Abstract
Organisms of Chlamydia spp. are obligate intracellular, gram-negative bacteria with a dimorphic developmental cycle that takes place entirely within a membrane-bound vacuole termed an inclusion. The chlamydial anomaly refers to the fact that cell wall-active antibiotics inhibit Chlamydia growth and peptidoglycan (PG) synthesis genes are present in the genome, yet there is no biochemical evidence for synthesis of PG. In this work, we undertook a genetics-based approach to reevaluate the chlamydial anomaly by characterizing MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase that catalyzes the first committed step of PG synthesis. The murA gene from Chlamydia trachomatis serovar L2 was cloned and placed under the control of the arabinose-inducible, glucose-repressible ara promoter and transformed into Escherichia coli. After transduction of a lethal ΔmurA mutation into the strain, viability of the E. coli strain became dependent upon expression of the C. trachomatis murA. DNA sequence analysis of murA from C. trachomatis predicted a cysteine-to-aspartate change in a key residue within the active site of MurA. In E. coli, the same mutation has previously been shown to cause resistance to fosfomycin, a potent antibiotic that specifically targets MurA. In vitro activity of the chlamydial MurA was resistant to high levels of fosfomycin. Growth of C. trachomatis was also resistant to fosfomycin. Moreover, fosfomycin resistance was imparted to the E. coli strain expressing the chlamydial murA. Conversion of C. trachomatis elementary bodies to reticulate bodies and cell division are correlated with expression of murA mRNA. mRNA from murB, the second enzymatic reaction in the PG pathway, was also detected during C. trachomatis infection. Our findings, as well as work from other groups, suggest that a functional PG pathway exists in Chlamydia spp. We propose that chlamydial PG is essential for progression through the developmental cycle as well as for cell division. Elucidating the existence of PG in Chlamydia spp. is of significance for the development of novel antibiotics targeting the chlamydial cell wall.
Chlamydiae, of which there are four species, are gram-negative, obligate intracellular eubacteria. Chlamydia psittaci and Chlamydia pecorum are important animal pathogens, while Chlamydia trachomatis and Chlamydia pneumoniae are important human pathogens that cause sexually transmitted ocular and respiratory infections. Chlamydiae have a unique biphasic developmental cycle that allows the bacteria to survive in two different habitats (38). The infectious elementary bodies (EBs) are small, osmotically stable, metabolically inactive particles that survive in the extracellular environment to attach to host epithelial cells and become internalized. Once inside, the bacteria reside in a specialized phagosome, termed the chlamydial inclusion, in the host cell cytoplasm. In the inclusion, the EBs differentiate into large, osmotically fragile, metabolically active reticulate bodies (RBs), which use host cell ATP and nutrients and divide by binary fission. The cycle remains synchronous for approximately the first 20 h postinfection (hpi), after which some RBs continue to divide while others begin to differentiate back to EBs. The host cells continue to support growth of the bacteria (30 to 72 hpi) before the cells lyse, releasing EBs, RBs, and intermediate forms. Released EBs initiate subsequent rounds of infection in surrounding cells.
While chlamydiae are classified as gram-negative bacteria, the chlamydial cell envelope differs from those of other eubacteria. Cell wall lipid and amino acid compositions are similar to those of other gram-negative organisms, and lipopolysaccharide is detected in the outer membrane (27). However, unlike most gram-negative as well as gram-positive bacteria, chlamydiae do not contain detectable levels of peptidoglycan (PG). PG is a polymer of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) units with an l- and d-amino acid-containing pentapeptide linked to MurNAc (26). This polymer layer determines the shape of the bacteria and protects the bacteria from osmotic shock. PG also plays a role in cell division. Several observations support the notion that chlamydiae lack PG. These include the inability to detect a PG layer by electron microscopy, the failure of antibodies against PG to react with chlamydiae, and the inability to detect muramic acid, a marker for PG, by using gas chromatography-mass spectrometry (5, 9). Disulfide cross-linking of the chlamydial major outer membrane protein and cysteine-rich proteins in the outer membrane of EBs are believed to substitute for PG and provide structural strength to the cell wall (14).
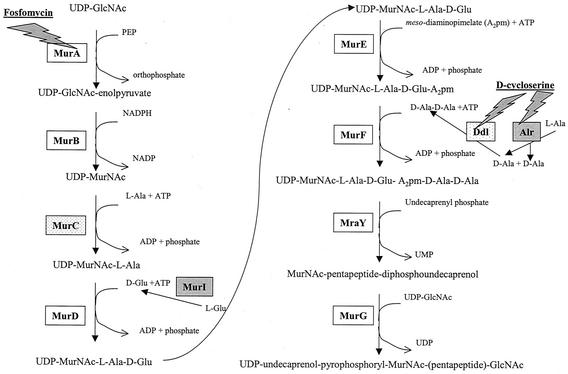
Paradoxically, unlike other bacteria that lack PG, chlamydiae contain penicillin-binding proteins and are sensitive to antibiotics that inhibit PG synthesis (2, 23). Matsumoto and Manire demonstrated by transmission electron microscopy that penicillin treatment of C. psittaci-infected L cells prevented RB binary fission and resulted in the formation of abnormally large RBs (21). These “penicillin forms,” which resume normal shape after penicillin removal, have been reported in every species of Chlamydia. Lin and Moulder reported similar observations for C. psittaci-infected cells treated with d-cycloserine (19). Correspondingly, genome sequencing of C. trachomatis serovars D and L2 and two strains of C. pneumoniae has revealed that both species contain a nearly complete pathway for PG synthesis similar to the pathway in Escherichia coli (Fig. 1). The paradox of phenotypic and genomic evidence for PG in Chlamydia and the absence of detectable PG has been termed the chlamydial anomaly (23).
FIG. 1.
Pathway of PG synthesis. In the cytoplasm of E. coli, eleven enzymes (MurA to MurG, MurI, MraY, Alr, and Ddl) catalyze the synthesis of the disaccharide-pentapeptide subunit of PG. Of these eleven enzymes, Chlamydia spp. encode nine; Alr and MurI (boxes shaded in gray) are missing (28, 34). MurC and Ddl (boxes with speckled gray) are encoded as a fused protein in Chlamydia spp. Antibiotics that inhibit cell wall synthesis are underlined, while gray-shaded bolts identify the enzymes targeted.
To date, there have been no reports demonstrating the functionality of an enzyme in the chlamydial PG pathway. We decided to take a genetic approach to demonstrate the functionality of the chlamydial MurA, the enzyme that commits UDP-GlcNAc to PG synthesis. MurA, a UDP-GlcNAc enolpyruvyl transferase, catalyzes the addition of enolpyruvate from phosphoenolypyruvate (PEP) to UDP-GlcNAc. In E. coli, murA is an essential gene. A single copy of murA exists in the genome, and deletion of this gene is lethal (4). MurA is also the site of action for the antibiotic fosfomycin, a structural analog of PEP that binds irreversibly to the active site of MurA (16).
