Abstract
SCF complexes are E3 ubiquitin-protein ligases that mediate degradation of regulatory and signaling proteins and control G1/S cell cycle progression by degradation of G1 cyclins and the cyclin-dependent kinase inhibitor, Sic1. Interchangeable F-box proteins bind the core SCF components; each recruits a specific subset of substrates for ubiquitylation. The F-box proteins themselves are rapidly turned over by autoubiquitylation, allowing rapid recycling of SCF complexes. Here we report a role for the UbL-UbA protein Ddi1 in the turnover of the F-box protein, Ufo1. Ufo1 is unique among F-box proteins in having a domain comprising multiple ubiquitin-interacting motifs (UIMs) that mediate its turnover. Deleting the UIMs leads to stabilization of Ufo1 and to cell cycle arrest at G1/S of cells with long buds resembling skp1 mutants. Cells accumulate substrates of other F-box proteins, indicating that the SCF pathway of substrate ubiquitylation is inhibited. Ufo1 interacts with Ddi1 via its UIMs, and Δddi1 cells arrest when full-length UFO1 is overexpressed. These results imply a role for the UIMs in turnover of SCFUfo1 complexes that is dependent on Ddi1, a novel activity for an UbL-UbA protein.
Ubiquitin-mediated proteolysis determines protein abundance and is a major pathway for regulating crucial biological processes (14). Proteins are degraded in the 26S proteasome that consists of a 20S catalytic chamber (CP) and a 19S regulatory particle (RP) (11). Target proteins are usually marked by phosphorylation, and this links substrate recruitment with intrinsic signal transduction pathways that are activated in response to environmental cues (39). Substrates targeted for degradation by the 26S proteasome are covalently modified with short ubiquitin chains by a cascade of E1 ubiquitin-activating, E2 ubiquitin-conjugating, and E3 ubiquitin-ligase activities (14). Substrate ubiquitylation usually occurs in macromolecular complexes, exemplified by the SCF (for Skp1-Cdc53/Cul1-F-box protein) and the APC (for anaphase promoting complex). These two complexes are the major engines that drive the cell cycle being responsible for degradation of cyclins and the cyclin kinase inhibitor (CKI), Sic1 inter alia (5). The SCF complex has a major role in G1/S progression. It consists of a Cdc53/Cul1 rodlike scaffold whose N terminus serves as a docking site for a substrate recognition module comprising a F-box protein and the Skp1 adaptor. The Cdc53 C-terminal domain forms a heterodimeric catalytic module with the RING protein, Rbx1, and this serves as a landing pad for the E2; in yeast it is Cdc34 (7). The SCF core elements Cdc53 and Skp1 are stable proteins, whereas F-box proteins are generally short-lived and undergo autoubiquitylation by Cdc34. The canonical assembly of different SCF complexes thus provides a rapid mechanism for regulating cell cycle progression and for checkpoint-mediated response to stalled DNA replication or DNA damage at different points of the cell cycle and in response to changes in the availability of nutrients in the cell environment (9, 46).
The F-box protein Ufo1 was identified during a study on the regulation of the activity of the mating switch endonuclease, Ho (22). Transcription of UFO1 is induced in response to DNA damage (20), and deletion of UFO1 has been observed to affect genome stability (40). Ufo1 is an unusual F-box protein in that besides the F-box and protein-protein interaction domains present in other F-box proteins, it has four copies of the ubiquitin-interacting motif (UIM). The UIM is a short N-terminal acidic box, followed by alternating long and short hydrophobic residues that terminates in an invariant signature sequence A-X-X-X-S (15). The UIM was originally identified as the ubiquitin chain-binding domain of subunit Rpn10 of the proteasome 19S RP (35); however, all other UIM proteins are components of the intracellular vesicular trafficking system (15). A comprehensive search of the protein databases indicates that it is only Ufo1 and its fungal orthologs that have UIMs. The UIM of the intracellular trafficking protein Epsin promotes ubiquitylation of glutathione S-transferase (GST) in the context of a GST-UIMEpsin fusion protein (34). Furthermore, during self-ubiquitylation of Hgs1, an adaptor protein that targets the EGF receptor for degradation, the Hgs1 UIM plays an essential role in recruitment of the E3, Nedd4 (25).
