Abstract
Spo0J (ParB) of Bacillus subtilis is a DNA-binding protein that belongs to a conserved family of proteins required for efficient plasmid and chromosome partitioning in many bacterial species. We found that Spo0J contributes to the positioning of the chromosomal oriC region, but probably not by recruiting the origin regions to specific subcellular locations. In wild-type cells during exponential growth, duplicated origin regions were generally positioned around the cell quarters. In a spo0J null mutant, sister origin regions were often closer together, nearer to midcell. We found, by using a Spo0J-green fluorescent protein [GFP] fusion, that the subcellular location of Spo0J was a consequence of the chromosomal positions of the Spo0J binding sites. When an array of binding sites (parS sites) were inserted at various chromosomal locations in the absence of six of the eight known parS sites, Spo0J-GFP was no longer found predominantly at the cell quarters, indicating that Spo0J is not sufficient to recruit chromosomal parS sites to the cell quarters. spo0J also affected chromosome positioning during sporulation. A spo0J null mutant showed an increase in the number of cells with some origin-distal regions located in the forespore. In addition, a spo0J null mutation caused an increase in the number of foci per cell of LacI-GFP bound to arrays of lac operators inserted in various positions in the chromosome, including the origin region, an increase in the DNA-protein ratio, and an increase in origins per cell, as determined by flow cytometry. These results indicate that the spo0J mutant produced a significant proportion of cells with increased chromosome content, probably due to increased and asynchronous initiation of DNA replication.
All dividing cells are faced with the challenges of accurately replicating and partitioning their chromosomes so that each daughter cell receives one complete copy of the genome. Spo0J is a DNA-binding protein that contributes to the fidelity of chromosome segregation in Bacillus subtilis. A spo0J null mutant produces 1 to 2% anucleate cells, a frequency approximately 100-fold greater than that of wild-type cells (25). In B. subtilis, Spo0J is also required for sporulation, and this requirement is alleviated by a null mutation in soj (suppressor of spo0J). That is, an soj spo0J double mutant is able to sporulate, although the double mutant still has a defect in chromosome partitioning (25). Soj (ParA) and Spo0J (ParB) homologs are found in many nonsporulating bacteria, and for those that have been tested, one or both are involved in accurate chromosome partitioning (15, 28, 36, 42).
Soj (ParA) and Spo0J (ParB) belong to the well-conserved Par family of partitioning proteins. Members of this family are involved in both plasmid and chromosome segregation. When placed on a plasmid, the chromosomal systems can stabilize the plasmid (15, 37, 62). Our understanding of this system comes largely from studies of partitioning of the low-copy P1 prophage and F plasmid of Escherichia coli: ParA is an ATPase, and ParB is a DNA-binding protein that interacts with ParA and parS, the binding site for ParB (references 2, 4, 7, 8, 9, 17, and 21 and references therein).
One model for the function of the Par system proposes that Par proteins recruit parS-containing DNA to specific intracellular locations. This model is supported by the observations that a variety of plasmids are positioned at the cell quarters (3, 10, 16, 22, 29, 44, 46, 48) and that the cognate Par system is required for this localization (3, 10, 29, 44). Par systems are required for stable inheritance of these plasmids (reviewed in references 2, 8, 17, and 21), leading to the suggestion that positioning at the cell quarters is important for plasmid segregation (10, 16, 29, 44). In further support of the model that Par systems are involved in positioning, inserting a sopC site in an oriC plasmid (that is otherwise not positioned at the cell quarters) is sufficient to cause positioning of this plasmid at the cell quarters in the presence of SopA and SopB (46). Remarkably, insertion of chromosomally encoded soj, spo0J, and parS from B. subtilis into a plasmid in E. coli causes positioning of sister copies of the plasmid at the cell quarters (62).
We were interested in determining whether Spo0J contributes positional information to the chromosomal origin region in B. subtilis. Spo0J binds at least eight parS sites located within the origin-proximal 20% of the chromosome, spread over a distance of ≈800 kbp (37). The subcellular position of Spo0J is indicative of origin position (14, 33, 37, 38, 52, 53, 55, 56). In both E. coli and B. subtilis, following duplication, sister origins are separated to opposite halves of the cell, where they remain for the majority of the cell cycle (14, 16, 33, 37, 38, 45, 50, 52, 53, 55, 56).
Spo0J is thought to contribute to chromosome orientation in sporulating cells (51). Early during sporulation, replicated origins become positioned at the extreme poles of the cell (14, 18, 38, 51, 56). Cell division occurs very close to one of the poles, producing a forespore and a larger mother cell. The polar septum closes around the nucleoid so that the origin-proximal 30% of a chromosome becomes trapped in the forespore (58). The remainder of that chromosome is pumped from the mother cell into the forespore by SpoIIIE, a DNA translocase located in the septum (59, 61). There is some disagreement over the contribution (or lack thereof) of spo0J to faithful origin trapping in the forespore (12, 51). Furthermore, inactivation of spo0J appears to cause increased chromosomal copy numbers in cells that have been induced to sporulate, suggesting that trapping of genes outside the origin-proximal 30% of the chromosome could be due to random trapping of regions from extra chromosomes rather than a direct involvement of spo0J in establishing chromosome orientation (56).
To examine the contributions of Spo0J to origin positioning, we measured origin position in wild-type and spo0J mutant cells during vegetative growth and sporulation. Our results indicate that Spo0J contributes to the normal positioning of sister origins at the cell quarters during vegetative growth. In addition, Spo0J appeared to affect chromosome positioning during sporulation, but probably indirectly. We also found that the subcellular position of Spo0J was dependent on the chromosomal location of its binding sites. That is, an array of parS sites inserted at various locations in the chromosome, in the absence of six of the eight known binding sites, recruited Spo0J away from the cell quarters. Finally, we found that spo0J mutant cells were longer than wild-type cells and had an increased DNA content, most likely due to increased and asynchronous initiation of DNA replication.
MATERIALS AND METHODS
Media and growth conditions.
For all microscopy experiments, cells were grown at 30°C in S7 defined minimal medium (with MOPS [morpholinepropanesulfonic acid] buffer at 50 mM rather than 100 mM) supplemented with 1% glucose and 0.1% glutamate (26, 54) and required amino acids (tryptophan, 40 μg/ml; phenylalanine, 40 μg/ml; and threonine, 120 μg/ml) as needed. Sporulation was induced by the addition of mycophenolic acid to 30 μg/ml (13). Cultures for flow cytometry were grown at 30°C in antibiotic medium 3 (Difco Laboratories, Detroit, Mich.) supplemented with adenine and guanosine (each at 20 μg/ml). Routine growth and strain constructions were done on Luria-Bertani plates, and antibiotics were used at standard concentrations.
Strains, alleles, and plasmids.
B. subtilis strains used in this study are listed in Table 1. Most are derivatives of AG174 (JH642) and contain the trpC and pheA mutations, unless indicated otherwise. Standard procedures were used for strain constructions (20).
TABLE 1.