Unlike most other bacteria of clinical significance, a system for genetic manipulation of Chlamydia is not available. Characterization of chlamydial genes is thus far limited to expression in a heterologous host system such as E. coli. In this report, we show that MurA from C. trachomatis functions as a UDP-GlcNAc enolpyruvyl transferase in vitro and in vivo. MurA from C. trachomatis complements a lethal deletion of the essential murA gene in E. coli. Genome sequencing revealed that the chlamydial MurA contains a cysteine-to-aspartate change in the active site of the enzyme. A similar change in the E. coli MurA renders the enzyme resistant to fosfomycin. We also demonstrate that C. trachomatis is resistant to high concentrations of fosfomycin. This innate resistance to fosfomycin appears to be due to the activity of the chlamydial MurA, as in vitro activity of the chlamydial MurA is also resistant to high concentrations of fosfomycin and fosfomycin resistance can be conferred upon E. coli expressing the chlamydial MurA. Expression of C. trachomatis murA mRNA and mRNA from other PG pathway genes is cell cycle dependent. Collectively, our data support the notion that Chlamydia organisms contain PG and suggest that PG in Chlamydia plays a role in RB development and division.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study are listed in Table 1. Strains were grown at 37°C in Luria-Bertani (LB) medium with aeration or on LB agar. When appropriate, medium was supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (15 μg/ml), arabinose (0.1%), or glucose (0.2%). Fosfomycin was obtained from the Sigma Chemical Company (St. Louis, Mo.).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| CodonPlus | E. coli BL21-CodonPlus-(DE3)-RIL [F−ompT hsdS(rB− mB−) dcm+ Tetrgal λ (DE3) endA Hte (argU ileY leuW Camr)] | Stratagene |
| ZK1746 | E. coli W3110 tna2 ΔlacU169 Δara ΔmurA::kan (pBAD30-Z) | 4 |
| ATM580 | CodonPlus transformed with pAJM5 | This work |
| ATM581 | ATM580 ΔmurAEc::kan | This work |
| ATM586 | CodonPlus transformed with pAJM6 | This work |
| ATM587 | ATM586 ΔmurACt::kan | This work |
| 2457T | S. flexneri 2a wild type | 8 |
| C. trachomatis | C. trachomatis serovar L2 | H. Caldwell |
| Plasmids | ||
| pBAD30 | Arabinose-inducible expression vector; Ampr; p15A origin; low copy number | 12 |
| pBAD33 | Arabinose-inducible expression vector; Cmr; p15A origin; low copy number | 12 |
| pBAD18 | Arabinose-inducible expression vector; Ampr; pMB1 origin; moderate copy number | 12 |
| pBAD30-Z | pBAD30::murAEc; coding sequence and RBSa of murA from E. coli (W3110) | 4 |
| pAJM5 | pBAD18::murAEc; coding sequence and RBS of murA from E. coli (W3110) | This work |
| pAJM6 | pBAD18::murACt; coding sequence and RBS of murA from C. trachomatis serovar L2 | This work |
RBS, ribosomal binding site.
pAJM5 and pAJM6 contain the −10 region and the coding sequence of the murA gene from E. coli K-12 and C. trachomatis serovar L2, respectively, under the control of the ara promoter of the pBAD vectors. pBAD was chosen for its arabinose-inducible, glucose-repressible regulation of gene expression. The E. coli murA gene was released from pBAD30-Z (4) as a 1.4-kb XbaI-KpnI fragment and ligated into XbaI-KpnI-digested pBAD33. This plasmid was then digested with SacI-SphI, and the liberated murA fragment was ligated into SacI-SphI-digested pBAD18 to create pAJM5. The chlamydial murA was PCR amplified, using Taq polymerase (Qiagen, Valencia, Calif.), from C. trachomatis serovar L2 genomic DNA. The upstream primer (5′-CGGAATTCGAAGGAACTGAAATGCCTG-3′) contained an EcoRI restriction site (underlined) and the −10 region and the start codon (bold), whereas the downstream primer (5′-GCGAATTCTTAAACATACACAGATAC-3′) contained an EcoRI restriction site (underlined) and the stop codon (bold). Both primers were designed on the basis of the genome sequence of C. trachomatis serovar D (http://chlamydia-www.berkeley.edu:4231/). The PCR fragment was gel purified and cloned into the EcoRI site of pBAD30. This plasmid was then digested with SacI-SphI, and the liberated murA fragment was ligated into the SacI-SphI-digested pBAD18 to create pAJM6. Sequencing of the cloned C. trachomatis serovar L2 murA gene was performed by the Biomedical Instrumentation Center at the Uniformed Services University of the Health Sciences.
Deletion of the chromosomal copy of murA.
The strategy used to delete the essential murA gene from the chromosome of E. coli has been described previously (4). BL21-CodonPlus-(DE3)-RIL (CodonPlus) was first transformed with pAJM5 or pAJM6 and selected on medium containing ampicillin and chloramphenicol to generate ATM580 or ATM586, respectively. Next, a generalized transducing lysate of P1L4 grown on ZK1746 was used to transduce these strains to kanamycin resistance. Transductants were selected on LB agar containing ampicillin, chloramphenicol, kanamycin, and arabinose. Potential ΔmurA::kan mutants were screened for arabinose-dependent growth by inoculating glucose-containing medium and observing growth after overnight (O/N) incubation at 37°C. Arabinose-dependent mutants were then screened for ΔmurA and the presence of the kan gene by PCR. Primers A (5′-CACTACTGGCGGAAGAACCGG-3′) and B (5′-ACAGAACGCAGTTGATGCGTAG-3′) amplify a 750-bp fragment from an internal site of the E. coli murA gene to an internal site in the 5′ upstream flanking gene (yrbA). Primers K (5′-CACCCCTTGTATTACTGTTTATGT-3′) and B amplify a 550-bp fragment to the yrbA gene from a site internal to the kan gene. Because the yrbA gene is not present in pAJM5 or pAJM6, any PCR product obtained from primers A and B results from amplification of the genomic murA.
Growth and viability curves.
Strains were grown O/N at 37°C with aeration. O/N cultures were diluted in 12.5 ml of LB medium containing appropriate antibiotics and arabinose (or glucose) to give an initial optical density at 600 nm (OD600) reading of 0.001. Cultures were grown at 37°C with aeration, and the OD600 of the samples was measured at various times to obtain the growth curve. For the viability curve, the same procedure was followed except that dilutions of the sample were plated on LB agar containing arabinose such that 30 to 300 CFU were obtained per plate.
Phase-contrast microscopy.
Cells were grown to mid-exponential (OD600 = 0.6) and stationary phase (O/N cultures) at 37°C and viewed with an Olympus BX60 system microscope. All images were obtained with a SPOT RT CCC digital camera (Diagnostic Instruments, Inc.).
Assay of MurA activity.
O/N cultures (3 ml) of ATM581 and ATM587 were harvested and washed three times with ice-cold 50 mM Tris, pH 7.5. Cells were resuspended in 1 ml of 50 mM Tris [pH 7.5]-2 mM dithiothreitol (DTT) and sonicated three times using a sonication probe for 15-s pulses. Cell debris was removed from the samples by centrifugation for 5 min, and the sample supernatant was desalted using a Pharmacia NAP-10 column equilibrated with 50 mM Tris [pH 7.5]-2 mM DTT. The protein concentration of the lysates was determined using the Bio-Rad protein assay (Bio-Rad, Hercules, Calif.).
