Abstract
The nucleosome core particle (NCP) is the fundamental building block of chromatin which compacts ∼146 bp of DNA around a core histone protein octamer. The effects of NCP packaging on long-range DNA charge transport reactions have not been adequately assessed to date. Here we study DNA hole transport reactions in a 157 bp DNA duplex (AQ-157TG) incorporating multiple repeats of the DNA TG-motif, a strong NCP positioning sequence and a covalently attached Anthraquinone photooxidant. Following a thorough biophysical characterization of the structure of AQ-157TG NCPs by Exonuclease III and hydroxyl radical footprinting, we compared the dynamics of DNA charge transport in ultraviolet-irradiated free and NCP-incorporated AQ-157TG. Compaction into a NCP changes the charge transport dynamics in AQ-157TG drastically. Not only is the overall yield of oxidative lesions decreased in the NCPs, but the preferred sites of oxidative damage change as well. This NCP-dependent attenuation of DNA charge transport is attributed to DNA–protein interactions involving the folded histone core since removal of the histone tails did not perturb the charge transport dynamics in AQ-157TG NCPs.
INTRODUCTION
Genomic DNA is under constant oxidative stress arising from agents like endogenous and exogenous free radicals, and γ- and X-ray irradiation. This challenge leads to DNA damage ranging from oxidized nucleobase products to single and double strand breaks (1). Understanding the origins of DNA oxidative damage is critical since increased levels of oxidative lesions have been implicated in the pathologies of adverse health conditions including cancer, atherosclerosis and diabetes (2–4). An analysis of the effects of oxidative stress on genomic DNA is complicated by the fact that DNA can act as an efficient carrier of excess charges. In other words, an excess electron or electron hole generated at a specific location in duplex DNA may not stay localized, but instead possess the ability to migrate long distances away from the initial damage site. Evidence for this electronic property of DNA comes primarily from in vitro experiments where the DNA nucleobases are either oxidized (5–8) or reduced (9,10) by a variety of exogenous agents. When DNA is oxidized, the resulting hole usually will localize at guanine to form a guanine radical cation (G•+), since the oxidation potential of this nucleobase is lower than those of A, T, C and the sugar phosphate backbone (11). Once formed, a G•+ has several fates available to it. First, it can simply recombine with the removed electron to return the system back to ground state. Second, G•+ can react with agents such as water to yield mutagenic guanine oxidative lesions (12). Third, the initially formed G•+ can undergo a hole hop, or a nearly isoenergetic electron exchange reaction with a nearby guanine (13,14), moving the electron deficient site >200 Å away from the site of initial oxidation (15,16).
While long-range DNA hole transport has been definitively established in vitro, the importance of this process in the eukaryotic nucleus has not. The primary reason for the lack of understanding of DNA hole migration in vivo arises from an incomplete picture of how chromatin structure impacts these reactions. The majority of the previous studies on DNA hole transport dynamics have been carried out using DNA substrates which were present naked in solution, and therefore they were free of any bound proteins. The reversible formation of a compact DNA–protein complex known as chromatin is a critical structural feature of the eukaryotic nucleus since 2 m of genomic DNA must fit into an ∼10 µm diameter human cell nucleus. The initial level of DNA compaction in chromatin is carried out by the formation of a nucleosome core particle (NCP). NCPs have been well characterized structurally through X-ray crystallography (17) and various additional thermodynamic and biophysical methodologies (18,19). A NCP consists of 146–147 bp of DNA wrapped 1.75 turns around an octamer built from two copies each of histones H2A, H2B, H3 and H4 (17–19). Between each NCP on a chromosome lies a linker region of DNA between 20 and 100 bp in length. The structure of the DNA packaged into a NCP is not the same as the Watson–Crick, B-form DNA found predominantly in naked DNA under intercellular conditions (17–19). For example, the DNA at the entrance and exit of the nucleosome is overwound and has a pitch of 10.0 bp/turn instead of the B-form 10.5 bp/turn. To help alleviate the strain placed upon the duplex in the overwound regions, the DNA near the pseudo-symmetry axis of the NCP is underwound and possesses a pitch of 10.7 bp/turn.
The effects of the changes in the DNA structure and/or local DNA environment associated with NCP formation on DNA hole transport reactions are of primary concern in the studies reported here. In the previous literature, the binding of proteins to DNA has been shown to have a variety of effects on the dynamics of DNA charge transport. Rajski et al. studied the effects of three different DNA binding proteins—restriction endonuclease PvuII, transcription factor ANTP and TATA-box binding protein (TBP)—on hole transport dynamics in DNA duplexes modified by a Rh-metallorganic photooxidant (20). Binding of PvuII and ANTP increased hole transport, whereas the binding of TBP decreased hole transport through the TBP-binding site. The authors attributed the different effects of these proteins to the extent of base stack structural perturbation in the DNA–protein complex. Consistent with their hypothesis that changes in DNA base stacking are responsible for the observed hole transport dynamics in DNA–protein complexes, Nunez et al. recently found that hole transport dynamics in a reconstituted NCP were nearly identical to those in the same DNA free in solution (21). This picture of base pair continuity being the primary factor controlling DNA hole transport in protein–DNA complexes is challenged by a report from Nakatani et al. (22). Here hole transport between two guanine oxidation sites flanking a BamHI binding site was greatly reduced by DNA–protein interactions, even though previous X-ray crystal structures indicated that BamHI does not perturb the integrity of the DNA base pair stacking upon binding (23). The authors attributed the attenuation of hole transport in the DNA–BamHI complex to the presence of a charged arginine residue making hydrogen bonding interactions with one of the guanine bases in the DNA major groove. Taken together, these studies indicate that an understanding of the effects of DNA–protein interactions on the dynamics of DNA hole transport is still in its infancy.
Even though Nunez et al. have studied DNA hole transport in the setting of a NCP, questions surrounding the effects of nucleosome formation on these long-range hole transport reactions are far from settled. For starters, NCPs form on a wide variety of genomic DNA sequences. This fact is reinforced by the observation that in the published X-ray structures electrostatic interactions are largely responsible for the stabilization of NCPs since there are no specific hydrogen bonds between the histone amino acid residues and the DNA nucleobases (17). Even though the histone octamer does not utilize a direct readout of DNA sequences, the thermodynamic stability of a NCP is directly related to the sequence of the bound DNA (18,19,24,25). With this large in vivo distribution of NCP thermodynamic stability in mind, we are initiating a strategic assessment of the hole transport dynamics in NCPs of varying DNA sequence in order to assess global trends. Our first efforts towards this endeavor are communicated in this paper for an artificial DNA sequence known as the TG-motif (26,27), a sequence which leads to the formation of structurally well-defined NCPs. For the selective initiation of oxidative damage, we utilize DNA duplexes covalently modified by an Anthraquinone photooxidant (Figure 1A) at one 5′-terminus. We show in this communication that NCP formation does in fact lead to drastic changes in both (i) the overall yield of DNA oxidative lesions and (ii) the observed distribution of oxidative damage sites in TG-motif NCPs. The results of this study are expected to contribute to our understanding of how DNA–protein interactions impact the dynamics of DNA-mediated charge transport reactions, as well as the impact of oxidative stress-induced DNA charge migration on global genomic integrity in eukaryotes.
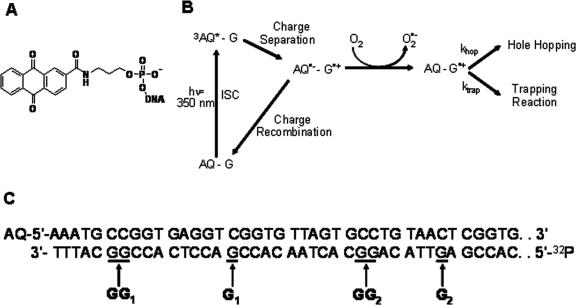
Figure 1.
