The city of Quebec, Canada, was a splendid setting for the ASM Conference on Prokaryotic Development, which was convened on 10 to 14 July 2002. Approximately 220 participants were treated to 39 talks and 128 poster presentations describing the latest insights into developmental processes in a variety of bacteria. Scientific program organizers Y. Brun (Indiana University) and L. Shimkets (University of Georgia), together with their advisory committee (M. Bibb, Diversa; J. Errington, University of Oxford; J. Golden, Texas A&M University; H. Kaplan, University of Texas-Houston; and L. Shapiro, Stanford University), planned seven sessions of talks on the topics of differential gene expression, positional information, checkpoints, signaling, cell cycle control, morphogenesis, and multicellularity. This topical format was effective in emphasizing common themes that have emerged from studies of different organisms. Each session featured two speakers invited prior to abstract submission and three to five speakers selected from among those who submitted abstracts. This format provided a lively mix of speakers that included established and young principal investigators, postdocs, and graduate students. Participants were also invited to submit manuscripts related to their abstracts for review and possible inclusion in this issue of the Journal of Bacteriology.
In this review, we describe highlights of the meeting, focusing mainly on the talks and to a lesser extent on related information from poster presentations. We generally follow the order of topics as presented at the meeting, but we have combined information from the sessions on signaling and multicellularity. Also, some talks presented in one session at the meeting are described here under a different topic. In keeping with the purpose of the meeting, we attempt to identify shared features of the molecular mechanisms that regulate and produce developmental change in different prokaryotes. Such features include networks of interacting regulators governing transcription, proteins that localize to a subcellular domain and then relocalize or rapidly oscillate to determine placement of a structure, proteolysis to activate or eliminate a key protein, self-assembling proteins that build structures inside cells to maintain shape or on the surface of cells for movement, adherence, or protection, and signaling between cells to coordinate movement and differentiation of the population. We also point out unique features of the developmental processes of different microbes and unanswered questions that make each worthy of continued study.
DIFFERENTIAL GENE EXPRESSION
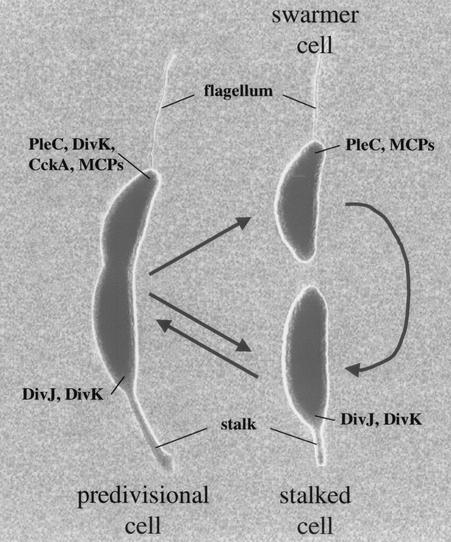
The meeting started with L. Shapiro (Stanford University) reminding us that the key to understanding microbial developmental relies on thinking about the bacterial cell in three dimensions. Recent studies of Caulobacter crescentus highlight how complex and tightly orchestrated developmental processes are. C. crescentus undergoes an obligatory developmental program. Cell division results in two dissimilar progeny, the motile swarmer cell and the sessile stalked cell. Whereas the stalked cell is capable of immediately initiating a new round of DNA replication and cell division, the swarmer cell must first differentiate into a stalked cell prior to progressing through the cell cycle (Fig. 1).
FIG. 1.
The C. crescentus cell cycle. The asymmetric predivisional cell divides to produce two distinct progeny: the flagellated swarmer cell and the sessile stalked cell. The stalked cell is competent to initiate DNA replication and cell growth to become a predivisional cell. The swarmer cell, however, first differentiates into a stalked cell. In addition to the external polar structures (stalk and flagellum), many membrane and membrane-associated proteins are found at specific cell poles during the cell cycle; some of these proteins are labeled.
The generation of asymmetric cells, as well as the progression through the cell cycle, is predominantly controlled by three distinct mechanisms. First, many genes are under transcriptional control and are expressed only when the protein product functions. Microarray analysis revealed that the transcription of approximately 20% of the genome is cell cycle controlled (72). In general, when a given function needs to be expressed, the genes are transcribed. Second, many proteins are specifically proteolysed when their function is no longer required. For example, McpA, a chemoreceptor protein found at the flagellated poles of swarmer cells and predivisional cells (Fig. 1), is specifically degraded during the swarmer-to-stalk differentiation (3). Finally, critical regulatory proteins are dynamically localized to specific locations, and this localization appears to be important for their function.
In C. crescentus, two-component signaling proteins play critical roles in coordinating cell cycle events with transcriptional events. CtrA is a response regulator involved in the control of 25% of the cell cycle-expressed genes (71) and is also involved in the control of DNA replication initiation (96). The initiation of replication depends on the proteolysis of CtrA, and recent work from the Shapiro group shows that DivK, another response regulator, is important for controlling CtrA proteolysis. The activity of CtrA is controlled by phosphorylation and involves a signal transduction network that includes the histidine kinases CckA (47), PleC (120), DivJ (120, 124), and DivL (125).
Many of the two-component signaling proteins involved in morphogenesis are dynamically localized during the cell cycle (Fig. 1). The interplay between these proteins results in changes in their localization, and most likely their activity, during the cell cycle. For example, DivK is distributed randomly in swarmer cells, assembles at the stalk pole in stalked cells, and is found at both poles of predivisional cells. PleC is responsible for the release of DivK from the cell poles (48). Consistent with this role, PleC localizes to the flagellated pole in swarmer cells and is dispersed in stalked cells and once again localized to the flagellated pole in predivisional cells (120).
Dynamic localization of proteins is not limited to regulatory proteins in C. crescentus. The nucleoid must also go to specific places at appropriate times. The origin of chromosome replication resides at the stalk cell pole, and the replisome assembles at that pole only during the G1-S transition, when the cell becomes competent for the initiation of replication (50). The newly replicated origin rapidly moves to the opposite pole, while the replisome complex, as an untethered replication factory, is pushed towards the division plane.
The lesson we learn from these and other studies is that it is not enough to know when an event happens; where proteins go is critical to understanding how they function. L. Shapiro drew an analogy between the cell and a department store with different areas of specialization and described the challenge of deciphering the entire regulatory network as being akin to playing three-dimensional chess. Clearly, unraveling the complexities of C. crescentus development will remain an exciting challenge for some years to come.
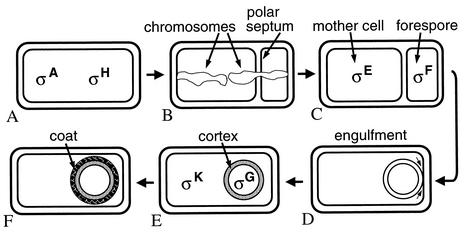
In contrast to the intrinsic asymmetry of the Caulobacter cell cycle, Bacillus subtilis divides to produce two indistinguishable cells during growth. This symmetry is broken upon starvation as cells begin the sporulation process. R. Losick (Harvard University) focused on the question of how the switch to asymmetric division occurs. The switch involves changes in gene expression directed by σA- and σH-containing RNA polymerases (Fig. 2A) and a change in nucleoid structure. Two copies of the chromosome form an axial filament in which DNA replication origin-proximal regions are at opposite cell poles (Fig. 2B). How the chromosomes become oriented is a mystery. Recently, cis-acting DNA sequences involved in this polar localization process have been shown by J. Errington's group (University of Oxford) to span a large region about 150 to 300 kbp away from the origin of replication, perhaps functioning like the large dispersed centromeres of some eukaryotic cells (127). Postdoc S. Ben-Yehuda of the Losick lab appears to have identified a trans-acting factor, RacA, involved in this process (7a).
FIG. 2.
Morphological changes during B. subtilis sporulation and the approximate time and location at which different σ factors become active. (A) σA and σH RNA polymerases transcribe genes whose products cause polar septation and axial filament formation. (B) The axial filament consists of two chromosomes extending the length of the cell with their replication origin-proximal regions attached at opposite ends of the cell. The polar septum forms around the axial filament, capturing one-third of one chromosome in the forespore. The remaining two-thirds of that chromosome is translocated into the forespore. (C) Upon completion of polar septation, σF becomes active in the forespore, and this leads to activation of σE in the mother cell. (D) Products of genes under σE control drive migration of the septal membranes around the forespore in the phagocytic-like process of engulfment. (E) Completion of engulfment pinches off the forespore as a free protoplast within the mother cell. Two membranes surround the forespore and separate its contents from the mother cell cytoplasm. σG becomes active in the forespore, leading to activation of σK in the mother cell. Primarily, genes under σE and σK control cause synthesis of a loosely cross-linked peptidoglycan termed cortex, between the two membranes surrounding the forespore, and synthesis of proteins that assemble on the surface of the forespore to produce the spore coat (F). Not shown are subsequent steps, which include spore maturation and release of the spore via lysis of the mother cell. Adapted from reference 67 with permission.
The switch to asymmetric division also involves a change in location of FtsZ. This key division protein normally forms a ring at midcell. The ring may actually be a very tight spiral, because upon entry into sporulation a relaxed spiral is observed transiently and is proposed to redeploy FtsZ to bipolar rings (or tight spirals) (7). One of the rings eventually becomes the polar septum that separates the larger mother cell from the smaller forespore (Fig. 2C). How one ring rather than the other matures into a polar septum is unclear, but recent studies show that polar septation involves at least two mechanisms that are partially redundant in function, one that boosts transcription of ftsZ and another that boosts transcription of spoIIE (7). SpoIIE is a membrane protein that associates with FtsZ. SpoIIE also dephosphorylates SpoIIAA, enabling it to bind to the anti-σ SpoIIAB, releasing active σF. SpoIIE plays a role in coupling polar septation to σF activation in the forespore (26, 65). A recent paper and two posters presented at the meeting describe mutations in spoIIE that uncouple σF activation from septum formation (25; K. Carniol, P. Eichenberger, and R. Losick, ASM Conf. Prokaryot. Dev., abstr. 40, 2002; D. Hilbert and P. Piggot, ASM Conf. Prokaryot. Dev., abstr. 34, 2002). These may provide a way to elucidate how coupling between the two functions of SpoIIE is achieved.
Activation of σF in the forespore also relies in part on transient genetic asymmetry created by polar septation (27). The polar septum forms around the axial filament and captures the replication origin-proximal one-third of one chromosome in the forespore (Fig. 2B) (126). The rest of the chromosome is translocated into the forespore by SpoIIIE in a matter of minutes, but the location of genes on the chromosome dictates when they are transferred from the mother cell to the forespore during this critical period for establishment of cell fate. In particular, the spoIIAB gene encoding the anti-σ that inhibits σF is located in the origin-distal region of the chromosome. This delays appearance of spoIIAB in the forespore transiently (21). With SpoIIIE acting as a barrier to diffusion of small proteins like SpoIIAB between mother cell and forespore (R. Agress, N. Liu, and K. Pogliano, ASM Conf. Prokaryot. Dev., abstr. 116, 2002), and because SpoIIAB is susceptible to degradation by ClpCP protease (90), the level of SpoIIAB would drop transiently in the forespore, causing activation of σF. In the mother cell, σF remains inactive, but the inactive precursor, pro-σE, is activated by proteolytic processing in response to a signal generated by σF-directed transcription in the forespore (39, 58, 76). Components of this signaling-processing pathway have been identified, but mechanistic details remain to be elucidated. Proteolytic degradation seems to ensure that any active σE produced in the forespore is eliminated (29), but the protease(s) has not been identified. With σF active in the forespore and σE active in the mother cell (Fig. 2C), differential gene expression ensues, driving engulfment of the forespore in a double membrane (Fig. 2D) and subsequent events discussed below.
