Abstract
In eukaryotes, the creation of ligatable nicks in DNA from flap structures generated by DNA polymerase δ-catalyzed displacement DNA synthesis during Okazaki fragment processing depends on the combined action of Fen1 and Dna2. These two enzymes contain partially overlapping but distinct endonuclease activities. Dna2 is well-suited to process long flaps, which are converted to nicks by the subsequent action of Fen1. In this report, we purified human Dna2 as a recombinant protein from human cells transfected with the cDNA of the human homologue of Saccharomyces cerevisiae Dna2. We demonstrated that the purified human Dna2 enzyme contains intrinsic endonuclease and DNA-dependent ATPase activities, but is devoid of detectable DNA helicase activity. We determined a number of enzymatic properties of human Dna2 including its substrate specificity. When both 5′ and 3′ tailed ssDNAs were present in a substrate, such as a forked-structured one, both single-stranded regions were cleaved by human Dna2 (hDna2) with equal efficiency. Based on this and other properties of hDna2, it is likely that this enzyme facilitates the removal of 5′ and 3′ regions in equilibrating flaps that are likely to arise during the processing of Okazaki fragments in human cells.
INTRODUCTION
Lagging strand DNA replication requires a series of complicated enzymatic steps including: (i) the synthesis of primer RNA coupled to the limited elongation of the RNA primer by DNA polymerase (pol) α-primase, (ii) a switch of the primer terminus from pol α to pol δ, (iii) elongation by pol δ and (iv) maturation of Okazaki fragments requiring the generation of ligatable nicks (1–3) and their ligation by DNA ligase. Generation of ligatable nicks is best studied in Saccharomyces cerevisiae using both genetic and biochemical approaches. Recently, it was shown that an exquisite coordination between the action of pol δ and Fen1 is required to produce a ligatable nick (4). In the absence of a functional Fen1, pol δ can contribute to generation of ligatable nicks by a process called ‘idling’ at nicks, which involves the reiterative incorporation of 2–3 nt into the double-stranded DNA, followed by degradation of the newly replicated DNA by the 3′ to 5′ exonuclease activity of pol δ to the nick position. In the presence of Fen1, however, idling is inhibited and the short flaps generated by pol δ are cleaved by Fen1. This process, identical to nick translation, occurs at least until all of the initiator RNA has been degraded. Both idling and nick translation can be terminated by DNA ligase I (5). Thus, it appears that Fen1 action, in conjunction with pol δ, can suppress the production of single-stranded DNA flaps.
Although Fen1 alone can efficiently generate ligatable nicks in vitro, it is not sufficient in vivo since Okazaki fragment maturation requires the endonuclease function of Dna2 (6–8). Based on its biochemical properties, Dna2 appears to act only when extensive strand displacement synthesis by pol δ results in the formation of flaps long enough (>20 nt) to bind replication protein A (RPA). The binding of RPA to flap structures inhibits their cleavage by Fen1 but stimulates Dna2-catalyzed cleavage activity (5,8). In addition, flaps which contain secondary structure are poor substrates for Fen1, necessitating the action of Dna2 helicase (9,10). Thus RPA-bound flaps are processed by Dna2, yielding shortened flaps (usually <6 nt), which are further processed by Fen1 into ligatable nicks. Thus, in vivo both Fen1 and Dna2 are required to generate ligatable nicks from flaps of varying sizes.
Dna2 is well conserved throughout eukaryotes, retaining the catalytic domains that are essential for both endonuclease and helicase activities. Despite their conserved motifs, the enzymatic activities associated with each protein vary. For example, S.cerevisiae Dna2 contains both endonuclease and helicase activities, while Xenopus laevis (11) and Schizosaccharomyces pombe Dna2 (H. Y. Kang and Y. S. Seo, unpublished data) possess endonuclease activity but lack helicase activity. In addition, there are notable differences in the primary structure of various eukaryotic Dna2 proteins (See Figure 1A); no significant homology is found in the N-terminal regions of the Dna2 proteins and they also vary in size. Dna2 enzymes from the two yeasts have additional 350–400 amino acids at their N-termini that are absent in the metazoan homologues. Though the N-terminal region of yeast Dna2 is dispensable for enzymatic activities, (12), yeast cells containing Dna2 devoid of this region show a temperature-dependent growth defect that is suppressed by a number of lagging strand enzymes, such as the subunits of pol δ, DNA ligase I, RPA (12–14). Most significantly, metazoan and single-cell organisms differ in their Dna2 requirement for viability. Deletion of Dna2 in Caenorhabditis elegans does not cause complete embryonic lethality since some mutants survive into the F2 generation (15), whereas it is absolutely essential in S.cerevisiae and S.pombe (6,7). This situation is reversed regarding the requirements for Fen1 (15,16); deletion of Fen1 results in embryonic lethality in mice and C.elegans. These observations suggest that the mechanism of Okazaki fragment processing in lower and higher eukaryotes may differ significantly. For example, the relative importance of Dna2 and/or Fen1 in each eukaryote may depend on the extent of displacement DNA synthesis carried out by pol δ during Okazaki fragment synthesis. As the length of the flap generated by displacement DNA synthesis increases, viability would become more dependent on Dna2. Therefore, in order to evaluate the in vivo role of Dna2 in eukaryotes, it is crucial to determine the frequency with which long flaps occur and their average size during Okazaki fragment synthesis. In order to address this issue, it is essential that an in vitro DNA replication system is reconstituted. For this purpose, we have isolated human Dna2 in order to evaluate its role in the reconstituted SV40 DNA replication system. In this report, we described the isolation and characterization of human Dna2. Based upon the enzymatic properties of human Dna2, we discuss how this influences Okazaki fragment processing.
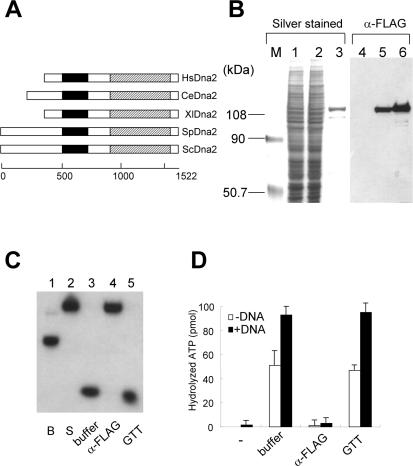
Figure 1.