(A) Chemical structure of the AQ Photooxidant. (B) Reaction mechanism of photoinduced DNA oxidation in AQ-labeled DNA. Details of each reaction step are provided in the Results section. (C) Sequence of the AQ-157TG duplex proximal to the AQ photooxidant. The full DNA sequence can be found in the Materials and Methods section.
MATERIALS AND METHODS
Reagents and chemicals
Monobasic sodium phosphate, phenylmethylsulfonic fluoride (PMSF), thiourea and sodium l-ascorbate were purchased from Sigma-Aldrich. Urea, phenol/chloroform and H2O2 were obtained from Fisher Scientific. Stock 40% acrylamide solutions were supplied by BioRad. T4 Polynucleotide Kinase (PNK), T4 DNA Ligase (T4 Lig) and Exonuclease III (ExoIII) were purchased from New England Biolabs. [γ-32P]ATP (5 mCi/mL, 3000 Ci/mmol specific activity) was supplied by Perkin Elmer. The 3500 MWCO Mini Slide-a-Lyzer Dialysis units were from Pierce. All phenol/chloroform-based separations of DNA and protein were performed using 0.5 mL Eppendorf Gel Phase Lock tubes. G-50 ProbeQuant micro spin columns were purchased from Amersham Biosciences.
Construction of DNA duplex AQ-TG157
AQ-157TG, a 157 bp DNA duplex covalently modified by AQ at one 5′-terminus and built around the strong NCP positioning TG DNA motif, was constructed in the following modular fashion. Six oligonucleotides AQ-A AQ-d(AAA TGC CGG TGA GGT CGG TGT TAG TGC CTG TAA CTC GGT GTT AGA GCC TGT AAC TCG G), B d(GTT ACA GGC ACT AAC ACC GAG TTA CAG GCA CTA ACA CCG ACC TCA CCG GCA TTT), C d(TGT TAG AGC CTG TAA CAC TCT CAG CCT TTG GC), D d(GAG TGC CAA AGG CTG AGA GTG TTA CAG GCT CTA ACA CCG A), E d(ACT CTC GGT GTT AGA GCC TGT AAC TCG GTG TTA GAG CCT GTA ACT CGG TGT TAG AGC CTG TAA CGA TA) and F d(TAT CGT TAC AGG CTC TAA CAC CGA GTT ACA GGC TCT AAC ACC GAG TTA CAG GCT CTA ACA GCG A) were synthesized by standard solid phase synthesis on an ABI Applied Biosystems 380B DNA synthesizer. Strand AQ-A was modified at its 5′-terminus by an AQ-phosphoramidite reagent generated using a previously reported synthetic scheme (28). Oligonucleotides B–F were synthesized Trityl-On, and all six strands were individually deprotected by heating them for 16 h at 55°C in concentrated ammonium hydroxide. After deprotection, the oligonucleotides were purified by high-performance liquid chromatography (HPLC) on a Waters 2690 system equipped with a Waters 996 Photodiode Array Detector and a Nucleosil C18 RP column (Hichrom). Following purification, oligonucleotides B–F were lyophilized, de-tritylated by treatment with 80% acetic acid for 30 min, and ethanol precipitated. Strand AQ-A was lyophilized, resuspended in H2O and ethanol precipitated. Oligonucleotide purity was ascertained using 20% polyacrylamide gel electrophoresis (PAGE), and the product yield was determined by A260 measurements on a Cary 100 Bio spectrophotometer (Varian).
After purification, the 5′-termini of strands B, C, D, and E (2 nmol each) were phosphorylated by incubation with 20 U of PNK and 10 mM ATP at 37°C for 45–60 min. The three duplexes AQ-AB, CD and EF were individually hybridized in a buffer composed of 100 mM NaCl, 10 mM Tris and 1 mM EDTA. Briefly, equimolar amounts of complementary oligonucleotides were mixed and placed in a temperature controlled spectrophotometer (Cary Bio 100), the samples heated to 90°C over 10 min, and the mixtures cooled at a rate of 0.5°C/min to 10°C. After hybridization, each duplex was ethanol precipitated. Next, the full length AQ-157TG duplex was produced by a step-wise DNA ligation protocol. First, duplexes CD and EF were ligated together in a reaction mixture containing 120 U of T4 Lig, 10 mM ATP, and 2 µM total DNA incubated at 16°C overnight. After verification of successful ligation (≥50% product yield) by electrophoresis on a 3% agarose gel, an equal concentration of duplex AQ-AB was added in situ, the ligation mixture supplemented with additional T4 Lig, and the reaction incubated overnight at 16°C. The desired 157 bp DNA product was purified by preparative (20 cm × 20 cm) 10% denaturing (7 M urea) PAGE, re-hybridized by the same protocol described previously for the individual duplexes, and precipitated by ethanol treatment. Following purification, the yield of AQ-157TG was calculated using A260 measurement. Before reconstitution, the 5′-terminus of strand BDF (counter to the AQ-labeled strand) was labeled with 32P by incubation of AQ-157TG with 20 U of T4 polynucleotide kinase and 2 µL (20 µCi) [γ-32P]ATP. Labeled samples were then extracted with phenol/chloroform, the excess [γ-32P]ATP removed using a G-50 ProbeQuant size exclusion column, and the DNA precipitated using ethanol.
Preparation of reconstituted nucleosome core particles
Chicken erythrocyte NCPs stripped of linker histones H1 and H5 (29) were a gift from the laboratory of Dr Michael J. Smerdon (Washington State University). The integrity of the core histone proteins was verified by 18% SDS–PAGE analysis on a routine basis. Reconstitution of the AQ-157TG DNA duplex onto NCPs was performed using the method of Moyer et al. (30). The 32P-labeled duplex (∼100 nM, ∼100k c.p.m.) was combined with NCPs (0.6 mg/mL) at a 1:50 molar ratio in 60 µL of a 1M NaCl, 10 mM sodium phosphate buffer (pH 7.0). Salt exchange dialysis was performed in 3500 MWCO Mini Slide-a-Lyzer Dialysis units at 4°C. Immediately prior to dialysis at each salt concentration, PMSF was added to the dialysis buffer to 0.2 mM final concentration. Initial dialysis was against buffered 1M NaCl for 45 min, followed by buffered 600 mM NaCl for 60 min, buffered 300 mM NaCl for 60 min, and finally 10 mM sodium phosphate (pH = 7.0) for 60 min. Evaluation of the reconstitution efficiency was performed by EMSA on 6% native acrylamide gels ran at 100 V and ∼0 W. The gels were dried, exposed to a Phosphorscreen (Amersham Biosciences) and visualized on a 445 SI Phosphorimager (Molecular Dynamics). All gel analysis was performed using ImageQuant software (Molecular Dynamics).
Footprinting of reconstituted nucleosome core particles
The translational setting of AQ-157TG rNCPs was assessed by ExoIII digestion. Briefly, separate samples of reconstituted nucleosomes and free AQ-157TG were digested by 1 U of ExoIII for 3 min at 37°C. The reaction was quenched by rapid phenol/chloroform extraction of the ExoIII from the mixture, followed by DNA ethanol precipitation. The DNA pellets were then resuspended in formamide loading buffer and ran on an 8% (7 M Urea) sequencing gel at 60 W for 120 min. To examine the rotational positioning of the DNA on the reconstituted NCPs, hydroxyl radical (OH•) footprinting was employed (31). Redox reactions between Fe(II)-EDTA (20 µM), H2O2 (0.3% v/v) and sodium l-ascorbate (1 mM) were used to generate OH• in situ. After 1 min at room temperature for free AQ-157TG, or 10 min at room temperature for rNCPs, the reactions were quenched by the addition of EDTA (40 mM) and thiourea (7 mM). The DNA was then treated by phenol/chloroform extraction and ethanol precipitation was carried out in the presence of glycogen. The DNA was resuspended in formamide loading buffer and was then separated on a 7M urea PAGE (6–8%) sequencing gel, followed by autoradiography.