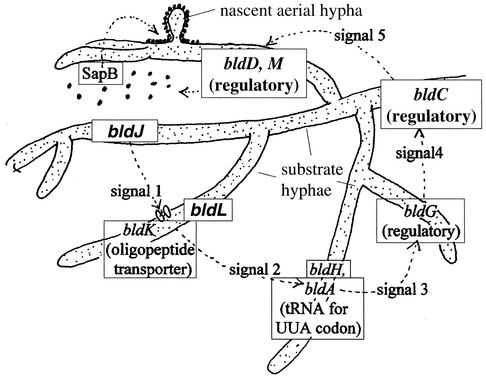
Streptomyces coelicolor differs from Caulobacter and Bacillus in that it grows as filaments called hyphae. A member of the actinomycetes, from which many important antibiotics are produced, S. coelicolor is a favorite model organism for studies of both secondary metabolism and development. Its genome sequence was reported recently (6). Growth on solid medium results in antibiotic-producing substrate hyphae (Fig. 3). In response to signals described below, the substrate hyphae switch to a different pattern of growth, giving rise to aerial hyphae that eventually form chains of spores. Aerial hyphae give the colony a hairy appearance. Bald (bld) mutants fail to produce aerial hyphae. The bld genes encode proteins needed for the switch to aerial-hypha formation. One such protein, σBldN, is a σ factor of the extracytoplasmic function (ECF) family (9). M. Elliot, a member of M. Buttner's group (John Innes Center), compared wild-type and bldN mutant cells at different stages of the life cycle using DNA microarrays, in order to identify genes downstream of bldN in the developmental pathway. Among the genes identified were some that also depend on bldM, which encodes a response regulator whose transcription is directed by σBldN (9). The downstream targets of σBldN and BldM include a family of secreted proteins found only in other Streptomyces organisms so far. These proteins may be linked to the cell wall and are very hydrophobic. The genes for two of these secreted proteins are adjacent to the genes specifying the rodlin proteins, which are themselves σBldN- and BldM-dependent, hydrophobic proteins that form the rodlet layer of the spore surface and can mediate attachment to a hydrophobic surface (18).
FIG. 3.
Extracellular signal exchange leading to aerial growth in S. coelicolor. The figure, based on data of Willey et al. (121) and Molle and Buttner (78), is modified from the work of Chater (16) with permission. The lower part depicts how substrate hyphae in different physiological states under bld gene control may produce signals that act in a cascade leading to production of SapB, which is thought to allow formation of aerial hyphae (one is illustrated near the top of the figure).
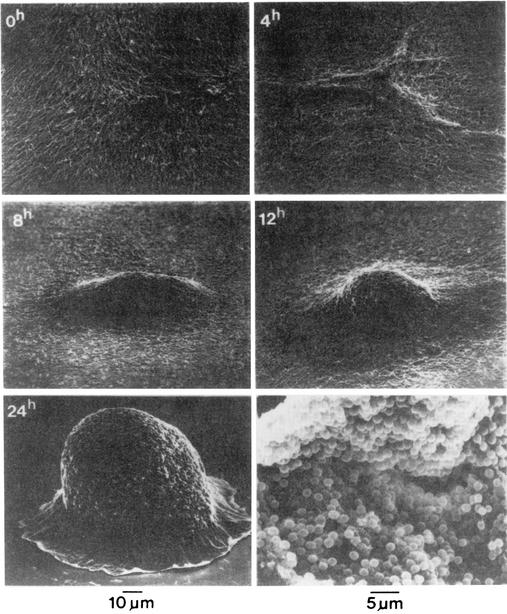
Like the actinomycetes, the myxobacteria produce a huge variety of secondary metabolites, some of which are used therapeutically. Like S. coelicolor, Myxococcus xanthus has a large (>9-Mbp), GC-rich (68%) genome and early on emerged as a favorite for studies of development, especially as genetic methods were devised to facilitate molecular analysis. But M. xanthus does not grow as filaments. Instead, the long, slender, rod-shaped cells glide over solid surfaces, secreting enzymes that lyse other bacteria and digest macromolecules for uptake as nutrients. Growth leads to binary fission. Starvation initiates a spectacular developmental process in which about 105 cells aggregate, forming a macroscopic mound roughly 0.1 mm wide and 0.1 mm tall (Fig. 4). The mound becomes a fruiting body as cells differentiate into ovoid spores. H. Kaplan (University of Texas—Houston) has used lacZ fusions to genes induced early during development in screens for mutants. This approach initially uncovered a two-component system (SasS/SasR) that senses and responds to A signal, a cell density signal composed of peptides and amino acids generated by extracellular M. xanthus proteases at the onset of development (35, 129). A third component, SasN, inhibits SasS/SasR during growth, providing a nutritional status input that does not depend on cell density (128). The nature of this input is unknown. Also, the Sas system does not regulate all early developmental genes. Using a different lacZ reporter, graduate student J. Rivera in Kaplan's lab recently discovered a parallel pathway that senses and responds to starvation and cell density signals early in development (J. Rivera and H. Kaplan, ASM Conf. Prokaryot. Dev., abstr. 31, 2002). This pathway involves an ECF σ factor and its cognate anti-σ. Interestingly, the two parallel pathways appear to be capable of sensing periplasmic stress brought about by a block in lipopolysaccharide (LPS) O-antigen biosynthesis. The Sas/ECF σ pathways are strikingly similar to the Cpx/σE envelope stress-responsive pathways of Escherichia coli (97). How disruption of LPS O-antigen biosynthesis generates envelope stress in M. xanthus and whether this is part of the normal developmental process are important questions for the future.
FIG. 4.
Fruiting body development in M. xanthus. Scanning electron micrographs of cells starved in submerged culture were made by Kuner and Kaiser (69) and are reprinted here with permission. Times poststarvation are indicated in the upper left corner of the first five frames, to which the 10-μm scale applies. Individual cells (about 5 μm long) begin to aggregate by 4 h and complete mound formation by 24 h, at which time some cells have begun to differentiate into spores. The lower right frame shows a mature fruiting body that has cracked open, revealing its spores (5-μm scale).
Some bacteria initiate a developmental program upon interaction with a plant or animal host. In some cases, a mutually beneficial symbiotic relationship is established. In others, pathogenesis occurs. Studies of nonpathogenic, nonsymbiotic model organisms, like those discussed above, dominated the meeting because the tractability of these systems allows rapid exploration in uncharted territory. The paradigms that emerge are not only fascinating but also extremely useful in guiding investigations of bacterium-host interactions, which are typically less tractable experimentally. One interaction that has yielded to molecular approaches is the symbiosis established between rhizobial bacteria and leguminous plants. After entry into plant cells, the bacteria differentiate into bacteroids, which are enlarged cells that can fix atmospheric nitrogen into ammonia for use by the plant. Bacteroids form in nodules on the plant roots. V. Oke (University of Pittsburgh) identified nodule-expressed (nex) bacterial genes (86) and implicated two pathways in the negative regulation of these genes. One pathway involves negative regulation by the transcription factor NodD3, one of several NodD proteins that positively activate genes early in the symbiosis process. The second pathway involves fadD, which encodes acyl coenzyme A synthetase. Although the reason for the connection to FadD is unclear, FadD affects gene expression in several pathogenic interactions.
POSITIONAL INFORMATION
At the heart of bacterial development is the asymmetric localization of macromolecules. For the most part, how these asymmetries are generated and how they are maintained is unknown. In this session, speakers provided insights into how specific proteins and protein complexes localize within the bacterial cell.
Bacterial two-component signal transduction systems control a wide array of physiological processes in response to a variety of environmental conditions. Although many of the C. crescentus two-component proteins are predicted to mediate responses to environmental conditions, others respond to internal cues and are central players in coupling morphogenesis with cell cycle progression.
C. Jacobs (Yale University) described recent studies on two histidine kinases, PleC and DivJ, and an essential single-domain response regulator, DivK, belonging to the CheY/Spo0F family (38). Using DivK-green fluorescent protein (GFP) fusions, she showed that these two histidine kinases play distinct roles in the polarity of DivK. DivJ is required for polar localization of DivK, whereas PleC is important for its release from the swarmer pole prior to cell division (48).
A. Newton (Princeton University) described two two-hybrid studies aimed at identifying the cognate sensor kinase and other components that interact with DivK. These studies revealed that a large number of the interacting clones encoded fragments of the catalytic domains of either DivJ or PleC, supporting the previous assignment of DivJ and PleC as cognate kinases of DivK (38, 124). A small number of DivL clones and two other novel kinase clones, designated CckN and CckO, were also isolated. Furthermore, DivK specifically binds a 72-amino-acid sequence of the DivJ kinase that contains the H box, corresponding to antiparallel α-helices I and II of the EnvZ kinase structure (113). Interestingly, the PleC and DivL kinase fragments also contain this 72-amino-acid sequence, which in addition to being responsible for dimerization, phosphorylation, and phosphotransfer appears to be sufficient to specify histidine kinase response regulator recognition. In collaboration with J.-P. Samama and colleagues (CNRS-IPBS, Toulouse, France), A. Newton and colleagues have solved the crystal structure of DivK (34), a critical step forward in delineating the relationships between DivK and its cognate histidine kinases.
How these critical two component proteins as well as other proteins are localized to the cell poles is an area of intense study. It was shown by P. Viollier, a postdoctoral fellow in L. Shapiro's group (Stanford University), that the pilus assembly protein, CpaE, required for the formation of the polar CpaC pilus secretion channel, is asymmetrically localized to the swarmer cell pole in predivisional cells and swarmer cells but not in stalked cells (116; P. H. Viollier, N. Sternheim, and L. Shapiro, ASM Conf. Prokaryot. Dev., abstr. 70, 2002). This pattern of CpaE localization parallels that of the histidine kinase PleC. In a mutant in which PleC cannot be autophosphorylated on its histidine residue (PleCH610A), CpaE and PleCH610A are not lost from the swarmer pole at the swarmer-to-stalked-cell transition. This results in predivisional cells in which CpaE and inactive PleC are localized to both cell poles instead of only one cell pole. While these results show that PleC activity controls the asymmetric location of CpaE and itself, the question of how polar localization is achieved remains unanswered. Part of the answer lies in a pleiotropic factor, PodJ, that directs PleC, CpaE, and, indirectly via CpaE, CpaC to the swarmer pole and is itself localized to the same cell pole. The localization of PodJ (also presented by A. Hinz of the Y. Brun lab [38a]) precedes that of CpaE, CpaC, and PleC. Thus, PodJ serves as a localization factor for the polar positioning of bacterial structural and regulatory proteins.
In C. crescentus, the establishment of cellular asymmetry in the predivisional cell and completion of the cell division cycle are prerequisites to flagellar pole differentiation into a stalked pole. It has been shown previously that the alternative sigma factor, σ54, is required for this critical differentiation step (13). E. coli σ54 requires a NtrC-like transcriptional activator in order to initiate the transcription of a subset of σ54-dependent genes (95). NtrC-like activators have a highly conserved central domain that E. Quardokus of the Y. Brun group (Indiana University) used to identify four σ54specific activators in C. crescentus. One of these, tacA, encodes an activator responsible for uncoupling differentiation of the new pole to the cell division cycle. Mutants with changes in both rpoN (encoding σ54) and tacA prematurely differentiate the new pole. Critical developmental regulators exhibit altered localization patterns or are not localized. These results suggest that cell cycle control of localization, which sets up asymmetry in the predivisional cell, may contribute to the proper timing of differentiation events.
One of the most exquisite examples of defining cellular position is observed in the placement of the cell division plane. Equipartitioning of cell components during division requires that Z-ring formation be accurately confined to a narrow zone around the midcell. Positioning of the Z ring in E. coli is determined by the cellular distribution of MinC. MinC inhibits Z-ring assembly (8) and undergoes an oscillatory localization cycle during which the protein alternately accumulates on the membrane at either cell end every 25 s (42, 99), effectively forcing proper positioning of the Z ring to the midcell (36, 94). The cellular location of MinC is directly dictated by that of MinD, which undergoes a similar oscillatory localization cycle (100). MinD is a self-interacting ATPase which associates with the cytoplasmic membrane in a peripheral manner. MinD also interacts directly with both MinC and MinE and recruits both to the membrane. MinE is required for MinD oscillation and itself undergoes a dynamic localization cycle that is coupled to that of MinD (36). The interplay among each of these proteins, the membrane, and potential topological anchors has been under intense study. Several talks and posters (described below and in the cell cycle section) shed light on critical features of these proteins in the cell division of E. coli.
Of fundamental importance is defining the role that cell polarity plays in the oscillation of MinCD. Several models for how the interactions between MinD, ATP, membrane, and MinE might culminate in the observed oscillatory behavior have been formulated. Contrary to previous expectations, it now appears that the cell poles do not play a role in this process other than serving as barriers that prevent MinD and MinE migration in a given direction. P. de Boer (Case Western University) presented an elegant computer simulation of how the placement of the cell division plane could occur (77). Surprisingly, in this simulation, no prelocalized determinant was needed for either the midcell placement of the FtsZ ring or the polar concentration of MinC. A similar conclusion was drawn by M. Howard (University of Leiden) using computer simulations of MinCDE localization with slightly different parameters (M. Howard, A. D. Rutenberg, and S. de Vet, ASM Conf. Prokaryot. Dev., abstr. 51, 2002). The exciting conclusion from these (40, 77) and other (68) studies is that MinCDE could oscillate between the cell poles, in principle, without any specific determinant to direct the proteins there.