The assay mixture (final volume, 50 μl) contained 50 mM Tris at various pH levels, 2 mM DTT, 10 mM UDP-GlcNAc, and 10 μg of bacterial extract protein. Samples were preincubated for 15 min at 37°C, and the reaction was started by the addition of 5 μl of 10 mM PEP. After 1 or 4 h of incubation for ATM581 and ATM587 lysates, respectively, 800 μl of color reagent (1% ammonium molybdate, 1 N HCl, 0.15% malachite green) was added to stop the reaction and measure the release of orthophosphate (Pi). Results are expressed as OD660 values corrected for the background reading in the absence of UDP-GlcNAc. To determine MurA activity in the presence of fosfomycin, the preincubation samples contained fosfomycin at concentrations ranging from 10 to 500 μg/ml (7).
MIC and MBC of fosfomycin.
MIC assays for C. trachomatis serovar L2 and Shigella flexneri 2a were performed with various concentrations of fosfomycin in a plaque assay (1, 24). Confluent monolayers of L2 mouse fibroblast cells were infected for 90 min at 37°C in 5% CO2 with 6 × 108 PFU of C. trachomatis serovar L2 or 6 × 106 CFU of S. flexneri 2a. After 90 min, the infection medium was removed. C. trachomatis- and S. flexneri-infected cells were incubated for 14 and 3 days, respectively, at 37°C in 5% CO2 after the addition of a 5-ml agarose overlay (0.75% low-melting-point agarose) containing Dulbecco's modified Eagle's medium, fetal bovine serum (10%), gentamicin (20 μg/ml), cycloheximide (20 μg/ml), and various concentrations of fosfomycin. For C. trachomatis-infected cells, a second 5-ml agarose overlay was added at day 7. To visualize plaques at the end of the incubation period, cells were stained with 0.5% neutral red for 2 h at 37°C in 5% CO2.
To determine the minimal bactericidal concentration (MBC) of fosfomycin for CodonPlus, ATM581, and ATM587, strains were grown O/N at 37°C with aeration and diluted 1:100 into fresh medium containing arabinose. Cultures were grown to mid-exponential phase and adjusted to an OD600 of 0.5 to normalize all cultures to the same initial OD. Dilutions (100 μl) of each culture were plated on LB agar containing arabinose and various concentrations of fosfomycin.
Reverse transcription-PCR (RT-PCR).
L2 mouse fibroblast cells were infected with C. trachomatis serovar L2 at a multiplicity of infection of 1 for 2 h at 37°C in 5% CO2. After the infection period, the infection medium was replaced with medium containing Dulbecco's modified Eagle's medium, fetal bovine serum (10%), and gentamicin (20 μg/ml). At various times postinfection, the cells were removed and total RNA from each sample was extracted using Trizol reagent (Invitrogen, Carlsbad, Calif.) according to the manufacturer's specifications. Total RNA was reverse transcribed using the Thermoscript reverse transcription system (Invitrogen), and the resulting cDNA was screened for murA and murB transcripts by PCR using Taq polymerase. Primers OAM2 (5′-CTTGTAGCCTCCTTACTTTCGGA) and OAM3 (5′-GTGATGTGTGCTCCAACAAGA) were expected to generate a 400-bp fragment from C. trachomatis murA mRNA. Primers OAM10 (5′-CGGGATTAGAATTTGCCGTA) and OAM11 (5′-ATATCTGCCCGCCTCCTATC) were expected to generate a 400-bp fragment from C. trachomatis murB mRNA.
Nucleotide sequence accession number.
The sequence of the C. trachomatis serovar L2 murA has been deposited in GenBank under accession number AY152390.
RESULTS
Cloning of murA from C. trachomatis.
To determine whether the chlamydial murA encoded a functional enzyme, we cloned the gene from C. trachomatis serovar L2. The −10 region and the complete coding sequence of murA from C. trachomatis serovar L2 were PCR amplified using primers designed from the published genome sequence of C. trachomatis serovar D (34). The resulting PCR product was cloned into pBAD18 to place the gene under the arabinose-inducible, glucose-repressible ara promoter. The resulting clone, pAJM6, was sequenced a minimum of four times on each strand to confirm that the insert contained the C. trachomatis murA. Amino acid alignment of the MurA of C. trachomatis serovar D and C. pneumoniae against our C. trachomatis serovar L2 sequence revealed 99 and 66% similarity, respectively. The sequence of E. coli MurA was 32% similar to that of MurA of C. trachomatis serovar L2.
Complementation of E. coli ΔmurA by murACt.
Previous studies in E. coli revealed that a single copy of murA exists in the genome. By cloning the wild-type E. coli murA under the control of the arabinose-inducible, glucose-repressible ara promoter, loss of murA from the E. coli chromosome was shown to be lethal, as the ΔmurA mutants are viable only in the presence of arabinose (i.e., when the cloned murA is expressed in trans) (4). To determine whether the chlamydial murA gene was able to complement a lethal murA deletion in E. coli, CodonPlus was transformed with pAJM6 (pBAD18::murACt [the murA gene of C. trachomatis]), and then a ΔmurA::kan mutation was introduced by transduction to kanamycin resistance. Transductants arose at a frequency of 7.1 × 10−7. One transductant was chosen as ATM587 (ctMurA). As a control, E. coli CodonPlus was transformed with pAJM5 (pBAD18::murAEc [the murA gene of E. coli]) and transduced to kanamycin resistance as described above. Transductants arose at a frequency of 4.3 × 10−6. One transductant was chosen as ATM581 (ecMurA). After 18 h of growth on LB arabinose medium at 37°C, a marked difference in colony size was observed between ATM581 and ATM587, with the strain expressing murACt being smaller. To test growth dependence on the expression of the cloned murA, ecMurA and ctMurA were inoculated into LB medium containing 0.2% glucose or 0.1% arabinose. Growth occurred for both strains only when murA expression was induced with arabinose, although there was some leaky expression of the E. coli murA.
We confirmed deletion of murA in ATM581 and ATM587 by PCR. To distinguish chromosomal murA from plasmid murA, we chose a primer that anneals upstream of murA to the yrbA gene and a primer that anneals internally to murA. Plasmid-encoded murA does not contain sequences from yrbA and therefore does not produce false-positive PCR data. Only the E. coli CodonPlus control generated a fragment by PCR using yrbA and murA primers. In contrast, PCR using the primer that anneals to yrbA and a primer that anneals to kan, ATM581, and ATM587 but not the E. coli CodonPlus control generated fragments. Together, these data confirmed the chromosomal deletion of murA and insertion of kan (data not shown).
In the presence of arabinose, the OD600 of O/N cultures of ctMurA was significantly lower than that of ecMurA (data not shown). This observation, as well as the fact that ctMurA grew slower than ecMurA on agar supplemented with arabinose, suggested that while chlamydial murA complemented the ΔmurA of E. coli, it affected the kinetics of growth differently than the cloned E. coli murA gene.
Growth and viability curves.