Purification of hDna2 endonuclease and ATPase. (A) Alignments of ORFs of Dna2 from different species. Endonuclease domain (a closed box); helicase domain (a hatched box). Hs; Homo sapiens, Ce; C.elegans, Xl; X.laevis, Sp; S.pombe, Sc; S.cerevisiae. The scales located at the bottom of the diagram indicate amino acid positions of S.cerevisiae Dna2. (B) Crude extracts prepared from 293T cells untransfected (lanes 1 and 4) and transfected (lanes 2 and 5) with a plasmid expressing FLAG-hDna2 as well as purified FLAG-hDna2 (lanes 3 and 6) were subjected to 7.5% SDS–PAGE. The gel was silver-stained (left panel) and analyzed by immuno-blotting with anti-FLAG monoclonal antibodies (right panel). (C and D) Immuno-depletion experiments demonstrating that FLAG-hDna2 is intrinsically associated with endonuclease (C) and ATPase (D). In brief, purified FLAG-hDna2 was depleted with anti-FLAG antibody or anti-GST control antibody and the supernatant was assayed for the endonuclease and ATPase activities. The nucleolytic products were analyzed by 10% native gel, dried and autoradiographed. ATPase assays were performed with or without ΦX174 sscDNA.
MATERIALS AND METHODS
Enzymes, antibodies, DNA and nucleotides
ΦX174 single-stranded circular (ssc) DNA was purchased from New England Biolabs (Beverly, MA). The oligonucleotides used were commercially synthesized from Genotech (Daejeon, Korea) and their sequences are summarized in Table 1. Nucleoside triphosphates were obtained from Sigma (St. Louis, MO). [γ-32P]ATP (>5000 Ci/mmol) was purchased from Amersham Biosciences (Piscataway, NJ). The following proteins were obtained commercially; T4 DNA ligase and terminal transferase were from New England Biolabs (Beverly, MA), restriction endonucleases and polynucleotide kinase were from KOSCO Inc. (Korea). Antibodies were obtained commercially; mouse monoclonal M2 anti-flag antibody from Sigma (St. Louis, MO). Secondary antibodies were from Amersham Biosciences. Human RPA (hRPA), human flap endonuclease 1 (hFen1) and human DNA ligase 1 were purified as described (17–19).
Table 1.
Oligonucleotide used in this study
| Oligo | Sequence (length of oligonucleotide) |
|---|---|
| 1 | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (55) |
| 2 | 5′-TGAAAACATTATTAATGGCGTCGAGCGTCCG-3′ (31) |
| 3 | 5′-TGAAAACATTATTAATGGCGTCGAGCGTCCGTTTTTTTTTTTTTTTTTTTTTTTTT-3′ (56) |
| 4 | 5′-CGGACGCTCGACGCCATTAATAATGTTTTC-3′ (30) |
| 5 | 5′-TGGGCTCACGTGGTCGACGCTGGAGGTGATCACCAGATGATTGCTAGGCATGCTTTCCGCAAGAGAACGGGCGTCTGCGTACCCGTGCAG-3′ (90) |
| 6 | 5′-CTGCACGGGTACGCAGACGCC-3′ (21) |
| 7 | 5′-CAGCGTCGACCACGTGAGCCC-3′ (21) |
| 8 | 5′-TGAAAACATTATTAATGGCGTCGAGCGTCCGTAGGCACAAGGCGAACTGCTAACGG-3′ (56) |
| 9 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTA-3′ (25) |
| 10 | 5′-CGGACGCTCGACGCCATTAATAATGTTTTC-3′ (30) |
| 11 | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (70) |
| 12 | 5′-GAAAACATTATTAATGGCGTCGAGCGTCCGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′ (70) |
| 13 | 5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (60) |
| 14 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTATTTTT-3′ (30) |
| 15 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTATTTTTTTTTT-3′ (35) |
| 16 | 5′-CCGTTAGCAGTTCGCCTTGTGCCTATTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′ (55) |
| 17 | 5′-TTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (36) |
| 18 | 5′-TTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC-3′ (43) |
| 19 | 5′-TCTCGACGACTAGATCTTGGCACAACTCTAGTCGTTGTTGTTCCACCCGTCCACCCGACGCCACGT-3′ (66) |
| 20 | 5′-ACGTGGCGTCGGGTGGACGGGTGGAACAACAACGACTAGAGTTGTGCCAAGATCTAGTCGTCGAGACCTATTGAAATTTAGGCTGGCACGGTCGCATAGC-3′ (100) |
| 21 | 5′-GCTATGCGACCGTGCCAGCCTAAATTTCAATAGGTCTCGACGACTAGATCTTGGACAAACTCTAGT-3′ (66) |
| 22 | 5′-GCTATGCGACCGTGCCAGCCTAAATTTCAATAGGTCTCGACGACTAGATCTTGGCACAACTCTAGTGCATAGCTAC-3′ (76) |
Preparation of substrates
The oligonucleotide–based partial duplex substrates, except for the equilibrating flap substrates, were prepared as described previously (20) using the synthetic oligonucleotides listed in Table 1. The oligonucleotides used to construct each substrate in individual experiments are indicated as circled numbers that are listed in Table 1. Equilibrating flap substrates were prepared as described (21). Briefly, a downstream oligonucleotide was labeled at its 3′ end with [α-32P]dideoxyATP and terminal transferase. The 3′-labeled oligonucleotide was annealed to a template oligonucleotide and an upstream oligonucleotide (molar ratio of 1:3:10, respectively). The annealed substrates were purified by polyacrylamide gel eletrophoresis as described (20), and their specific activities (∼2500 c.p.m./fmol) were determined by scintillation counting (Beckman).
Cloning of cDNA encoding human Dna2 and construction of expression vector
The cDNA encoding the human homologue of S.cerevisiae Dna2 was amplified by RT–PCR using 5′ human Dna2 (hDna2) expressed-sequence tag (EST) primer (5′-CCGGTACCTGGTGTTGGCAGTCAATACT-3′) and 3′ hDna2 EST primer (5′-AATAGTGTTTTATTCTCTTTGAAAGTCACCCAA-3′) derived from an EST clone (GenBank™ accession no. D42046). An amplified band was excised from an agarose gel and purified using QIAquick gel extraction kit (Qiagen), and subcloned into pCR2.1 TOPO vector (Invitrogen). For cloning of full-length cDNA, the 5′ end of the cDNA was extended by a 5′ RACE-PCR. An M13F primer (5′-GGAAACAGCTATGACCATGATTACGC-3′) complementary to a flanking sequence of inserts in the library plasmid (Uni-ZAP™ XR; Stratagene) and a hDna2-RACE primer (5′-TACAGTATTGACTGCCAACACCAGGTACCGG-3′) specific to the hDna2 cDNA were used for 5′ RACE-PCR. The longest reaction products were subcloned into pCR2.1 TOPO. The sequences of several independent clones were determined with an automated DNA sequencer (ABI PRISM 310 Genetic Analyzer, Perkin-Elmer). The longest hDna2 cDNA was identical to the EST clone (GenBank™ accession no. D42046). After analysis of other EST clones, we concluded that the first start codon of hDna2 is the 52–54 bp of GenBank™ accession no. D42046 clone. The full-length cDNA of hDna2 encodes 1060 amino acids and also contains the UvrD superfamily helicase domain and the RecB ribonuclease domain. Amino acid sequences of yeast and human Dna2 are 31% identical and 49% similar. Later, the expected full-length cDNA of hDna2 was updated as an accession no. XP_166103 in Entrez (http://www.ncbi.nlm.nih.gov/Entrez). This newly predicted cDNA has additional 86 amino acids at its N-terminus.