The mechanism by which integral membrane proteins are positioned at a specific location in a bacterial cell is also under intensive study. Three general models predominate. Proteins could be inserted randomly into the membrane and diffuse to their specific position, where they are captured (lateral sorting). Alternatively, randomly inserted proteins could be eliminated by proteolysis from sites other than their proper destination. Finally, membrane proteins could be inserted directly at their final location. The Losick group has previously demonstrated that the B. subtilis membrane protein, SpoIVFB, is localized to the outer forespore membrane by lateral sorting after first being inserted in the cytoplasmic membrane of the mother cell (104). New insight into how mother cell proteins could be specifically localized was presented in a poster by C. van Ooij in R. Losick s lab (Harvard University) (C. van Ooij, P. Eichenberger, and R. Losick, ASM Conf. Prokaryot. Dev., abstr. 81, 2002). Like SpoIVFB, SpoVM-GFP becomes localized to the outer forespore membrane. This localization depends on the mother cell-specific sigma factor σE; SpoVM-GFP is localized uniformly along all membranes in a mutant without σE activity. Thus, there must be a gene under σE control whose protein product recruits SpoVM to the mother cell membrane of the polar septum, which becomes the outer forespore membrane during engulfment (Fig. 2C and D). Interestingly, the localization of SpoVM does not rely on gene expression in the forespore. The SpoVM anchor is absent in the forespore, since SpoVM expressed exclusively in the forespore from a forespore-specific promoter was not recruited to the polar septum. Finally, based on the analysis of specific alanine substitutions, it appears that the N terminus of SpoVM is critical for its interaction with the σE controlled gene product.
The rules governing the localization of integral membrane proteins in the forespore appear to be different. A. Rubio of K. Pogliano's group (University of California, San Diego) described a variation of the direct targeting mechanism. The targeting determinants for localization to the sporulation septum were studied by using both heterologous and native GFP fusion proteins synthesized in the forespore of B. subtilis. Surprisingly, all GFP fusion proteins were found to first target to the septum. Membrane-spanning segments within a protein seem sufficient for septal localization of proteins produced in the forespore. After initial insertion into the forespore septal domain, however, the heterologous GFP fusions and native forespore membrane protein, SpoIIQ, show remarkably different localization patterns. Nonnative fusion proteins quickly diffused around the forespore membrane. In contrast, GFP-SpoIIQ tracked with the engulfing mother cell membrane, became punctate late in engulfment, and was observed throughout the forespore after engulfment was complete. Based on localization studies in mutant backgrounds, it was determined that movement of GFP-SpoIIQ required engulfment and its retention in the septal membrane domain required an unknown mother cell-expressed protein, while a heterologous GFP fusion protein was not retained in any background. These data led to a catch-and-release model for membrane proteins synthesized in the forespore: all membrane proteins are first directly inserted into the forespore septal membrane domain and are released to diffuse freely throughout the membrane if they are not caught in the septal domain by a specific interaction.
The mechanisms responsible in bacteria for faithful segregation of replicated chromosomes to daughter progeny, until recently, were largely unknown. Recent evidence indicates that in B. subtilis a stationary DNA replication factory may be responsible for supplying some of the force necessary to partition duplicated DNA toward the cell poles (74). One of the most exciting findings in bacterial architecture has been the discovery of actin-like filaments that also play key roles in the segregation of replicons to daughter progeny. J. Moller-Jensen (University of Southern Denmark) presented evidence that polymerization of actin-like filaments is important for the directional movement of plasmids to the cell poles in E. coli (79). Plasmids encode partitioning genes (par) that are required for faithful plasmid segregation at cell division. The plasmid R1 par locus encodes a prokaryotic centromere-like system that mediates plasmid stabilization. The locus encodes two proteins, ParM and ParR, and contains a centromere-like DNA region (parC) to which the ParR protein binds. Immunofluorescence microscopic images of ParM filaments reveal that these filaments undergo dynamic changes that are dependent on the ATPase activity. ATP binding by ParM is required for polymerization, whereas the hydrolysis of ATP is required for filament depolymerization. ParM filament formation is strictly dependent on the presence of both ParR and parC DNA in vivo. In addition, the ParR/parC complex was able to induce ParM polymerization (below its critical concentration) in vitro, confirming that the complex formed between replicated plasmids and ParR serves as a nucleation point for ParM polymerization. In this way filament formation is restricted to occur at the right time and place.
ParM belongs to a superfamily of ATPases that includes MreB (11). J. Moller-Jensen presented structural data (obtained in collaboration with F. van den Ent, J. Löwe, and L. Amos, Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom) showing that filaments formed by ParM are structurally very similar to F actin. Moreover, the structure of the ParM protein in the empty and ADP-bound forms was solved by X-ray crystallography to resolutions of 2.3 and 2.0 Å, respectively. The structures show that ParM obtains a fold that is very similar to those of MreB and actin. The structural similarity is remarkable, since these proteins are very different in terms of their amino acid sequence (less than 15% identity).
The finding that ParM, an actin homologue in bacteria, is involved in plasmid partitioning prompts the question of whether MreB, another member of the family of actin-like proteins, could be playing a similar role in chromosome partitioning. Two posters addressed the localization of chromosomal DNA in E. coli and C. crescentus mreB cells (R. Figge, J. J. Easter, and J. W. Gober, presented at the ASM Conference on Prokaryotic Development, 2002; T. Kruse, A. Lobner-Olesen, and K. Gerdes, ASM Conf. Prokaryot. Dev., abstr. 48, 2002). Both of these studies show that in mreB mutants, origins of replication are mislocalized and that the origin copy number was increased. Thus, it appears that actin-like filaments are important for both plasmid and chromosome segregation in bacteria.
CHECKPOINTS
Biological checkpoints ensure the completion of one event before the initiation of another. Eukaryotic cells employ several checkpoints to regulate progression through their cell cycle. Likewise, bacteria link DNA replication control to cell division, and some of the proteins and regulatory pathways involved are being defined by studies of the Caulobacter cell cycle, as described elsewhere in this review. Bacteria also employ checkpoints at the transition between growth and development (e.g., A signaling early in Myxococcus development serves as a cell density checkpoint; see above). B. subtilis surveys the replication status of its chromosome and DNA damage prior to the initiation of sporulation gene expression via a small polypeptide (Sda) that inhibits phosphorylation of the key transcription factor Spo0A (14).
Some checkpoints couple completion of a morphological event to further gene expression. A classic example is secretion of FlgM, an anti-σ, upon completion of the hook-basal body in E. coli or Salmonella enterica serovar Typhimurium (2). This relieves FlgM inhibition of σ28, allowing transcription of flagellar genes. In Caulobacter, transcription of class III/IV flagellar genes is coupled to assembly of a class II basal body structure. Previous studies in J. Gober's lab (University of California, Los Angeles [UCLA]) showed that this coupling involves control of the activity of an NtrC-like response regulator, FlbD, by FliX (82). These proteins also regulate the completion of cell division, thus coupling the cell division event to progression of flagellar assembly (80). Recent work suggests that FliX does not affect FlbD activity by regulating its phosphorylation (81). Rather, FliX appears to interact directly with FlbD to regulate its activity. How FliX senses assembly of a class II flagellar structure and how this affects its proposed interaction with FlbD are questions whose answers will reveal the molecular details of morphological coupling in this system.
Morphological coupling is also employed during B. subtilis sporulation. One instance, mentioned above, couples completion of the asymmetric septum to activation of σF in the forespore, and efforts to understand this coupling are focused on SpoIIE (25; Carniol et al., ASM Conf. Prokaryot. Dev.; Hilbert and Piggot, ASM Conf. Prokaryot. Dev.). Another instance of morphological coupling occurs later, upon completion of forespore engulfment (Fig. 2D). It governs activation of σG in the forespore (Fig. 2E). The sigG gene, encoding σG, is transcribed in the forespore by σF RNA polymerase. However, there is a delay in transcription of sigG until engulfment is complete or nearly complete (91). An unidentified signal under σE control in the mother cell is necessary for sigG transcription in the forespore. New results from J. Errington's lab (University of Oxford) suggest that a repressor may bind between the −35 and −10 elements of the sigG promoter and that the σE-dependent signal relieves this repression (L. Evans, A. Feucht, and J. Errington, ASM Conf. Prokaryot. Dev., abstr. 83, 2002). Once σG is made, the available evidence suggests it is held inactive by SpoIIAB, the same anti-σ that inhibits σF (62). Relief from this inhibition seems to be coupled to the completion of engulfment by a putative complex of eight membrane proteins (encoded by the spoIIIA operon) that appear to localize to the outermost of the two membranes surrounding the engulfed forespore (102). In a poster presented at the meeting, M. Serrano of A. Henriques's lab (Universidade Nova de Lisboa) showed evidence that SpoIIIJ is also required to relieve SpoIIAB inhibition of σG (M. Serrano, L. Corte, J. Opdyke, C. Moran, and A. Henriques, ASM Conf. Prokaryot. Dev., abstr. 112, 2002). The mechanism by which SpoIIIA proteins and SpoIIIJ couple completion of engulfment to activation of σG is unknown.
The two instances of morphological coupling just described are checkpoints that link septum formation and engulfment to σF and σG activation in the forespore, respectively. In these cases, as well as in the cases of morphological coupling in flagellar biosynthesis discussed above, the proteins believed to sense morphogenesis affect the activity of a transcription factor via direct interaction (FlgM and FliX), via dephosphorylation (SpoIIE), or by an unknown mechanism (SpoIIIA and SpoIIIJ). Although in the last case the mechanism is still unknown, a theme that emerges from the other three examples is that morphological coupling is accomplished without new gene expression.
Other checkpoints link morphogenesis to activation of a transcription factor indirectly. These checkpoints do involve expression of a new gene(s), which produces a signal that is passed between cells to coordinate subsequent gene expression. For example, σF directs transcription of spoIIR in the B. subtilis forespore, and SpoIIR signals activation of σE in the mother cell via proteolytic processing of pro-σE (39, 58, 76). Hence, activation of σE is indirectly coupled to septum formation due to dependence on σF. This coupling ensures that active σE is produced during a critical period following septum formation (102).
Likewise, the σK checkpoint links proteolytic processing of pro-σK in the B. subtilis mother cell to the completion of engulfment, through dependence on a signaling pathway that begins with activation of σG in the forespore (19, 102). σG directs transcription of spoIVB in the forespore, and SpoIVB signals by an unknown mechanism to a complex of three proteins located in the outermost membrane that surrounds the engulfed forespore (103). The complex includes SpoIVFB, a founding member of a novel family of putative metalloproteases that cleave their substrates within or near the surface of membranes (101, 133). SpoIVFB is thought to be the protease that processes pro-σK. In support of this idea, R. Zhou and L. Kroos (Michigan State University) presented evidence that E. coli strains engineered to produce a SpoIVFB-GFP fusion protein efficiently and accurately process pro-σK in vivo (R. Zhou and L. Kroos, ASM Conf. Prokaryot. Dev., abstr. 24, 2002). In complex with SpoIVFB are SpoIVFA and BofA, which inhibit processing (103). L. Kroos presented studies showing that the level of SpoIVFA decreases in response to SpoIVB signaling from the forespore (66). The SpoIVFA decrease coincides with the appearance of σK, as if the SpoIVFA decrease relieved inhibition of the SpoIVFB protease, allowing pro-σK processing. However, a decrease in the level of SpoIVFA was not sufficient to allow processing if oxidative phosphorylation or translation was blocked. The requirements for oxidative phosphorylation and translation were bypassed by mutations that uncouple processing from dependence on forespore signaling. Hence, pro-σK processing can occur in the absence of an energized membrane and without ongoing translation, but during the normal course of sporulation, these are necessary for synthesis of SpoIVB, and it activates the pro-σK processing complex in a way that is not mimicked by loss of SpoIVFA upon translational arrest. These studies, together with recently published results of D. Rudner and R. Losick (103), shift the focus from SpoIVFA to BofA as the likely target of SpoIVB signaling in the activation of pro-σK processing. To investigate the substrate requirements for pro-σK processing, graduate student H. Prince of Kroos's lab fused GFP or six histidine residues to the C terminus of pro-σK truncated at various points (H. Prince and L. Kroos, ASM Conf. Prokaryot. Dev., abstr. 23, 2002). In contrast to pro-σE, for which 55 N-terminal amino acids was sufficient for processing (55), pro-σK required more than 109 amino acids (through region 2.3). Constructs with 126 N-terminal amino acids (through region 2.4) were sufficient for processing, indicating that region 2.4 has one boundary of a determinant necessary for processing. This determinant may allow proper folding of the N-terminal domain, targeting to membranes (134), and/or recognition by SpoIVFB or another protein.
