAS |
| Amino acid exported | OAS, Cys, Gln, Asn | OAS, Cys |
| Growth phase of export (OAS) | Exponential | Exponential |
| Growth phase of export (Cys) | Stationary | Exponential |
Apart from excretion of amino acids.
What are the selective values and the connected physiological functions of two exporters of cysteine and OAS? Although we are far from having a definite answer, there are a few characteristics which may lead the way. First, extrusion of cysteine and OAS via the YfiK system requires an upregulated biosynthetic pathway, which is not a prerequisite for YdeD. The affinity for cysteine and OAS of YfiK therefore appears to be lower than that of YdeD. This matches the observation that YdeD but not YfiK overproduction drains the cellular level of OAS (and therefore the level of the CysB activator N-acetylserine [NAS]) so much that activation of the cysteine regulon by NAS (13) can no longer take place (9). Therefore, YdeD might be involved in some fine-tuning process in balancing the synthesis of the carbon skeleton of cysteine and the formation of the reduced sulfur source, e.g., by exporting OAS or NAS when it is no longer needed for activation of the regulon. There is no information in the literature on how NAS is removed under nonactivation conditions. YfiK, on the other hand, could be an emergency valve, pumping out cysteine and its precursor (and activator) when they attain toxic levels.
YdeD and YfiK, when overproduced singly, also exert different effects on the timing of OAS and cysteine extrusion. In a YdeD overproducer, OAS is pumped out during the exponential growth phase; later, it is taken up again and serves as a precursor for the subsequent cysteine formation and secretion (8). In a strain expressing yfiK from a plasmid, OAS and cysteine are exported side by side in the exponential phase. This again would be consistent with an emergency function of YfiK and a modulation of the OAS (and NAS) concentration by YdeD during exponential growth.
The results described here and reported in the literature (12, 13) emphasize the key role of SAT in the control of the cysteine regulon, both kinetically and genetically. This is highlighted by the finding that addition of OAS to the culture of a wild-type strain results in massive cysteine secretion. In view of the toxicity of OAS (Fig. 3), it is mandatory that this level must be strictly controlled, such as under conditions of sulfur limitation. In such a situation, cysteine cannot be made in sufficient amounts to feedback-inhibit SAT, and the only measure to overcome the accumulation of toxic levels of OAS might be extrusion (12).
Both ydeD and yfiK are expressed at very low levels (reference 9 and our unpublished results). No regulation could be detected for ydeD expression. Although it is unknown whether expression of yfiK is also constitutive, its chromosomal organisation indicates that it may be regulated. yfiK is separated by a 57-bp spacer from an inversely oriented gene, yfiE, whose derived amino acid sequence indicates that it belongs to the LysR family of regulators. This fact, the positioning relative to yfiK, and the unusually short spacer segment could indicate that YfiE regulates the expression of yfiK, possibly in concert with some metabolites.
Acknowledgments
We thank Renate Flinspach for competent technical assistance.
I.F., A.R., and A.B. thank the Consortium für Elecktrochemische Industrie and the Fonds der Chemischen Industrie for generous financial support.
REFERENCES
- 1.Aleshin, V. V., N. P. Zakataeva, and V. A. Livshits. 1999. A new family of amino-acid-efflux proteins. Trends Biochem. Sci. 24:133-135. [DOI] [PubMed] [Google Scholar]
- 2.Bellmann, A., M. Vrljic, M. Patek, H. Sahm, R. Krämer, and L. Eggeling. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765-1774. [DOI] [PubMed] [Google Scholar]
- 3.Bost, S., F. Silva, and D. Belin. 1999. Transcriptional activation of ydeA, which encodes a member of the major facilitator superfamily, interferes with arabinose accumulation and induction of the Escherichia coli arabinose PBAD promoter. J. Bacteriol. 181:2185-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bröer, S., and R. Krämer. 1991. Lysine excretion by Corynebacterium glutamicum. 1. Identification of a specific secretion carrier system. Eur. J. Biochem. 202:131-135. [DOI] [PubMed] [Google Scholar]
- 5.Bröer, S., and R. Krämer. 1991. Lysine excretion by Corynebacterium glutamicum. 2. Energetics and mechanism of the transport system. Eur. J. Biochem. 202:137-143. [DOI] [PubMed] [Google Scholar]
- 6.Burkovski, A., and R. Krämer. 2002. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biotechnol. 58:265-274. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daßler, T. 2001. Ph.D. thesis. Universität München, Munich, Germany.
- 9.Daßler, T., T. Maier, C. Winterhalter, and A. Böck. 2000. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol. Microbiol. 36:1101-1112. [DOI] [PubMed] [Google Scholar]
- 10.Gaitonde, M. K. 1967. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, and B. Magasanik (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, D.C.
- 13.Kredich, N. M. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747-2753. [DOI] [PubMed] [Google Scholar]
- 14.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Leinfelder, W., and P. Heinrich. April 1995. Process for preparing O-acetylserine, l-cysteine-related products. German patent WO97/15673.
- 17.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, J. Y., P. F. Miller, M. Gosink, and E. R. Olson. 1999. The identification of a new family of sugar efflux pumps in Escherichia coli. Mol. Microbiol. 31:1845-1851. [DOI] [PubMed] [Google Scholar]
- 19.Luntz, M. M., N. I. Zhdanova, and G. I. Bourd. 1986. Transport and excretion of l-lysine in Corynebacterium glutamicum. J. Gen. Microbiol. 132:2137-2146. [Google Scholar]
- 20.Lyons, L. B., and N. D. Zinder. 1972. The genetic map of the filamentous bacteriophage f1. Virology 49:45-60. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Rouanet, C., and W. Nasser. 2001. The PecM protein of the phytopathogenic bacterium Erwinia chrysanthemi, membrane topology and possible involvement in the efflux of the blue pigment indigoidine. J. Mol. Microbiol. Biotechnol. 3:309-318. [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 25.Simic, P., H. Sahm, and L. Eggeling. 2001. l-Threonine export: use of peptides to identify a new translocator from Corynebacterium glutamicum. J. Bacteriol. 183:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vrljic, M., J. Garg, A. Bellmann, S. Wachi, R. Freudl, M. J. Malecki, H. Sahm, V. J. Kozina, L. Eggeling, and M. H. Saier, Jr. 1999. The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradigm for a novel superfamily of transmembrane solute translocators. J. Mol. Microbiol. Biotechnol. 1:327-336. [PubMed] [Google Scholar]
- 28.Vrljic, M., H. Sahm, and L. Eggeling. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol. Microbiol. 22:815-826. [DOI] [PubMed] [Google Scholar]
- 29.Winterhalter, C., and W. Leinfelder. June 1997. Mikroorganismen und Verfahren zur fermentativen Herstellung von l-Cystein, l-Cystin, N-Acetylserin oder Thiazolidinderivaten. European patent 0885962A1.
- 30.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 32.Zakataeva, N. P., V. V. Aleshin, I. L. Tokmakova, P. V. Troshin, and V. A. Livshits. 1999. The novel transmembrane Escherichia coli proteins involved in the amino acid efflux. FEBS Lett. 452:228-232. [DOI] [PubMed] [Google Scholar]