Purification of recombinant human Dna2
The cDNA of hDna2 with a FLAG epitope (FLAG-hDna2) at its N-terminus was subcloned into the BamHI and NotI sites of pIRESpuro2 (BD Biosciences). This plasmid was transfected into 293T cells using lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Transfected cells were grown and selected in DMEM containing 10% (v/v) fetal bovine serum and 1 µg/ml of puromycin for at least two weeks. Selected cells were analyzed for the expression of FLAG-hDna2 by western blot analyses. Approximately 6 × 108 puromycin resistant cells were collected and suspended with 150 ml of Buffer H500 [50 mM HEPES-KOH (pH 7.4), 500 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM phenylmethlysulfonyl fluoride (PMSF), 0.1 mM benzamidine, 1.25 µg/ml leupeptin and 0.625 µg/ml pepstatin A; the subscript number indicates NaCl in mM] and lyzed by sonication (three cycles of a 30 s pulse and a 2 min cooling interval). Cell lysates were incubated with 600 µl of FLAG M2 beads (Sigma) pre-equilibrated with buffer H500. After 6 h incubation, beads were washed with buffer H100 (more than 100 vol of beads) used and then incubated with buffer H100 containing 1 mg/ml 3× FLAG peptide (Sigma) overnight. Eluted proteins were subjected to a Hitrap-heparin column (1 ml; Amersharm-Phamacia Biotech) chromatography pre-equilibrated with buffer H100. After extensive washing with buffer H200, bound proteins were eluted with a linear gradient (200–800 mM) of NaCl in buffer H (10 ml). The eluted fractions were analyzed for proteins and activities.
ATPase and endonuclease assays
Standard ATPase assays were carried out in reaction mixtures (20 µl) containing 25 mM Tris–HCl (pH 7.8), 2 mM DTT, 2 mM MgCl2, 0.25 mg/ml BSA, 100 µM cold-ATP, 20 nM [γ-32P]ATP (>5000 Ci/mmol) and 50 ng of ΦX174 sscDNA or other polynucleotides as indicated. After incubation at 37°C for 10 min, aliquots (2 µl) were spotted onto a polyethyleneimine-cellulose plate (J. T. Baker Inc.) and developed in 0.5 M LiCl/1.0 M formic acid. The products were analyzed using a Phosphorimager (Molecular Dynamics, Inc.).
Standard endonuclease assays were performed in reaction mixtures (20 µl) containing 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 2 mM DTT, 50–100 mM NaCl, 0.25 mg/ml BSA and a partial duplex DNA substrate (15 fmol). The omission and addition of other components were as indicated in each experiment. Reactions were incubated at 37°C for 10 min, followed by the addition of 4 µl of 6× stop solution [60 mM EDTA (pH 8.0), 40% (w/v) sucrose, 0.6% SDS, 0.25% bromophenol blue and 0.25% xylene cyanol]. The reaction products were subjected to electrophoresis for 1 h at 150 V (10 V/cm) through 10% polyacrylamide gels containing 0.1% SDS in 0.5× TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA). The gels were dried on DEAE-cellulose paper and autoradiographed. Labeled DNA products were quantitated with the use of a PhosphoImager. For high resolution gel, reactions were stopped by addition of 2× stop solution (95% formamide, 20 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanol). The nucleolytic products were boiled for 1 min and subjected to electrophoresis for 1.5 h at 35 W in 1× TBE through 12 or 20% denaturing gel containing 7 M urea as described previously.
All the assays were repeated in multiple times and error bars were indicated where applicable.
Immuno-depletion experiments
To establish that endonuclease and ATPase activities are associated intrinsically with the purified recombinant protein, immuno-depletion experiments were carried out using M2 anti-FLAG beads. A mixture (200 µl) containing the beads (20 µl) and purified FLAG-hDna2 (800 ng) was incubated for 4 h at 4°C with occasional rocking in binding buffer [50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 2 mM DTT, 300 mM NaCl, 0.25 mg/ml BSA]. The beads were spun down in a microcentrfuge. Endonuclease and ATPase assays were carried out in standard reaction mixtures as described above with 0.2 and 5 µl, respectively, of the supernatant as enzyme source. Glutathione–Sepharose 4B beads (GTT) were used as a negative control.
RESULTS
Purification of FLAG-hDna2
To determine whether hDna2 is also a DNA endonuclease/helicase, we purified and characterized recombinant hDna2. Initial efforts to express hDna2 in a number of organisms, including bacteria, yeast and insect cells failed due to poor expression and extensive degradation. In contrast, expression of the protein in human cells yielded full-length soluble hDna2. In order to facilitate its production and isolation, we established stable 293T cell lines that expressed FLAG-hDna2. Recombinant FLAG-hDna2 protein was purified from these cells as described in Materials and Methods. When examined by western analysis using anti-FLAG antibodies, extracts from the transfected cells contained a ∼120 kDa protein, the expected size for the FLAG-hDna2 fusion protein, whereas it was not detected in untransfected control extracts (Figure 1B, compare lanes 1 and 4 with lanes 2 and 5). Using the purification procedure described in Materials and Methods, we obtained ∼20 µg of FLAG-hDna2 from a 10 l culture of transfected cells. Both the affinity-purified FLAG-hDna2 and protein present in crude extracts were detected at the same position using anti-FLAG antibodies (Figure 1B, compare lanes 5 and 6).