The σK checkpoint works differently in Clostridium difficile, a pathogenic cousin of B. subtilis. Graduate student J. Haraldsen of L. Sonenshein's group (Tufts University) has determined that the C. difficile sigK gene lacks the prosequence. Also, the genome lacks genes for the processing complex (i.e., spoIVFB, spoIVFA, and bofA). Regulation instead appears to involve excision of a DNA element inserted in the sigK gene. C. difficile strains lacking the DNA element sporulate poorly. B. subtilis has a different DNA element inserted at a different position in its sigK gene, and strains engineered to lack the element sporulate normally (70), because the σK checkpoint ensures that pro-σK is not processed to active σK prematurely. On the other hand, B. subtilis with a sigK gene lacking the prosequence sporulates poorly (19), perhaps owing to a feedback loop initiated by σK RNA polymerase activity that negatively regulates early mother cell gene expression (135). Similarly, in C. difficile strains lacking the sigK insertion element, sporulation may be impaired by a premature switch from early to late mother cell gene transcription. The comparison raises the intriguing possibility that in C. difficile strains with the DNA element, excision may depend on a signal from the forespore. Evolutionarily, pro-σK processing, rather than DNA excision, seems to have prevailed, because for other Bacillus and Clostridium species that have been studied the sigK gene has a prosequence and lacks a DNA insertion element.
Endospore formation presents an opportunity to understand signaling checkpoints between the mother cell and forespore. In the section on signaling and multicellularity below, we see that signaling also governs B. subtilis entry into the sporulation pathway, its exit from a spore (germination), and presumably the multicellular behaviors observed in wild isolates. Signal transduction systems can be thought of as checkpoint mechanisms that monitor intercellular and environmental signals and respond appropriately. In multicellular eukaryotes, new patterns of gene expression and cellular differentiation are commonly linked to morphogenesis by signaling interactions between cells that serve as developmental checkpoints. One such checkpoint in prokaryotes is C signaling during M. xanthus development (see Signaling and Multicellularity below).
CELL CYCLE CONTROL
The coordinated oscillatory behavior of the three Min proteins provides regulation for midcell positioning of the division septum in E. coli, as described in the section on positional information above. J. Lutkenhaus's group (University of Kansas) is investigating the biochemical basis for the oscillatory behavior by examining the interactions among the Min proteins and the membrane. They have observed that MinE stimulates MinD's ATPase activity but only in the presence of phospholipid vesicles (41). Talks by both J. Lutkenhaus and P. de Boer revealed that the ATP-bound form of MinD binds phospholipid vesicles and recruits MinC. MinE interacts with the MinCD complex bound to the bilayer and stimulates the dissociation of MinD and MinC from the vesicles. The release of MinC does not require ATP hydrolysis, indicating that MinE and MinC compete for binding to MinD bound to the bilayer. Taken together, these results show that MinD can bind membrane by direct interactions with phospholipids and support models wherein MinE induces Min protein dynamics by stimulating the conversion of the membrane-bound form of MinD (MinD-ATP) to the cytoplasmic from (MinD-ADP). The results further suggest that, in vivo, MinE-stimulated dissociation of MinC from the MinC-MinD-ATP membrane complex may occur prior to the hydrolysis of the nucleotide.
Details of the association of MinD with both MinC and the membrane were revealed by H. Zhou in J. Lutkenhaus's laboratory (University of Kansas). She described mutant forms of MinD that were unable to bind to MinC (H. Zhou and J. Lutkenhaus, ASM Conf. Prokaryot. Dev., abstr. 108, 2002). Among these, minD44 and minD130 were mapped to the switch I region of MinD. Yeast two-hybrid assays showed that the self-association was not affected but the interaction with MinC was abolished. When fused to GFP and examined by fluorescence microscopy, these mutant proteins display a halo-like appearance, indicating membrane interaction. Further, in vitro biochemical assays showed that they still have ATPase activity that can be stimulated by MinE. Thus, it appears that the switch I region of MinD is involved in MinC interaction. Truncation and site-directed mutagenesis of the carboxyl terminus of MinD indicated that it is involved in membrane interaction, a conclusion also reached by T. Szeto in G. King’s group (Drake University) (T. H. Szeto, S. L. Rowland, S. Kobsa, and G. F. King, ASM Conf. Prokaryot. Dev., abstr. 39, 2002).
MinE has two functions that can be assigned to different domains: the N-terminal domain is involved in antagonizing MinCD inhibition, and the C-terminal domain is involved in topological specificity. Y.-L. Shih in L. Rothfield's laboratory (University of Connecticut Health Science Center) described progress toward identifying the functional sites within the topological specificity domain of MinE (MinETSD) (Y. Shih, G. F. King, and L. I. Rothfield, ASM Conf. Prokaryot. Dev., abstr. 25, 2002). Based upon the structure of MinETSD (64), solvent-exposed residues of MinETSD were targeted for site-directed mutagenesis followed by phenotypic assays. The study demonstrated that the two α-helical and β-sheet faces of MinETSD provide topological specificity using different mechanisms. Asp45 and Val49 form a contiguous patch near the center of the α-helical face of MinETSD. This patch is required for the formation of the MinE ring. Data obtained from a D45A/V49A double mutant provided evidence for the involvement of the MinE ring in arresting MinD polar zone assembly near midcell and triggering the disassembly of MinD polar zones.
The Min proteins are also involved in the determination of cell division site placement in cocci, such as Neisseria gonorrhoeae (98, 108). Previously, J. Szeto and other members of the Dillon group (University of Ottawa) showed that MinD self-associates and that MinD from N. gonorrhoeae (MinDNg) is active in E. coli (108). In a poster presentation, they correlated MinDNg structure and function using a variety of assays that examined the abilities of different MinDNg mutants to influence cell division in gonococcal and E. coli backgrounds, to oscillate intracellularly, to interact with themselves and with other Min proteins, and to hydrolyze ATP (J. Szeto, S. R. M. Costford, S. Ramirez-Arcos, and J. R. Dillon, ASM Conf. Prokaryot. Dev., abstr. 13, 2002). These results highlight the complexity of MinD function, showing that discrete mutations in MinDNg can produce nonfunctional protein by simultaneously affecting its localization, interactions, and enzyme activity. In addition, their data suggest that self-interaction alone is insufficient for MinD function.
In B. subtilis, polar division is also blocked by the activities of the division inhibitor MinCD. In this case, these proteins are stably maintained at the cell poles through interaction with DivIVA (23). DivIVA has a second role in sporulating cells. It interacts with the chromosome segregation machinery to position the replication origin near the cell pole (110). DivIVA also plays a critical role in Streptomyces, as discussed by K. Flardh (Uppsala University). S. coelicolor grow in a highly polarized fashion. New cell wall material is preferentially incorporated at the hyphal tips. A DivIVA-EGFP fusion protein localized at growing hyphal tips but not at the cell poles generated by cell division (i.e., at the hyphal cross walls). DivIVA is essential in S. coelicolor but repression of divIVA expression resulted in a phenotype reminiscent of that in many fungal mutants with defects in tip growth or nuclear migration (i.e., irregular and crooked cell shape, apical branching, and poor growth). On the other hand, overexpression of DivIVA gave rise to short, swollen, and rounded cells instead of hyphae. These results indicate that, in contrast to the cell division role found for B. subtilis DivIVA, S. coelicolor DivIVA plays a critical role in hyphal tip growth and cell shape determination.
In C. crescentus, cell division results in asymmetric progeny that follow different developmental fates. Whereas the stalked cell is capable of immediately initiating a new round of DNA replication, the motile swarmer cell must differentiate into a sessile stalked cell in order for chromosome replication to occur. The C. crescentus origin has five CtrA binding sites (96). CtrA represses chromosome replication in swarmer cells (96), but CtrA is degraded in stalked cells, allowing chromosome replication (20). In C. crescentus, as in most eubacteria, chromosome replication is initiated by the essential DnaA protein. Repression of dnaA expression results in a block in the initiation of DNA replication. Interestingly, cell division also stops, but not swarmer-to-stalked-cell differentiation (32). In E. coli, DnaA is subject to tight regulation to ensure that replication occurs once and only once per cell cycle. This regulation is provided through titration of DnaA, regulated inactivation of DnaA, and sequestration of the origin. Graduate student B. Gorbatyuk (G. Marczynski laboratory, McGill University) provided evidence that in C. crescentus, DnaA is subject to a fourth level of control: selective cell cycle proteolysis. He showed that DnaA is naturally unstable in C. crescentus but is stabilized in the presence of azide, suggesting that its turnover requires ATP-dependent proteases. Preliminary evidence suggests that ClpP is the protease involved in this degradation. The majority of DnaA protein synthesized during each of the stages of the cell cycle is degraded just prior to cell division. Therefore, newly divided progeny rely on de novo-synthesized DnaA to initiate replication.
Development of M. xanthus involves a portion of the rod-shaped cells in a population differentiating into spherical spores within fruiting bodies. Other cells remain outside fruiting bodies and are called peripheral rods. L. Tzeng in M. Singer’s group (University of California, Davis) examined the possibility that the cell cycle state of M. xanthus governs progression through the developmental program. Vegetative cultures of M. xanthus are asynchronous with respect to the cell cycle; therefore, when development is initiated the population has a heterogeneous chromosome number. Using flow cytometry and fluorescence microscopy, it was shown that after development, myxospores and peripheral rod cells are homogeneous populations containing 2n and 1n chromosome complements, respectively. The benefit of having two copies of the chromosome in a spore is unknown. Addition of DNA replication inhibitors within the first 6 h of development caused a dramatic decrease in both fruiting body formation and sporulation, while addition of these same inhibitors after 12 h had no effect on development. These data suggest that chromosome number is controlled early during the developmental program, leading to the intriguing possibility that cell cycle cues influence the ultimate fate of developing M. xanthus cells.
MORPHOGENESIS
As discussed earlier, the actin-like proteins (MreB and ParM) appear to play critical roles in the partitioning of DNA. However, several proteins of the MreB family were first predicted to play a cytoskeletal-type role in bacterial cell morphogenesis (54). J. Errington (University of Oxford) described recent progress in characterizing the B. subtilis actin homologue, Mbl. B. subtilis has two actin-like proteins, MreB and Mbl, that were shown previously to form helical filamentous structures running just under the surface of the cell and which are required for cell shape determination (54). Like actin, Mbl was able to polymerize in vitro and to bind and hydrolyze ATP. In vivo experiments with a fully functional GFP-Mbl fusion showed that the filaments are dynamic, undergoing continuous remodeling during growth of the cell. Several lines of evidence suggest that Mbl, together with its paralogue, MreB, directs the localization of penicillin-binding proteins and peptidoglycan precursor synthetic enzymes to specific sites of cell wall synthesis (“peptidoglycan factories”). Y. Wei and D. Popham (Virginia Tech) presented evidence that penicillin binding proteins PBP2 and YkuA play redundant roles in directing the peptidoglycan synthetic machinery to produce a rod-shaped, rather than an ovoid, B. subtilis cell (Y. Wei and D. L. Popham, ASM Conf. Prokaryot. Dev., abstr. 117, 2002).
R. Figge and J. Gober (UCLA) presented evidence for a similar role for MreB in C. crescentus. MreB forms spirals that assemble and disassemble in a dynamic manner. During most of the cell cycle, small spirals are distributed over the cell. After cell division is initiated, a predominant spiral forms at midcell in an FtsZ-dependent manner and disassembles at the end of cell division. Interestingly, PBP2, which is involved in cell shape determination in E. coli, forms small MreB-dependent spirals in C. crescentus. One attractive model is that MreB spirals serve as tracks for the peptidoglycan-synthesizing machinery. The small spirals would track the cell elongation complex and the large spirals at midcell would track the FtsI/FtsW complex required for septation. Based on the segregation defect of the mreB mutants, a third function of MreB filaments would be in chromosome partitioning, as discussed above.
One of the most impressive morphological feats is the biogenesis of a spore coat. The outermost proteinaceous layer of many bacterial spores, called the coat, is critical for spore survival, germination, and, for some species, pathogenesis. The coat is composed of over 30 structural components, and J. Maddock (University of Michigan) presented a poster (E. Lai, N. Phadke, A. Driks, and J. R. Maddock, ASM Conf. Prokaryot. Dev., abstr. 118, 2002) describing a proteomics approach that revealed additional spore coat proteins. The proteins that comprise the coat form a series of layers that are evident in thin-section electron micrographs. In B. subtilis, this layered architecture is directed, in part, by the coat protein CotE. A. Driks and coworkers (Loyola University) have identified multiple functional modules within CotE. One of these is responsible for guiding CotE to a specific location within the coat layers. Other modules direct the assembly of specific subsets of coat proteins. Preliminary protein-protein interaction studies suggest that CotE directly interacts with most of the proteins whose assembly it controls. Work described by T. Costa of A. Henriques's lab (Universidade Nova de Lisboa) revealed that CotE is critical for spore resistance to lysozyme, even under conditions where most of the outer coat proteins are missing (T. Costa, M. Serrano, A. J. Ozin, C. P. J. Moran, and A. O. Henriques, ASM Conf. Prokaryot. Dev., abstr. 102, 2002).