UV-irradiation and assessment of DNA charge transport reactions
Irradiation of free AQ-157TG (∼100 nM) and rNCPs were separately performed in a Luzchem photoreactor (Luzchem Research) with 6 UV-A lamps (∼0.3 mW/cm2 per lamp) centered at 350 nm for 30–60 min at 25°C. After irradiation, the AQ-157TG duplex was dissociated from the core histones by heating the samples at 90°C for 5 min. The DNA was then immediately subjected to a phenol/chloroform extraction and ethanol precipitation with 2 µL glycogen (20 mg/mL) as a carrier. Both irradiated and non-irradiated DNA were treated with hot 10% piperidine (90°C) for 30 minutes, followed by lyophilization and two washes with 30 µL ddH2O. The dried DNA pellets were resuspended in formamide loading buffer and samples (∼10 k c.p.m.) were loaded and ran on 7M urea, 8% acrylamide sequencing gels at 60 W constant power for 2 h. The gels were dried, exposed to a Phosphorscreen (Amersham Biosciences), and visualized on a 445 SI Phosphorimager (Molecular Dynamics). All gel analysis was performed using ImageQuant software (Molecular Dynamics).
RESULTS
Design and construction of AQ-157TG DNA
N-alkyl-Anthraquinone-2-amide (AQ; Figure 1A) is a well characterized photooxidant used in many previous studies to initiate DNA charge transport (CT) reactions (6,32,33). A reaction scheme for AQ-initiated DNA hole transport is shown in Figure 1B. Briefly, irradiation of AQ at ∼350 nm generates the lowest singlet excited state of AQ (1AQ*) which quickly undergoes intersystem crossing (ISC; Figure 1B) to the lowest triplet state (3AQ*) (6). 3AQ* oxidizes a nearby G base, thus forming the AQ radical anion (AQ•−) and guanine radical cation (G•+). To liberate the electron hole in the DNA base pair stack, O2 acts as a scavenger to remove the excess electron from AQ•− (6). Once liberated, the hole is free to move between guanines in the DNA duplex. The interplay of hole hopping rates (khop) between guanine sites and site-dependent charge trapping rates by agents like water, O2 (34) and O2 •− (35) (ktrap) leads to a final steady state distribution of piperidine-labile oxidized guanine products.
In order to study DNA CT reactions in NCPs possessing a well-defined structure, we chose to utilize DNA duplexes containing multiple repeats of the non-natural TG-motif NCP binding sequence d(TCGGTGTTAGAGCCTGTAAC) (27). The phasing of the alternating G/C and A/T rich blocks (underlined above) was designed to ensure that DNA duplexes built around the TG-motif will possess only one structural conformation upon reconstitution to form a NCP (27). Our 157 bp synthetic oligonucleotide duplex, AQ-157TG (Figure 1C), incorporates a total of six TG motifs. The AQ photooxidant was covalently attached at one 5′ end of the duplex, while the other 5′ end was labeled by 32P for the quantization of DNA oxidative damage yields using autoradiography. Previous NMR structures (36) of 5′-covalent DNA modifications, and molecular modeling of AQ-DNA complexes (28), lead to the expectation that in an aqueous environment the AQ photooxidant will preferentially end-cap, or stack onto the base pairs at the end of the DNA duplex. However, we initially had uncertainties about whether the AQ would end-cap onto the DNA duplex in a reconstituted NCP (rNCP), as intended, or instead associate with hydrophobic regions on the core histone proteins. We therefore designed AQ-157TG such that the AQ would be placed at the end of 10 bp of linker DNA, and thus one full duplex turn away from the expected entrance of a rNCP. In addition, the sequence d(AAATGCC) was placed proximal to the AQ in order to ensure that DNA oxidative damage yields would not be limited by charge injection efficiency in these studies (37).
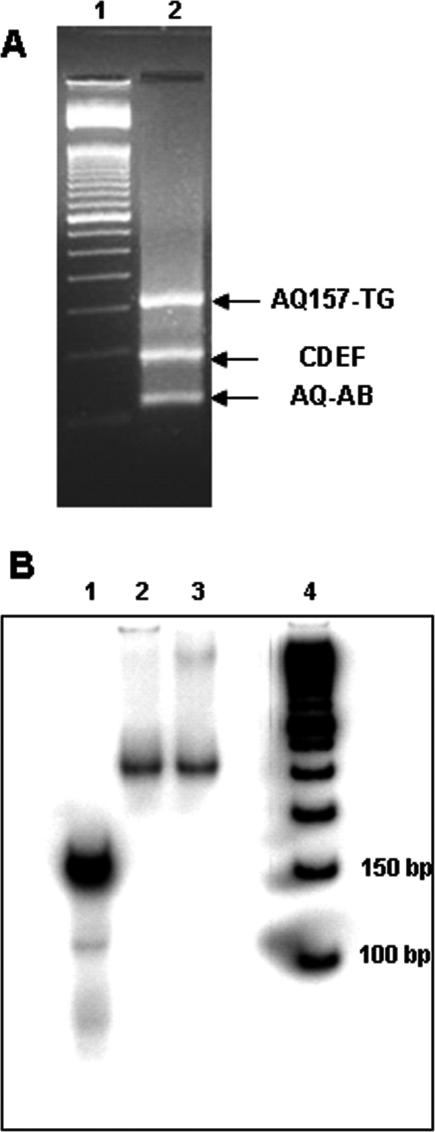
The production of AQ-157TG utilized a modular system involving the solid phase synthesis of six oligonucleotides, labeled AQ-A through F, through standard phosphoramidite chemistry. Following reverse phase purification by HPLC, oligonucleotides B–E were phosphorylated at their 5′-termini. Duplexes AQ-AB, CD and EF were individually hybridized, then ligated together in a stepwise, one-pot ligation reaction. The results of a typical multistep ligation are shown inFigure 2A. AQ-157TG was purified from the ligation reaction by electrophoresis on a 10% denaturing PAGE gel. The AQ-157TG single strands were then extracted from the appropriate acrylamide fragments, rehybridized and the yield of AQ-157TG calculated from A260 measurements. This methodology does not produce high yields of full length AQ-157TG (<10%); however it does result in a very pure (>95%) single parent band when labeled with 32P and electrophoresed on a DNA sequencing gel. In particular, we note that there are little to no extra non-ligated DNA fragments present in any of the experiments reported here.
Figure 2.
(A) Ethidium-stained 3% Agarose gel showing the results of a typical ligation reaction used to produce AQ-157TG. Lane 1 is a 50 bp DNA ladder, and Lane 2 contains the ligation mixture. Assignment of the bands in Lane 2 is indicated in the figure. (B) A native 6% acrylamide electrophoretic mobility shift assay (EMSA) showing the reconstitution efficiency of AQ-157TG. Lanes: (1) Free AQ-157TG, (2) and (3) separate rNCP preparations, and (4) 50 bp DNA ladder.
Reconstitution of AQ-157TG nucleosome core particles and footprinting
The incorporation of AQ-157TG into NCPs was carried out by process known as reconstitution (19). NCP reconstitution is initiated by mixing, in the presence of 1M NaCl, native NCPs with DNA that forms thermodynamically stable NCPs. The high salt concentration will disrupt the electrostatic interactions between the negatively charged genomic DNA and the basic histones. Lowering the ionic strength of the solution by stepwise dialysis leads to the formation of NCPs, only now the exogenous DNA duplex out-competes some of the endogenous DNA to yield reconstituted nucleosome core particles (rNCPs) (29). Reconstitution was initiated by mixing an ∼1:50 molar ratio of 32P labeled AQ-157TG and chicken erythrocyte NCPs in a 1 M NaCl, 10 mM NaPi, pH 7.0 buffer, followed by slow dialysis down to 10 mM NaPi. The reconstitution products were analyzed by an electrophoretic mobility shift assay (EMSA) (Figure 2B). This EMSA shows that (i) from autoradiography, the reconstitution efficiency of AQ-157TG was >95%, (ii) AQ-157TG reconstitutes onto NCPs to form predominantly mononucleosomes since there is only (a) one slow migrating band and (b) no evidence of higher order DNA–protein aggregates in the gel (Supplementary Figure 2) and (iii) the presence of the 5′-AQ label does not perturb the reconstitution of AQ-157TG onto NCPs since this EMSA is identical to an EMSA of NCPs formed via the reconstitution of a 157TG duplex with a 5′-aminopropyl linker in place of the AQ chromophore (data not shown).