B. subtilis strains
| Strain | Relevant genotype or characteristics | Reference |
|---|---|---|
| DCL629 | spo0J-gfp (parS mutation in spo0J) ΔparS6 | |
| DCL631 | cgeD (181°)::pDL141 (parS16 cat) spo0J-gfp (parS mutation in spo0J) ΔparS6 | |
| DCL696 | yyaC (359°)::pDL175 (lacO cassette cat) thrC::(lacI-gfp mls) | |
| DCL705 | Δspo0J::spc yyaC (359°)::pDL175 (lacO cassette cat) thrC::(lacI-gfp mls) | |
| IRN424 | rtp::pGK97 (rtp-yfp spc) | 32 |
| KPL686 | cgeD (181°)::pAT14 (lacO cassette cat) thrC::(lacI-cfp mls) | 32 |
| KPL716 | cotS (270°)::pAT25 (lacO cassette cat) thrC::(lacI-cfp mls) | 31 |
| KPL718 | yheH (90°)::pAT26 (lacO cassette cat) thrC::(lacI-cfp mls) | |
| PSL10 | spo0J::pDL50B (spo0J-gfp cat) | |
| PSL23 | cotS (270°)::pPSL1A (parS16 cat) spo0J-gfp (parS mutation in spo0J) ΔparS6 | |
| PSL25 | yheH (90°)::pPSL2A (parS16 cat) spo0J-gfp (parS mutation in spo0J) ΔparS6 | |
| PSL27 | yqkF (210°)::pPSL3A (parS16 cat) spo0J-gfp (parS mutation in spo0J) ΔparS6 | |
| PSL62 | yyaC (359°)::pDL175 (lacO cassette cat) thrC::(lacI-gfp mls) ΔspoIIIE::tet | |
| PSL64 | cgeD (181°)::pAT14 (lacO cassette cat) thrC::(lacI-cfp mls) ΔspoIIIE::tet | |
| PSL66 | cotS (270°)::pAT25 (lacO cassette cat) thrC::(lacI-cfp mls) ΔspoIIIE::tet | |
| PSL73 | Δ(soj-spo0J)::spc yyaC (359°)::pDL175 (lacO cassette cat) thrC::(lacI-gfp mls) ΔspoIIIE::tet | |
| PSL74 | Δ(soj-spo0J)::spc cgeD (181°)::pAT14 (lacO cassette cat) thrC::(lacI-cfp mls) ΔspoIIIE::tet | |
| PSL76 | Δ(soj-spo0J)::spc cotS (270°)::pAT25 (lacO cassette cat) thrC::(lacI-cfp mls) ΔspoIIIE::tet | |
| PSL101 | Δspo0J::spc cotS (270°)::pAT25 (lacO cassette cat) thrC::(lacI-cfp mls) | |
| PSL110 | Δspo0J::spc cgeD (181°)::pAT14 (lacO cassette cat) thrC::(lacI-cfp mls) | |
| CRK6000 | purA16 metB5 hisA3 guaB | 43 |
| NIS6074 | CRK6000 spo0J::ermC |
(i) spo0J and rtp.
The spo0J-gfp (38) and rtp-yfp (32) fusions were described previously. In each case, the fusion is driven from the endogenous promoter, is the only expressed copy of the gene, and is mostly functional. pDL152 contains mls (encoding erythromycin and lincomycin resistance) and the 3′ end of spo0J with the parS site inactivated (37) fused to the gene for green fluorescent protein (gfp). It was integrated into the chromosomal spo0J to generate the spo0J-gfp fusion.
Δspo0J::spc and Δ(soj-spo0J)::spc are deletion-insertion mutations from strains AG1468 and AG1505, respectively (25). spo0J::ermC is a null allele with ermC inserted into the BglII site in spo0J.
(ii) ΔspoIIIE::tet.
A spoIIIE null allele was constructed by replacing all of the spoIIIE coding region with a tetracycline resistance cassette (tet). A ≈1,000-bp fragment immediately upstream of spoIIIE was amplified by PCR with oligonucleotides LEE-19 and LEE-20, which added NheI and XhoI restriction sites at the ends of the fragment. A ≈1,000-bp fragment immediately downstream of spoIIIE was amplified with oligonucleotides LEE-21 and LEE-22, which added PstI and BamHI sites at the ends of the fragment. These fragments were cloned into the corresponding restriction sites on either side of the tet gene in pDG1513 (19), creating the spoIIIE knockout plasmid pPSL5. ΔspoIIIE::tet was introduced by transformation into the B. subtilis chromosome, with selection for resistance to tetracycline and a defect in sporulation. PCR was used to confirm that the ΔspoIIIE::tet allele was integrated by double crossover.
(iii) lac operator system.
LacI-green fluorescent protein (GFP) and LacI-cyan fluorescent protein (CFP) fusions were used to visualize regions of the chromosome marked with arrays of lac operators. The lacI-gfp and lacI-cfp fusions are driven by a constitutive promoter, are integrated into thrC, and have been described in detail (31, 32). Briefly, lacI is missing the C-terminal 11 codons (to inhibit tetramerization) and is fused to either gfpmut2 (lacI-gfp) or cfpw7 (lacI-cfp).
The lacO cassette was inserted into several regions of the chromosome (90°, 181°, 270°, and 359°) by homologous recombination. To insert the lacO cassette into the 359° region of the chromosome, oligonucleotides LIN-145 and LIN-146 were used to amplify a ≈660-bp fragment from yyaC (immediately downstream from spo0J). The PCR product was digested with AatII and BglII, and the resulting ≈380-bp fragment was inserted into the lacO cassette-containing plasmid pAT12 (56), creating pDL175. Plasmid pDL175 (which cannot replicate in B. subtilis) was integrated into the B. subtilis chromosome by single crossover, selecting for resistance to chloramphenicol. Chromosomal DNA from a transformant was used to introduce the array into additional strains. The lacO cassettes at 90°, 181°, and 270° were transformed into appropriate strains with chromosomal DNA from AT53, AT54, and AT52, respectively (53). The cassettes were amplified by selecting for resistance to chloramphenicol (25 μg/ml) as described previously (53, 56).
(iv) parS arrays.
pDL139 contains 16 tandemly repeated parS sites (parS16) and was used as the parent plasmid for targeting the parS array into various chromosomal sites. It was constructed by a strategy similar to that used to construct the lacO cassette (49), taking advantage of the compatible cohesive ends of SalI and XhoI and loss of both sites upon ligation. First, a plasmid (pDL135) was constructed with an approximately 60-bp fragment from spo0J that contains the 16-bp parS site internal to spo0J. Single-stranded oligonucleotides LIN116 (65 nucleotides) and LIN117 (61 nucleotides) were annealed, giving a double-stranded oligonucleotide with a single parS site approximately in the middle, one blunt end with an XhoI site, and a SalI overhang at the other end. This largely double-stranded oligonucleotide was ligated into pGEMcat (20) that had been digested with SmaI and SalI to generate pDL135. An EcoRI-SalI fragment from this plasmid was then cloned back into pDL135 that had been digested with EcoRI and XhoI, generating pDL136, containing two parS sites. This cycle was repeated three more times, each time with the new plasmid as the source of the insert (EcoRI to SalI) and vector (cloning between EcoRI and XhoI) to generate pDL137 (four parS sites), pDL138 (eight parS sites), and finally pDL139 (parS16). The parS16 array was able to stabilize an otherwise unstable low-copy plasmid (data not shown) to an extent indistinguishable from that previously determined for a single parS site (37), and plasmid stability was dependent on both soj and spo0J (data not shown).
The parS16 array was targeted to specific chromosomal sites by cloning fragments from the location of interest into a plasmid that had the parS16 array. Plasmids were then integrated into the chromosome by single-crossover homologous recombination into the targeted location. The positions targeted were identical to or within few hundred base pairs of the regions in which the lac operator arrays had been inserted.
For the terminus region, the parS16 array was targeted downstream of cgeD at 181° with pDL141, a derivative of pET21(+) (Novagen), which contains a selectable marker for B. subtilis (cat), a fragment from cgeD, and the parS16 array. The chloramphenicol acetyltransferase (cat) gene was excised from pMI1101 (63) and inserted into the SphI site of pET21(+), generating pET21cat. An approximately 490-bp fragment from cgeD was amplified by PCR (with oligonucleotides LIN118 and LIN119, containing AatII sites) and cloned into the AatII site of pET21cat, generating pDL124. The parS16 array was excised from pDL139 by digestion with EcoRI and HindIII and ligated into EcoRI- and HindIII-digested pDL124 to create plasmid pDL141.
Derivatives of pDL139 were made to target the parS16 array to 90° (pPSL2A), 210° (pPSL3A), and 270° (pPSL1A). In each case, a fragment of chromosomal DNA was amplified by PCR with primers containing EcoRI sites near the ends and cloned into the EcoRI site of pDL139. pPSL1A (parS16 array at 270°) contains a 994-bp fragment from the 3′ end of cotS, generated by PCR with primers LEE-8 and LEE-9. pPSL2A (parS16 array at 90°) contains a 673-bp fragment from the 3′ end of yheH, generated by PCR with primers LEE-10 and LEE-11. pPSL3A (parS16 array at 210°) contains a 510-bp fragment from the 3′ end of yqkF, generated by PCR with primers LEE-12 and LEE-13.
(v) ΔparS6 mutant.