The current models of coat assembly address how the architecture of the coat layers are formed but not how assembly within a layer is controlled. To identify proteins critical for assembly within each layer, A. Driks and colleagues used atomic force microscopy to characterize the coat surfaces from wild-type and coat protein mutant spores. This analysis showed that CotA and CotB are needed for normal surface morphology, although not for establishment of the coat layers. Therefore, they constitute a novel class of morphogenetic factors.
The bacterial flagellum is a remarkable molecular propeller that mediates movement of many different types of bacteria. Flagellum assembly requires ordered export and polymerization of small numbers of hook and hook-associated protein (HAP) subunits and about 20,000 filament subunits (flagellin, FliC). The FliC polymerizes onto the distal end of the growing filament following export and passage through the channel running through the center of the growing flagellum. Since the channel diameter is small (105), it is likely that FliC traverses the channel in a partially unfolded state. Export of FliC and the other axial proteins (hook, HAPs, and cap) is facilitated by dimeric substrate-specific chaperones (FlgN, FliT, and FliS) in the cytosol. Previous work in the laboratory of C. Hughes (Cambridge University) showed that the FlgN protein binds to the HAPs FlgK and FlgL, while the FliT protein binds FliD (28). At this meeting, C. Hughes detailed the interactions between the chaperone FliS and FliC. FliS binds to the C-terminal domain of FliC and prevents premature polymerization of the newly synthesized flagellin subunits in the cytoplasm (5). Thus, it appears that all three chaperones act as substrate-specific bodyguards that pilot their substrates to the export channel for secretion.
Export of the axial proteins occurs through a specialized type III export apparatus in the membrane located at the base of the basal body. The FliI ATPase is thought to couple ATP hydrolysis with secretion. FliH binds to FliI and negatively regulates its activity. Recent studies from the Hughes (4) and Macnab (31) laboratories have shown that the C-terminal domain of FliH is essential for FliH dimerization and interaction with FliI. Furthermore, membrane association of FliI appears to be critical for its control as FliI ATPase activity was stimulated tenfold by bacterial phospholipids. Thus, association of FliI with the membrane, perhaps mediated by interactions with FlhA and FlhB (136), may provide topological cues that allow secretion at the correct location.
In addition to flagella, bacteria express a variety of surface appendages that are important for adherence and invasion. Curli fimbriae are extracellular fibers produced by E. coli and Salmonella that are involved in host colonization and biofilm formation. Interestingly, curli are a bacterial amyloid (nonbranching, aggregated fibrils consisting of proteins), sharing distinguishing features with the medically important pathological fibers that are the hallmark of many neurodegenerative diseases. Eukaryotic amyloid formation is thought to be a biological accident—the result of misbehaving proteins that adopt nonnative yet stably folded structures. However, curli formation is clearly not a mistake. It is the end product of an elaborate and finely tuned amyloid assembly system. Curlin assembly requires the products of at least five genes located on two divergently transcribed operons. M. Chapman, a postdoctoral fellow with S. Hultgren (Washington University), described recent work on the role of different csg gene products on the assembly of curli fibers (15). He found that the major curlin subunit, CsgA, was secreted from bacteria in a CsgG- and CsgE-dependent fashion. CsgG oligomerizes in the outer membrane into a barrel-like structure with an apparent central pore of 2 nm. This structure is the backbone of the curli subunit secretion apparatus, and without it CsgA is confined to the periplasmic space, where it is proteolytically degraded. Secretion of CsgA is guided by the 22 N-terminal amino acids on mature CsgA, which form a specific CsgG-dependent secretion signal. CsgE is a type III-like chaperone that is required for full CsgG stability and activity. Once on the cell surface, CsgA interacts with the nucleator protein, CsgB. In the absence of CsgB, no curli are formed and CsgA remains in a soluble, monomeric state. When purified, CsgA can spontaneously assemble into amyloid fibers, demonstrating that amyloid formation is an inherent property of the CsgA protein. Teasing apart the details of curli biogenesis will continue to elucidate a model of amyloid fiber formation, and it will also give insights into such fundamental bacterial processes as protein secretion, folding, and assembly.
In order to gain insight into prokaryotic development and to determine the interplay with evolution, U. Jenal's lab (University of Basel) has been carrying out experimental evolution studies in C. crescentus. M. Ackermann described studies aimed at investigating how development and differentiation of C. crescentus would change in response to long-term evolution in a homogenous environment. Under such conditions, selection for asymmetry is released since there is no longer an advantage in producing motile, division-incompetent swarmer cells. Furthermore, the experimental conditions impose a strong selection for rapid growth. Three populations of cells were serially propagated for 7,000 generations in liquid culture. In all populations, growth rates increased rapidly in the first 1,000 generations, primarily due to a shortening of the swarmer phase. At later stages, each population acquired unique morphological alterations such that they differed from each other and from their ancestor. These later morphological changes were dependent on mutations that were fixed in earlier generations. Thus, the tight connections between morphogenesis and replication that are characteristic of the C. crescentus cell cycle initially constrained the evolution of morphology, but these constraints were released by changes that occurred during the first 1,000 generations. Likewise, M. xanthus organisms that evolved in nutrient liquid rapidly lost social motility and development behaviors (115).
SIGNALING AND MULTICELLULARITY
Entry into the B. subtilis sporulation pathway is governed by a phosphorelay that integrates a variety of signals and controls the phosphorylation of the key transcription factor Spo0A. This checkpoint includes five histidine kinases that respond to positive signals for sporulation by phosphorylating Spo0F (51). The checkpoint also involves members of two families of protein phosphatases that respond to negative signals for sporulation by dephosphorylating phosphorylated Spo0A (Spo0A∼P) or Spo0F∼P (93). M. Perego (Scripps Research Institute) has focused on biochemical and structural characterization of members of the Rap family of proteins, at least three of which are phosphatases that dephosphorylate Spo0F∼P (45). One of these, RapA, is inhibited by a pentapeptide derived from the 44-amino-acid product of phrA. The PhrA precursor is secreted and cleaved extracellularly, and the pentapeptide inhibitor is imported. Whether this serves as a timing device or a means to sense cell density or some other environmental factor is unclear. The PhrA pentapeptide inhibits RapA by disrupting its interaction with Spo0F∼P. Very recent work indicates that another Rap family member, RapC, regulates the development of competence to take up DNA by a different mechanism. RapC does not induce dephosphorylation of the key response regulator ComA∼P. Instead, it inhibits the ability of ComA∼P to bind to its target promoters. Hence, not all Rap proteins are phosphatases. More generally, they are proteins that interact directly with response regulators to inhibit their activity. FliX, of the C. crescentus flagellar hierarchy, is thought to interact directly with the FlbD response regulator (81), as noted above. This may be a case of functional, but not structural, homology, because Rap proteins are characterized by the presence of six tetratricopeptide repeats known to promote protein-protein interactions (92), and such repeats have not been recognized in FliX.
Exit from sporulation is a complex process, known as germination, that includes loss of spore properties and resumption of metabolism. Germination is triggered by nutrients that are sensed by receptors, resulting in a number of biochemical and physical changes, including the release of dipicolinic acid (DPA) as a calcium chelate. Ca2+-DPA can trigger germination of spores lacking nutrient receptors. Studies in P. Setlow's lab (University of Connecticut) have shown that CwlJ is essential for the germination response to Ca2+-DPA (89). CwlJ is an enzyme involved in the lysis of the peptidoglycan cortex that surrounds the spore (Fig. 2E). K. Ragkousi of the Setlow lab has performed a genetic screen that identified another component of the Ca2+-DPA response pathway. The gene identified, ywdL, is transcribed by σE RNA polymerase in the mother cell and is necessary for localization and/or stability of CwlJ.
Wild isolates of B. subtilis exhibit robust multicellular behaviors not seen with domesticated strains used in most labs. In standing (rather than shaking) culture, wild isolates form a floating biofilm with aerial projections (12). Spores form preferentially at the tips of the projections, so they have been called fruiting bodies, by analogy with myxobacterial fruiting bodies. Genes involved in B. subtilis biofilm and fruiting body formation are being identified by using mutational (S. Branda, E. Gonzalez-Pastor, R. Losick, and R. Kolter, ASM Conf. Prokaryot. Dev., abstr. 61, 2002) and microarray (N. Stanley, R. Britton, A. Grossman, and B. Lazazzera, ASM Conf. Prokaryot. Dev., abstr. 26, 2002) approaches. The genes encode transcriptional regulators that control entry into sporulation, biosynthetic enzymes for a surfactant and for extracellular matrix material, and proteins potentially involved in signaling. Postdoc D. Kearns of the Losick lab has discovered another behavior in the B. subtilis repertoire. A wild isolate exhibits swarming motility. This strain, unlike lab strains, can move on a solid surface (0.7% agar medium). Cells spotted on agar medium, after a delay, become hyperflagellated and swarm outward in groups. This type of movement requires flagella, lipopeptide production, surfactin, and a subset of the chemotaxis genes. The involvement of chemotaxis genes, together with the observation that the lag between spotting and swarming varies with cell density, suggests that signaling initiates swarming behavior. Based on studies of M. xanthus swarming motility (see below), it is likely that B. subtilis cells interact extensively during swarming.
M. xanthus has long been known to glide over solid surfaces using two different motility systems. System A (adventurous) controls movement of cells as individuals, and system S (social) governs movement of cells in groups (123). S motility is related to twitching motility in other bacteria in that it requires type IV pili. These pili are thought to extend from the end of an M. xanthus cell and then retract, pulling the cell along. S motility also requires extracellular fibril material, composed of about equal amounts of protein and carbohydrate. Evidence from W. Shi's lab (UCLA) supports the idea that fibril material stimulates pilus retraction (Y. Li, H. Sun, and W. Shi, ASM Conf. Prokaryot. Dev., abstr. 44, 2002). System A motility may be related to that in certain cyanobacteria, where hydration of polysaccharide in nozzle structures at the cell surface is believed to result in slime extrusion that propels the bacterium (123). Consistent with this idea, genes likely to be involved in polysaccharide synthesis have recently been identified in a screen for A motility mutants (R. Yu and D. Kaiser, ASM Conf. Prokaryot. Dev., abstr. 21, 2002).
P. Hartzell (University of Idaho) is investigating how the activity of M. xanthus's two motility systems is coordinated. mgl is a locus that affects both systems. It encodes MglA, a novel GTPase, and MglB, a putative GDP release factor (37). An mglA mutant appears to be nonmotile, based on colony phenotype, but microscopic examination reveals rapid reversal of gliding direction, resulting in no net movement (107). The Hartzell lab isolated a second-site mutation that partially restores a motile colony phenotype to an mglA8 missense mutant. The second-site mutation is in a gene near mgl that encodes a membrane tyrosine kinase of the serine-threonine-tyrosine (STY) family, which has not been recognized in prokaryotes previously (111). The mutation restores only S motility and results in greatly increased levels of extracellular fibril material. The STY kinase interacts directly with MglA in a yeast two-hybrid assay. This assay also identified AglZ, a protein with response regulator and coiled-coil domains, as an MglA-interacting partner. Disruption of aglZ blocks A motility. Hence, MglA interacts directly with components of both the A and S systems, and further studies should reveal how activity of the two systems is coordinated.
The activity of each motility system is also regulated by interactions between M. xanthus cells. Certain mutants can be stimulated to move transiently after contact with wild-type cells. Graduate student E. Nudleman of D. Kaiser's lab (Stanford University) has demonstrated that lipoproteins Tgl and CglB are transferred between cells to stimulate S and A motility, respectively (E. Nudleman, D. Wall, and D. Kaiser, ASM Conf. Prokaryot. Dev., abstr. 101, 2002). Contact dependent transfer of lipoproteins is proposed to coordinate the swarming behavior of large groups of cells.
Certain lipids can also affect the motility behavior of M. xanthus cells, altering the frequency with which they reverse their direction of gliding, to elicit a chemotactic response (61). Extracellular fibril material, which is now implicated in pilus retraction thought to drive S motility (see above), is necessary for excitation by dilauroyl phosphatidylethanolamine. Moreover, a fibril protein, FibA, has been shown to be essential for this chemotactic response (60). FibA is similar to a family of zinc metalloproteases that are secreted as inactive zymogens and are activated by autoproteolysis. Graduate student P. Bonner of L. Shimkets's lab (University of Georgia) has analyzed fibA mutants for the putative self-cleavage and for the lipid chemotactic response. Changing a glutamic acid residue to aspartic acid in the putative active site abolished the lipid response but not processing of FibA, as if FibA protease activity is needed for the chemotactic response but not for self-cleavage. Perhaps another protease processes FibA to its active form. Deleting one of FibA's two C-terminal repeats still allowed the lipid response, but deleting both repeats eliminated it. The role of the repeat, the identity of FibA's substrate, the issue of whether FibA directly senses lipid, and the reason for placing this novel signal transduction system in the extracellular fibril material are some of the interesting questions that remain.