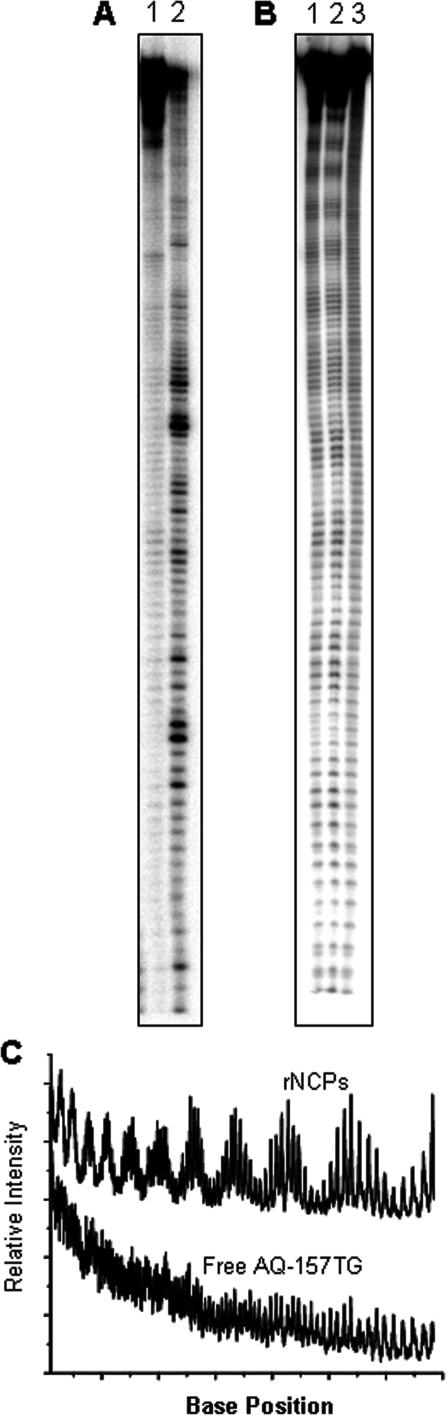
In order to characterize the conformational structure of the AQ-157TG rNCPs, DNA footprinting experiments were performed. These experiments allow a full characterization of both the rotational and the translational settings of a DNA duplex on the surface of a NCP. To investigate the perturbation of DNA CT by NCP formation, ideally a homogeneous mixture of rNCPs containing only one DNA rotational and one DNA translational setting would be used. When a NCP possesses a single translational setting, there is no sliding of the DNA on the protein surface and there are defined nucleobases at the entrance and exit to the core particle (38). Exonuclease III (ExoIII) is a 3′–5′ DNA exonuclease whose activity is retarded by the presence of DNA–protein contacts (39). Hence, a limited ExoIII digestion can determine if there is a single, well-defined, nucleobase at the entrance of a rNCP. Figure 3A shows the results of ExoIII digestion of both free and rNCP AQ-157TG DNA. Free AQ-157TG is easily digested along its entire length by ExoIII, with some regions of preferred cutting apparent. After reconstitution onto NCPs, AQ-157TG is vigorously digested in the ∼10 bp region next to the AQ photooxidant, whereas the rest of the DNA is highly protected from ExoIII cutting. These experiments indicate that (i) the extended 10 bp overhang with the AQ cap is outside of the NCP, (ii) AQ does not interact with the histone proteins since we observe ExoIII activity on the surrounding nucleobases, and (iii) there appears to be only one major translational setting of AQ-157TG on the nucleosome surface since only the ∼10 bp of DNA proximal to the AQ-label are digested.
Figure 3.
Structural characterization of AQ-157TG rNCPs. (A) Exonuclease III footprinting of AQ-157TG rNCPs (lane 1) and free AQ-157TG (lane 2). The restriction of ExoIII activity to the ∼10 bp proximal to AQ in the AQ-157TG rNCPs is evident. (B) Autoradiogram of hydroxyl radical footprinting on AQ-157TG rNCPs (lanes 1 and 2) and free AQ-157TG (lane 3). (C) Partial scan of the footprint in B of both free AQ-157TG (bottom) and AQ-157TG rNCPs (top). The 10 bp periodic cutting in the rNCPs is apparent.
The term rotational setting refers to how the minor groove of the DNA duplex at each individual base pair is oriented with respect to the histone octamer (31). On an rNCP with only one rotational setting, there are specific regions where the minor groove rotates inward to face the histone protein core interspersed with regions where the minor groove rotates outward towards solution. Hydroxyl radical (OH•) footprinting is a very sensitive method to probe the DNA rotational setting in rNCPs (31). Generation of OH• in the presence of DNA results in the induction of DNA strand breaks in regions where the phosphodiester backbone, and hence DNA minor groove, is exposed to solvent. The results of OH• footprinting experiments on free and rNCP AQ-157TG are shown in Figure 3B. When free in solution, there is nearly equal intensity of strand breaks at every base as ascertained by autoradiography (Figure 3C). Conversely, AQ-157TG rNCPs show the expected periodical phasing of damaged/protected regions associated with a single DNA rotational setting (Figure 3C).
Taken together, the footprinting experiments and EMSAs indicate that there is one predominant structure of AQ-157TG in an rNCP and that any measurements of DNA CT come from a nearly homogenous source.
DNA charge transport in free AQ-157TG and rNCPs
Irradiation of both free and rNCP AQ-157TG was achieved by placing the samples in the rotating carrousel of a Luzchem photoreactor. The photoreactor was equipped with six ultraviolet-A (UV-A) lamps whose emission was centered at 350 nm and cutoff at 300 nm in order to selectively irradiate the AQ-photooxidant but not the DNA nucleobases or aromatic amino acid side chains of the histones. After irradiation at room temperature, the oxidative lesions arising from G•+ trapping reactions were treated with hot piperidine and the ensuing strand cleavage products visualized by autoradiography of a 8% PAGE/7 M Urea DNA sequencing gel. Separate time courses for both free and rNCP AQ-157TG DNA were conducted to ensure that single turnover conditions, as defined by the relative ratios of the oxidized guanine nucleobases remaining unchanged, were fulfilled in both samples (data not shown).