Immuno-depletion experiments were carried out to confirm that the purified FLAG-hDna2 intrinsically contained ATPase and endonuclease activities. When purified FLAG-hDna2 was incubated with anti-FLAG antibody beads, the endonuclease activity of FLAG-hDna2 disappeared from supernatants upon removal of the beads by centrifugation (Figure 1C, lanes 4). In contrast, this activity remained in the supernatant when buffer alone or glutathione beads were used in place of anti-FLAG antibody beads (Figure 1C, lanes 3 and 5). Both ATPase and endonuclease activities behaved similarly upon immuno-depletion (Figure 1D). The recombinant FLAG-hDna2 displayed a substantial level of ATPase activity in the absence of cofactor DNA, which was depleted along with the DNA-stimulated ATPase activity (Figure 1D). These results indicate that both ATPase and endonuclease activities are intrinsic to the FLAG-hDna2. We confirmed this further by analyzing fractions obtained from a glycerol gradient sedimentation of the affinity-purified FLAG-hDna2 (data not shown). We noted that the DNA-dependent ATPase activity of FLAG-hDna2 was ∼10-fold lower than that of yeast or C.elegans Dna2 (data not shown). Though FLAG-hDna2 contained substantial ATPase activity, it did not unwind duplex DNA substrates including those less than 20 bp long (data not shown). Thus, human Dna2 is similar to Xenopus Dna2. They both contained robust endonuclease activity but do not unwind duplex DNA.
Properties of hDna2 endonuclease activity
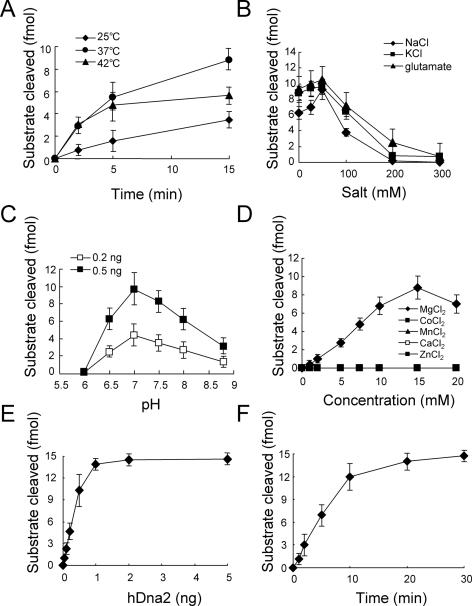
We next examined the biochemical properties of the FLAG-hDna2 endonuclease activity. For this purpose, its endonuclease activity was measured in standard reaction mixtures (described in Materials and Methods) containing a double-flap DNA substrate (15 fmol, substrate h in Figure 3). As shown in Figure 2A, endonuclease activity was most active at 37°C. Product accumulation at 42°C ceased after 5 min of incubation, indicating that FLAG-hDna2 was rapidly inactivated at 42°C. The endonuclease activity of FLAG-hDna2 was slightly stimulated in the presence of low concentrations (50 mM) of salts (NaCl, KCl and potassium glutamate), but was inhibited at higher concentrations (Figure 2B). Endonuclease activity was optimal at pH 7 when we tested two levels of enzyme (Figure 2C). Mg2+ was the only divalent metal ion that supported endonuclease activity with an optimal concentration of Mg2+ relatively high (10–20 mM) (Figure 2D). Other metal ions examined (Co2+, Ca2+, Zn2+ and Mn2+) were virtually inactive (Figure 2D).
Figure 2.
Determination of biochemical properties of endonuclease activity of hDna2. (A) Determination of an optimal temperature for endonuclease activity of hDna2. The reaction mixture (80 µl) containing 0.5 ng of hDna2 was incubated at 25, 37 or 42°C. Aliquots (20 µl) were withdrawn at the indicated times. The reaction products were analyzed by native 10% PAGE in 1× TBE as described under Materials and Methods. (B) Influence of salt on endonuclease activity of hDna2. The standard endonuclease assays were carried out as described under Materials and Methods except concentration of added salt. (C) Determination of optimal pH. The standard endonuclease assays were performed as described under Materials and Methods except pH. MES-NaOH (2-Morpholinoethanesulfonic acid) was used in pH 6.0 and pH 6.5 buffer. Tris–HCl was used in other pH buffer. (D) Requirement of divalent ions. The standard endonuclease assays were carried out as described under Materials and Methods using various divalent ions. (E) Enzyme titration. Increasing amounts of hDna2 were added to the standard reaction mixture. Nucleolytic products were analyzed under Materials and Methods. (F) Kinetic analysis of endonuclease activity of hDna2. The reaction mixture (140 µl) contained 3.5 ng of hDna2 (0.5 ng per reaction); aliquots (20 µl) were withdrawn at the indicated times and the reaction products were analyzed under Materials and Methods.
Using the optimal conditions determined above, we examined the rate of substrate cleavage as a function of enzyme concentration. In the presence of 15 fmol of the substrate used above, endonuclease activity increased linearly up to 0.5 ng (5 fmol) of enzyme during the 10 min of incubation period (Figure 2E), and plateaued upon addition of more than 1 ng (10 fmol) of enzyme. In the presence of a fixed amount (0.5 ng, 5 fmol) of hDna2, the rate of cleavage increased linearly up to ∼10 min; ∼1.2 fmol of the substrate was cleaved per min up to 10 min (Figure 2F). The turnover rate calculated (0.24/min/enzyme) could be underestimated in our assays since only initial cleavage of the labeled 5′ end was scored. With similar assays, however, human Dna2 has a turnover rate which is not far slower than those of other eukaryotic Dna2, such as C.elegans [0.82/min/enzyme; (22)] and yeast [0.33/min/enzyme; (23)].
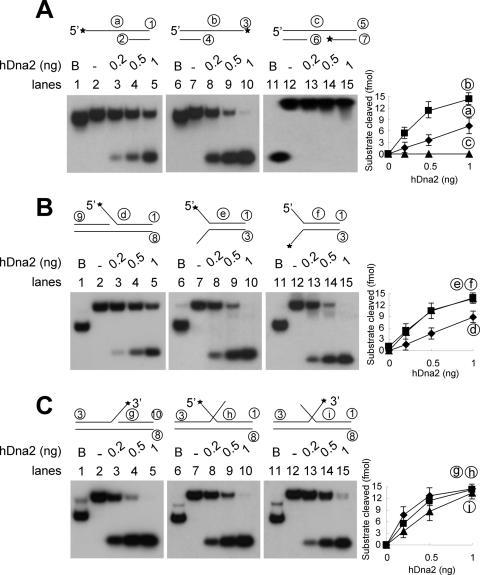
Substrate specificity of hDna2
To determine the substrate specificity, a number of different substrates, shown in Figure 3, were examined. To avoid sequence preferences, all single-stranded regions in the substrates contained homopolymeric dT. As shown in Figure 3A, hDna2 cleaved both 5′ and 3′ ssDNA overhang regions (substrates a and b), but not ssDNA flanked by duplex DNA (substrate c). The enzyme cleaved 3′ overhang regions more efficiently (3- to 5-fold) than 5′ overhang substrates. We also examined the cleavage of ssDNA tailed substrates in a variety of different flap structures as shown in Figure 3B and C. The 5′ flap substrate was cleaved as efficiently as the 5′ overhang substrate (compare substrate a and d). Unlike simple overhang ssDNAs, both 5′ and 3′ tails in fork-structured substrates were cleaved equally efficiently by hDna2 (Figure 3B, substrates e and f). These observations suggest that hDna2 can more efficiently cleave the 5′ tail when it is present at a fork. Alternatively, the presence of a 3′ tail at the fork can significantly stimulate hDna2's cleavage of the 5′ tail. In either case, it appears that hDna2 can recognize structural features in the substrate.