Plasmids containing the parS16 array were introduced into strain DCL629, which contains a spo0J-gfp fusion created by integrating pDL152 into the spo0J locus and has six of the eight known endogenous parS sites inactivated, referred to as ΔparS6 (Table 1). DCL629 was constructed in a manner similar to that described previously for strain DCL484, with six of the eight known Spo0J binding sites inactivated (37). Five of the parS mutations are deletion-insertions and are marked with resistance cassettes for spectinomycin (spc), kanamycin (kan), phleomycin (phl), tetracycline (tet), and erythromycin and lincomycin (mls). The parS site in spo0J was inactivated by changing 7 bp of the 16-bp binding site without altering the gene product (37).
Fluorescence microscopy.
Where indicated, cells were stained with the vital dye FM4-64 (200 ng/ml; Molecular Probes) to visualize membranes and 4′,6′-diamidino-2-phenylindole (DAPI) (40 to 80 ng/ml) to visualize nucleoids. Microscopy was performed essentially as described previously (30, 31). Briefly, cells were immobilized on pads of 1% agarose in 1× T'base-1 mM MgSO4 (20), and images were captured with a Nikon E800 microscope equipped with a Hamamatsu digital camera and filters tetramethylrhodamine isothiocyanate for FM4-64 and the chroma filter set at 31044 for CFP and 41012 for GFP. Improvision OpenLabs 2.0 software was used to process images.
Measurement of focus position in cells with two foci.
Focus positions in cells with two foci were measured with a strategy designed to eliminate any unintentional visual bias in scoring. For each cell, the focus closest to a pole was designated focus A, and the other focus was designated focus B. Three measurements were made: (i) a, the distance from the center of focus A to the closest pole; (ii) b, the distance from that same pole to the center of focus B; and (iii) l, the cell length. This created a systematic bias in scoring so that in every cell, focus A was closer to a pole than focus B. In order to remove this bias, a random number generator was used to assign each focus an approximately 50% chance of being counted as closest to a pole. The corresponding distance of each focus from the same cell pole (the a and b measurements) was then recalculated.
The focus position as a percentage of cell length was determined with the measurements described above. We determined the distance of each focus from the nearest cell pole (a for focus A; l minus b for focus B), divided by cell length, and multiplied by 100 to give the focus position as a percentage of cell length. These numbers were averaged for all the cells in a sample. The 95% confidence intervals for the mean were calculated with the approximation that, for large samples (n > 30), which cannot be assumed to be normally distributed, the 95% confidence interval is approximately equal to the sample mean ± 1.96 times the standard error of the mean (23). This is an indication of confidence in the mean and not an indication of the breadth of the distribution.
Interfocal distance was calculated by subtracting a from b. The correlation coefficient between cell length and interfocal distance was calculated with the least-squares method of linear regression (23).
Flow cytometry.
Cells were grown at 30°C in antibiotic medium 3 (Difco Laboratories, Detroit, Mich.) supplemented with adenine and guanosine (both at 20 μg/ml). Flow cytometry was performed essentially as described previously (47). Briefly, chloramphenicol (200 μg/ml) was added to cells to inhibit protein synthesis and block initiation of new rounds of replication. Cells were incubated for 5 h to allow ongoing rounds of replication to finish. Next, cells were collected, fixed with ethanol, and treated with a mixture of fluorescent dyes, mithramycin A, and ethidium bromide. The amount of DNA per cell was measured with a Bryte HS flow cytometer (Bio-Rad Laboratories).
Measurement of DNA-protein ratios.
The DNA-protein ratio was determined essentially as described previously (27). Briefly, 30 ml of exponentially growing cells was collected by centrifugation, resuspended, lysed, and separated into nucleic acid and protein fractions. DNA and protein concentrations in the fractions were measured with colorimetric methods as described before (6, 39). Reported values are averages from three to four experiments, followed by the standard deviations from the mean.
RESULTS AND DISCUSSION
Replicated origins are positioned at or near the cell quarters.
We measured the intracellular position of origin regions (0°/360° on the circular chromosome) along the length of cells during exponential growth. The origin region was visualized with either Spo0J-GFP bound to endogenous origin-proximal parS sites (Fig. 1A) or LacI-GFP bound to an array of lac operators inserted at 359°. Under the growth conditions used, the majority of cells (≈84%) had two spatially resolved foci of the origin region, as visualized with LacI-GFP bound to lac operators at 359° (Table 2). When origin position was plotted against cell length for cells with two foci, origins appeared to be distributed around the cell quarters with both Spo0J-GFP and the lacO/LacI-GFP systems (Fig. 2A and 2B; Table 3). Average origin positions, relative to the nearest pole (± the 95% confidence interval for the mean) were at or near the cell quarters: 27.9% ± 1.4% and 26.1% ± 1.0% of cell length for foci of Spo0J-GFP and LacI-GFP, respectively (Table 3). There was no significant difference in the average positions determined by the two methods, consistent with the previous finding that Spo0J usually colocalizes with LacI-GFP bound to lac operators inserted at 359° (53).
FIG. 1.
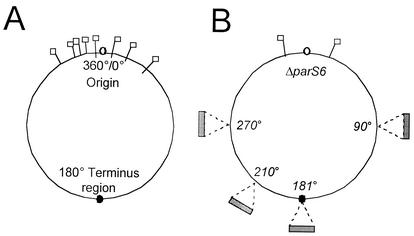
(A) B. subtilis chromosome with parS sites shown. B. subtilis has a circular chromosome with the origin of replication at 0°/360° and the terminus region at 180°. Spo0J binds eight known parS sites located in the origin-proximal 20% of the chromosome (37). The parS sites are located at 330°, 354°, 355°, 356°, 359°, 4°, 15°, and 40° and are indicated by flags. (B) Strains with an array of 16 parS sites inserted in different regions of the chromosome (90°, 181°, 210°, and 270°), indicated by the shaded rectangles, were constructed. In each strain, six of the eight known endogenous parS sites were inactivated. The remaining parS sites were located at 355° and 15°, indicated by flags.
TABLE 2.
Number of foci per cell of various chromosomal regions during exponential growth in wild-type and spo0J mutant cellsa
| Strain | Insertion region | spo0J alleleb | % of cells with indicated no. of foci
|
No. of cells analyzed | |||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | >4 | ||||
| DCL696 | 359° | + | 1.2 | 1.9 | 83.8 | 2.9 | 10.0 | 0.2 | 421 |
| DCL705 | − | 4.4 | 15.3 | 44.9 | 20.1 | 11.2 | 4.1 | 517 | |
| KPL716 | 270° | + | 0.2 | 31.8 | 64.5 | 2.1 | 1.2 | 0.2 | 515 |
| PSL101 | − | 3.5 | 20.4 | 60.5 | 9.0 | 6.1 | 0.6 | 491 | |
| KPL686 | 181° | + | 0.6 | 74.2 | 24.9 | 0.4 | <0.2 | <0.2 | 511 |
| PSL110 | − | 1.4 | 50.0 | 46.4 | 0.9 | 1.4 | <0.5 | 222 | |
An array of lac operators were inserted in the indicated regions of the chromosome and visualized with LacI-GFP or LacI-CFP. The strains were grown in defined minimal medium, and samples were taken during exponential growth. The number of foci of LacI-GFP (or LacI-CFP) per cell was determined, and the percentage of cells with the indicated number of foci was calculated.
+, wild type; −, null mutation.
FIG. 2.