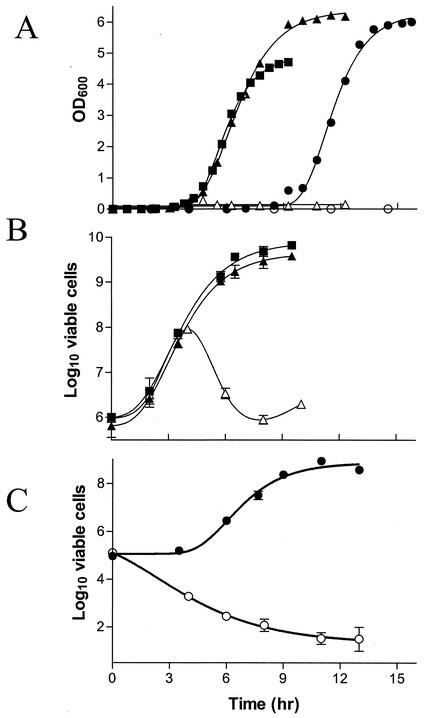
The growth rate of ecMurA and ctMurA was measured by OD and compared to that of the parent strain (murA+). Figure 2A depicts the time course of growth for each strain in the presence of arabinose or glucose. As expected, under glucose-repressing conditions neither ΔmurA mutant was able to grow. When the cloned murA strain was induced by the addition of arabinose to the culture, the growth rate of ecMurA paralleled the growth rate of wild-type E. coli CodonPlus over time. ctMurA also exhibited a similar growth rate, but a much longer lag phase occurred before the culture entered the logarithmic stage. This lag phase might be the result of the chlamydial MurA being less active than the E. coli MurA, a poor ribosomal binding site recognition in the chlamydial coding sequence, poor expression due to codon usage differences in E. coli, or fewer viable cells in the O/N culture of the E. coli strain expressing the chlamydial MurA. Both complemented mutants reached similar plateaus when grown with arabinose, demonstrating higher OD600 values than that of the wild-type parent. This result suggested that either more cells were present or the cell morphology had been altered.
FIG. 2.
Growth and viability of ecMurA and ctMurA under arabinose-inducing and glucose-repressing conditions. E. coli CodonPlus (squares), ecMurA (triangles), and ctMurA (circles) were inoculated into medium containing arabinose (filled symbols) or glucose (open symbols). At various times, the OD600 was measured to monitor the growth of the cultures (A) or dilutions were plated on LB medium containing arabinose to assess the viability of the cultures (B and C). Viability platings were performed in triplicate.
To determine the viable numbers of ctMurA and ecMurA under inducing and repressing conditions, bacteria were grown in LB medium containing arabinose or glucose and plated on agar containing arabinose. In the presence of arabinose or glucose, the number of viable cells of ecMurA was the same as that for the wild-type parent during the first 4 h of growth (Fig. 2B). After 4 h, however, ecMurA grown in glucose-containing medium quickly lost viability. The initial increase in bacterial numbers of ecMurA under glucose-repressing conditions was probably due to residual activity of MurA remaining in the cells from O/N growth with arabinose. Viability decreased as turnover of MurA occurred because no new MurA was being made under the repressing conditions. The number of viable ecMurA cells in the presence of arabinose paralleled the number of viable E. coli CodonPlus cells throughout the course of the experiment (Fig. 2B). Consistent with the OD600 readings in Fig. 2A, the viability of ctMurA in the presence of arabinose was similar to that of E. coli CodonPlus and ecMurA except for an initial lag period (Fig. 2C). However, the initial and final viable counts of ctMurA were each 1 order of magnitude lower than those of E. coli CodonPlus and ecMurA even though the OD600 values for each culture were identical at time zero. In contrast to what was observed for ecMurA in the presence of glucose, no initial growth of ctMurA occurred and viability of the culture gradually diminished to approximately 100 to 1,000 CFU/ml after 10 h.
Cellular morphology.
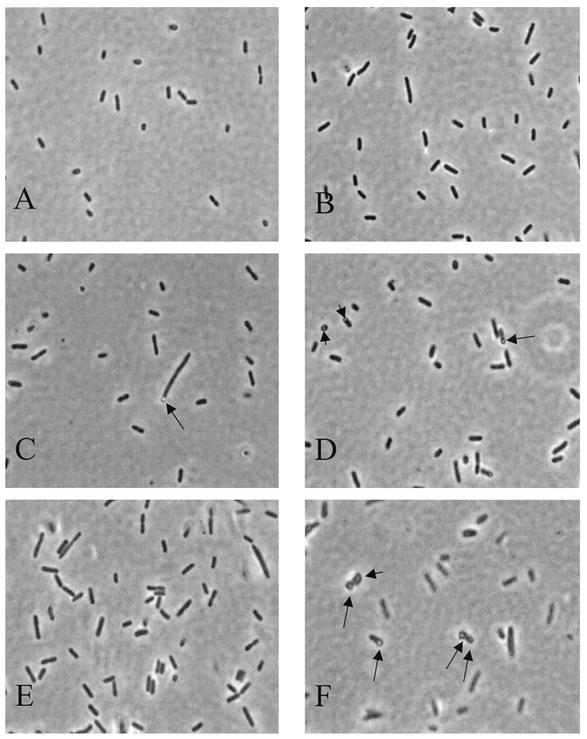
Cell morphology was examined using phase-contrast microscopy. Compared to the uniformly small rod shape of E. coli CodonPlus from O/N cultures (Fig. 3A), the morphology of both complemented mutants appeared longer and more filamentous (Fig. 3B to D). One striking characteristic of approximately 25% of the ctMurA cells was the presence of bulbs polarized at one end of the bacterium (Fig. 3C to D). Similar bulbs have been observed in cell wall mutants of E. coli (6, 17, 30, 33). During logarithmic growth, these bulbs were observed as transparent holes in the center of both daughter cells as they were dividing (Fig. 3F). Similar bulbs were not seen in logarithmic cultures of ecMurA (Fig. 3E).
FIG. 3.
Morphology of ecMurA and ctMurA cells. O/N cultures of E. coli CodonPlus (A), ecMurA (B), and ctMurA (C and D) and logarithmic phase cultures of ecMurA (E) and ctMurA (F) were visualized by phase-contrast microscopy. Black arrows point to bulbs, which were present in approximately 25% of the ctMurA cells.
In vitro activity of Chlamydia MurA.
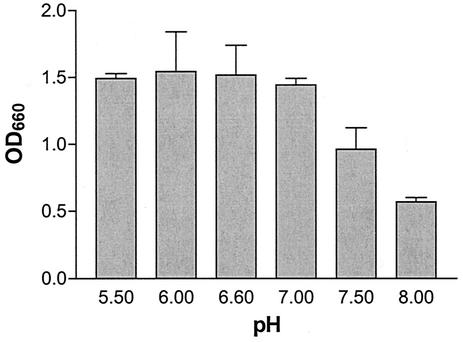
The addition of enolpyruvate to UDP-GlcNAc by MurA releases Pi. This phosphate release can be measured optically and used to indirectly assess the activity of MurA. DNA sequence analysis predicted that the C. trachomatis serovar L2 MurA contains a cysteine-to-aspartate (C → D) change at amino acid 119 in the active site of the enzyme. Kim et al. previously showed that an E. coli MurA mutant containing the same C → D substitution in the active site alters the pH optimum for the enzyme (18). The activity of the wild-type E. coli MurA remains relatively constant over a pH range of 5 to 9. In contrast, the mutant MurA has greater activity than the wild-type MurA at pH values less than 7 and dramatically loses activity at pH values greater than 7 (18). To test whether the activity of the Chlamydia MurA was also pH sensitive, the release of Pi from a crude lysate of ctMurA was measured over a pH range of 5.5 to 8.0. Because measurement of Pi release required saturation kinetics, we determined that saturation was reached by 1 h for ecMurA, whereas it took 4 h to reach saturation for ctMurA (data not shown). Figure 4 represents the chlamydial MurA activity at different pH values. As predicted from the E. coli mutant data, Pi release was maximal at pH values lower than 7.0. Further in vitro characterization of the chlamydial MurA activity was performed at pH 6.6.