Figure 3.
Determination of substrate specificity. (A) The schematic structures of partial duplex DNA substrates used are shown at the top of each figure. The asterisks indicate 32P-labeled ends. The oligonucleotides used to construct each substrate in individual experiments are indicated as circled numbers that are listed in Table 1. Reaction mixtures (20 µl) containing the indicated amounts of hDna2 and 15 fmol of each substrate were incubated at 37°C for 10 min, and the reaction products were analyzed as described under Materials and Methods. B denotes the boiled substrate. Graphs on right indicate quantitation of the results in each row. (A) 5′-overhang (a), 3′-overhang (b) and flush (c) substrates. (B) 5′-flap (d), 5′-labeled fork (e) and 3′-labeled fork (f) substrates. (C) 3′-flap (g), 5′-labeled double-flap (h) and 3′-labeled double-flap (i) substrates.
To further confirm this notion, we examined the cleavage of a 3′ single flap and double-flapped oligonucleotide substrates structurally related to the forked structure shown in Figure 3C. Like the fork-structure substrates, hDna2 cleaved both flaps in the double-flapped substrate efficiently (Figure 3C). There was a significant increase in the extent of cleavage of the 5′ flap of the double-flap substrate (Figure 3B and C, compare substrates d and h), similar to the result observed with the fork-structured substrate. However, the cleavage efficiency of the 3′ flap in the double-flap substrate was reduced reproducibly about 2-fold (Figure 3C, compare substrates g and i).
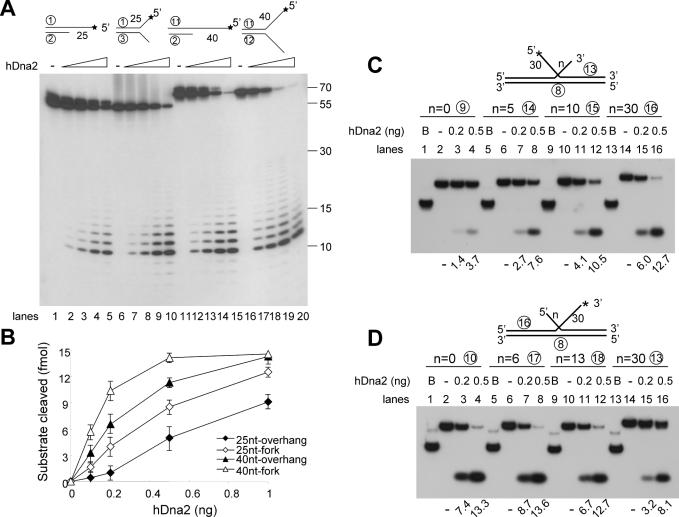
Effects of ssDNA flap lengths on hDna2-catalyzed cleavage
Since hDna2 displayed enhanced cleavage of the 5′ ssDNA tail in forked and double-flapped substrates, we investigated the influence of the 3′ tail length on the cleavage of the 5′ tail. In the presence of low levels (<0.2 ng) of enzyme, the cleavage of the 25 nt 5′ tailed substrate was stimulated significantly by the presence of a 25 nt 3′ tail (Figure 4A, lanes 1–10; Figure 4B). When both tails were increased to 40 nt, the overall efficiency of cleavage of the 5′ tailed substrate increased, but the stimulation by the presence of the 3′ tail was reduced (Figure 4A, lanes 11–20). We then examined the influence of the length of one tail on the cleavage of the other tail of double-flap substrates (Figure 4C and D). For this purpose, two types of double-flap substrates were prepared; one contained a 5′ flap of a fixed length (30 nt) with varying 3′ flap lengths (0, 5, 10 and 30 nt), while the other contained a 3′ flap of a fixed length (30 nt) with varying 5′ flap lengths (0, 6, 13, 30 nt). As shown in Figure 4C, cleavage of the 5′ flap by hDna2 increased in response to the increase in length of the 3′ flap. In contrast, cleavage of the 3′ tail was inhibited slightly (∼2-fold) in response to the lengthening of the 5′ flap (Figure 4D), consistent with the results shown in Figure 3C. These findings indicate that the presence of two tails at forks or in double-flap substrates mutually affect the processing by hDna2. They also show that 5′ tails are cleaved more efficiently by hDna2 in the presence of 3′ tails that are longer than 5 nt.
Figure 4.
The influence of one flap on the hDna2-catalyzed cleavage of the other flap in a two-flapped substrate. The four partial duplex substrates used are shown at the top of figure (* indicates 32P-labeled end). The 5′-ssDNA tail and 3′-ssDNA consist of homopolymer dT. The number in the schematic diagram of substrates used indicates the length of ssDNA tail. In case of the fork substrates, the length of 5′-ssDNA and 3′-ssDNA are identical. (A) Each substrate was incubated with increasing amounts of hDna2 at 37°C for 10 min and the reaction products formed were analyzed on 20% denaturing polyacrylamide gel. The numbers shown on the right of the figure indicate the size of the markers. (B) Quantitation of the results obtained in (A). The double-flap substrates used are shown at the top of figure (* indicates 32P-labeled end). n = number indicates length of 5′ or 3′ ssDNA in double-flap substrate. B denotes the boiled substrate. The numbers at the bottom of figure indicate fmol of nucleolytic products. (C) The double-flap substrates containing a 30 nt 5′ flap and a 3′ flap of various lengths (0, 5, 10 or 30 nt) were prepared. Each substrate was incubated with increasing amounts of hDna2 at 37°C for 10 min. The products formed were analyzed as described under Materials and Methods. (D) The double-flap substrates containing a 30 nt 3′ flap and a 5′ flap of various length (0, 6, 13 or 30 nt) were prepared. Each substrate was incubated with increasing levels of hDna2 at 37°C for 10 min. The reaction products formed were analyzed as described under Materials and Methods.