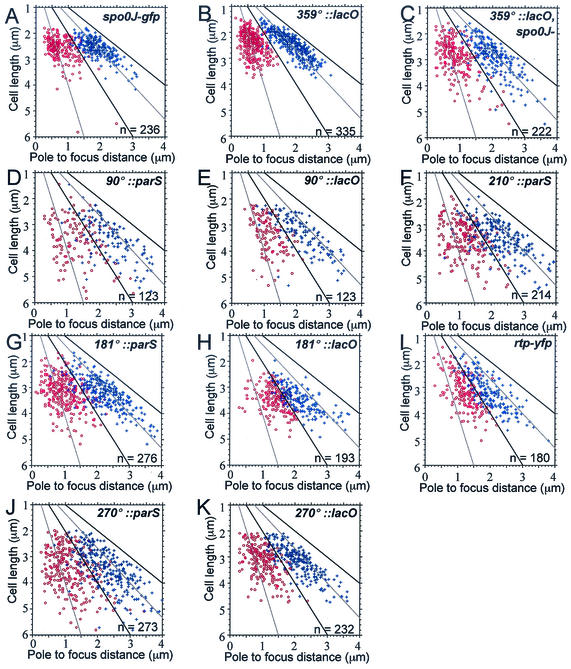
Locations of sister copies of various regions of the chromosome. Strains producing Spo0J-GFP, LacI-GFP, LacI-CFP, or Rtp-YFP were grown at 30°C in defined minimal medium. Only cells with two foci of the region of interest were analyzed. The distance from each focus to the same cell pole was measured from images of live cells in exponential growth (Materials and Methods) and is plotted on the x axis, and cell length is plotted on the y axis. Cell length and the midcell positions are indicated by solid lines. Cell quarter positions are indicated by dotted lines. The number of cells analyzed (n) is indicated in the lower right corner of each panel. One focus is indicated with red open circles and the other with blue crosses. (A) Position of Spo0J-GFP bound to endogenous parS sites (strain PSL10). (B, C) Position of sister origin regions (359°) visualized with LacI-GFP bound to lac operator arrays in spo0J+ (strain DCL696) and spo0J mutant (strain DCL705) cells. (D, E) Position of Spo0J-GFP bound to a parS array (D, strain PSL25) or LacI-CFP bound to a lac operator array (E, strain KPL718) inserted at 90°. (F) Position of Spo0J-GFP bound to a parS array inserted at 210° (strain PSL27). (G, H) Position of Spo0J-GFP bound to a parS array (G, strain DCL631) or LacI-CFP bound to a lac operator array (H, strain KPL686) inserted at 181°. (I) Position of Rtp-YFP bound to endogenous ter sites (strain IRN424). (J, K) Position of Spo0J-GFP bound to a parS array (J, strain PSL23) or LacI-CFP bound to a lac operator array (K, strain KPL716) inserted at 270°.
TABLE 3.
Subcellular positions of sister copies of various regions of the chromosomea
| Strain | Region and genotype | % of cell length ± 95% CI
|
No. of cells analyzedb | |
|---|---|---|---|---|
| Focus position | Interfocal distance | |||
| PSL10 | spo0J-gfp | 27.9 ± 1.4 | 44.3 ± 1.5 | 236 |
| DCL696 | 359°::lacO | 26.1 ± 1.0 | 47.8 ± 1.2 | 335 |
| DCL705 | 359°::lacO Δspo0J | 30.1 ± 1.5 | 39.7 ± 1.8 | 222 |
| PSL23 | 270°::parS | 33.5 ± 1.6 | 33.0 ± 1.6 | 273 |
| KPL716 | 270°::lacO | 32.6 ± 1.3 | 34.8 ± 1.6 | 232 |
| PSL25 | 90°::parS | 34.5 ± 2.5 | 31.1 ± 2.4 | 123 |
| KPL718 | 90°::lacO | 34.9 ± 1.9 | 30.2 ± 2.0 | 123 |
| PSL27 | 210°::parS | 34.5 ± 1.4 | 31.0 ± 1.7 | 214 |
| DCL631 | 181°::parS | 31.5 ± 1.5 | 37.0 ± 1.6 | 276 |
| KPL686 | 181°::lacO | 38.6 ± 1.4 | 22.8 ± 1.7 | 193 |
| IRN424 | rtp-yfp | 38.3 ± 1.5 | 23.4 ± 1.7 | 180 |
The indicated strains were grown in defined minimal medium, and samples were taken during exponential growth. Spo0J-GFP was used to visualize the subcellular location of parS arrays, and LacI-GFP (or LacI-CFP) was used to visualize lac operator arrays. The arrays were integrated into the chromosome in the indicated regions. For strains PSL10 (spo0J-gfp) and IRN424 (rtp-yfp), there were no ectopic arrays, and Spo0J-GFP was bound to the endogenous parS sites in the origin region (37, 38) and Rtp-YFP was bound to endogenous ter sites in the terminus region (32). The distance of each focus from the nearest cell pole was measured in cells with two foci and is presented as a percentage of cell length ± the 95% confidence intervals (CI). The distance between foci (interfocal distance) was determined as a percentage of cell length in cells with two foci.
The total number of cells with two foci that were analyzed.
We also analyzed the distance between foci (interfocal distance) and compared the interfocal distance to cell length. With either Spo0J-GFP (Fig. 2A) or LacI-GFP (Fig. 2B) bound to lac operators at 359° to visualize origin regions, the interfocal distance between replicated origins increased as cell length increased (Fig. 3A and 3B), consistent with the notion that sister origin regions are positioned relative to cell length. This indicates that origins are not positioned by a segregation apparatus of fixed length and contrasts with a previous report indicating that the distance between Spo0J-GFP foci is fixed in B. subtilis (52). We do not understand the reasons for this discrepancy, but it could be due to differences in the Spo0J-GFP fusions, strain backgrounds, or growth conditions.
FIG. 3.
Relationship between cell length and interfocal distance. Data from Fig. 2A to C were used to determine the distance between the two foci in each cell (the interfocal distance). This is plotted as a function of cell length. (A) Spo0J-GFP from Fig. 2A. (B) LacI-GFP in spo0J+ cells from Fig. 2B. (C) LacI-GFP from spo0J mutant cells from Fig. 2C. The correlation coefficient, r, which measures how strongly the interfocal distance and cell length are correlated, is shown for each plot (r = 1 for two perfectly, positively correlated variables). An ellipse was drawn around the distribution of wild-type cells (B), and this ellipse was superimposed on the corresponding plots in panels A andC. The interfocal distances in most of the spo0J mutant cells were within this ellipse; a subset (≈15%) fell below and to the right of the wild-type distribution. These cells had replicated origins that were closer together than the origins in wild-type cells of similar length.
Replicated origins are closer together in spo0J null cells.
We compared the positions of replicated origins in a large number of spo0J mutant cells to those of wild-type cells. Origin regions were visualized with LacI-GFP bound to a lacO cassette inserted at 359°. The average position of the origin regions in spo0J null mutants was 30.1% ± 1.5% of cell length from the nearest pole (Fig. 2C; Table 3). This gives an average interfocal distance of 39.7% ± 1.8% of cell length in the spo0J mutant (Table 3). In contrast, the average distance between sister origin regions in wild-type cells was 47.8% ± 1.2% of cell length (Table 3).
The interfocal distance was not as tightly correlated with cell length in the spo0J mutant (Fig. 3C) as in wild-type cells (Fig. 3B). A subset of spo0J null cells had replicated origins that were closer together than origins in wild-type cells of similar lengths. The smaller interfocal distance in this subset of cells probably accounts for the smaller average interfocal distance in the spo0J mutant. The subset of cells having aberrantly closely spaced origins was approximately 15% of the spo0J null cells, estimated from the number of cells falling below the wild-type distribution (Fig. 3B, C). This is greater than the proportion (1 to 2%) of anucleate cells produced by a spo0J null mutant, indicating that most of the cells with mispositioned origin regions can still successfully partition sister chromosomes. In fact, the vast majority of spo0J mutant cells still manage to separate sister origins to opposite halves of the cell (Fig. 2C).
Following duplication, origin regions move toward opposite halves of the cell in both B. subtilis and E. coli (14, 16, 33, 37, 38, 45, 50, 52, 53, 55, 56). Our data indicate that in B. subtilis, the sister origin regions are then positioned at or near the cell quarters. The defect in positioning in the spo0J null mutant could reflect a defect in: (i) recruiting the origins to the quarters, (ii) separating newly duplicated sister origins, or (iii) maintaining origin regions at the cell quarters after they are initially positioned there. These possibilities are not mutually exclusive. Results presented below indicate that Spo0J is not sufficient to recruit chromosomal binding sites to the cell quarters, leading us to favor a role for Spo0J in separation of sister origin regions or in maintenance of sister origins at the cell quarters.
parS arrays at 90°, 181°, 210°, and 270° are not recruited to the cell quarters.