Here we report that deletion of the Ufo1 UIMs creates a dominant-negative phenotype of inactivation of the SCF pathway of protein degradation. The cells arrest with long buds resembling skp1-3 mutants at the restrictive temperature, corresponding to a G1/S arrest. Furthermore, we show that there is a specific interaction between Ufo1 and the UbL-UbA protein Ddi1. This interaction requires the presence of the UIMs of Ufo1 and is important for the turnover of SCFUfo1 complexes and cell cycle progression.
MATERIALS AND METHODS
Strains.
W303 (MATa his3 leu2 trp1 ura3-52) and isogenic wild-type and Δrad23 Δdsk2 Δddi1 mutant strains were purchased from Euroscarf. The wild type and Δufo1 mutant are Research Genetics BY4730 and #142, respectively. The G1/S skp1-3 mutant is described elsewhere (26). Temperature-sensitive (ts) cdc34-2 and cdc53-1 were obtained from M. Tyers and are in a W303 background with the genotypes MATa can1-100 ade2-1 his3-11,15 leu2,3,112 trp1-1 ura3-1 GAL+ cdc34-2 and MATa ura3-1 can1-100 GAL+ leu2-3,112 trp1-1 his3-11,15 ade2-1 cdc53-1, respectively. FY56 and its isogenic rsp5 mutant, Fw1808 (MATa his4-912ΔΡ5 lys2-128Δ ura3-52 rsp5-1 [L7335]), were obtained from R. Haguenauer-Tsapis. The atg7 mutant was generated by one-step gene replacement in Euroscarf BY4741 by using the primers ATG7-URAF (ATAACTAAAGTTCATTATATTTCAACAAATATAAGATAATCAAGAATAAAATGTCGAAAGCTA CATATAAGG) and ATG7-URAR (CCGCAAGTTCTGCCTTTGGCTTTCCACAGTCCTCAAAATTATATAACGCT TTAGTTTTGCTGGCCGCATCTTC) to introduce URA3 at the genomic ATG7 locus. Cells expressing hemagglutinin (HA)-tagged Cln2 were prepared by integrating plasmid pRD114a from R. Deshaies at the LEU2 locus. SIC1 was deleted in the cdc53 ts strain above by using the primers SicHIS3F (AAATCAATCAACCAAACCTCTACGGAATTTTGACCCTTGAAGCAGGGACTATTACACGAAAATGACAGAGCAGAAAGCCC) and SicHIS3R (ATGTAGAATAAGTAAGTAAATAAAATATAATCGTTCCAGAAACTTTTTTTTTTCATTTCTCTACATAAGAACACCTTTGG) to substitute HIS3 for the genomic SIC1 gene.
Plasmids.
Construction of pGAL-GFP-UFO1 is described in reference 21; construction of of pTET-HO-LACZ is described in reference 22. pGFP-UFO1ΔUIM was cloned in plasmid pYCPGAL (41) by using the primers Ufo1BamF (CGGGATCCATGGAGCGGCCTGGCTTGGTATTGC) and Ufo1SalR (CCGACAGGATCCCCCGCTTAAGCTTGATTCCG) to amplify genomic UFO1. The K134A K141A mutations were introduced by primer overlap extension by using the external primers described above and the mutagenic primers NLSHindF (CACGCAAGGCAGAAGAAATTTAAAGCTTTGATTTATATTCC) and NLSHindR (GGAATATAAATCAAAGCTTTAAATTTCTTCTGCCTTGCGTG). The GFP-UFO1-UIMs was subcloned into the same plasmid by using the primers UfoUIMBamF (GCACTCTTAGGATCCCAGGAGGCGCAAGC) and UfoUIMBamR (GCGGATCCTCAATTGATTTCACTCAATGAC). The genomic UIM domain was replaced with a TAP tag (36) by using the primers UfoΔUIMTAPF (CGGAATCAAGCTGAAGCGGGGGAGCCTGTCGGAGATGATGTCCATGGAAA AGAGAAG) and UfoΔUIMTAPR (CTATAAATAAAATATTTAACATATGCTCTTCCAAATGTACATACTTTCATACGACTCACTATAGGG) to amplify the tag from plasmid pBS1479 (Euroscarf). The clone for overexpressing GFP-F-box residues 1 to 233 was made by deleting all residues between the HindIII site at bp 700 of the UFO1 open reading frame and a similar site in the vector. Ufo1 deleted for its F-box was cloned into pOBD using primers WDF (GGATTCAATATTAATGCTGCAGTG) and UIMR (GATCCCCGGGAATTGCCATGTCAATTGATTTCACTCAATGACAACGCAAT). The fusion of the Ufo1 WD40 domain in pOAD is described in reference 21.