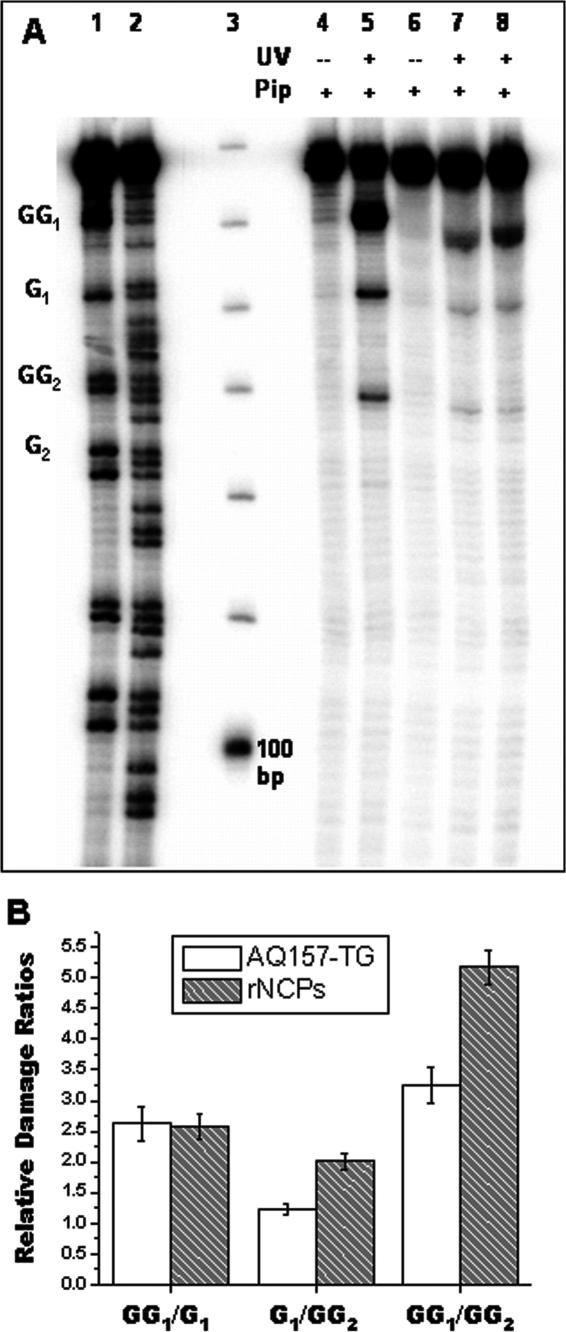
In the AQ-157TG sequence chosen for study, there are three guanine-containing steps in the first 28 bp of the 5′-32P labeled counterstrand (BDF). In order of increasing distance from AQ, these sites are labeled GG1, G1 and GG2 (Figure 1C). In free AQ-157TG DNA CT is observed over 95 Å (28 bp) to the 5′-Guanine of the GG2 step (Figure 4A, Lane 5). The yield of CT to the next single G in strand BDF (G2; 34 bp, 116Å from AQ) is not significantly above background. In addition, Figure 4A also displays the results using non-irradiated, piperidine treated AQ-157TG (Lane 4) indicating (i) the minimal background inherent in these experiments and (ii) the lack of piperidine-labile lesions in the absence of AQ photooxidation. Using autoradiography, the ratios of the strand break yields at each guanine step (GG1/G1, G1/GG2 and GG1/GG2) in free AQ-157TG were determined (Table 1, Figure 4B). These ratios give an approximate steady state snapshot of the guanine damage distribution resulting from the interplay between khop and ktrap (Figure 1B). In addition, the overall yield of guanine oxidative damage (Ydam, Table 1) was obtained by calculating the ratio of total piperidine-sensitive guanine oxidative damage to the intensity of all bands in that lane.
Figure 4.
(A) Visualization of DNA charge transport products in free AQ-z157TG, rNCPs, and tailless rNCPs as a function of 60 min. of UV-A irradiation and hot piperidine treatment. Lanes: (1) Maxam-Gilbert G ladder, (2) Maxam-Gilbert G+A ladder, (3) 10 bp DNA ladder, (4) free AQ-157TG, (5) free AQ-157TG, (6) AQ-157TG rNCPs, (7) AQ-157TG rNCPs, (8) tailless AQ-157TG rNCPs. (B) Graphical representation of the damage ratios GG1/G1, G1/GG2, and GG1/GG2 from Table 1.
Table 1.
Quantitative values for the site damage distribution ratios GG1/G1, G1/GG2 and GG1/GG2 for free AQ157-TG and rNCPs
| DNA sample | Ydam | GG1/G1 | G1/GG2 | GG1/GG2 |
|---|---|---|---|---|
| Free AQ-157TG | 0.64 | 2.6 | 1.2 | 3.2 |
| AQ-157TG rNCP | 0.21 | 2.6 | 2.0 | 5.2 |
Ydam values for AQ157-TG and the AQ-157TG rNCPs were determined by normalizing for total damage in a single lane, i.e. Ydam=1 − (parent band/total lane counts). All values are averages over five independent experiments.
The rNCP samples were irradiated and treated with piperidine in exactly the same manner as the free AQ-157TG. A typical DNA sequencing gel resulting from these reactions is shown in Figure 4A. Once again, the non-irradiated samples (Lane 6) show the lack of piperidine-labile nucleobase oxidation products in the absence of photoinduced charge injection. A minor shift of the piperidine cleavage bands and the 157 bp parent band in the rNCP samples was repeatedly observed on the DNA sequencing gels. This shift was attributed to an rNCP-related gel artifact, and it does not interfere with the analysis of the oxidative damage product ratios or Ydam. A cursory examination of the sequencing gel in Figure 4A indicates that while the damage spectrum in free AQ-157TG (Lane 5) and rNCPs (Lane 7) is similar, there is an attenuation of the total yield of piperidine-labile guanine oxidation products in the rNCPs. This qualitative observation was quantified by autoradiography and it was found that the value of Ydam decreases by ∼60% in the rNCPs (Table 1).
Not only is the overall yield of oxidative damage decreased in a NCP, but the distribution of oxidative damage is also perturbed by packaging DNA into chromatin structures (Table 1, Figure 4B). The site damage ratio GG1/G1 does not differ significantly between the free AQ-157TG and the rNCPs, however significant increases in the ratios G1/GG2 (46% increase), and GG1/GG2 (59% increase) were reproducibly observed in the rNCPs.
As a control reaction, we mixed free AQ-157TG with chicken erythrocyte NCPs at a 1:50 molar ratio in a 10 mM sodium phosphate buffer without performing a reconstitution and performed similar analyses as those carried out on the AQ-157TG rNCPs. An EMSA showed that incubation of AQ-157TG and native NCPs at low salt results in the formation of aggregates (Supplementary data) of uncharacterized structure. Irradiation of these aggregates followed by piperidine treatment resulted in a 30% reduction in Ydam as compared with free AQ-157TG, but no changes in the damage ratios G1/GG2 and GG1/GG2 (data not shown).
Effect of histone tail removal on DNA charge transport in rNCPs
From X-ray crystallography and biophysical investigations, it is known that there are two distinct features of the core histones in a NCP. The first are the α-helical regions which form the histone surface which DNA wraps around (17). The second are the largely unstructured histone N-terminal and C-terminal tails which contain the majority of sites involved in generating the ‘histone code’ arising from posttranslational modifications such as acetylation and methylation (40). The histone tails are basic in nature, but do not make contact with the DNA wrapped around the NCP. However, there are reports indicating that the histone tails do make contact with the DNA linker regions between two NCPs in chromatin, regions similar to the AQ-labeled 10 bp overhang (41). Therefore, we investigated if the histone tails in the AQ-157TG rNCPs are responsible for the observed changes in the DNA CT dynamics by removing the N-terminal tails of native NCPs via a limited trypsin digestion prior to reconstitution (Supplementary data). OH• footprinting verified no change in the rotational setting in the tailless AQ-157TG rNCPs (Supplementary data). Irradiation of the tailless rNCPs yielded DNA CT results indistinguishable from those of the native rNCPs (Figure 4A). We infer from this observation that the histone tails play no role in regulating the dynamics of CT in mononucleosomes.
DISCUSSION
Progress in the field of DNA CT has been strongly driven by the possibility of novel applications of this phenomenon in materials science and biosensors (42). However, consideration of the consequences of efficient, long range DNA CT on fundamental processes in biology has lagged behind. In this paper we have chosen to interrogate the feasibility of intracellular DNA CT by modeling these reactions in a NCP, the fundamental building block of chromatin. The NCP presents a dramatically different DNA structure and environment compared with the typical Watson–Crick B-form structure (43). For starters, in the regions of the NCP where we observe CT the DNA is overwound and possesses an average 10.0 bp/turn rise instead of the typical 10.5 bp/turn. Also, the accessibility of the DNA grooves is perturbed by NCP formation. For instance, there are alternating regions where the DNA backbone, and hence minor groove, is facing away from the histone core surface and accessible from bulk solvent, interspersed with regions where the minor groove is rotated inwards towards the histone protein surface. The effects of these structural changes and solvent accessibility on DNA CT reactions are just starting to receive the attention they deserve (44,45).