A simple model for the function of ParB proteins is that they recruit their cognate binding sites to a characteristic subcellular position (29). This model can account for many observations regarding the plasmid Par systems and the fact that chromosomal systems can function to stabilize a plasmid (15, 37, 62) and position plasmids at the cell quarters (62). In this view, Spo0J could be positioned at the cell quarters and recruit or tether its binding sites there. To test this model, we measured the subcellular position of Spo0J-GFP bound to an array of 16 parS sites (parS16) inserted into different regions of the chromosome (90°, 181°, 210°, and 270°) in cells with six of the eight known endogenous parS sites inactivated (Fig. 1B) (37).
In cells with the parS16 array inserted in the 270° region of the chromosome, the subcellular position of Spo0J-GFP was different from that of the origin region (Fig. 2A, B, and J; Table 3). Spo0J-GFP foci in cells with the parS16 array at the 270° region showed a broader distribution than origin foci and were positioned 33.5% ± 1.6% of cell length from the nearest pole (Table 3). The results indicate that Spo0J is not sufficient to recruit parS sites at the 270° region to the cell quarters.
We also measured the position of Spo0J-GFP bound to ectopic parS sites inserted at three other regions in the chromosome, 90°, 181°, and 210° (Fig. 1B). None of these regions were recruited to the cell quarters (Fig. 2D, G, and F; Table 3), and in each case, the subcellular position of Spo0J-GFP was statistically distinguishable from that of the origin at the cell quarters. Again, these results indicate that Spo0J is not sufficient to recruit its binding sites to the cell quarters.
Subcellular position of Spo0J-GFP bound to parS16 arrays at 90° and 270° is a reflection of the normal position of each chromosomal region.
For the 270°, 90°, and 181° regions, we compared the positioning of Spo0J-GFP bound to the parS16 array to that of the normal position of these regions as visualized with LacI-CFP bound to an array of lac operators inserted at the same chromosomal region (see Materials and Methods).
The subcellular position of Spo0J-GFP bound to parS sites at 90° and 270° appeared to be a reflection of the normal subcellular position of those regions. The average position of LacI-CFP bound to an array of lac operators inserted at 90° was 34.9% ± 1.9% of cell length (Fig. 2E; Table 3). This was statistically indistinguishable from the average position of Spo0J-GFP bound to the parS16 array inserted at 90° (Fig. 2D; Table 3). Similarly, the average position of LacI-CFP bound to an array of lac operators at 270° was 32.6% ± 1.3% of cell length (Fig. 2K; Table 3), statistically indistinguishable from the average position of Spo0J-GFP bound to the parS16 array at 270° (Fig. 2J; Table 3). These results indicate that not only is Spo0J insufficient to recruit its binding sites to the cell quarters, but that the subcellular position of Spo0J is largely determined by the subcellular position of its binding sites, at least for the 90° and 270° regions.
In contrast to our findings with the 90° and 270° regions, the subcellular position of Spo0J-GFP bound to the parS array at 181° was significantly different from the subcellular position of LacI-CFP bound to lac operators at that position. In cells with two foci, the foci of Spo0J-GFP bound to the parS16 array inserted at 181° were farther apart than those of LacI-CFP bound to lac operators inserted at the same position (Fig. 2G, H; Table 3). The average focus positions were 31.5% ± 1.5% versus 38.6% ± 1.4% of cell length for Spo0J-GFP and LacI-CFP, respectively. We also measured the position of the terminus region with a fusion of yellow fluorescent protein (YFP) to the replication termination protein Rtp. Rtp binds to multiple sites in the terminus region (34, 35) and has been used to visualize the subcellular position of this region (32).
We found that the subcellular position of Rtp-YFP was similar to that determined for the 181° region determined with the lac system (Fig. 2H, I; Table 3), consistent with previous findings (32). These results indicate that the position of the terminus region is perturbed in cells with Spo0J bound to an array of parS sites at 181°. The terminus and origin regions are different from other regions of the chromosome in that they both appear to interact, either directly or indirectly, with factors associated with the bacterial membrane (11, 24, 57, 59). It is possible that Spo0J bound to the parS array inserted at 181° interferes with endogenous positional information or that the parS array titrates Spo0J away from other sites that affect, either directly or indirectly, the positioning of the terminus region.
Effects of spo0J on positioning various regions of the chromosome in the forespore.
During sporulation, the origin-proximal 30% of one chromosome is initially positioned in the forespore, with the remaining ≈70% in the larger mother cell (58). The mother cell also contains an intact chromosome, which is essential for sporulation. Loss-of-function mutations in spoIIIE trap the sporangium (forespore plus mother cell) in this situation by preventing the translocation of the remainder of the chromosome into the forespore (59, 61). Previous findings were interpreted to indicate that Spo0J might provide a “centromere-like” function and play a role in positioning origin regions in the forespore during the early stages of sporulation (51). The experiments involved monitoring expression of genes that are induced only in the forespore and placing the reporter gene in different chromosomal locations. The findings were interpreted to indicate that a spo0J null mutation causes a defect in orienting the duplicated chromosomes so that origin regions were sometimes excluded from the forespore and other regions were inappropriately positioned in the forespore (51).
These results and interpretations were called into question by a somewhat different type of analysis investigating the effects on sporulation of the transient genetic asymmetry between the forespore and the mother cell (12). These results indicated that the 174° and 192° regions of the chromosome were not positioned in the forespore in a spo0J mutant, apparently contradicting the earlier report (51). Finally, it has been suggested that during sporulation, some spo0J mutant cells have an increased number of chromosomes, and parts of these might be randomly trapped in the forespore and not reflect a role for Spo0J in positioning origin regions to the pole (56).
In order to measure the effects of Spo0J on positioning of the origin region in the forespore, we visualized the 359° (Fig. 4A to E) and the 181° and 270° regions of the chromosome in sporulating cells. Regions of the chromosome were visualized with LacI-GFP or LacI-CFP bound to an array of lac operators inserted in the region of interest. Deleting spo0J causes a sporulation defect that is suppressed by deleting soj (25). Therefore, as in previous reports (12, 51, 56), comparisons were made between wild-type and soj spo0J double mutants. In addition, we used strains that contained a null mutation in spoIIIE, the gene necessary for transport of the origin-distal 70% of the chromosome into the forespore (59, 61). A spoIIIE null mutation prevents translocation of the entire chromosome into the forespore and allows visualization of the region that is trapped. Images of live cells were captured 4 h after the induction of sporulation, and cells that had undergone polar septation were analyzed. The number of foci located in the forespore and mother cell was determined for each chromosomal region of interest (Table 4).
FIG. 4.
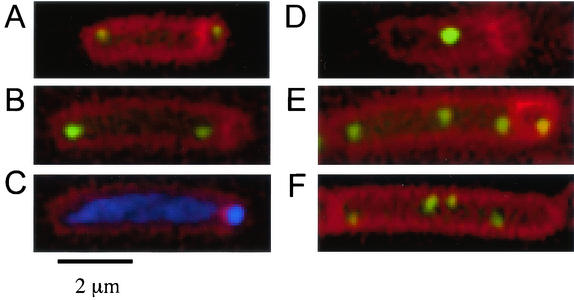
Visualization of chromosomal regions during sporulation and growth. (A to E) Cells were grown in defined minimal medium and induced to sporulate by the addition of mycophenolic acid. Samples were taken 4 h after the initiation of sporulation, and membranes were stained with FM4-64. The positions of origin regions were visualized with LacI-GFP bound to an array of lac operators inserted at 359°. Note that for cells with two or more foci, there was no detectable effect of spo0J on the frequency of positioning an origin region in the forespore (see text). Images illustrate some of the different types of sporangia observed. (A) Typical origin position in spo0J+ cells (strain PSL62 spo0J+ ΔspoIIIE::tet). (B to E) spo0J mutant cells [strain PSL73 Δ(soj-spo0J)::spc ΔspoIIIE::tet] with two origin foci located in the mother cell (B) and the chromosomal DNA visualized with DAPI (C), indicating that there is DNA in the forespore. (D) A spo0J mutant cell with a single focus of the origin region that is excluded from the forespore. (E) A spo0J mutant cell with four copies of the origin region, one of which is in the forespore. (F) Increased copies of the terminus region, visualized with LacI-CFP bound to an array of lac operators inserted at 181°, in a spo0J mutant (strain PSL110) during exponential growth.