CLN2 and SIC1 were cloned as GFP fusion proteins in YCPGAL by using the primers YCpGALCLN2BamF (CCATTCATTCATTAAATTTAAGGATCCACAATGGCTAGTGC) and YCpGALCLN2BamR (GGTACGTTTGGCAAATTGGGATCCATTTATCATGAAAAGAACAGG) and the primers YCpGAL-SIC1BamF (CGGAATTTTGACCCTGGATCCAGGGACTATTACACGAAAATG) and YCpGAL-SIC1BamR (GTAAATAAAATATAATGGATCCAGAAACTTTTTTTTTTCATTTC), respectively.
Subclones of Ddi1 with the UbL or UbA domain deleted were LexA fusions (3). 2μm plasmids with SKP1 from a genomic library, or CDC4 cloned as a PCR product amplified from genomic DNA, were a gift from D. Kornitzer. GST-Ddi1 was a gift of D. Skowyra and GST-Rad23 and GST-Dsk2 were obtained from S. Elsasser (8).
Yeast two-hybrid system.
UFO1 was fused to the Gal4 activation domain of pOAD (4). Rad23, Dsk2, and Ddi1 fusions to the Gal4 DNA-binding domain were from Ito et al. (17). The plasmids were cotransformed into yeast strain PJ694a, where three reporter genes are under the control of the GAL promoter (19) and cells in which the two proteins interact can grow on selective plates lacking histidine and adenine. Transformations of yeast cells were performed by using lithium acetate (1).
Protocols for metabolic labeling, immunoprecipitation, and pulse-chase are described in reference 21, as are the methods for coimmunoprecipitation and immunoblotting. In these experiments, GFP-UFO1 was induced from the GAL promoter by overnight growth in minimal medium with 2% galactose. In the experiment with G1-arrested cells, overnight cultures were diluted into medium with 2 μg of α-factor/ml and 2% galactose, followed by incubation for 3 h before promoter shutoff by the addition of 3% glucose. An overnight 50-ml culture of 108 cells/ml served as the source of a 300-μl extract that had 80 μg of protein/μl. A total of 190 μl (15 mg of protein) was taken for immunoprecipitation (IP) with anti-Ddi1 or anti-GFP, and the immunoprecipitate was run in a single lane for Western blotting (WB) with the appropriate antisera. Anti-Ddi1 was a gift from J. Gerst and was used at dilutions of 1:1,000 for IP and 1:5,000 for WB; anti-GFP antiserum from Roche Molecular Biochemicals and anti-myc (mouse monoclonal antibody 9E10) from Santa Cruz Biotechnology were used at 1:200 for IP and at 1:1,000 for WB. Goat anti-mouse and goat anti-rabbit antibodies were used at 1:1,000 and were from Santa Cruz Biotechnology. Protein A-Sepharose was purchased from Amersham and was used at 50%; 30-μl portions were added to each sample.
Microscopy.
Cells expressing green fluorescent protein (GFP)-tagged proteins were observed with a Nikon fluorescence microscope fitted with GFP-specific filters (dichromic 505 nm, excitation 450 to 490 nm, emission 515 nm). Images were captured with a Micromax 512 BFT camera (Roper Scientific) by using WinView32 imaging software.
RESULTS
Deletion of the Ufo1 UIMs leads to cell cycle arrest.