The FibA pathway, as well as other signal receptors, may provide input into the frizzy (frz) system of chemotaxis (che)-like genes discovered in D. Zusman's lab (University of California, Berkeley) (118). Classical frz mutants exhibit a frizzy colony morphology due to reduced cell reversal frequency. A C-terminal truncation of FrzCD causes the opposite phenotype. Cells change direction so often that little net movement occurs, resulting in a colony that appears almost nonmotile (similar to the mglA mutant described above). Postdoc J. Kirby of the Zusman lab has searched the partial M. xanthus genome sequence in the Cereon Microbial Sequence Database (http://microbial.cereon.com) and discovered at least nine clusters of che-like genes. Mutations in the che3 locus cause early aggregation of starved cells, even at low densities. Several developmental genes are overexpressed, and the fruiting bodies contain few spores. The locus does not encode a CheY-like response regulator, which typically mediates motility responses. However, immediately upstream and divergently transcribed is a gene, crdA (for “chemosensory regulator of development”), encoding an NtrC-like transcription factor. CrdA interacts with the CheA homolog of the che3 locus in a yeast two-hybrid assay. Therefore, the che3 locus is proposed to encode a Che-like signal transduction system that regulates developmental gene transcription. This theme of using che-like genes to regulate development was revisited in a talk by J. Berleman of C. Bauer's lab (Indiana University). Mutations in che-like gene clusters other than the one shown previously to control chemotaxis and phototaxis of Rhodospirillum centenum (52) caused cyst formation under nutrient conditions that would normally promote growth. In both R. centenum and M. xanthus, the broad sensitivity range and adaptation features of chemosensory systems may be employed for nutrient sensing in the decision whether to initiate development.
Other che-like gene clusters in M. xanthus impact development in a manner more like frz, by affecting motility. The che4 (H. Vlamakis, J. Kirby, and D. Zusman, ASM Conf. Prokaryot. Dev., abstr. 123, 2002) and dif (130) clusters specifically affect S motility. Mutational (W. Black, K. Bellenger, and Z. Yang, ASM Conf. Prokaryot. Dev., abstr. 128, 2002; H. Vlamakis et al., ASM Conf. Prokaryot. Dev.) and yeast two-hybrid (H. Lancero and W. Shi, ASM Conf. Prokaryot. Dev., abstr. 1, 2002) analyses are being used to discern the function and interactions of proteins in these systems. The theme of using more than one che-like cluster to control motility is also observed for the phototactic response of the gliding cyanobacterium Synechocystis sp. strain PCC6803 (D. Bhaya and A. Grossman, ASM Conf. Prokaryot. Dev., abstr. 55, 2002). Integration of signals transduced by more than one che-like system likely involves the target of the CheY-like proteins, which remains to be identified.
Starvation changes the gliding movements of M. xanthus cells. A population of starved cells self-organizes in patterns called ripples, streams, and aggregates. Postdoc R. Welch of D. Kaiser's lab (Stanford University) has developed clever experimental systems to observe the movement of cells as they form these patterns. Ripples are sets of parallel traveling waves in which the wave “crests” are areas of higher cell density and the “troughs” between waves are areas of lower cell density. Conditions were established under which a population of M. xanthus produced opposing ripples (119). Movement of individual cells tagged with GFP was monitored in the rippling population. The characteristics of cell movement during rippling were used to successfully model rippling behavior in one dimension (44), and more recently in two dimensions. Under different conditions, designed to evoke a strong stringent response, cells move together in streams and gradually form aggregates that become fruiting bodies. While streaming cells are frequently oriented toward an aggregate, cells do not always stream to the nearest aggregate and remain there. Streams frequently travel past the nearest aggregation center to one that is more distant, and individual cells move in and out of aggregates at the same speed. These observations suggest complexity in the relationship between streaming and aggregation. Further investigation of this relationship promises to provide new insight into the mechanics of building a fruiting body.
C signaling plays a key role in the temporal and spatial coordination of rippling, streaming, aggregation, developmental gene expression, and sporulation during M. xanthus development. The C signal is encoded by the csgA gene. The full-length CsgA protein (24.5 kDa) is similar to the short-chain alcohol dehydrogenase family of proteins and has an N-terminal NAD(P)+ binding pocket (73). There is disagreement about whether full-length CsgA can rescue the C signaling defect of csgA mutant cells (M. Avadhani and L. Shimkets, ASM Conf. Prokaryot. Dev., abstr. 29, 2002; S. Lobedanz and L. Sogaard-Andersen, ASM Conf. Prokaryot. Dev., abstr. 4, 2002), but it is clear that a shortened 17-kDa form (p17) is sufficient for rescue (63). Graduate student S. Lobedanz of L. Sogaard-Andersen's lab (University of Southern Denmark) has found that p17 corresponds to the C-terminal 17-kDa portion of CsgA and lacks the ability to bind NAD+ (Lobedanz and Sogaard-Andersen, ASM Conf. Prokaryot. Dev.). p17 can be produced from full-length CsgA in vitro by an unidentified protease with an inhibition profile that suggests that it is a serine protease.
The C signal remains associated with the cell surface, and C signaling requires contact between cells. Early in development, cells produce a low level of CsgA, and this induces rippling behavior (75). When a cell makes end-to-end contact with a countermigrating cell in a neighboring ripple, C signaling causes both cells to reverse their direction of movement (119). Hence, two colliding waves reflect from each other. This signaling leads to a rise in csgA expression due to a positive feedback loop elucidated in D. Kaiser's lab (Stanford University). The loop involves the products of the act operon, which regulate csgA transcription to ensure a steady rise of C signal as development proceeds (33). The act operon encodes a compound response regulator (ActA) similar to PleD of C. crescentus and an NtrC-like transcription factor (ActB). In-frame deletion of actA or actB eliminates positive feedback, so the level of C signal does not rise, resulting in prolonged rippling rather than progression to the streaming pattern of motility. In-frame deletion of actC causes C-signal levels to rise rapidly and speeds the course of development, whereas deletion of actD (encoding a protease whose substrate is unknown) has the opposite effect. Determining how the Act proteins sense early C signaling and respond by increasing csgA transcription is a key challenge for the future.
The rising level of C signal causes a change in the cellular response to C signaling. Cells glide in the same direction (streaming), rather than reversing direction (rippling), upon contact (49). Cells also increase their average speed. Streaming eventually leads to aggregation. Within aggregates, cells align and make many contacts. C signaling in the aggregate causes further C-signal production and triggers expression of some developmental genes (57). Other developmental genes and the sporulation differentiation program itself exhibit a still higher threshold for induction by C signaling.
How do cells respond differently to different levels of C signal during the course of development? Little is known. A receptor(s) for C signal has not yet been identified. A response regulator, FruA, is likely phosphorylated (24), but the cognate kinase(s) has been elusive. The immediate targets of FruA∼P are unknown, but it is clear that fruA is needed for changes in motility and gene expression. Nearly all genes induced after the early aggregation stage at about 6 h of M. xanthus development exhibit partial or absolute dependence on C signaling for expression. P. Viswanathan of L. Kroos's lab (Michigan State University) has characterized the cis-acting DNA elements necessary for expression of a promoter that depends absolutely on C signaling (117). For comparison, D. Yoder of Kroos's lab has studied promoters that depend partially on C signaling (D. Yoder and L. Kroos, ASM Conf. Prokaryot. Dev., abstr. 18, 2002). Different patterns of mutational effects in sequences that are conserved among the promoters suggest that they are recognized by different proteins. Identifying these proteins would be an important step toward understanding differential gene expression in response to C signaling.
The switch of S. coelicolor from growth as substrate hyphae to formation of aerial hyphae also appears to be governed by extracellular signals (Fig. 3). Extracellular complementation between different bld mutants, which alone fail to form aerial hyphae, suggests that a cascade of at least five signals regulates the production of SapB, a secreted, hydrophobic peptide that acts as a surfactant, thought to allow hyphae to overcome surface tension at the colony-air interface and project upward (112, 121). J. Willey (Hofstra University) reported that two other streptomycetes, Streptomyces tendae and Streptomyces hygroscopicus, use a similar strategy to solve the surface tension problem. Complete structural characterization of these small (8- to 18-amino-acid) peptides has been difficult due to their hydrophobicity. Isolation of the bld-derived signals that precede SapB in the S. coelicolor signaling cascade has been even more problematic, but persistent efforts in the Willey lab have recently led to the extraction of signaling molecules from spent agar on which bldC or bldG mutant strains had been grown. The extracts restored the capacity of several other bld mutants to undergo complete morphological differentiation. This is the first step toward further purification and structural elucidation of bld-derived signal molecules.
The bld signaling cascade can be interpreted as a series of checkpoints, each involving a signal produced by hyphae undergoing physiological change under the control of bld genes (Fig. 3). K. Chater (John Innes Center) focused on the checkpoint regulated by bldA and bldH. The bldA gene product is the only tRNA that recognizes the leucine codon UUA, which is the rarest codon in S. coelicolor. It seems that bldA exerts its effect on morphological development (but not on secondary metabolism) through a UUA codon in bldH mRNA. In terms of controlling aerial-hypha formation, bldH appears to be the S. coelicolor equivalent of the homologous Streptomyces griseus adpA gene (85). AdpA is an AraC-like regulatory protein through which the γ-butyrolactone A factor of S. griseus exerts its effects on both secondary metabolism and normal morphological development. A γ-butyrolactone signal affects secondary metabolism but not development of S. coelicolor (109). This signal is communicated to antibiotic biosynthetic genes via bldA but not through the UUA codon in bldH mRNA. Rather, the target of the bldA tRNA is antibiotic pathway-specific regulatory gene mRNAs that contain a UUA codon. These studies show that there are differences in developmental decision-making at this stage among Streptomyces spp., both in the signals that activate developmental regulatory genes and in the targets of action of these genes.
How are signals perceived by S. coelicolor? The ram locus encodes five proteins, at least two of which play a key role in signal transduction governing aerial hyphae formation. RamR is a response regulator that when overproduced allows bld mutants to form aerial hyphae (K. Nguyen, J. Willey, L. Nguyen, P. Viollier, and C. Thompson, ASM Conf. Prokaryot. Dev., abstr. 3, 2002). J. Nodwell's group (McMaster University) has shown that RamR binds to the promoter region of the ramCSAB operon (84). Unphosphorylatable mutants of RamR fail to support morphogenesis, but a cognate kinase has not been found. Mutational analysis of ramC supports the idea that it encodes a membrane-associated serine/threonine kinase, which may serve as a receptor for an extracellular signal (43). The ramCSAB operon is expressed in substrate hyphae but not aerial hyphae, and the RamC protein is absent from spores. Expression of ramCSAB depends not only directly on RamR but also indirectly on BldD and CprA, suggesting a further connection in the signaling network, since CprA may be a γ-butyrolactone receptor and a transcription factor (87).
Cyanobacteria resemble streptomycetes in that both grow to produce chains of cells. The cyanobacterium Anabaena sp. strain PCC7120 grows as filaments with incomplete separation between adjacent cells at the level of the outer membrane. Hence, a signal that crosses the inner membrane of one cell can diffuse along the filament and affect other cells. Several years ago J. Golden (Texas A&M University) and his colleagues discovered that this type of signaling helps produce the developmental pattern of single nitrogen-fixing heterocysts separated by 10 to 15 vegetative cells upon nitrogen limitation (131). The signal is thought to be a 13- or 17-amino-acid peptide encoded by patS, which is expressed in clusters of cells early in development and by lateral inhibition is believed to resolve the clusters into single preheterocysts spaced approximately equidistantly along the filament (132). A synthetic peptide representing the C-terminal five amino acids of PatS can inhibit heterocyst development. Continued studies in J. Golden's lab show that a minigene engineered to express only the last five codons of patS inhibits heterocyst formation. Since the pentapeptide expressed in this way is unlikely to be secreted, the results suggest that the PatS receptor is located in the cytoplasm rather than on the cell surface. To identify components of the PatS signaling pathway, mutations that can bypass the heterocyst inhibition caused by patS overexpression have been sought. Some lie in hetR, which encodes a key regulator of early heterocyst-specific gene expression (1). Others lie in a new gene, hetL, which when overexpressed stimulates heterocyst formation. HetL is predicted to be an unusual protein consisting almost entirely of nearly 40 copies of a pentapeptide-repeat motif. While clearly important, PatS signaling is not the only determinant of heterocyst patterning. Analysis of a patS null mutant indicates that products of nitrogen fixation are involved (132).