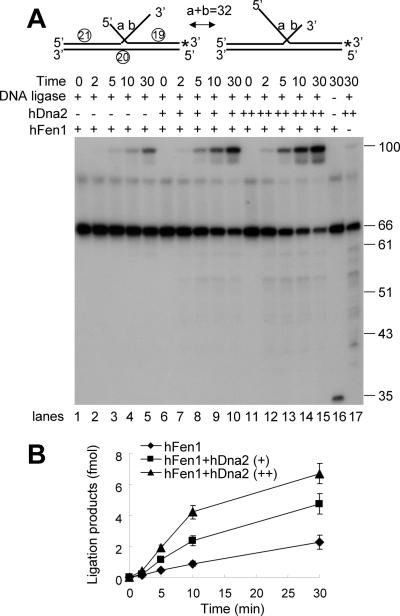
Human Dna2 promotes formation of ligatable nick by Fen1 in the presence of equilibrating flap substrates
It was previously shown that Dna2 facilitates Fen1-catalyzed nick formation by shortening long 5′ flaps (8). When the flap structures generated by displacement DNA synthesis catalyzed by pol δ are not processed rapidly, equilibrating flap structures can form. Since the nucleotide sequences of these single-stranded flap regions are identical, their annealing to the template DNA is competitive. This competition results in the production of two flap intermediates of different sizes (see Figure 5A). Since cleavage of 5′ tail substrates by hDna2 is optimal in the presence of a 3′ tail, it is likely that equilibrating flaps are physiological substrates of hDna2 in vivo. If this were the case, the processing of equilibrating flaps should require Dna2 for the efficient formation of ligatable nicks by Fen1. We tested this possibility by measuring the rate of formation of ligated product from a 32 nt equilibrating flap in the presence and absence of hDna2. The reaction mixtures used contained a fixed amount of human Fen1 and human DNA ligase I. As expected from our previous results, the addition of hDna2 increased both the rate and efficiency of formation of ligated products (Figure 5A, compare lanes 1–5, 6–10 and 11–15; Figure 5B), suggesting that hDna2 plays a critical role in the maturation of Okazaki fragments particularly when an equilibrating flap is formed. The addition of RPA neither stimulated nor inhibited the formation of ligated products in this reaction (data not shown). Fen1 alone generated a 34 nt cleavage product from the equilibrating flap substrate (Figure 5A, lane 16). In the absence of Fen1, ligated products were hardly detected (Figure 5A, lane 17), indicating that Fen1 is the primary enzyme responsible for the generation of a ligatable nick.
Figure 5.
hDna2 stimulates the formation of ligatable nicks with equilibrating flap substrates. The equilibrating flap substrate used are shown at the top of figure (* indicates 32P-labeled end). The combined length of the two equilibrating flaps (a and b) is 32 nt as shown. (A) The reaction mixtures containing 0.5 ng of Fen1 and 30 ng of human DNA ligase 1 were incubated with increasing amounts of hDna2 (0, 1 or 3 ng) at 37°C for the indicated time period. The products formed were analyzed on 12% of denaturing polyacrylamide gel. The numbers shown on the right of the figure indicate the size of the markers. (B) Quantitation of the results obtained in (A).
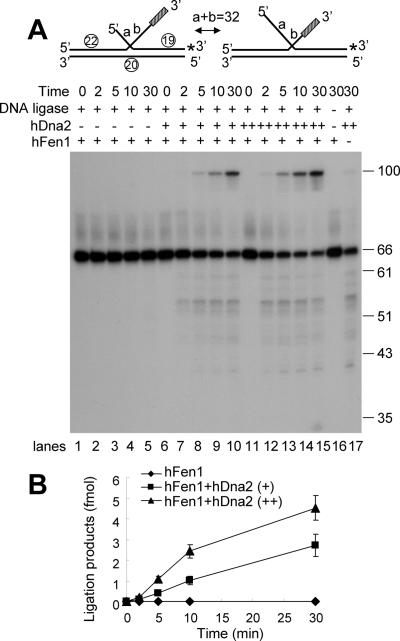
The above results do not exclude the possibility that a subset of equilibrating flaps (e.g. those only with a 5′ flap) is cleaved by hDna2 to promote formation of ligation products. If this were the case, removal of the 3′ tail would not contribute to the enhanced rate of ligation. In order to confirm that cleavage of the 3′ flap by hDna2 is crucial for maturation of an equilibrium flap, we prepared a DNA substrate containing 32 nt region complementary to the template oligonucleotide and a 10 nt random sequence located terminally at the 3′ flap which should interfere in the equilibration of the double-flaps. This equilibrating flap substrate contains an unannealed 10 nt 3′ flap that cannot be processed by Fen1 alone, and cannot produce ligatable nicks unless it is removed (Figure 6A, lanes 1–5). As predicted, formation of ligated products was totally dependent on the addition of hDna2 (Figure 6A, lanes 6–15). Increased levels of hDna2 resulted in the concomitant increase in formation of ligated products (Figure 6B), indicating that hDna2 contributes importantly in the formation of Fen1-catalyzed ligatable nick because of its ability to remove the bulk of the 3′ ssDNA flap. This result confirms that cleavage of both 5′ and 3′ ssDNA flap structures by hDna2 is critical for the processing of equilibrating double-flaps and the generation of ligatable nicks by Fen1.
Figure 6.
Cleavage of the 3′ flap by hDna2 also contributes to efficient formation of ligatable nicks. The equilibrating flap substrate used are shown at the top of figure (* indicates 32P-labeled end). The substrate is identical to that used in Figure 5 except that the 3′ tail contains an additional 10 nt random sequence (indicated as a shaded box). (A) The reaction mixtures containing 0.5 ng of Fen1 and 30 ng of human DNA ligase 1 were incubated with increasing amounts of hDna2 (0, 1 or 3 ng) at 37°C for indicated time period. The products formed were analyzed on a 12% denaturing polyacrylamide gel. The numbers shown on the right of the figure indicate the size of the markers. (B) Quantitation of the results obtained in (A).