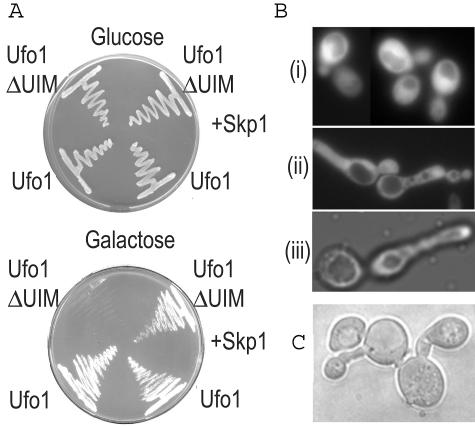
Ufo1 has an F-box domain (residues 5 to 51) attached via a linker to a WD40 repeat region (residues 190 to 420) that binds substrates for degradation. The unique UIMs are between residues 547 and 668. At least two canonical UIMs are present in most fungal Ufo1 orthologs, with many species, among them Saccharomyces cerevisiae, showing up to four motifs. To determine the functional significance of the UIMs of Ufo1, we expressed from the GAL promoter in wild-type yeast, full-length pGFP-UFO1, and truncated UFO1 without the UIMs (pGFP-UFO1ΔUIM). pGFP-UFO1 transformants grew normally on galactose plates, whereas those that expressed pGFP-UFO1ΔUIM were growth inhibited. Growth could be restored by ectopic expression of SKP1 (Fig. 1A). pGFP-UFO1ΔUIM cells in liquid culture arrested with long buds, similar to the G1/S arrest phenotype of skp1-3 mutants at 37°C, implying a lack of SCF function (Fig. 1B). Quantification showed about half the GFP-UFOΔUIM-expressing cells arrested with long buds. These experiments relied on high-level expression of UFO1 and its truncated form. We therefore investigated the effect of deleting the UIMs in genomic UFO1. This was done by integrating a PCR product encoding a TAP tag and the TRP1 marker gene (36) either just prior to the stop codon of UFO1 (UFO11-668::TAP) or in place of its UIMs (UFO11-538uim::TAP). Cells expressing UFO11-668::TAP grew normally, whereas those expressing UFO11-538uim::TAP formed microcolonies with cells arrested with long buds. These cells did not continue to grow, and the transformants died shortly after reaching the stage shown in Fig. 1C. This is in contrast to Δufo1 mutants that have no obvious growth phenotype.
FIG. 1.
Deletion of the Ufo1 UIMs leads to G1/S cell cycle arrest. (A) pGFP-UFO1 or pGFP-UFO1ΔUIM galactose-inducible transformants without (left side of plate) or with (right side of plate) ectopic expression of SKP1 were plated on glucose and galactose plates and photographed after 36 h growth. (B) Cells expressing pGFP-UFO1 show a normal morphology (i), whereas transformants expressing pGFP-UFO1ΔUIM arrest with long buds (ii) and resemble the skp1-3 G1 mutant at the restrictive temperature (iii). (C) Terminal arrest phenotype of cells deleted for the UIMs of genomic UFO1.
Ufo1ΔUIM is active in Ho degradation.
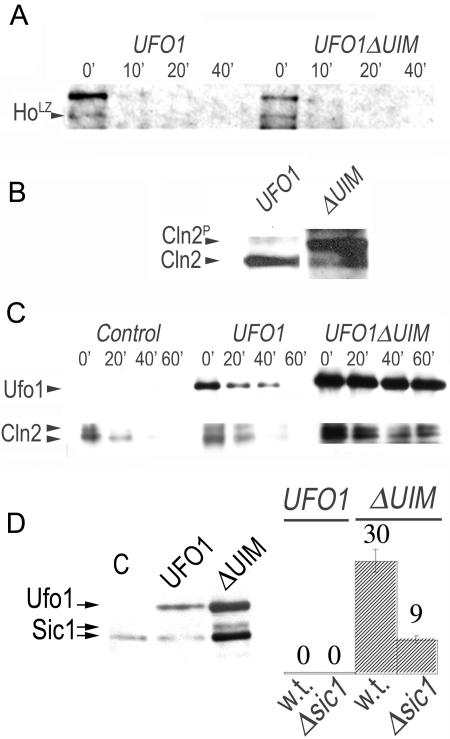
The G1/S arrest could be due to lack of degradation of a substrate of Ufo1 that has a role in cell cycle progression. However, we found that the Ufo1 substrate, Ho endonuclease, was degraded with a similar half-life in the presence of full-length Ufo1 or Ufo1ΔUIM, indicating that Ufo1ΔUIM is capable of forming active SCF complexes. This was done by cotransforming Δufo1 cells with pGFP-UFO1 or pGFP-UFO1ΔUIM together with pTET-HO-LACZ and measuring the half-life of Ho-LacZ by pulse-chase and IP (Fig. 2A).
FIG. 2.