The DNA CT reactions reported here come from protein–DNA complexes whose molecular structure is well-characterized. Neither the extra 10 bp of DNA beyond the 147 in direct contact with the histone octamer nor the covalently tethered AQ photooxidant perturbed the ability of the TG-motif to generate stable rNCPs (27). From the EMSAs, AQ-157TG associates with the core histones to form mononucleosomes in >95% yield, without the formation of higher order protein–DNA aggregates. The OH• experiments indicate that AQ-157TG possesses only one rotational setting on the surface of a NCP. Structurally, this implies that there are specific contact points between the DNA backbone and the core histone proteins which do not vary from NCP to NCP. The ExoIII footprinting reactions show that AQ-157TG has only one translational setting in a NCP. This means that as counted from the 5′-AQ label, base pair 11 always resides near the entrance to the NCP, and there is no evidence for sliding of the DNA along the protein surface. In addition, the AQ-label and the proximal ∼10 bp of the duplex are out in solution and not associated with the histone octamer.
A comparison of the oxidative damage yields and distributions between AQ-157TG free and in rNCPs shows that there are several significant effects of chromatin packaging on DNA CT. The first effect is an overall decrease in piperidine-labile guanine oxidation products in the rNCPs when both free and rNCP samples are irradiated and subsequently treated under similar conditions. The decrease in Ydam in the rNCPs (Table 1) arises solely from interactions between AQ-157TG and the folded histone core since the removal of the histone tails by trypsin digestion had no effect on the rNCP CT reactions. From the reaction scheme illustrated in Figure 1B we can propose and discuss three hypotheses which could account for the observed attenuation of oxidative damage in the rNCPs. Hypothesis 1: the yield of charge injection into the NCP is decreased because AQ preferentially associates with the histone proteins. This hypothesis can be ruled out because (i) full activity of both ExoIII and OH• in the DNA proximal to AQ means AQ is not associating with the folded histone core and (ii) similar CT dynamics in both the native and tailless AQ-157TG rNCPs implies that AQ is not associating with the histone tails. Hypothesis 2: charge liberation is inefficient because O2 is restricted from oxidizing AQ•-. We reject this hypothesis because the DNA near the AQ is accessible to OH• generated in solution, and it is difficult to envision a scenario where OH• and O2 accessibility would differ so dramatically. Hypothesis 3: the rates of charge trapping at G•+ to form piperidine-labile lesions (ktrap) and/or the rates of hole transport between neighboring guanines (khop) are different between the rNCP and free DNA environments. The perturbation of either ktrap or khop by the formation of a rNCP is plausible for two reasons. First, khop may decrease because the DNA in the entrance to the nucleosome is overwound, and thus the bases are stacked closer together and have a slightly larger helical twist angle as compared to B-form DNA. From quantum chemical calculations (46,47), changes in either of these DNA structural features can perturb the relative energetics of the GG and G oxidation sites in AQ-157TG. According to Marcus theory, these changes in energetics could lead directly to changes in khop (48). Additionally, the change in the energetics of G•+ could also perturb ktrap (49). A second possibility would be changes in ktrap in the rNCPs arising from DNA–protein contacts restricting the accessibility of trapping reagents to G•+.
With just the Ydam values, we would be unable to ascertain whether the changes in structure or solvent accessibility were responsible for the changes in CT dynamics in the rNCPs. However, the differences in the site-specific relative yields of piperidine-labile damage in AQ-157TG rNCPs allow us to propose a clearer picture of how DNA packaging perturbs DNA CT in AQ-157TG. In the non-AQ counterstrand of AQ-157TG (BDF), the three sites of guanine oxidation are labeled GG1 (7 bp from AQ), G1 (16 bp from AQ) and GG2 (28 bp from AQ) (Figure 1C). While the damage ratio GG1/G1 is insensitive to DNA compaction, there are significant increases in the ratios GG1/GG2 and G1/GG2 when AQ-157TG is incorporated into a NCP (Table 1, Figure 4B). These results imply that there might be something specific occurring to GG2 in the rNCP which changes the relative khop and/or ktrap values associated with this site, but not sites GG1 or G1. Coupling together the ExoIII and OH• footprinting data in this paper with the previously reported high resolution crystal structure of a core particle (17), we can map out the structural environment of GG1, G1 and GG2 in AQ-157TG rNCPs (Figure 5). First GG1 is located in a DNA region resembling naked DNA since it does not associate with the histone proteins in the rNCPs. While both G1 and GG2 lie within the DNA region compacted into the NCP structure, both oxidation sites are located in regions where the DNA major groove is rotated out towards bulk solvent (Figure 5B). Since the solvent accessibility of G1 and GG2 appear to be so similar, we contend that changes in ktrap due to nucleobase accessibility plays a relatively minor role in the differences in the site damage yields observed between free and rNCP AQ-157TG.
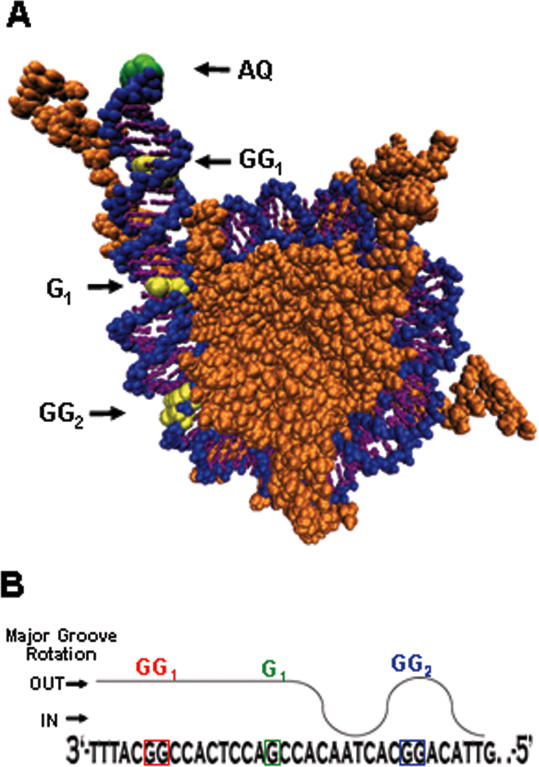
Figure 5.
(A) Crystal structure of the NCP (Protein Data Bank Accession no. 1KX5) with 147 bp of DNA (nucleobases = purple, sugar-phosphate backbone = blue) modified to include a 10 bp extension and a covalently attached AQ (green) end-capped onto the DNA. The histone octamer is in orange. The relative positions of the guanine oxidation sites GG1, G1, and GG2 of AQ-157TG are highlighted in yellow. (B) Illustration of the rotational position of the DNA major groove in the AQ-157TG rNCPs as deduced from Part A. The relative position of damage sites GG1, G1 and GG2 with respect to the histone protein surface is indicated.
There is a specific DNA–protein interaction in the AQ-157TG rNCPs which could be a possible explanation for the decrease in CT to site GG2. In NCP crystal structures, the histone H2A Arg77 guanidinium side chain is inserted into the DNA minor groove and causes a local kink in the DNA structure by making hydrogen bonds with phosphate groups from the base pairs 16 and 19 nt away from the NCP entrance (17). Mapping AQ-157TG onto the NCP crystal structure shows that GG2 lays dead center of this region, occupying base pairs 17 and 18 from the entrance of the NCP. This local structural change in the DNA near GG2 could perturb the energetics of this oxidation site and thus perturb either khop or ktrap. While the charge transfer attenuation here is not as dramatic as the Arg-induced diminution of DNA CT in the BamHI experiments of Saito et al. (22) it suggests that protein–DNA interactions can play subtle, but significant roles in controlling the observed distribution of DNA oxidative damage in protein–DNA complexes. Clearly, additional experimental and theoretical research is needed to address these structural effects further.