DISCUSSION
In this report, we expressed and purified recombinant hDna2 from 293T cells transfected with a cDNA encoding the human homologue of yeast Dna2 in an attempt to define its role in lagging strand synthesis in mammalian cells. The cDNA of hDna2 used in our studies was cloned based upon EST information (NCBI accession no. CAI17238) and contained an open reading frame (ORF) of 1040 amino acids. Computational analysis of the human genome, however, revealed the presence of an additional 86 amino acids at the N-terminus of the EST-hDna2 (NCBI accession no. XP166103). However, our attempts to isolate a cDNA that included this additional 5′ region encoding the predicted 86 amino acids using a 5′ race technique were unsuccessful. Thus, it remains unclear whether the full-length ORF of hDna2 includes the additional 86 amino acids that are not encoded by the EST-hDna2. Since expression of hDna2 in bacteria, insects and yeasts were unsuccessful, we established a human cell line that stably expressed hDna2, which was used to purify the recombinant FLAG-hDna2. The level of hDna2 expression in such established cell lines, however, gradually diminished and was barely detectable after culturing for one month (data not shown). These findings suggest that the continued overexpression of hDna2 in human cells may be deleterious. Similar findings have been reported with yeast Dna2 (24).
As shown above, the purified recombinant hDna2 contains ssDNA-specific endonuclease and ATPase activities as observed with Dna2 enzymes isolated from other sources. Although hDna2 contained ATPase activity, it was devoid of DNA unwinding activity. Nonetheless, it may be able to translocate along ssDNA. If this were the case, hDna2 could yield different sized cleavage products from a ssDNA in the presence of ATP. The addition of ATP, however, did not alter the size or pattern of cleavage products formed from these substrates (data not shown). These findings contrast to those obtained with the yeast Dna2; the addition of ATP resulted in the formation of longer cleavage products from 5′ overhang substrates (23). Our results indicate that hDna2 lacks the ability (or has marginal activity) to unwind duplex or translocate along single-stranded DNA despite its DNA-dependent ATPase activity. In this regard, hDna2 is similar to Xenopus Dna2 which also contains robust endonuclease but lacks duplex DNA unwinding activity (11). In contrast, both the yeast and C.elegans Dna2 proteins displayed robust helicase activity (12,22). Our results, however, do not exclude the possibility that hDna2 contains low levels of helicase activity that are obscured by its nuclease activity. We also noted that hDna2 endonuclease activity was inhibited by high levels of RPA unlike the yeast or C.elegans Dna2 (data not shown). These distinct enzymatic properties of Dna2 isolated from different eukaryotes may influence the mechanism by which they contribute to Okazaki fragment maturation. For example, C.elegans Dna2 utilizes short flap structures more efficiently than S.cerevisiae Dna2, raising the possibility that the C.elegans pol δ catalyzes a more limited displacement of DNA than the S.cerevisiae pol δ. If this were the case, the roles of Dna2 and Fen1 in C.elegans may be substantially redundant. In keeping with this possibility, C.elegans Dna2 is less critical for viability than the budding or fission yeast homologues. Furthermore, Fen1 can suppress the loss of Dna2 function to a substantial extent in C.elegans.
The difference in enzymatic properties of Dna2 isolated from single and multicellular eukaryotes could be attributed to the primary structure of the protein; metazoan Dna2 enzymes, including those present in human, frog and C.elegans, possess a shorter N-terminus than those from single-cell eukaryotes. Deletion of the N-terminal 405 amino acids of yeast Dna2 (dna2Δ405N) resulted in cells that exhibit a temperature-sensitive phenotype and a mutant enzyme that is stimulated less efficiently by RPA. In addition, the N-terminal 405 amino acids of yeast Dna2 play a role in binding to and resolving secondary structures present in the flap, which ultimately assists Dna2-catalyzed cleavage of the secondary-structured flap (S. H. Bae and Y. S. Seo, unpublished data). However, C.elegans or hDna2 is not able to resolve secondary structures present in a flap (data not shown). The role played by the N-terminal domain found in lower eukaryotes may be carried out by other proteins. For example, Werner and Bloom helicases (WRN and BLM, respectively) can suppress the temperature-dependent growth defects caused by the presence of the yeast dna2-1 mutant allele (25,26). Both helicases stimulate the endonuclease activity of Fen1 as well as resolve secondary structures in flaps, which in wild-type yeast Dna2 are attributed to the N-terminal 405 amino acid domain and its helicase activity. In addition, it has been shown that WRN is required for lagging strand synthesis of telomeres in human cells (27).
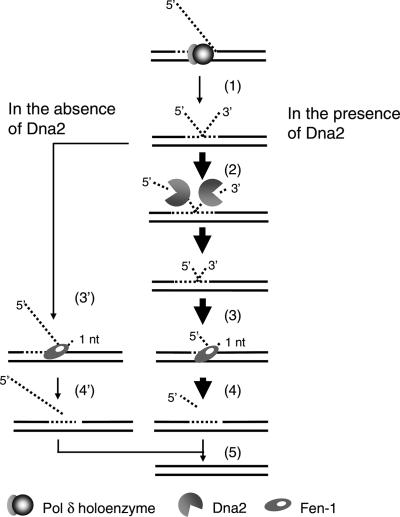
Significantly, our results show that hDna2 cleaves 3′ ssDNA substrates more efficiently than those containing a 5′ ssDNA tail, a specificity that is not observed with the yeast or C.elegans Dna2. In addition, the stimulation of cleavage of one tail due to the presence of a second tail (in forked- or double-flap substrates) by hDna2 was more pronounced than that observed with the yeast and C.elegans Dna2 (data not shown). If this unique property of human Dna2 described here also occurred in vivo, what additional role would be added to the action of hDna2? We suggest that this property is well-suited for the more rapid processing of equilibrating long flaps. As shown in Figure 7, the failure to rapidly process long flap structure generated by pol δ holoenzyme would result in an equilibrating flap. The simultaneous cleavage of both 5′ and 3′ flaps in the equilibrating flap by hDna2 would reduce the size of each flap and expedite the formation of a substrate for Fen1, leading to the production of a juxtaposed nick that is sealed by DNA ligase. In the absence of Dna2, however, due to the reduced efficiency of Fen1-catalyzed cleavage of a long 5′ flap (28), the rate of formation of ligatable nicks would be decreased.
Figure 7.
A model for a role of hDna2 in processing equilibrating flaps. (1) The failure of timely processing of a long flap generated by pol δ holoenzyme results in an equilibrating flap. (2) Cleavage of both 5′ and 3′ flaps by hDna2 reduces the size of each flap, (3) which expedites the formation of a substrate for Fen1. (4) Subsequent cleavage by Fen1 generates a ligatable nick. (5) The nick is sealed by DNA ligase. In the absence of Dna2, the slow formation of substrates for Fen1 (3′) and the reduced efficiency of Fen1-catalyzed cleavage of the long 5′ flap (4′) decrease the rate of production of ligatable nicks. The enzymes involved are as indicated.