SCFUfo1ΔUIM is active but prevents alternative SCF complex formation. (A) The Ufo1 UIMs are not required for turnover of Ho. Half-life of Ho-LacZ (HoLZ) ubiquitylated by SCFUfo1 or by SCFUfo1ΔUIM in vivo determined by pulse-chase and immunoprecipitation. (B) Cln2HA accumulates when pGFP-UFO1ΔUIM is overexpressed. Cln2HA was immunoprecipitated with anti-HA antiserum from cells expressing pGFP-UFO1 or pGFP-UFO1ΔUIM and was detected by Western blotting with anti-HA antiserum. The top band is phosphorylated Cln2HA. (C) Turnover of GFPCln2 in cells is retarded in the presence of Ufo1ΔUIM. Turnover of GFPCln2 is shown in cells without ectopic UFO1 compared to cells expressing pGFP-UFO1 or pGFP-UFO1ΔUIM. Aliquots of cells were collected after promoter shutoff at the times indicated above each lane in minutes, and the presence of GFPUfo1 and GFPCln2 was determined by anti-GFP immunoprecipitation and Western blotting. (D) Accumulation of the CKI, Sic1 in cells expressing pGFP-UFO1ΔUIM. Suppression of the cell cycle arrest in cells deleted for SIC1. The percentage of cells arrested with long buds was calculated when full-length pGFP-UFO1 or pGFP-UFO1ΔUIM were expressed in either wild type or Δsic1 mutants.
Cln2 accumulates in the presence of Ufo1ΔUIM.
The lethality caused by deletion of the genomic UFO1 UIMs implies that Ufo1 is exerting a dominant-negative effect on the SCF pathway. If the deletion of the Ufo1 UIMs is interfering directly with SCFUfo1 turnover, we would expect cells producing Ufo1ΔUIM to accumulate SCF degradation substrates of other F-box proteins such as Cln2 (ubiquitylated by SCFGrr1), and the Cdk inhibitor Sic1 (ubiquitylated by SCFCdc4) (38). Indeed, Western blotting demonstrated significant accumulation of Cln2 in cells expressing pGFP-UFO1ΔUIM but not in cells expressing full-length UFO1 (Fig. 2B). Furthermore, in an anti-GFP immunoprecipitation and Western blot experiment, we could show directly that the turnover of Cln2 is retarded when GFP-UFO1ΔUIM, but not GFP-UFO1, is expressed (Fig. 2C). These data are further supported genetically by the observation that the cell cycle arrest upon GFP-UFO1ΔUIM expression was ameliorated in a Δsic1 mutant and by the accumulation of Sic1 in these cells (Fig. 2D).
Suppression of the arrest caused by deletion of the UIMs of Ufo1.
The data presented above show that Ufo1ΔUIM has a dominant-negative effect on SCF activity, suggesting that it may sequester core subunits of the SCF. We therefore tested whether the arrest can be suppressed by coexpression of subunits of the SCF. F-box proteins are recruited to the SCF by binding the Skp1 adaptor; however, we showed previously that the Ufo1 F-box domain binds both Skp1 and Cdc53 and that a WD40 domain subclone binds Cdc53 (21). Thus, Ufo1 may be sequestering both core components of the SCF and preventing them from recycling. In accordance with this we found that overexpression of the F-box protein, CDC4, suppressed the phenotype of cells with a deletion of the genomic UFO1 UIMs, UFO11-538uim::TAP. Presumably, the large amounts of Cdc4 could compete for binding the SCF core subunits, implying that they are indeed limiting for substrate degradation (Fig. 3A and B). Similarly, when we coexpressed SKP1 with UFO1ΔUIM we found marked restoration of growth with only 16% arrested cells with long buds. Likewise, coexpression of CDC53 suppressed the cell cycle arrest induced by UFOΔUIM by half. The Ufo1 WD40 domain alone fused to the Gal4 DNA activating domain did not cause cell cycle arrest, and high-level expression of an Ufo1 F-box clone and putative linker region (residues 1 to 233) led to only 13% arrested cells compared to ca. 50% when Ufo1ΔUIM was produced from the same plasmid (Fig. 3A and B). A mutant version of Ufo1ΔUIM in which we introduced two point mutations into a putative nuclear localization sequence of basic residues between the F-box and the WD40 domains, Ufo1ΔUIMK134A,K141A, was not affected in nuclear import of Ufo1 but did lead to suppression of the cell cycle arrest induced by Ufo1ΔUIM (Fig. 3C). Taken together, the results confirm that it is the truncated Ufo1ΔUIM protein itself that is directly inhibiting cell cycle progression and that contacts between the Ufo1 F-box domain and both Skp1 and Cdc53 are involved in the dominant-negative effect on function of other SCF complexes. Moreover, this experiment emphasizes the difference between the inhibitory effect on the cell cycle of Ufo1ΔUIM, which consists of the F-box and WD40 domains, compared to the rescue by Cdc4 comprising the same two domains.
FIG. 3.