TABLE 4.
Effect of spo0J on positioning chromosomal regions in the foresporea
| Strain | Region | soj spo0Jb | % (no.) of sporangia with region in foresporec | No. of sporangia analyzed |
|---|---|---|---|---|
| PSL66 | 270° | + | <0.28 (0) | 361 |
| PSL76 | − | 3.8 (21) | 551 | |
| PSL64 | 181° | + | <0.24 (0) | 420 |
| PSL74 | − | 0.25 (1) | 402 | |
| PSL62 | 359° | + | 92 (331) | 360 |
| PSL73 | − | 83 (287) | 346 |
The indicated strains (all containing a null mutation in spoIIIE) had an array of lac operators inserted at the indicated region of the chromosome. The strains were grown in defined minimal medium, and sporulation was induced by the addition of mycophenolic acid. Samples were taken 4 h after the initiation of sporulation and stained with the membrane dye FM4-64 to allow identification of cells that had an asymmetric septum.
+, wild-type soj and spo0J; −, soj spo0J double mutant.
Percentage (and total number in parentheses) of sporangia (mother cell plus forespore) with a focus of LacI-GFP (or LacI-CFP) positioned entirely in the forespore.
Origin (359°) region.
Our results indicate that parS and Spo0J do not provide a centromere-like function for positioning origin regions to opposite poles during sporulation. In cells that had undergone asymmetric septation and had two foci of LacI-GFP bound to arrays of lac operators in the origin (359°) region (Fig. 4A to C), there was no noticeable effect of spo0J on positioning of foci in the forespore. Ninety-three percent (297 of 319) of the sporangia from spo0J+ cells and 92% (224 of 243) of the sporangia from soj spo0J mutant cells had one focus of LacI-GFP in the forespore and one in the mother cell (Fig. 4A). This finding is not consistent with the notion that Spo0J functions as an anchor to hold or recruit sister origins to opposite cell poles during sporulation.
There was an effect of spo0J on trapping the origin region in the forespore (Table 4), but only in cells with one focus of the origin region. Only seven sporangia (1.9% of the total) from the spo0J+ cells had a single focus of the origin region, and in five of those the focus was in the forespore. In striking contrast, 43 sporangia (12% of the total) from the soj spo0J mutant had a single focus of the origin region (Table 5). Only seven of these had the focus in the forespore, and the remaining 36 had the focus in the mother cell (Fig. 4D). Most of the soj-spo0J null cells that failed to trap origins still had chromosomal DNA trapped in the forespore, as visualized by DAPI staining (Fig. 4C and data not shown). soj single mutants trapped the origin at a frequency similar to wild-type cells, indicating that the trapping defect was due to the absence of spo0J (data not shown). These results indicate that Spo0J does influence positioning of the origin region in the forespore but probably because it affects the number of sporangia that have only a single focus of the origin region. The soj spo0J null mutant also had an increase in the number of sporangia that had more than two foci of the origin region (Table 5), although almost all of these had a focus positioned in the forespore (Fig. 4E). These effects of spo0J on the number of origin regions per sporangium are probably due to asynchronous replication and defects in coordinating cell division with replication (see below).
TABLE 5.
Effect of spo0J on chromosome content during sporulationa
| Strain | Region | soj spo0Jb | % of cells with indicated no. of focia
|
No. of sporangia analyzed | |||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | >4 | ||||
| PSL62 | 359° | + | 0.28 | 1.9 | 89 | 7.2 | 1.9 | <0.28 | 360 |
| PSL73 | − | 0.87 | 12 | 70 | 10 | 6.1 | <0.29 | 346 | |
| PSL66 | 270° | + | 0.55 | 1.7 | 94 | 2.8 | 1.4 | <0.28 | 361 |
| PSL76 | − | 1.6 | 6.5 | 62 | 17 | 12 | 1.6 | 551 | |
| PSL64 | 181° | + | 0.71 | 32 | 63 | 1.7 | 0.9 | <0.24 | 420 |
| PSL74 | − | 1.0 | 21 | 65 | 6.0 | 5.7 | 0.7 | 402 | |
The indicated strains (all containing a null mutation in spoIIIE) were grown in defined minimal medium and induced to sporulate by the addition of mycophenolic acid. Samples were taken 4 h after the initiation of sporulation and stained with the membrane dye FM4-64 to allow identification of cells that had divided asymmetrically. The number of foci per sporangium (mother cell plus forespore) of LacI-GFP (or LacI-CFP) was determined for cells that had undergone an asymmetric septation. The percentage of sporangia with the indicated number of foci for each strain is presented. An array of lac operators was inserted in the indicated region of the chromosome and visualized with LacI-GFP or LacI-CFP.
+, wild-type soj and spo0J; −, soj spo0J double mutant.
The 270° and 181° (terminus) regions.
We also examined the effects of loss of spo0J on trapping the 270° and 181° regions in the forespore. In spo0J+ cells that had undergone asymmetric septation, we never (among 361 sporangia examined) observed positioning of the 270° region in the forespore (Table 4). However, in the soj spo0J mutant, 3.8% of the sporangia (21 of 551) had the 270° region positioned in the forespore (Table 4). Of these, 10 had one focus in the mother cell and one focus in the forespore, and one sporangium had a focus only in the forespore. The remaining 10 sporangia had one focus in the forespore and multiple foci in the mother cell. These observations indicate that approximately half the time, mispositioning of the 270° region in the forespore is probably due to cells with extra copies of this region. Many of the sporangia that did not have the 270° region trapped in the forespore had extra foci of the region in the mother cell (see below), consistent with the previous observation that some cells in a population of spo0J mutants have extra copies of the chromosome (56).
In contrast to the 270° region, only 0.25% of the soj spo0J mutant sporangia (1 of 402) had the 181° region positioned in the forespore, virtually indistinguishable from the spo0J+ sporangia (0 of 420) (Table 4). Thus, it appears that even in the absence of spo0J, the terminus region (181°) is rarely trapped in the forespore. These findings probably explain part of the apparent discrepancy between previous reports (12, 51). If the terminus region is not significantly positioned in the forespore in the soj spo0J mutant, then, as reported (12), experiments to measure expression in the forespore of genes in the terminus region would indicate little or no expression.
spo0J null mutants have more origin foci and increased chromosomal copy number.
Cells containing the spo0J null mutation had phenotypes in addition to the origin-positioning defect. Inactivating spo0J affected the number of origin foci per cell: there were fewer cells with two foci and more cells with one, three, or more foci relative to the wild type, as visualized with LacI-GFP bound to lac operators at 359° (Table 2). Additionally, the mutant cells had more foci of the 270° and 181° regions of the chromosome, visualized by LacI-CFP bound to lac operators inserted in these regions (Table 2; Fig. 4F). These results indicate that a subset of the spo0J mutant cells have an increase in chromosomal copy number. Similar results were observed in sporulating cells (Table 5), consistent with a previous observation (56).
The increased number of chromosomes in some cells could be due to a defect in the timing of replication initiation: if origins fired more often, this could increase the chromosomal copy number, and if origins fired asynchronously within the same cell, this could create cells with three foci of the origin region. The increase in cells with one focus of the origin could be due, in part, to asynchronous initiation (for example, a cell with three foci of the origin could divide to produce a one-focus cell and a two-focus cell). We would also expect that the spo0J mutant would have an increase in the percentage of cells with a single focus of other regions. However, this was not the case (Table 2), indicating that the increase in the percentage of cells with a single focus of the origin region might be due to something other than or in addition to asynchronous replication. A defect in separation of sister origins could also contribute to an increase in cells that appear to have a single focus of the origin. The finding that sister origins are on average closer together in the spo0J mutant is also consistent with the notion that Spo0J is somehow facilitating separation.