FIG. 4.
pH dependence of C. trachomatis serovar L2 MurA activity in vitro. Whole-cell lysates of ctMurA were preincubated with 10 mM UDP-GlcNAc, 2 mM DTT, and 50 mM Tris at the pH indicated. 10 mM PEP was added to each sample to start the reaction. After 4 h of incubation, release of inorganic phosphate was measured by adding color reagent and measuring the OD660 of the sample (described in Materials and Methods). Inorganic phosphate release was measured in triplicate.
Previous studies have also demonstrated that the C → D conversion in the MurA active site renders the enzyme resistant to fosfomycin (7, 18). To determine whether the same phenotype was true for the chlamydial MurA, crude lysates of ecMurA and ctMurA were tested for their ability to release Pi in the presence of various concentrations of fosfomycin. As expected, wild-type E. coli MurA activity was nearly abolished in the presence of concentrations of fosfomycin as low as 0.5 μg/ml (Fig. 5A) and no further reduction in activity was seen at concentrations up to 10 μg/ml. Incubation of ecMurA for 4 h in the presence of fosfomycin did not increase the amount of Pi release from that seen at 1 h of incubation (data not shown), suggesting that the E. coli enzyme is completely inactivated at these concentrations of fosfomycin. Unlike ecMurA, ctMurA was strikingly resistant to fosfomycin, and MurA activity was detected in the presence of concentrations of fosfomycin up to 500 μg/ml (Fig. 5B). Taken together, these data suggest that the chlamydial MurA is a functional enzyme that can catalyze the addition of enolpyruvate to UDP-GlcNAc and is resistant to fosfomycin.
FIG. 5.
In vitro MurA activity in the presence of fosfomycin. Whole-cell lysates of ecMurA (A) and ctMurA (B) were preincubated with a mixture containing 10 mM UDP-GlcNAc, 2 mM DTT, the indicated fosfomycin concentration, and 50 mM Tris at pH 7.5 (A) or pH 6.6 (B). 10 mM PEP was added to start the reaction. After 1 h (A) or 4 h (B) of incubation, release of inorganic phosphate was measured by adding color reagent and determining the OD660 of the sample. Inorganic phosphate release was measured in triplicate.
In vivo resistance of C. trachomatis MurA to fosfomycin.
Chlamydiae are obligate intracellular eubacteria. Because chlamydiae require host eukaryotic cells for growth, MIC assays cannot be performed in broth or on L agar. To determine whether resistance of ctMurA to fosfomycin in vitro correlated to resistance in vivo, we examined growth of Chlamydia in a plaque assay (1). In this assay, Chlamydia is allowed to infect confluent monolayers of mouse fibroblast cells. After a 120-min infection period, the infection medium is replaced by an agarose overlay. This overlay contains nutrients to support the viability of the eukaryotic cells. When newly replicated EBs are released from infected cells, the agarose overlay keeps the released EBs localized such that they can reinfect only neighboring cells. As infection, release, and reinfection of the eukaryotic cells by Chlamydia proceeds, a plaque is formed in the monolayer of eukaryotic cells that can be observed after the cells are stained with neutral red. Each plaque arises from initial infection of a single eukaryotic cell. Effectiveness of any inhibitor of chlamydial growth can be assessed by addition of the compound to the agarose overlay and comparison of the number of plaques that form in the presence and absence of the compound.
In our laboratory, we also use the plaque assay to determine the effects of compounds on the intracellular growth of S. flexneri, a facultative intracellular bacterial pathogen closely related to E. coli. Like Chlamydia spp., S. flexneri infects mouse fibroblast cells and spreads to neighboring cells, forming plaques in the monolayer (24). However, Shigella is sensitive to fosfomycin when grown on L agar. Therefore, as a control for drug penetration into fibroblast cells, plaque formation by S. flexneri 2a was determined in the presence and absence of fosfomycin. At a fosfomycin concentration of 1 μg/ml, plaque formation by S. flexneri 2a was completely abolished (data not shown), suggesting that the drug can be taken up efficiently by eukaryotic cells and can inhibit plaque formation by fosfomycin-sensitive organisms.
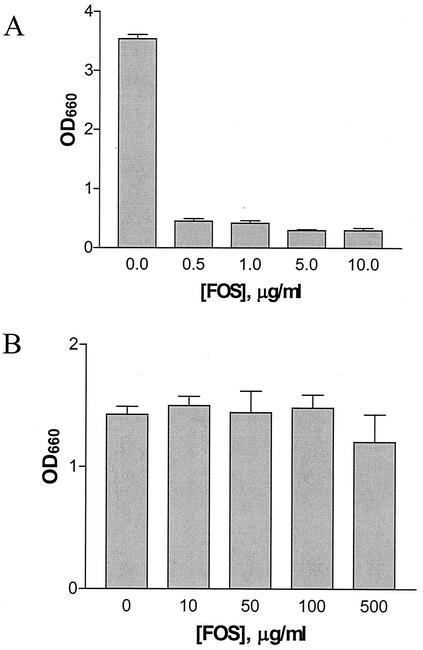
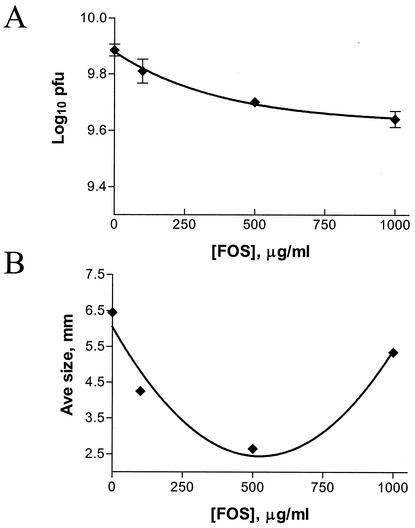
Next, we tested the effect of fosfomycin on C. trachomatis serovar L2 in the plaque assay. As seen in Fig. 6A, concentrations of fosfomycin up to 1,000 μg/ml did not significantly inhibit plaque formation of C. trachomatis in the plaque assay. Although no dose-dependent inhibition of the ability of C. trachomatis serovar L2 to form plaques was observed, a dose-dependent decrease in the average plaque size was observed at concentrations up to 500 μg/ml (Fig. 6B). At higher concentrations of fosfomycin, the average plaque size returned to sizes similar to that seen in the absence of fosfomycin. It is not understood why the plaque sizes increased at higher concentrations of the antibiotic. It is unlikely that toxic effects of fosfomycin on the fibroblast cells account for the increase in plaque size, as uninfected fosfomycin-treated controls displayed normal morphology in this assay. In this same assay, C. trachomatis serovar D and C. psittaci strains Cal10 and GPIC were also resistant to fosfomycin and formed plaques at concentrations of fosfomycin up to 500 μg/ml (data not shown).
FIG. 6.
MIC of fosfomycin for C. trachomatis serovar L2. Confluent mouse fibroblast monolayers were infected with C. trachomatis serovar L2 as described in Materials and Methods. At 2 h postinfection, a 0.5% agarose overlay containing the indicated concentration of fosfomycin was added. (A) The log10 of the number of PFU per milliliter was calculated after 14 days. (B) Plaque size was measured for 10 individual plaques, and the average was plotted against the fosfomycin concentration.