Like many eukaryotes, cyanobacteria possess internal timing systems called circadian clocks. S. Golden (Texas A&M University) described the clock of Synechococcus elongatus (53). Although the mechanism of 24-h timekeeping by the three products of the kai locus remains a mystery, the clock shares with other developmental processes the properties of responding to environmental cues and regulating expression of many genes. Cues from the environment can reset the clock via CikA (circadian input kinase), which has a plant phytochrome-like domain that could conceivably respond to light, and govern the activity of its kinase domain, perhaps directly or indirectly leading to phosphorylation of a Kai protein (106). In this issue, Katayama et al. (59) describe another gene involved in light-dependent modulation of the circadian period. Output from the clock involves SasA, a histidine protein kinase that interacts directly with KaiC (46), and a family of four group 2 σ factors (83). Knocking out one of the σ genes lengthened the circadian period of expression from one promoter but not two others, demonstrating that different timing circuits can coexist in a cell. How the σ factors coordinate their activities to convey circadian information to downstream genes in this system is relevant to the long-standing question of how σ factor activity is coordinated in cascades during phage and bacterial development (56, 135).
Many species of bacteria utilize a type of signaling called quorum sensing to regulate their gene expression and multicellular behavior in response to changes in population density (122). Acyl-homoserine lactone-based or peptide-based intraspecies quorum sensing plays a role in the formation of bacterial biofilms. In medical and industrial settings, biofilms have a significant impact and are often composed of multiple species. For example, dental plaque is a major cause of tooth decay and contains Streptococcus mutans in association with many other bacteria. Interspecies quorum sensing often involves the luxS-dependent signal known as autoinducer 2, recently shown to be a furanosyl borate diester (17). An ortholog of luxS was identified in S. mutans (J. Merritt, S. Goodman, F. Qi, M. Anderson, and W. Shi, ASM Conf. Prokaryot. Dev., abstr. 45, 2002). Disruption of this gene eliminated autoinducer 2 production and changed the structure of the S. mutans biofilm, providing the first evidence for this type of signaling in S. mutans biofilm formation. The oral pathogen Actinobacillus actinomycetemcomitans also forms biofilms. Transposon mutations that prevent synthesis of the O-antigen component of LPS alter biofilm development (J. Kaplan, M. Meyenhofer, and D. Fine, ASM Conf. Prokaryot. Dev., abstr. 42, 2002). Secretion of exopolysaccharide to form the extracellular matrix plays an important role in the early stages of biofilm formation by Vibrio cholerae (88). Other early steps require extracellular appendages: flagella enable movement to a suitable surface, and pili allow attachment. N. Bomchil of R. Kolter's lab (Harvard Medical School) has identified a gene that affects a late stage of biofilm development (10). Normally, pillars of cells encased in matrix material are separated by channels that permit fluid passage. A knockout of mbaA (for “maintenance of biofilm architecture”) results in a biofilm with few or no channels. Interestingly, the predicted MbaA protein contains GGDEF and EAL domains, which are thought to be characteristic structural features of a novel family of signal transduction proteins (30). This family includes PleD of C. crescentus, a key regulator of cell cycle events. MbaA may regulate production of extracellular matrix material so that pillars and channels are maintained in the mature biofilm. In the soil bacterium and plant pathogen Agrobacterium tumefaciens, biofilm maturation is controlled by sinR, which is predicted to encode a transcriptional regulator of the FNR superfamily (B. Ramey and C. Fuqua, ASM Conf. Prokaryot. Dev., abstr. 12, 2002). Not surprisingly, it is becoming increasingly clear that the behavior of bacteria in biofilms relies on molecular mechanisms similar to those discovered in studies of the development of model organisms like C. crescentus, B. subtilis, M. xanthus, S. coelicolor, and E. coli.
CONCLUDING REMARKS
In our opinion, and based on conversations with other participants, this was a fabulous meeting. The pace of prokaryotic development research is quickening with the availability of complete genome sequences for most of the major model organisms. M. xanthus will soon join the club, as H. Kaplan (University of Texas—Houston) announced that funding has been obtained to complete its sequence. Genomic approaches are providing a more comprehensive view of the immense complexity of gene regulation and protein function, which bring about morphological change during development. The realization that a protein's function is often carried out at a specific location in the cell continues to grow, thanks to widespread use and continual improvements of techniques for visualizing GFP. Some of the rules for localizing proteins to particular subcellular addresses are beginning to emerge, and this promises to be a fruitful area for further investigation. Likewise, understanding how phospholipids affect the activity of peripheral and integral membrane proteins poses an important challenge. There is already evidence of an interplay between phospholipids and protein-protein interactions controlling ATPase activity of MinD in cell division site selection and of FliI in secretion of flagellar components. Do phospholipids, as well as protein-protein interactions, influence the activity of actin-like ATPases involved in plasmid (ParM) and chromosome (MreB) segregation or involved in cell shape determination (MreB and Mbl)? These questions should be amenable to biochemical approaches. More daunting, though, is the challenge of biochemical reconstruction when it involves integral membrane proteins. Yet it is tempting to speculate that interactions with phospholipids, as well as with proteins, influence the kinases that localize to the swarmer or stalk cell pole of C. crescentus to establish asymmetry, or that they influence the activity of SpoIIE phosphatase in the B. subtilis polar septum to ensure coupling between morphogenesis and gene expression. This theme of membrane-associated proteins playing pivotal regulatory roles extends to signaling interactions between cells. For example, a membrane protease cleaves a membrane-associated σ factor (pro-σK) in the mother cell of sporulating B. subtilis, in response to a signal from the forespore. This was one of the first examples of regulated intramembrane proteolysis, which is now emerging as a widely conserved mechanism for controlling diverse signaling pathways (114). In M. xanthus, contact between cells can result in transfer of lipoproteins to stimulate motility or in C signaling to orchestrate cell movement (rippling, streaming, and aggregation) and differentiation (sporulation within a fruiting body). M. xanthus fibrils act like antennae (22) to sense particular lipids and modulate gliding reversals, a chemotactic response. And chemotaxis-like genes in M. xanthus regulate not only motility but also developmental gene expression. Still more surprises await those who venture to work with wild isolates rather than domesticated lab strains, as exemplified by the discovery of B. subtilis fruiting bodies and swarming motility. Conversely, laboratory evolution of C. crescentus is yielding mutants with novel developmental defects. Stay tuned for more intriguing developments, because it is clear from this meeting that studies of prokaryotic development are well into the exponential phase!
Acknowledgments
We thank all the conference participants for making this a stimulating scientific meeting. We apologize to those whose work we were not able to review or cite due to space constraints. We thank the speakers that provided input on sections that discuss their research and we are especially grateful to L. Shimkets, Y. Brun, and K. Chater for critical reading of the entire manuscript.
Research in the laboratory of L.K. is supported by NIH grant GM43585, NSF grant MCB-0090478, a Strategic Partnership Grant from the Michigan State University Foundation, and the Michigan Agricultural Experiment Station. Research in the laboratory of J.R.M. is supported by NIH grant GM55133 and American Cancer Society grant RSG-01-090-01-MCB.
REFERENCES
- 1.Adams, D. G. 2000. Heterocyst formation in cyanobacteria. Curr. Opin. Microbiol. 3:618-624. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 3.Alley, M. R., J. R. Maddock, and L. Shapiro. 1993. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science 259:1754-1757. [DOI] [PubMed] [Google Scholar]
- 4.Auvray, F., A. J. Ozin, L. Claret, and C. Hughes. 2002. Intrinsic membrane targeting of the flagellar export ATPase FliI: Interaction with acidic phospholipids and FliH. J. Mol. Biol. 318:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auvray, F., J. Thomas, G. M. Fraser, and C. Hughes. 2001. Flagellin polymerization control by a cytosolic export chaperone. J. Mol. Biol. 308:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 7a.Ben-Yehuda, S., D. Rudner, and R. Losick. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science, in press. (First published 19 December 2002; http://www.sciencemag.org/cgi/content/abstract/1079914v1.) [DOI] [PubMed]
- 8.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase σ factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384. [DOI] [PMC free article] [PubMed]
- 11.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brun, Y. V., and L. Shapiro. 1992. A temporally controlled σ factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 6:2395-2408. [DOI] [PubMed] [Google Scholar]
- 14.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269-279. [DOI] [PubMed] [Google Scholar]
- 15.Chapman, M. R., L. S. Robinson, J. S. Pinkner, R. Roth, J. Heuser, M. Hammar, S. Normark, and S. J. Hultgren. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chater, K. 1998. Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465-1478. [DOI] [PubMed] [Google Scholar]
- 17.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 18.Claessen, D., H. A. Wosten, G. van Keulen, O. G. Faber, A. M. Alves, W. G. Meijer, and L. Dijkhuizen. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 44:1483-1492. [DOI] [PubMed] [Google Scholar]
- 19.Cutting, S., V. Oke, A. Driks, R. Losick, S. Lu, and L. Kroos. 1990. A forespore checkpoint for mother-cell gene expression during development in Bacillus subtilis. Cell 62:239-250. [DOI] [PubMed] [Google Scholar]
- 20.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 21.Dworkin, J., and R. Losick. 2001. Differential gene expression governed by chromosomal spatial asymmetry. Cell 107:339-346. [DOI] [PubMed] [Google Scholar]
- 22.Dworkin, M. 1999. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. Bioessays 21:590-595. [DOI] [PubMed] [Google Scholar]
- 23.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905-915. [DOI] [PubMed] [Google Scholar]
- 24.Ellehauge, E., M. Norregaard-Madsen, and L. Sogaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30:807-817. [DOI] [PubMed] [Google Scholar]
- 25.Feucht, A., L. Abbotts, and J. Errington. 2002. The cell differentiation protein SpoIIE contains a regulatory site that controls its phosphatase activity in response to asymmetric septation. Mol. Microbiol. 45:1119-1130. [DOI] [PubMed] [Google Scholar]
- 26.Feucht, A., R. A. Daniel, and J. Errington. 1999. Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis. Mol. Microbiol. 33:1015-1026. [DOI] [PubMed] [Google Scholar]
- 27.Frandsen, N., I. Barak, C. Karmazyn-Campelli, and P. Stragier. 1999. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 13:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser, G. M., J. C. Bennett, and C. Hughes. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569-580. [DOI] [PubMed] [Google Scholar]
- 29.Fujita, M., and R. Losick. 2002. An investigation into the compartmentalization of the sporulation transcription factor σE in Bacillus subtilis. Mol. Microbiol. 43:27-38. [DOI] [PubMed] [Google Scholar]
- 30.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Pedrajo, B., G. M. Fraser, T. Minamino, and R. M. Macnab. 2002. Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol. Microbiol. 45:967-982. [DOI] [PubMed] [Google Scholar]
- 32.Gorbatyuk, B., and G. T. Marczynski. 2001. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 40:485-497. [DOI] [PubMed] [Google Scholar]
- 33.Gronewold, T. M., and D. Kaiser. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40:744-756. [DOI] [PubMed] [Google Scholar]
- 34.Guillet, V., N. Ohta, S. Cabantous, A. Newton, and J. P. Samama. 2002. Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus. J. Biol. Chem. 277:42003-42010. (First published 10 August 2002; http://www.jbc.org/cgi/content/abstract/M204789200v1.) [DOI] [PubMed] [Google Scholar]
- 35.Guo, D., Y. Wu, and H. B. Kaplan. 2000. Identification and characterization of genes required for early Myxococcus xanthus developmental gene expression. J. Bacteriol. 182:4564-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hale, C. A., H. Meinhardt, and P. A. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartzell, P. L. 1997. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc. Natl. Acad. Sci. USA 94:9881-9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hecht, G. B., T. Lane, N. Ohta, J. M. Sommer, and A. Newton. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Hinz, A. J., D. E. Larson, C. S. Smith, and Y. V. Brun. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol. Microbiol., in press. [DOI] [PubMed]
- 39.Hofmeister, A. E. M., A. Londono-Vallejo, E. Harry, P. Stragier, and R. Losick. 1995. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 83:219-226. [DOI] [PubMed] [Google Scholar]
- 40.Howard, M., A. D. Rutenberg, and S. de Vet. 2001. Dynamic compartmentalization of bacteria: accurate division in E. coli. Phys. Rev. Lett. 87:278102.. [DOI] [PubMed] [Google Scholar]
- 41.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99:6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu, Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82-90. [DOI] [PubMed] [Google Scholar]