This notion is supported by the demonstration that Dna2, in the presence of Fen1, enhanced the generation of ligatable nicks from equilibrating flaps (Figures 5 and 6). We showed that the removal of a 3′ tail in an equilibrating flap also contributed to generation of ligatable nicks. Since a physiological substrate for Fen1 is a double-flap substrate with a 1 nt 3′ flap and a 5′ flap of varying length (20), Fen1 can generate ligatable nicks only after an equilibrating flap is converted to this structure. Therefore, hDna2 shortens the two flaps in the equilibrating flaps and by doing so accelerates the conversion of the equilibrating flap into a physiological substrate for Fen1. Why is hDna2 most suited for this function? One speculation is that it may have something to do with displacement DNA synthesis carried out by human pol δ; if the human DNA pol δ is capable of extensive displacement DNA synthesis, the equilibrating flaps formed in humans may be longer than those produced in other organisms. This would necessitate the presence of a Dna2 with an enhanced ability to cleave both flaps simultaneously and provide Fen1 with a substrate that can be converted into a ligatable nick. This possibility is supported by observations that human DNA pol δ efficiently carries out displacement DNA synthesis in vitro (29).
Acknowledgments
This work was supported by a grant from the Creative Research Initiatives of the Korea Science Foundation (KOSEF) give to Y. -S. S. Funding to pay the Open Access publication charges for this article was provided by Korea Advanced Institute of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Garg P., Burgers P.M. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.Kao H.I., Bambara R.A. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol. 2003;38:433–452. doi: 10.1080/10409230390259382. [DOI] [PubMed] [Google Scholar]
- 3.Hubscher U., Seo Y.S. Replication of the lagging strand: a concert of at least 23 polypeptides. Mol. Cells. 2001;12:149–157. [PubMed] [Google Scholar]
- 4.Garg P., Stith C.M., Sabouri N., Johansson E., Burgers P.M. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 6.Lee K.H., Kim D.W., Bae S.H., Kim J.A., Ryu G.H., Kwon Y.N., Kim K.A., Koo H.S., Seo Y.S. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 2000;28:2873–2881. doi: 10.1093/nar/28.15.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budd M.E., Choe W., Campbell J.L. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000;275:16518–16529. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- 8.Bae S.H., Bae K.H., Kim J.A., Seo Y.S. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 9.Bae S.H., Kim D.W., Kim J., Kim J.H., Kim D.H., Kim H.D., Kang H.Y., Seo Y.S. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 2002;277:26632–26641. doi: 10.1074/jbc.M111026200. [DOI] [PubMed] [Google Scholar]
- 10.Kao H.I., Campbell J.L., Bambara R.A. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J. Biol. Chem. 2004;279:50840–50849. doi: 10.1074/jbc.M409231200. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q., Choe W., Campbell J.L. Identification of the Xenopus laevis homolog of Saccharomyces cerevisiae DNA2 and its role in DNA replication. J. Biol. Chem. 2000;275:1615–1624. doi: 10.1074/jbc.275.3.1615. [DOI] [PubMed] [Google Scholar]
- 12.Bae S.H., Kim J.A., Choi E., Lee K.H., Kang H.Y., Kim H.D., Kim J.H., Bae K.H., Cho Y., Park C., et al. Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res. 2001;29:3069–3079. doi: 10.1093/nar/29.14.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budd M.E., Campbell J.L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang H.Y., Choi E., Bae S.H., Lee K.H., Gim B.S., Kim H.D., Park C., MacNeill S.A., Seo Y.S. Genetic analyses of Schizosaccharomyces pombe dna2(+) reveal that dna2 plays an essential role in Okazaki fragment metabolism. Genetics. 2000;155:1055–1067. doi: 10.1093/genetics/155.3.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K.H., Lee M.H., Lee T.H., Han J.W., Park Y.J., Ahnn J., Seo Y.S., Koo H.S. Dna2 requirement for normal reproduction of Caenorhabditis elegans is temperature-dependent. Mol. Cells. 2003;15:81–86. [PubMed] [Google Scholar]
- 16.Kucherlapati M., Yang K., Kuraguchi M., Zhao J., Lia M., Heyer J., Kane M.F., Fan K., Russell R., Brown A.M., et al. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl Acad. Sci. USA. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henricksen L.A., Umbricht C.B., Wold M.S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 18.Dianova I.I., Bohr V.A., Dianov G.L. Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision repair. Biochemistry. 2001;23:12639–12644. doi: 10.1021/bi011117i. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson Z.O., Hindges R., Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae S.H., Choi E., Lee K.H., Park J.S., Lee S.H., Seo Y.S. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 21.Kao H.I., Henricksen L.A., Liu Y., Bambara R.A. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J. Biol. Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 22.Kim D.H., Lee K.H., Kim J.H., Ryu G.H., Bae S.H., Lee B.C., Moon K.Y., Byun S.M., Koo H.S., Seo Y.S. Enzymatic properties of the Caenorhabditis elegans Dna2 endonuclease/helicase and a species-specific interaction between RPA and Dna2. Nucleic Acids Res. 2005;33:1372–1383. doi: 10.1093/nar/gki255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae S.H., Seo Y.S. Characterization of the enzymatic properties of the yeast dna2 Helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 24.Parenteau J., Wellinger R.J. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 1999;19:4143–4152. doi: 10.1128/mcb.19.6.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura O., Campbell J.L. The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc. Natl Acad. Sci. USA. 2003;100:8193–8198. doi: 10.1073/pnas.1431624100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S., Sommers J.A., Brosh R.M., Jr In vivo function of the conserved non-catalytic domain of Werner syndrome helicase in DNA replication. Hum. Mol. Genet. 2004;13:2247–2261. doi: 10.1093/hmg/ddh234. [DOI] [PubMed] [Google Scholar]
- 27.Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 28.Chai Q., Zheng L., Zhou M., Turchi J.J., Shen B. Interaction and stimulation of human FEN-1 nuclease activities by heterogeneous nuclear ribonucleoprotein A1 in alpha-segment processing during Okazaki fragment maturation. Biochemistry. 2003;42:15045–15052. doi: 10.1021/bi035364t. [DOI] [PubMed] [Google Scholar]
- 29.Maga G., Villani G., Tillement V., Stucki M., Locatelli G.A., Frouin I., Spadari S., Hubscher U. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl Acad. Sci. USA. 2001;98:14298–14303. doi: 10.1073/pnas.251193198. [DOI] [PMC free article] [PubMed] [Google Scholar]