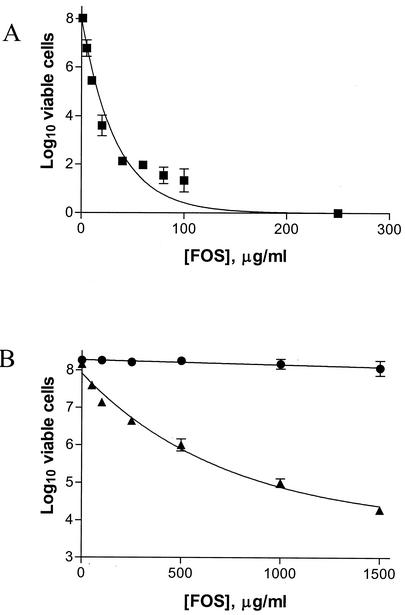
Because the in vitro and in vivo data suggested that the chlamydial MurA was responsible for the resistance of Chlamydia to fosfomycin, we wanted to verify this resistance by asking whether the chlamydial MurA was able to impart resistance to the sensitive E. coli strain. Therefore, the MBC of fosfomycin was determined for E. coli CodonPlus, ecMurA, and ctMurA. Figure 7A shows the MBC of fosfomycin for E. coli CodonPlus. A dramatic decrease in viability was observed at a concentration as low as 5 μg/ml, and growth of this strain was completely inhibited at a fosfomycin concentration of 200 μg/ml. When the Chlamydia murA gene was expressed in trans (ctMurA), the strain displayed complete resistance to fosfomycin at concentrations up to 1,500 μg/ml (Fig. 7B). This resistance was due to enzyme activity of Chlamydia MurA and not to high-level expression of murACt from a moderate-copy-number vector. Some resistance to fosfomycin was also observed for ecMurA (murAEc cloned on the same moderate-copy-number plasmid as murACt), but this strain exhibited increased sensitivity as the fosfomycin concentration increased (Fig. 7B). The difference in levels of bacterial growth between ecMurA and ctMurA was 4 orders of magnitude at a fosfomycin concentration of 1,500 μg/ml. These data suggest that C. trachomatis serovar L2 resistance to the cell wall inhibitor fosfomycin is due to synthesis of a resistant form of MurA.
FIG. 7.
MBC of fosfomycin for E. coli CodonPlus, ecMurA, and ctMurA. Logarithmic phase cultures were diluted in LB medium and plated on LB agar containing the indicated fosfomycin concentrations. The log10 of the number of viable cells per milliliter was calculated for E. coli CodonPlus (squares) (A) and ecMurA (triangles) and ctMurA (circles) (B) and plotted against fosfomycin concentration.
RT-PCR of murA from C. trachomatis serovar L2.
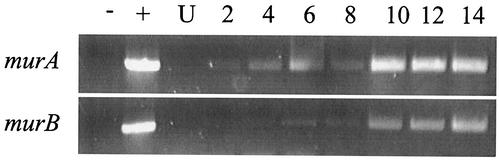
The genome sequences of C. trachomatis and C. pneumoniae contain homologues of genes in the PG biosynthesis pathway, including murA. While we were able to demonstrate in vitro activity of Chlamydia MurA and in vivo activity in E. coli, we wanted to determine whether murA mRNA was expressed in Chlamydia bacteria at any time during the chlamydial developmental cycle. L2 mouse fibroblast cells were infected with C. trachomatis serovar L2, and total RNA was extracted from Chlamydia-infected cells at various times postinfection. Figure 8 depicts the time course of expression for C. trachomatis murA mRNA. murA mRNA transcripts were detected as early as 2 hpi and expressed throughout the time course of the chlamydial infection. This expression profile was similar to that of C. trachomatis hsp60 (reference 31 and data not shown). We also examined the expression of C. trachomatis murB mRNA. murB encodes a UDP-GlcNAc enolpyruvyl reductase, which reduces UDP-GlcNAc enolpyruvate to UDP-MurNAc (Fig. 1). MurNAc is the hallmark constituent of PG. We were able to detect murB mRNA transcripts; however, expression lagged behind the expression of murA mRNA (Fig. 8).
FIG. 8.
Detection of C. trachomatis serovar L2 gene transcripts by RT-PCR during infection. RT-PCR analysis was performed for murA and murB mRNA isolated from infected cells at the indicated time points after infection. −, RNA-DNA negative control; +, genomic DNA positive control; U, uninfected cells.
DISCUSSION
Despite the lack of evidence for the presence of PG in Chlamydia, a nearly complete E. coli PG synthesis pathway is encoded in the genomes of each Chlamydia species (Fig. 1). Until now, functional assessment of any enzyme in the PG biosynthesis pathway in Chlamydia had not been performed. Here, we have demonstrated that the chlamydial MurA functions as an UDP-GlcNAc enolpyruvyl transferase and commits UDP-GlcNAc to PG synthesis. Using the strategy of Brown et al. (4), we demonstrated the activity of the chlamydial MurA by assessing its ability to restore viability to an E. coli ΔmurA mutant. Growth and viability of the E. coli mutant was dependent on the arabinose-induced expression of the cloned chlamydial murA (Fig. 2A and C).
The altered morphology observed in ctMurA cells is quite interesting. Approximately 25% of the cells formed bulbs. These bulbs were polarized when the bacterial cultures were in stationary phase yet were found in the centers of both daughter cells as they were dividing. It is unclear what caused these bulbs to form or why they occurred in only 25% of the population. A similar phenotype has been reported in other cell wall mutants of E. coli, including htrB, pbpA, and pbpB mutants (6, 17, 30, 33). HtrB is a lauroyl transferase, a late-functioning acyltransferase of lipid A biosynthesis, and is required for growth above 32°C. The presence of bulbs in htrB mutants depends on the initial OD of the culture when the culture is shifted to higher temperatures. At OD600 values below 0.05, the cells form bulbs and bulges, but when the cells are shifted to 42°C at higher OD600 values, the phenotype shifts from bulbs and bulges in the cells to a filamentous phenotype (17). We have not explored whether the 25% of ctMurA cells that form bulbs shift to a filamentous phenotype when the OD of the cultures is higher than 0.05. Regardless of the initial OD of the htrB mutant cultures, the cells eventually ceased dividing and lost viability (17). It is possible that this 25% of ctMurA cells also cease dividing, and this might explain the order-of-magnitude difference in total viable cells between ctMurA and ecMurA cultures.
In E. coli, PBP2 and PBP3 are classified as high-molecular-mass penicillin-binding proteins and are essential to E. coli development (10). PBP2 and PBP3, which are encoded by pbpA (mrdA) and pbpB (ftsI), respectively, are transpeptidases involved in establishing the cell shape and in cell division. PBP2 maintains the cell's rod shape. This has been demonstrated by the formation of round cells in pbpA(Ts) mutants and in E. coli treated with mecillinam, a β-lactam antibiotic that specifically targets PBP2 (3, 33). PBP3 is involved in the formation of septal PG and is essential for cell division. In pbpB(Ts) mutants and in E. coli cultures treated with cephalexin, a β-lactam antibiotic that specifically targets PBP3, cell division ceases and filamentation occurs (3, 33). Bulbs and bulges have not been identified in Chlamydia treated with β-lactam antibiotics, although Chlamydia genomes encode homologues of PBP2 and PBP3. However, abnormalities in morphology and cell division are observed. It should be noted that chlamydial RBs are not rod shaped but, rather, are round. Recently, it was shown that mecillinam specifically binds to the chlamydial PBP2 homologue in vitro and inhibits chlamydial development in vivo, although the effect of mecillinam on cell morphology was not studied (35). It is not known whether the chlamydial PBPs have transpeptidase activity, as these enzymes have to date been characterized only by their ability to bind radiolabeled β-lactams (2, 35). It is possible that the roles of PBP2 and PBP3 in the formation of chlamydial PG differ from their roles in E. coli.