There is only one other report of photoinduced DNA CT reactions in the setting of a NCP (21). In contrast to the results obtained here, Nunez et al. found that there was little to no difference in DNA CT dynamics in a rNCP vs. naked DNA. There are several critical differences between these two studies which should be noted. First, the photooxidant used in each experiment is different in nature. Nunez et al. utilized an intercalating Rh-photoinjection system, whereas we have used the smaller end-capping AQ. We note that the absence of reported DNA footprinting experiments on the Rh-labeled rNCPs raises the possibility that their large organometallic photooxidant may have led to changes in rNCP structure, e.g. changes related to DNA aggregation phenomena known to arise from Rh-labeled DNA (32,50). A second difference is that the DNA sequences used in both studies are not the same. The prior report utilized the palindromic DNA sequence from the crystal structures of Luger et al. (51) whereas we have used DNA duplexes built around the TG motif. It may be that the differences between their studies and ours are due to differences in the thermodynamic stability and DNA rotational and/or translational freedom inherent in the two different DNA sequences utilized. If true, this would imply that the effects of NCP formation on DNA CT may vary between different genomic regions based upon the thermodynamic stability of the NCPs present. We are currently investigating this hypothesis by performing DNA CT studies on rNCPs formed from DNA sequences of varying thermodynamic stability and sequence. We also note that our observation of decreased DNA damage in both the AQ-157TG-NCP aggregates and the AQ-157TG rNCPs is similar to the results in a recent report by Das and Schuster (52) on DNA CT reactions in DNA–spermidine condensates. The similarities between these two studies indicates that one of the constant effects of in vivo DNA packaging, in all of its guises, may be to decrease DNA oxidative damage arising from long-range CT reactions.
The observation that the NCP can attenuate DNA CT is important from a biological standpoint because it indicates that chromatin may act as a protective barrier against long-range charge migration in genomic DNA sequences. In addition, it implies that oxidative damage may not occur randomly in genomic DNA during times of oxidative stress, but that it might arise from preferential trapping at guanines located in inter-nucleosomal linker regions and near the entrance to the nucleosome (53). This scenario would have important implications for DNA repair processes like Base Excision Repair (BER) which must find and repair nucleobase-derived oxidative lesions buried in the vast sea of chromatin in the eukaryotic nucleus. If oxidative damage occurs preferentially in the DNA linker regions, these sites may be easily repaired since they only make contact with the relatively unstructured tail regions of the core histones (41). Also, oxidative lesions in the first turns of the NCP may be easily repaired as well. Recent FRET measurements on reconstituted NCPs have shown that there is a periodic unwinding of the first couple of turns of DNA near the entrance of a nucleosome (53,54). This limited dissociation of the DNA–histone complex may be adequate to allow repair proteins access to any damage sites which are present in the entrance region of the nucleosome. Contrast these scenarios to the case of oxidative damage in the DNA more tightly associated with the histone octamer. Recent experiments by Beard et al. (55) on the efficiency of the removal of uracil residues in nucleosomal DNA by the BER repair protein Uracil deglycosylase have shown a 10 fold decrease in repair efficiency when a uracil is packaged into a rNCP. Therefore, keeping charge migration from leading to oxidative lesions deep in a NCP might be advantageous to the cell, since BER repair processes appear to be hindered by nucleosome packaging.
In conclusion, we have found significant differences in the dynamics of DNA-mediated hole transport in the presence and absence of packaging into a NCP. The NCPs utilized in this work are structurally well-characterized and this has allowed us to strongly correlate many of the changes in the guanine oxidative damage yields in the rNCP with the observed changes in DNA structure and environment arising from interaction with the core histone proteins. CT attenuation arises solely from interactions between the DNA and the folded core of the histone octamer since removal of the histone tails has no effect on DNA CT reactions in rNCPs. Clearly, future experimental and theoretical work will need to focus upon the origin of these changes in DNA hole transport dynamics, as well as focus upon the effects of higher order chromatin structure, such as the formation of the 30 nm fiber, on intracellular DNA hole transport reactions.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
Supplementary Material
Acknowledgments
The authors wish to thank Prof. Mick Smerdon and Drs. Brian Beard and Zeljko Svedruzic for supplying purified chicken erythrocyte NCPs, and their advice and assistance on the reconstitution and handling of NCPs. This material is based upon work supported by the National Science Foundation under CAREER Award #0347270 (WBD) and startup funds provided by Washington State University. Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Halliwell B., Gutteridge J.M.C. Free Radicals in Biology and Medicine. 3rd edn. Oxford: Oxford University Press; 1999. pp. 86–95. [Google Scholar]
- 2.Ames B.N., Shigenaga M.K., Hagen T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feig D.I., Reid T.M., Loeb L.A. Reactive oxygen species in tumorigenesis. Cancer Res. 1994;54(Suppl 7):1890–1894. [PubMed] [Google Scholar]
- 4.Olinski R., Gackowski D., Foksinski M., Rozalski R., Roszkowski K., Jaruga P. Oxidative DNA damage: assessment of the role in carcinogenesis, atherosclerosis, and acquired immunodeficiency syndrome. Free Radic. Biol. Med. 2002;33:192–200. doi: 10.1016/s0891-5849(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 5.Boon E.M., Barton J.K. Charge transport in DNA. Curr. Opin. Struct. Biol. 2002;12:320–329. doi: 10.1016/s0959-440x(02)00327-5. [DOI] [PubMed] [Google Scholar]
- 6.Joy A., Schuster G.B. Long-range radical cation migration in DNA: Investigation of the mechanism. Chem. Commun. 2005;14:2778–2784. doi: 10.1039/b500412h. [DOI] [PubMed] [Google Scholar]
- 7.Lewis F.D., Letsinger R.L., Wasielewski M.R. Dynamics of photoinduced charge transfer and hole transport in synthetic DNA hairpins. Acc. Chem. Res. 2001;34:159–170. doi: 10.1021/ar0000197. [DOI] [PubMed] [Google Scholar]
- 8.Giese B. Electron transfer in DNA. Curr. Opin. Chem. Biol. 2002;6:612–618. doi: 10.1016/s1367-5931(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 9.Behrens C., Cichon M.K., Grolle F., Hennecke U., Carell T. Excess electron transfer in defined donor-nucleobase and donor-DNA-acceptor systems. Topics. Curr. Chem. 2004;236:187–204. [Google Scholar]
- 10.Ito T., Rokita S.E. Reductive electron injection into duplex DNA by aromatic amines. J. Am. Chem. Soc. 2004;126:15552–15559. doi: 10.1021/ja045637n. [DOI] [PubMed] [Google Scholar]
- 11.Steenken S., Jovanovic S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 12.Shukla L.I., Adhikary A., Pazdro R., Becker D., Sevilla M.D. Formation of 8-oxo-7,8-dihydroguanine-radicals in gamma-irradiated DNA by multiple one-electron oxidations. Nucleic Acids Res. 2004;32:6565–6574. doi: 10.1093/nar/gkh989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jortner J., Bixon M., Langenbacher T., Michel-Beyerle M.E. Charge transfer and transport in DNA. Proc. Natl Acad. Sci. USA. 1998;95:12759–12765. doi: 10.1073/pnas.95.22.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C.-S., Hernandez R., Schuster G.B. Mechanism for radical cation transport in duplex DNA oligonucleotides. J. Am. Chem. Soc. 2004;126:2877–2884. doi: 10.1021/ja0378254. [DOI] [PubMed] [Google Scholar]