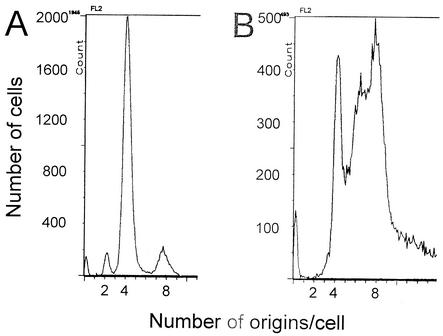
Flow cytometry results confirmed that inactivating spo0J caused increased chromosome content as well as asynchronous initiation of replication. Replication runoff assays were performed to count the number of origins per cell. Ongoing rounds of replication were allowed to finish, while reinitiation and cell division were inhibited. Under the growth conditions used (see Materials and Methods), the majority of wild-type cells had four origins per cell, and origin numbers reflected synchronous replication initiation in most of the population (Fig. 5A). Inactivating spo0J caused a shift towards six to eight origins per cell (Fig. 5B), consistent with the observation that a spo0J null mutation caused increased numbers of foci per cell of several chromosomal regions visualized by the lac system. These results could be due to (i) replication initiation occurring asynchronously and at a smaller cell mass in the absence of spo0J, and/or (ii) effects of spo0J on growth rate or cell division.
FIG. 5.
Early and asynchronous initiation of replication in spo0J mutant cells. spo0J+ (A, strain CRK6000) and spo0J mutant (B, strain NIK6074) cells were grown in Difco antibiotic medium 3, and samples were taken during exponential growth and treated for analysis by flow cytometry (see Materials and Methods). Histograms show the number of cells with a given amount of DNA per cell after completion of rounds of replication, as determined by flow cytometry. The numbers on the x axis represent the number of origins per cell at the time the sample was taken. More than 20,000 cells were analyzed in each experiment.
Under the growth conditions used, inactivating spo0J did not cause a detectable change in doubling time (54 ± 2 min for the wild type and the spo0J mutant). Although spo0J mutant cells were found to be 15 to 20% larger (1.17-fold) than wild-type cells by light scattering, the origin-mass ratio was 1.6 times higher in a spo0J null mutant than in the wild type, indicating that the increased chromosomal content was due to early initiation rather than a cell division defect alone. Furthermore, the DNA-protein ratio was higher in a spo0J null mutant (0.060 ± 0.001) than in the wild type (0.041 ± 0.002), indicating that there is overreplication in the spo0J mutant.
In addition to the increased chromosome content in some of the spo0J mutant cells, the average length of spo0J null mutant cells, measured from micrographs of exponentially growing cells, was ≈20% greater than that of wild-type cells, consistent with the light-scattering results obtained by flow cytometry. The average length of wild-type cells was 2.45 ± 0.07 μm, whereas the spo0J null mutant had an average length of 2.93 ± 0.15 μm (expressed as the 95% confidence intervals for the means). This difference is indicative of a defect or delay in cell division in the spo0J mutants.
Interestingly, depletion of ParB in Caulobacter crescentus causes a severe defect in cell division (41), perhaps indicating a role for many of the ParB family members in regulating cytokinesis. We suspect that the defect in cell division in the spo0J mutants could partly contribute to the increased chromosome copy number. However, this alone cannot account for the presence of cells with asynchronous numbers of origin foci or the increased origin-mass and DNA-protein ratios observed in the spo0J mutant. Taken together, the results indicate that spo0J is a negative regulator, either directly or indirectly, of replication initiation in B. subtilis.
Phenotypes of spo0J mutants.
Experiments presented here indicate that duplicated origin regions are typically found approximately at the cell quarters and that Spo0J contributes to this positioning. Sister origins were closer together in a subpopulation of spo0J mutant cells. However, binding of Spo0J to an array of parS sites was not sufficient to recruit these sites to the cell quarters. It seems likely that Spo0J contributes indirectly to positioning duplicated origins at the cell quarters.
Rather than providing positional information per se, it is possible that Spo0J contributes to separation of sister origin regions. If Spo0J is helping to compact the origin region, as suggested previously (1, 5), then perhaps this compaction facilitates movement of the sister origin regions away from each other. Compaction could also enhance the function of other proteins that might bind to the origin region and contribute to positioning. A recent report identified a site in the origin region that appears to be the main determinant of origin positioning (27). Whereas Spo0J does not appear to bind to this site, perhaps Spo0J bound to its sites in the origin region contributes to the function of this positioning site. A broad region of the chromosome that appears to contribute to positioning of the origin region in the forespore has also been identified (60). However, Spo0J function appears to override the effects of this region, and the interactions between Spo0J function and this large partitioning region are not understood.
In addition to the alteration in positioning of sister origin regions, a subpopulation of the spo0J null mutant cells had extra copies of the chromosome and appeared to have asynchronous replication. These phenotypes are somewhat similar to those caused by seqA mutations of E. coli (references 21 and 40 and references therein) and may indicate that binding of Spo0J to endogenous parS sites sequesters origins and prevents inappropriate initiation of replication.
Apparent discrepancy between plasmid and chromosomal par systems.
Our results highlight an apparent discrepancy between plasmid and chromosomal Par systems. Spo0J was not sufficient to position ectopic chromosomal parS sites at the cell quarters. However, plasmid Par proteins are required and appear to be sufficient for positioning duplicated plasmids at the cell quarters. In fact, when placed on a derivative of the E. coli F plasmid, the B. subtilis Par system (soj, spo0J, and parS) confers positional information, so that duplicated plasmids are now found predominantly at the cell quarters and the plasmid is significantly stabilized in E. coli (62). In B. subtilis, both soj and spo0J are necessary for plasmid stability (37).
ParA/ParB/parS partitioning systems are widespread. First characterized in the low-copy plasmid F and the prophage P1, homologous systems have been found in numerous plasmids and on many bacterial chromosomes (reviewed in references 2, 8, 17, and 21). It would be comforting to think that the mechanisms by which the plasmid and chromosomal systems contribute to partitioning are similar, and the fact that a chromosomal system can function on a plasmid is consistent with this view. However, there appear to be significant differences between the plasmid and chromosomal systems. First, as indicated above, the Par system from B. subtilis does not appear to be sufficient to provide positional information to ectopic sites in the chromosome. Furthermore, while loss of the B. subtilis Par system causes some perturbation in the positioning of sister origin regions, there still appears to be significant positioning. In addition, in the plasmid systems, and for chromosomal systems to function on a plasmid, both the parA and parB products are required (10, 37, 44). In contrast, in B. subtilis, loss of parB has a much more severe effect on chromosome partitioning than loss of parA (25).
We do not understand the nature of these differences between plasmids and chromosomes. They could reflect differences in the mechanisms by which the Par systems function on plasmids and chromosomes. However, we think it more likely that they reflect differences between plasmids and chromosomes (size, time required for replication, proximity of parS sites to the origin) and that the biochemical functions of the homologous Par systems are quite similar. For example, if the primary positional information comes from another site and a protein that binds to that site, as appears to be the case for the chromosome (27) and could be the case for plasmids, then the Par system could facilitate function of the primary system, but not by providing positional information per se. For plasmids (and the chromosome too), the system providing positional information could be related to the origin of replication and replication initiation proteins. Further analysis of and comparisons between plasmid and chromosomal Par systems should help determine the mechanisms by which they act.
Acknowledgments
We are grateful to Richard M. Dudley (Department of Mathematics, MIT) for enthusiastic help with statistical analyses. We thank Chris Webb, Aurelio Telemann, and Richard Losick for providing the strains and plasmids from which the lacO cassette strains were derived and Katherine Lemon for constructing the lacO cassette strains. We thank K. Kobayashi in N. Ogasawara's laboratory (Nara Institute of Science and Technology) for constructing the spo0J::ermC mutation. We also thank William Burkholder, Nicholas Houstis, Janet Lindow, Megan Rokop, Melanie Barker, and Frank Solomon for helpful comments on the manuscript and members of the Grossman laboratory for helpful and engaging discussions.
P.S.L. was supported in part by a predoctoral training grant from NIH. This work was also supported in part by a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Science to S.M. and by Public Health Services grant GM41934 to A.D.G.