We found that ctMurA displayed altered growth kinetics compared to the wild-type parent (murA+) and ecMurA. A 5- to 6-h lag occurred before ctMurA cultures entered the exponential phase of growth (Fig. 2A). One possible explanation for this lag is the viability difference observed in ctMurA cultures (Fig. 2B and C). Although the initial OD600 values of the ctMurA, ecMurA, and wild-type E. coli CodonPlus cultures were identical, the viability of ctMurA was 1 order of magnitude lower than that of ecMurA and wild-type E. coli CodonPlus. Nevertheless, this explanation cannot account for the lag in time before the cells began to divide (Fig. 2C). A difference in the activity of the chlamydial MurA is more likely to be responsible for the altered growth and viability kinetics. The optimal enzyme activity of the chlamydial MurA is at pH values lower than 7, and a significant decrease in activity is seen at pH 7.5. In E. coli, the cytoplasmic pH is maintained at 7.5 over an extracellular pH range of 5 to 9 (25, 32).
The correlation between the pH optimum of chlamydial MurA activity and the pH of the chlamydial inclusion is also interesting. Schramm et al. found that at 2 hpi, the pH of inclusions formed by C. trachomatis serovar L2 is neutral and drops to a pH of 6.26 at 4 hpi before stabilizing at pH 6.6 at 12 hpi (29). Because of the method used to measure the pH, they were unable to measure the pH after 12 h. Recently, Grieshaber et al. were able to measure the pH of the chlamydial inclusion at 20 hpi and reported a pH of 7.25 (11). We suggest that the C → D change in the active site of the chlamydial MurA is the result of evolutionary pressure to select a variant enzyme with optimal activity within the unique environment of the chlamydial inclusion. Our data suggest that the chlamydial MurA is required during early stages of the chlamydial life cycle, when EBs convert to RBs and RBs begin to expand in size and divide. At these times, the pH of the inclusion is such that MurA is optimally active. Later in the developmental cycle, the environment of the inclusion potentially changes, which may render the chlamydial MurA less active, to accommodate other events. Perhaps Chlamydia is able to recycle PG and therefore needs less MurA during late stages of the developmental cycle.
Fosfomycin is a broad-spectrum antibiotic that irreversibly binds to the active site of MurA (16, 37). Fosfomycin forms a covalent adduct with a cysteine residue in the active site of MurA, thus rendering the enzyme inactive (16, 20). In E. coli, replacement of the cysteine in the active site of MurA with aspartate renders the bacteria resistant to fosfomycin (18). Mycobacterium tuberculosis is innately resistant to fosfomycin, and M. tuberculosis MurA is resistant to fosfomycin in vitro. Sequence analysis of the M. tuberculosis murA revealed a cysteine-to-aspartate change in the MurA active site. Engineering an aspartate-to-cysteine change in the active site of M. tuberculosis MurA renders the enzyme sensitive to fosfomycin in vitro (7). Our sequence analysis of C. trachomatis serovar L2 MurA predicted a similar C → D change in the active site. C. trachomatis serovar D and C. pneumoniae MurA sequences also contain an aspartate residue instead of cysteine in the active site of the enzyme (28, 34). In this work, we have demonstrated that, similar to M. tuberculosis, Chlamydia spp. are innately resistant to fosfomycin. Although we have shown that fosfomycin can enter the eukaryotic cell in a plaque assay and effectively kill the intracellular pathogen Shigella, we cannot rule out the possibility that Chlamydia is innately resistant to fosfomycin due to the inability of the drug to enter the inclusion. However, it has been determined that the pore size of the chlamydial inclusion is between 45 and 520 Da (11, 15). Fosfomycin, which is approximately 185 Da in size, is small enough to pass through this pore. Therefore, we do not believe that the innate resistance to fosfomycin is due to an inability of the drug to reach Chlamydia. Rather, fosfomycin resistance in Chlamydia can be attributed to the activity of the chlamydial MurA. Correlating with our in vivo resistance data, the in vitro chlamydial MurA activity was uninhibited at high concentrations of fosfomycin. Resistance to high concentrations of fosfomycin was also conferred to an E. coli ΔmurA mutant expressing the chlamydial murA. Although overexpression of the fosfomycin-sensitive E. coli MurA in ecMurA increased resistance to fosfomycin (Fig. 7B), this strain was still sensitive to high concentrations of fosfomycin. However, ctMurA was resistant to high concentrations of fosfomycin, suggesting that the activity of the chlamydial MurA accounted for the resistance of ctMurA. Fosfomycin is generally not used to treat chlamydial infections. Therefore, it is reasonable to conclude that the C → D conversion in the active site of the chlamydial MurA is not the result of antibiotic pressure to select for a fosfomycin-resistant variant of Chlamydia. Rather, evolutionary selective pressure for an enzyme better adapted to the environment of the chlamydial inclusion is the likely explanation for this alteration in the active-site residue.
Within 2 h after infection by C. trachomatis, EBs begin to convert to RBs. A size and volume increase in the RBs becomes evident at 8 to 12 hpi. This increase continues over several hours. Around 12 hpi, RBs begin dividing by binary fission. After 18 hpi, increasing numbers of RBs begin differentiating back to EBs while others continue to divide. At approximately 36 to 72 hpi, the cells lyse and EBs are released to initiate another round of infection (13). Our RT-PCR data suggest that PG is involved in size expansion of RBs and chlamydial cell division. Chlamydia murA mRNA transcripts are expressed shortly after the eukaryotic cells are infected with EBs, while there is a 6-h lag before murB mRNA transcripts are expressed. Shaw et al. reported that murG (Fig. 1) was not expressed at 6 hpi but was expressed at 12 hpi (31). We propose that the expression of mRNA transcripts from each PG pathway gene differs in relation to its place in the pathway and the developmental cycle. Chlamydia murA mRNA is expressed as soon as EBs begin converting to RBs, possibly generating a pool of UDP-GlcNAc enolpyruvate. Approximately 6 h later, murB mRNA and potentially other PG genes are expressed, allowing UDP-MurNAc and all or part of the attached pentapeptide to be generated. Between 6 and 12 hpi, murG and possibly other PG genes are expressed and the disaccharide-pentapeptide subunit is synthesized and flipped to the periplasm. In the periplasm, penicillin-binding proteins cross-link the subunits, giving rise to PG layers that expand the size and volume of RBs and facilitate cell division. Further support for this model includes the expression of pbp2 mRNA transcripts at 6 to 8 hpi (data not shown) and the effectiveness of cell wall antibiotics, which interfere with RB development and cell division (19, 21, 35).