- 43.Hudson, M. E., D. Zhang, and J. R. Nodwell. 2002. Membrane association and kinase-like motifs of the RamC protein of Streptomyces coelicolor. J. Bacteriol. 184:4920-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Igoshin, O. A., A. Mogilner, R. D. Welch, D. Kaiser, and G. Oster. 2001. Pattern formation and traveling waves in myxobacteria: theory and modeling. Proc. Natl. Acad. Sci. USA 98:14913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishikawa, S., L. Core, and M. Perego. 2002. Biochemical characterization of aspartyl phosphate phosphatase interaction with a phosphorylated response regulator and its inhibition by a pentapeptide. J. Biol. Chem. 277:20483-20489. [DOI] [PubMed] [Google Scholar]
- 46.Iwasaki, H., S. B. Williams, Y. Kitayama, M. Ishiura, S. S. Golden, and T. Kondo. 2000. A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101:223-233. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs, C., I. J. Domian, J. R. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111-120. [DOI] [PubMed] [Google Scholar]
- 48.Jacobs, C., D. Hung, and L. Shapiro. 2001. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 98:4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jelsbak, L., and L. Sogaard-Andersen. 1999. The cell surface-associated intercellular C-signal induces behavioral changes in individual Myxococcus xanthus cells during fruiting body morphogenesis. Proc. Natl. Acad. Sci. USA 96:5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen, R. B., S. C. Wang, and L. Shapiro. 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 20:4952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 52.Jiang, Z. Y., H. Gest, and C. E. Bauer. 1997. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J. Bacteriol. 179:5720-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson, C. H., and S. S. Golden. 1999. Circadian programs in cyanobacteria: adaptiveness and mechanism. Annu. Rev. Microbiol. 53:389-409. [DOI] [PubMed] [Google Scholar]
- 54.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 55.Ju, J., T. Luo, and W. Haldenwang. 1997. Bacillus subtilis pro-σE fusion protein localizes to the forespore septum and fails to be processed when synthesized in the forespore. J. Bacteriol. 179:4888-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ju, J., T. Mitchell, H. Peters III, and W. G. Haldenwang. 1999. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 181:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Julien, B., A. D. Kaiser, and A. Garza. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 97:9098-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karow, M. L., P. Glaser, and P. Piggot. 1995. Identification of a gene, spoIIR, that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 92:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katayama, M., T. Kondo, J. Xiong, and S. Golden. 2003. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC7942. J. Bacteriol. 185:1415. [DOI] [PMC free article] [PubMed]
- 60.Kearns, D. B., P. J. Bonner, D. R. Smith, and L. J. Shimkets. 2002. An extracellular matrix-associated zinc metalloprotease is required for dilauroyl phosphatidylethanolamine chemotactic excitation in Myxococcus xanthus. J. Bacteriol. 184:1678-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kearns, D. B., and L. J. Shimkets. 2001. Lipid chemotaxis and signal transduction in Myxococcus xanthus. Trends Microbiol. 9:126-129. [DOI] [PubMed] [Google Scholar]
- 62.Kellner, E. M., A. Decatur, and C. P. Moran. 1996. Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol. Microbiol. 21:913-924. [DOI] [PubMed] [Google Scholar]
- 63.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 64.King, G. F., Y. L. Shih, M. W. Maciejewski, N. P. Bains, B. Pan, S. L. Rowland, G. P. Mullen, and L. I. Rothfield. 2000. Structural basis for the topological specificity function of MinE. Nat. Struct. Biol. 7:1013-1017. [DOI] [PubMed] [Google Scholar]
- 65.King, N., O. Dreesen, P. Stragier, K. Pogliano, and R. Losick. 1999. Septation, dephosphorylation, and the activation of σF during sporulation in Bacillus subtilis. Genes Dev. 13:1156-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kroos, L., Yu, D. Mills, and S. Ferguson-Miller. 2002. Forespore signaling is necessary for pro-σK processing during Bacillus subtilis sporulation despite the loss of SpoIVFA upon translational arrest. J. Bacteriol. 184:5393-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kroos, L., B. Zhang, H. Ichikawa, and Y.-T. N. Yu. 1999. Control of σ factor activity during Bacillus subtilis sporulation. Mol. Microbiol. 31:1285-1294. [DOI] [PubMed] [Google Scholar]
- 68.Kruse, K. 2002. A dynamic model for determining the middle of Escherichia coli. Biophys. J. 82:618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuner, J. M., and D. Kaiser. 1982. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J. Bacteriol. 151:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunkel, B., R. Losick, and P. Stragier. 1990. The Bacillus subtilis gene for the developmental transcription factor σK is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 4:525-535. [DOI] [PubMed] [Google Scholar]
- 71.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. USA 99:4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 73.Lee, B.-U., K. Lee, J. Mendez, and L. Shimkets. 1995. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev. 9:2964-2973. [DOI] [PubMed] [Google Scholar]
- 74.Lemon, K. P., and A. D. Grossman. 2000. Movement of replicating DNA through a stationary replisome. Mol. Cell 6:1321-1330. [DOI] [PubMed] [Google Scholar]
- 75.Li, S.-F., B. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 76.Londono-Vallejo, J. A., and P. Stragier. 1995. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 9:503-508. [DOI] [PubMed] [Google Scholar]
- 77.Meinhardt, H., and P. A. de Boer. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Molle, V., and M. J. Buttner. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36:1265-1278. [DOI] [PubMed] [Google Scholar]
- 79.Moller-Jensen, J., R. B. Jensen, J. Lowe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muir, R. E., and J. W. Gober. 2001. Regulation of late flagellar gene transcription and cell division by flagellum assembly in Caulobacter crescentus. Mol. Microbiol. 41:117-130. [DOI] [PubMed] [Google Scholar]
- 81.Muir, R. E., and J. W. Gober. 2002. Mutations in FlbD that relieve the dependency on flagellum assembly alter the temporal and spatial pattern of developmental transcription in Caulobacter crescentus. Mol. Microbiol. 43:597-615. [DOI] [PubMed] [Google Scholar]
- 82.Muir, R. E., T. M. O'Brien, and J. W. Gober. 2001. The Caulobacter crescentus flagellar gene, fliX, encodes a novel trans-acting factor that couples flagellar assembly to transcription. Mol. Microbiol. 39:1623-1637. [DOI] [PubMed] [Google Scholar]
- 83.Nair, U., J. L. Ditty, H. Min, and S. S. Golden. 2002. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J. Bacteriol. 184:3530-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45:45-57. [DOI] [PubMed] [Google Scholar]
- 85.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 86.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 87.Onaka, H., T. Nakagawa, and S. Horinouchi. 1998. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28:743-753. [DOI] [PubMed] [Google Scholar]
- 88.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 89.Paidhungat, M., K. Ragkousi, and P. Setlow. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pan, Q., D. A. Garsin, and R. Losick. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis. Mol. Cell 8:873-883. [DOI] [PubMed] [Google Scholar]
- 91.Partridge, S. R., and J. Errington. 1993. Importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol. Microbiol. 8:945-955. [DOI] [PubMed] [Google Scholar]
- 92.Perego, M., and J. A. Brannigan. 2001. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22:1541-1547. [DOI] [PubMed] [Google Scholar]
- 93.Perego, M., and J. A. Hoch. 1996. Protein aspartate phosphatases control the output of two-component signal transduction systems. Trends Genet. 12:97-101. [DOI] [PubMed] [Google Scholar]
- 94.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Popham, D. L., D. Szeto, J. Keener, and S. Kustu. 1989. Function of a bacterial activator protein that binds to transcriptional enhancers. Science 243:629-635. [DOI] [PubMed] [Google Scholar]
- 96.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 98.Ramirez-Arcos, S., J. Szeto, T. Beveridge, C. Victor, F. Francis, and J. Dillon. 2001. Deletion of the cell division inhibitor MinC results in lysis of Neisseria gonorrhoeae. Microbiology 147:225-237. [DOI] [PubMed] [Google Scholar]
- 99.Raskin, D. M., and P. A. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raskin, D. M., and P. A. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rudner, D., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA 96:14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rudner, D. Z., and R. Losick. 2001. Morphological coupling in development: lessons from prokaryotes. Dev. Cell 1:733-742. [DOI] [PubMed] [Google Scholar]
- 103.Rudner, D. Z., and R. Losick. 2002. A sporulation membrane protein tethers the pro-σK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 16:1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rudner, D. Z., Q. Pan, and R. M. Losick. 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. USA 99:8701-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ruiz, T., N. R. Francis, D. G. Morgan, and D. J. DeRosier. 1993. Size of the export channel in the flagellar filament of Salmonella typhimurium. Ultramicroscopy 49:417-425. [DOI] [PubMed] [Google Scholar]
- 106.Schmitz, O., M. Katayama, S. B. Williams, T. Kondo, and S. S. Golden. 2000. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289:765-768. [DOI] [PubMed] [Google Scholar]
- 107.Spormann, A. M., and D. Kaiser. 1999. Gliding mutants of Myxococcus xanthus with high reversal frequencies and small displacements. J. Bacteriol. 181:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szeto, J., S. Ramirez-Arcos, C. Raymond, L. D. Hicks, C. M. Kay, and J. A. Dillon. 2001. Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol. 183:6253-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 110.Thomaides, H. B., M. Freeman, M. El Karoui, and J. Errington. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15:1662-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomasson, B., J. Link, A. Stassinopoulos, N. Burke, L. Plamann, and P. Hartzell. 2002. MglA, a small GTPase, interacts with a tyrosine kinase to control type IV pili-mediated motility and development of Myxococcus xanthus. Mol. Microbiol. 46:1399-1413. [DOI] [PubMed] [Google Scholar]
- 112.Tillotson, R. D., H. A. Wosten, M. Richter, and J. M. Willey. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30:595-602. [DOI] [PubMed] [Google Scholar]
- 113.Tomomori, C., T. Tanaka, R. Dutta, H. Park, S. K. Saha, Y. Zhu, R. Ishima, D. Liu, K. I. Tong, H. Kurokawa, H. Qian, M. Inouye, and M. Ikura. 1999. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 6:729-739. [DOI] [PubMed] [Google Scholar]
- 114.Urban, S., and M. Freeman. 2002. Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Curr. Opin. Genet. Dev. 12:512.. [DOI] [PubMed] [Google Scholar]
- 115.Velicer, G. J., L. Kroos, and R. E. Lenski. 1998. Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat. Proc. Natl. Acad. Sci. USA 95:12376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Viollier, P. H., N. Sternheim, and L. Shapiro. 2002. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 21:4420-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Viswanathan, P., and L. Kroos. 2003. cis elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 1405. 185: [DOI] [PMC free article] [PubMed]
- 118.Ward, M. J., and D. R. Zusman. 1999. Motility in Myxococcus xanthus and its role in developmental aggregation. Curr. Opin. Microbiol. 2:624-629. [DOI] [PubMed] [Google Scholar]
- 119.Welch, R., and D. Kaiser. 2001. Cell behavior in traveling wave patterns of myxobacteria. Proc. Natl. Acad. Sci. USA 98:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wheeler, R. T., and L. Shapiro. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4:683-694. [DOI] [PubMed] [Google Scholar]
- 121.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 122.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wolgemuth, C., E. Hoiczyk, D. Kaiser, and G. Oster. 2002. How myxobacteria glide. Curr. Biol. 12:369-377. [DOI] [PubMed] [Google Scholar]
- 124.Wu, J., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. USA 95:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu, J., N. Ohta, J. L. Zhao, and A. Newton. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. USA 96:13068-13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu, L. J., and J. Errington. 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol. 27:777-786. [DOI] [PubMed] [Google Scholar]
- 127.Wu, L. J., and J. Errington. 2002. A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. EMBO J. 21:4001-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu, D., C. Yang, and H. B. Kaplan. 1998. Myxococcus xanthus sasN encodes a regulator that prevents developmental gene expression during growth. J. Bacteriol. 180:6215-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang, C., and H. B. Kaplan. 1997. Myxococcus xanthus sasS encodes a sensor histidine kinase required for early developmental gene expression. J. Bacteriol. 179:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoon, H. S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 132.Yoon, H. S., and J. W. Golden. 2001. PatS and products of nitrogen fixation control heterocyst pattern. J. Bacteriol. 183:2605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu, Y.-T. N., and L. Kroos. 2000. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 182:3305-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang, B., A. Hofmeister, and L. Kroos. 1998. The pro-sequence of pro-σK promotes membrane association and inhibits RNA polymerase core binding. J. Bacteriol. 180:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang, B., P. Struffi, and L. Kroos. 1999. σK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis. J. Bacteriol. 181:4081-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu, K., B. Gonzalez-Pedrajo, and R. M. Macnab. 2002. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry 41:9516-9524. [DOI] [PubMed] [Google Scholar]