REFERENCES
- 1.Autret, S., R. Nair, and J. Errington. 2001. Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA-binding and interactions with Soj protein. Mol. Microbiol. 41:743-755. [DOI] [PubMed] [Google Scholar]
- 2.Bignell, C., and C. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1-34. [DOI] [PubMed] [Google Scholar]
- 3.Bignell, C. R., A. S. Haines, D. Khare, and C. M. Thomas. 1999. Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol. Microbiol. 34:205-216. [DOI] [PubMed] [Google Scholar]
- 4.Bouet, J.-Y., and B. E. Funnell. 1999. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 19:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britton, R. A., and A. D. Grossman. 1999. Synthetic lethal phenotypes caused by mutations affecting chromosome partitioning in Bacillus subtilis. J. Bacteriol. 181:5860-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton, K. 1956. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of DNA. Biochem. J. 62:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davey, M. J., and B. E. Funnell. 1997. Modulation of the P1 plasmid partition protein ParA by ATP, ADP, and P1 ParB. J. Biol. Chem. 272:15286-15292. [DOI] [PubMed] [Google Scholar]
- 8.Draper, G. C., and J. W. Gober. 2002. Bacterial chromosome segregation. Annu. Rev. Microbiol. 56:567-597. [DOI] [PubMed] [Google Scholar]
- 9.Easter, J., Jr., and J. W. Gober. 2002. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol. Cell 10:427-434. [DOI] [PubMed] [Google Scholar]
- 10.Erdmann, N., T. Petroff, , and B. Funnell. 1999. Intracellular localization of P1 ParB protein depends on ParA and parS. Proc. Natl. Acad. Sci. USA 96:14905-14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firshein, W. 1989. Role of the DNA/membrane complex in prokaryotic DNA replication. Annu. Rev. Microbiol. 43:89-120. [DOI] [PubMed] [Google Scholar]
- 12.Frandsen, N., I. Barak, C. Karmazyn-Campelli, and P. Stragier. 1999. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 13:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freese, E., J. M. Lopez, and K. Ochi. 1981. Role of guanine nucleotides and of the stringent response to amino acid deprivation in the initiation of bacterial sporulation, p. 11-16. In D. Schlessinger (ed.), Microbiology—1981. American Society for Microbiology, Washington, D.C.
- 14.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 15.Godfrin-Estevenon, A. M., F. Pasta, and D. Lane. 2002. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol. Microbiol. 43:39-49. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 18.Graumann, P. L., and R. Losick. 2001. Coupling of asymmetric division to polar placement of replication origin regions in Bacillus subtilis. J. Bacteriol. 183:4052-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Genes Dev. 167:335-336. [DOI] [PubMed] [Google Scholar]
- 20.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus, p. 27-35. John Wiley & Sons, Chichester, England.
- 21.Hiraga, S. 2000. Dynamic localization of bacterial and plasmid chromosomes. Annu. Rev. Genet. 34:21-59. [DOI] [PubMed] [Google Scholar]
- 22.Ho, T. Q., Z. Zhong, S. Aung, and J. Pogliano. 2002. Compatible bacterial plasmids are targeted to independent cellular locations in Escherichia coli. EMBO. J. 21:1864-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogg, R., and E. Tanis. 1997. Probability and statistical inference, p. 58-62, 294-297. Prentice-Hall, Inc., Upper Saddle River, N.J.
- 24.Hoshino, T., T. McKenzie, S. Schmidt, T. Tanaka, and N. Sueoka. 1987. Nucleotide sequence of Bacillus subtilis dnaB: a gene essential for DNA replication initiation and membrane attachment. Proc. Natl. Acad. Sci. USA 84:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ireton, K., N. W. Gunther IV, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 171:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadoya, R., A. K. Hassan, Y. Kasahara, N. Ogasawara, and S. Moriya. 2002. Two separate DNA sequences within oriC participate in accurate chromosome segregation in Bacillus subtilis. Mol. Microbiol. 45:73-87. [DOI] [PubMed] [Google Scholar]
- 28.Kim, H.-J., M. J. Calcutt, F. J. Schmidt, and K. F. Chater. 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182:1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S. K., and J. C. Wang. 1998. Localization of F plasmid SopB protein to positions near the poles of Escherichia coli cells. Proc. Natl. Acad. Sci. USA 95:1523-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemon, K. P., and A. D. Grossman. 1998. Localization of the bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 31.Lemon, K. P., and A. D. Grossman. 2000. Movement of replicating DNA through a stationary replisome. Mol. Cell 6:1321-1330. [DOI] [PubMed] [Google Scholar]
- 32.Lemon, K. P., I. Kurtser, and A. D. Grossman. 2001. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis, P. J., and J. Errington. 1997. Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the Spo0J partitioning protein. Mol. Microbiol. 25:945-954. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, P. J., G. B. Ralston, R. I. Christopherson, and R. G. Wake. 1990. Identification of the replication terminator protein binding sites in the terminus region of the Bacillus subtilis chromosome and stoichiometry of the binding. J. Mol. Biol. 214:73-84. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, P. J., M. T. Smith, and R. G. Wake. 1989. A protein involved in termination of chromosome replication in Bacillus subtilis binds specifically to the terC site. J. Bacteriol. 171:3564-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, R. A., C. R. Bignell, W. Zeng, A. C. Jones, and C. M. Thomas. 2002. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology 148:537-548. [DOI] [PubMed] [Google Scholar]
- 37.Lin, D. C.-H., and A. D. Grossman. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675-685. [DOI] [PubMed] [Google Scholar]
- 38.Lin, D. C.-H., P. A. Levin, and A. D. Grossman. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 94:4721-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 40.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 41.Mohl, D. A., J. J. Easter, and J. W. Gober. 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 42:741-755. [DOI] [PubMed] [Google Scholar]
- 42.Mohl, D. A., and J. W. Gober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 43.Moriya, S., K. Kato, H. Yoshikawa, and N. Ogasawara. 1990. Isolation of a dnaA mutant of Bacillus subtilis defective in initiation of replication: amount of DnaA protein determines cells' initiation potential. EMBO J. 9:2905-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 45.Niki, H., and S. Hiraga. 1998. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 12:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niki, H., and S. Hiraga. 1999. Subcellular localization of plasmids containing the oriC region of the Escherichia coli chromosome, with or without the sopABC partitioning system. Mol. Microbiol. 34:498-503. [DOI] [PubMed] [Google Scholar]
- 47.Ogura, Y., Y. Imai, N. Ogasawara, and S. Moriya. 2001. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 183:3833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinett, C. C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A. S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization with Lac operator/repressor recognition. J. Cell Biol. 135:1685-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roos, M., A. B. van Geel, M. E. Aarsman, J. T. Veuskens, C. L. Woldringh, and N. Nanninga. 1999. Cellular localization of oriC during the cell cycle of Escherichia coli as analyzed by fluorescent in situ hybridization. Biochimie 81:797-802. [DOI] [PubMed] [Google Scholar]
- 51.Sharpe, M. E., and J. Errington. 1996. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol. Microbiol. 21:501-509. [DOI] [PubMed] [Google Scholar]
- 52.Sharpe, M. E., and J. Errington. 1998. A fixed distance for separation of newly replicated copies of oriC in Bacillus subtilis: implications for coordination of chromosome segregation and cell division. Mol. Micro. 28:981-990. [DOI] [PubMed] [Google Scholar]
- 53.Teleman, A. A., P. L. Graumann, D. C.-H. Lin, A. D. Grossman, and R. Losick. 1998. Chromosome arrangement within a bacterium. Curr. Biol. 8:1102-1109. [DOI] [PubMed] [Google Scholar]
- 54.Vasantha, N., and E. Freese. 1980. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J. Bacteriol. 144:1119-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webb, C. D., P. L. Graumann, J. A. Kahana, A. A. Teleman, P. A. Silver, and R. Losick. 1998. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol. 28:883-892. [DOI] [PubMed] [Google Scholar]
- 56.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C.-H. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 57.Winston, S., and N. Sueoka. 1980. DNA-membrane association is necessary for initiation of chromosomal and plasmid replication in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 77:2834-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 59.Wu, L. J., and J. Errington. 1997. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 16:2161-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, L. J., and J. Errington. 2002. A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. EMBO J. 21:4001-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, L. J., P. J. Lewis, R. Allmansberger, P. M. Hauser, and J. Errington. 1995. A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev. 9:1316-1326. [DOI] [PubMed] [Google Scholar]
- 62.Yamaichi, Y., and H. Niki. 2000. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:14656-14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]