Chlamydial genomes are relatively small and for the most part encode pathways to synthesize compounds that the organism cannot acquire from the host. Therefore, from an evolutionary point of view, it is highly unlikely that Chlamydia would retain a nearly complete pathway for PG synthesis if these genes did not encode functional products. Because nearly all the PG synthesis homologues are present in Chlamydia, it is unlikely that the PG synthesis enzymes are involved in other cellular processes. Chlamydia lacks both the alanine and glutamine racemases (Alr and MurI, respectively) required for a complete PG synthesis pathway. It has been speculated that Chlamydia can acquire these d-amino acids from the host. However, there does not appear to be conclusive evidence to support this idea. More likely, Chlamydia bacteria encode a racemase(s) that is not similar to racemases in other bacteria. The presence of d-alanine in chlamydial PG is supported by the sensitivity of Chlamydia to d-cycloserine, which inhibits the d-alanine ligase.
Many recent publications have indirectly suggested that the question regarding the presence of PG in Chlamydia needs to be reevaluated. For example, synthesis of MurC/DdlA was identified in a proteome analysis of C. pneumoniae EBs, while MurG was detected in a combined genomic-proteomic analysis of surface-exposed proteins in C. pneumoniae EBs (22, 36). It should be noted that previous attempts to detect PG have been performed with purified EBs. Our data strongly suggest not only that PG is capable of being synthesized in Chlamydia but also that the presence of PG in Chlamydia should be reanalyzed in RBs. Biochemical evidence of PG in Chlamydia would be of significance in that it would open up a new avenue to the treatment of chlamydial infections through the application of preexisting and novel drugs that target chlamydial PG synthesis.
Acknowledgments
We thank Harlan Caldwell for the gift of C. trachomatis serovar L2, Roberto Kolter for the gift of E. coli strain ZK1746, Michael N. Flora and the USUHS Biomedical Instrumentation Center for DNA sequencing and oligonucleotide synthesis services, and Rachel Binet for thoughtful discussions. We also thank Priscilla Wyrick for helping us get started in Chlamydia research and for her continuing support.
This work was supported by grant AI44033 from the National Institute of Allergy and Infectious Diseases.
The opinions or assertions contained herein are those of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
REFERENCES
- 1.Banks, J., B. Eddie, J. Schachter, and K. F. Meyer. 1970. Plaque formation by Chlamydia in L cells. Infect. Immun. 1:259-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G., K. Amano, T. Hackstadt, L. Perry, and H. D. Caldwell. 1982. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J. Bacteriol. 151:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begg, K. J., and W. D. Donachie. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, E. D., E. I. Vivas, C. T. Walsh, and R. Kolter. 1995. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 177:4194-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopra, I., C. Storey, T. J. Falla, and J. H. Pearce. 1998. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology 144:2673-2678. [DOI] [PubMed] [Google Scholar]
- 6.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeSmet, K. A., K. E. Kempsell, A. Gallagher, K. Duncan, and D. B. Young. 1999. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology 145:3177-3184. [DOI] [PubMed] [Google Scholar]
- 8.Formal, S. B., G. J. Dammin, E. H. LaBrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, A., J. C. Rogers, J. Gilbart, S. Morgan, C. H. Davis, S. Knight, and P. B. Wyrick. 1990. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect. Immun. 58:835-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grieshaber, S., J. A. Swanson, and T. Hackstadt. 2002. Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell. Microbiol. 4:273-283. [DOI] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackstadt, T. 1999. Cell biology, p. 101-138. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, D.C.
- 14.Hatch, T. P. 1996. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 178:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzen, R. A., and T. Hackstadt. 1997. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect. Immun. 65:1088-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahan, F. M., J. S. Kahan, P. J. Cassidy, and H. Kropp. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 235:364-386. [DOI] [PubMed] [Google Scholar]
- 17.Karow, M., O. Fayet, A. Cegielska, T. Ziegelhoffer, and C. Georgopoulos. 1991. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J. Bacteriol. 173:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, D. H., W. J. Lees, K. E. Kempsell, W. S. Lane, K. Duncan, and C. T. Walsh. 1996. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolypyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35:4923-4928. [DOI] [PubMed] [Google Scholar]
- 19.Lin, H. S., and J. W. Moulder. 1966. Patterns of response to sulfadiazine, d-cycloserine and d-alanine in members of the psittacosis group. J. Infect. Dis. 116:372-376. [DOI] [PubMed] [Google Scholar]
- 20.Marquardt, J. L., E. D. Brown, W. S. Lane, T. M. Haley, Y. Ichikawa, C. H. Wong, and C. T. Walsh. 1994. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry 33:10646-10651. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto, A., and G. P. Manire. 1970. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J. Bacteriol. 101:278-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montigiani, S., F. Falugi, M. Scarselli, O. Finco, R. Petracca, G. Galli, M. Mariani, R. Manetti, M. Agnusdei, R. Cevenini, M. Donati, R. Nogarotto, N. Norais, I. Garaguso, S. Nuti, G. Saletti, D. Rosa, G. Ratti, and G. Grandi. 2002. Genomic approach for analysis of surface proteins in Chlamydia pneumoniae. Infect. Immun. 70:368-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moulder, J. W. 1993. Why is Chlamydia sensitive to penicillin in the absence of peptidoglycan? Infect. Agents Dis. 2:87-99. [PubMed] [Google Scholar]
- 24.Oaks, E. V., M. E. Wingfield, and S. B. Formal. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padan, E., D. Zilberstein, and S. Schuldiner. 1981. pH homeostasis in bacteria. Biochim. Biophys. Acta 650:151-166. [DOI] [PubMed] [Google Scholar]
- 26.Park, J. T. 1996. The murein sacculus, p. 48-57. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 27.Raulston, J. E. 1995. Chlamydial envelope components and pathogen-host cell interactions. Mol. Microbiol. 15:607-616. [DOI] [PubMed] [Google Scholar]
- 28.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schramm, N., C. R. Bagnell, and P. B. Wyrick. 1996. Vesicles containing Chlamydia trachomatis serovar L2 remain above pH 6 within HEC-1B cells. Infect. Immun. 64:1208-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwarz, U., A. Asmus, and H. Frank. 1969. Autolytic enzymes and cell division of Escherichia coli. J. Mol. Biol. 41:419-429. [DOI] [PubMed] [Google Scholar]
- 31.Shaw, A. C., K. Gevaert, H. Demol, B. Hoorelbeke, J. Vandekerckhove, M. R. Larsen, P. Roepstorff, A. Holm, G. Christiansen, and S. Birkelund. 2002. Comparative proteome analysis of Chlamydia trachomatis serovar A, D and L2. Proteomics 2:164-186. [DOI] [PubMed] [Google Scholar]
- 32.Slonczewski, J. L., R. M. Macnab, J. R. Alger, and A. M. Castle. 1982. Effects of pH and repellent tactic stimuli on protein methylation levels in Escherichia coli. J. Bacteriol. 152:384-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 35.Storey, C., and I. Chopra. 2001. Affinities of β-lactams for penicillin binding proteins of Chlamydia trachomatis and their antichlamydial activities. Antimicrob. Agents Chemother. 45:303-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandahl, B. B., S. Birkelund, H. Demol, B. Hoorelbeke, G. Christiansen, J. Vandekerckhove, and K. Gevaert. 2001. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 22:1204-1233. [DOI] [PubMed] [Google Scholar]
- 37.Wu, H. C., and P. S. Venkateswaran. 1974. Fosfomycin-resistant mutant of Escherichia coli. Ann. N. Y. Acad. Sci. 235:587-592. [DOI] [PubMed] [Google Scholar]
- 38.Wyrick, P. B. 2000. Intracellular survival by Chlamydia. Cell. Microbiol. 2:275-282. [DOI] [PubMed] [Google Scholar]