- 15.Henderson P.T., Jones D., Hampikian G., Kan Y., Schuster G.B. Long-distance charge transport in DNA: the phonon-assisted polaron-like hopping mechanism. Proc. Natl Acad. Sci. USA. 1999;96:8353–8358. doi: 10.1073/pnas.96.15.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunez M.E., Hall D.B., Barton J.K. Long-range oxidative damage to DNA: effects of distance and sequence. Chem. Biol. 1999;6:85–97. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 17.Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 angstrom resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 18.Widom J. Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct. 1998;27:285–327. doi: 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe A.P. Chromatin Structure and Function. San Diego: Academic Press, Inc.; 1992. [Google Scholar]
- 20.Rajski S.R., Barton J.K. How different DNA-binding proteins affect long-range oxidative damage to DNA. Biochemistry. 2001;40:5556–5564. doi: 10.1021/bi002684t. [DOI] [PubMed] [Google Scholar]
- 21.Nunez M.E., Noyes K.T., Barton J.K. Oxidative charge transport through DNA in nucleosome core particles. Chem. Biol. 2002;9:403–415. doi: 10.1016/s1074-5521(02)00121-7. [DOI] [PubMed] [Google Scholar]
- 22.Nakatani K., Dohno C., Ogawa A., Saito I. Suppression of DNA-mediated charge transport by BamHI binding. Chem. Biol. 2002;9:361–366. doi: 10.1016/s1074-5521(02)00119-9. [DOI] [PubMed] [Google Scholar]
- 23.Newman M., Strzelecka T., Domer L.F., Schildkraut I., Aggarwal A.K. Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 24.Thastrom A., Bingham L.M., Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone—DNA interactions and nucleosome positioning. J. Mol. Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 25.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc. Natl Acad. Sci. USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowary P.T., Widom J. New DNA sequence rules for high affinity binding to histone ocatmer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 27.Shrader T.E., Crothers D.M. Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasper S.M., Schuster G.B. Intramolecular photoinduced electron transfer to anthraquinones linked to duplex dna: the effect of gaps and traps on long-range radical cation migration. J. Am. Chem. Soc. 1997;119:12762–12771. [Google Scholar]
- 29.Libertini L.J., Small E.W. Salt-induced transitions of chromatin core particles studied by tyrosine fluorescence anisotropy. Nucleic Acids Res. 1980;8:3517–3534. doi: 10.1093/nar/8.16.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moyer R., Marien K., van Holde K., Bailey G. Site-specific aflatoxin B1 adduction of sequence-positioned nucleosome core particles. J. Biol. Chem. 1989;264:12226–12231. [PubMed] [Google Scholar]
- 31.Tullius T.D., Dombroski B.A., Churchill M.E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein–DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- 32.Fahlman R.P., Sharma R.D., Sen D. The charge conduction properties of DNA Holliday junctions depend critically on the identity of the tethered photooxidant. J. Am. Chem. Soc. 2002;124:12477–12485. doi: 10.1021/ja020495n. [DOI] [PubMed] [Google Scholar]
- 33.Williams T.T., Dohno C., Stemp E.D.A., Barton J.K. Effects of the photooxidant on DNA-mediated charge transport. J. Am. Chem. Soc. 2004;126:8148–8158. doi: 10.1021/ja049869y. [DOI] [PubMed] [Google Scholar]
- 34.Kasai H., Yamaizumi Z., Berger M., Cadet J. Photosensitized formation of 7,8-dihydro-8-Oxo-2′-deoxyguanosine (8-hydroxy-2′-deoxyguanosine) in DNA by riboflavin—a nonsinglet oxygen mediated reaction. J. Am. Chem. Soc. 1992;114:9692–9694. [Google Scholar]
- 35.Misiaszek R., Uvaydov Y., Crean C., Geacintov N.E., Shafirovich V. Combination reactions of superoxide with 8-Oxo-7,8-dihydroguanine radicals in DNA: kinetics and end products. J. Biol. Chem. 2005;280:6293–6300. doi: 10.1074/jbc.M412253200. [DOI] [PubMed] [Google Scholar]
- 36.Norman D.G., Grainger R.J., Uhrin D., Lilley D.M.J. Location of cyanine-3 on double-stranded DNA: importance for fluorescence resonance energy transfer studies. Biochemistry. 2000;39:6317–6324. doi: 10.1021/bi992944a. [DOI] [PubMed] [Google Scholar]
- 37.Sanii L., Schuster G.B. Long-distance charge transport in DNA: sequence-dependent radical cation injection efficiency. J. Am. Chem. Soc. 2000;122:11545–11546. [Google Scholar]
- 38.Negri R., Buttinelli M., Panetta G., De Arcangelis V., Di Mauro E., Travers A. Sequence dependence of translational positioning of core nucleosomes. J. Mol. Biol. 2001;307:987–999. doi: 10.1006/jmbi.2001.4546. [DOI] [PubMed] [Google Scholar]
- 39.Neubauer B., Horz W. Analysis of nucleosome positioning by in vitro reconstitution. Methods Enzymol. 1989;170:630–644. doi: 10.1016/0076-6879(89)70069-0. [DOI] [PubMed] [Google Scholar]
- 40.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 41.Hayes J.J., Tullius T.D., Wolffe A.P. The structure of DNA in a nucleosome. Proc. Natl Acad. Sci. USA. 1990;87:7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhalla V., Bajpai R.P., Bharadwaj L.M. DNA electronics—DNA and electronics seem to be two different things, but a series of events has highlighted the unusual ability of DNA to form electronic components. Embo Rep. 2003;4:442–445. doi: 10.1038/sj.embor.embor834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bloomfield V.A., Crothers D.M., Tinoco I., Jr . Nucleic Acids: Structures, Properties and Functions. Sausalito, CA, USA: University Science Books; 2000. [Google Scholar]
- 44.Cao H., Schuster G.B. Complex formation between a DNA duplex and lipid-like spermine derivatives: hydrophobic protection of DNA from one-electron oxidation. Bioconjugate Chem. 2005;16:820–826. doi: 10.1021/bc0500185. [DOI] [PubMed] [Google Scholar]
- 45.Wagenknecht H.-A., Stemp E.D.A., Barton J.K. DNA-bound peptide radicals generated through DNA-mediated electron transport. Biochemistry. 2000;39:5483–5491. doi: 10.1021/bi992897m. [DOI] [PubMed] [Google Scholar]
- 46.Voityuk A.A., Jortner J., Bixon M., Rosch N. Energetics of hole transfer in DNA. Chem. Phys. Lett. 2000;324:430–434. [Google Scholar]
- 47.Voityuk A.A., Michel-Beyerle M.E., Rosch N. Energetics of excess electron transfer in DNA. Chem. Phys. Lett. 2001;342:231–238. [Google Scholar]
- 48.Barbara P.F., Meyer T.J., Ratner M.A. Contemporary issues in electron transfer research. J. Phys. Chem. 1996;100:13148–13168. [Google Scholar]
- 49.Giese B., Spichty M. Long distance charge transport through DNA: quantification and extension of the hopping model. Chem. Phys. Chem. 2000;1:195–198. doi: 10.1002/1439-7641(20001215)1:4<195::AID-CPHC195>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 50.Olson E.J.C., Hu D.H., Hormann A., Barbara P.F. Quantitative modeling of DNA-mediated electron transfer between metallointercalators. J. Phys. Chem. B. 1997;101:299–303. [Google Scholar]
- 51.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 52.Das P., Schuster G.B. Effect of condensate formation on long-distance radical cation migration in DNA. Proc. Natl Acad. Sci. USA. 2005;102:14227–14231. doi: 10.1073/pnas.0506778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li G., Levitus M., Bustamante C., Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 54.Li G., Widom J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 55.Beard B.C., Wilson S.H., Smerdon M.J. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proc. Natl Acad. Sci. USA. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.