Cell cycle arrest by deletion of the Ufo1 UIMs and its suppression by coexpression of CDC4, SKP1, and CDC53. (A) Diagram of Ufo1 and the subclones used in the experiments. Asterisks mark the K134A K141A mutations. (B) Quantitation of percentage of arrested cells, median of 300 cells, and the standard deviation. The clone expressed is listed below the histogram. ΔgUIM designates deletion of the genomic UFO1 UIMs ufo1:uimTAP; ΔUIM is the expression of pGFP-UFO1ΔUIM; UFO1 is the expression of pGFP-UFO1; WD40 is this domain fused to the Gal4-activating domain expressed from the ADH promoter; F-box is a truncated GFP-UFO1 gene that encodes 233 N-terminal residues. Genes above the specific columns with a plus sign were coexpressed with either pGFP-UFO1 or pGFP-UFO1ΔUIM or the F-box clone as indicated. (C) Phenotype of cells expressing UFO1, UFO1ΔUIM, or UFO1ΔUIMK134AK141A (UFO1ΔUIM**) that no longer arrest.
Ufo1 is a long-lived F-box protein, and its degradation requires SCF activity.
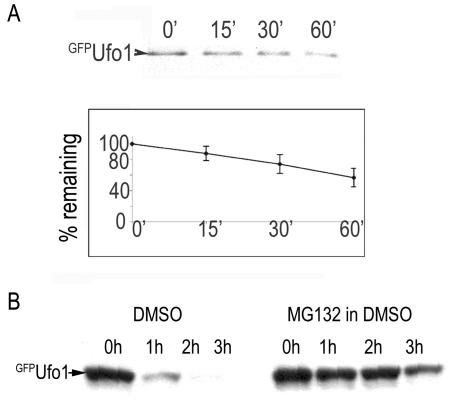
Most F-box proteins are short-lived and undergo autoubiquitylation by the SCF and degradation by the proteasome (43). Ufo1 has an unusually long half-life in a pulse-chase and IP experiment with >50% still present after 1 h (Fig. 4A). We confirmed the role of the proteasome in Ufo1 degradation by comparing the half-life of Ufo1 in the presence or the absence of the proteasome inhibitor, MG132 (27). The rate of Ufo1 turnover is severely retarded in the presence of MG132 (Fig. 4B). Moreover, Ufo1 degradation shows a requirement for a functional SCFUfo1 complex: Ufo1 deleted of its FB domain, which cannot be incorporated into SCF complexes, is not degraded in wild-type cells (Fig. 5A). Likewise, Ufo1 degradation is severely retarded in both cdc34 and cdc53 mutants at the restrictive temperature (Fig. 5B). Both cdc34 and cdc53 mutants arrest at the G1/S interphase at 37°C; however, Ufo1 was degraded normally in α-factor-arrested G1 cells, excluding the possibility that Ufo1 degradation requires progression into S phase. Furthermore, Ufo1 was stabilized in cdc53 mutants in which SIC1 was deleted to enable cell cycle progression (Fig. 5C). These results indicate that the main pathway for ubiquitylation of Ufo1 involves SCFUfo1, followed by degradation in the proteasome.
FIG. 4.
Ufo1 has an unusually long half-life and is degraded in the proteasome. (A) Ufo1 half-life is ca. 60 min determined by pulse-chase and immunoprecipitation. Degradation graph is the mean of three different experiments. (B) Half-life of Ufo1 in the presence of the proteasome inhibitor MG132 or its dimethyl sulfoxide solvent.
FIG. 5.
Half-life of GFPUfo1 in E2 and E3 mutants. (A) Half-life of GFPUfo1 compared to that of N-terminal truncated Ufo1 that has its F-box domain replaced in frame with the Gal4-binding domain (BDΔFB-Ufo1). In this experiment GFPUfo1 and BDΔFB-Ufo1 were immunoprecipitated with anti-GFP and anti-BD antisera, respectively, and detected by Western blotting with the same antisera (IP-WB). (B) Half-life of GFPUfo1 in wild type and mutants of the SCF scaffold, Cdc53, and its associated E2, Cdc34. The experiment was done at 37°C. (C) Half-life of GFPUfo1 in G1 α-factor-arrested cells and in cdc53 sic1 double mutants. (D) Half-life of GFPUfo1 in functions of the vacuolar degradation pathway in the isogenic wild type and rsp5 mutant and in the wild type and atg7 mutant. (E) The vacuolar protease inhibitor phenylmethylsulfonyl fluoride and its solvent control were also tested. Transformants were grown overnight in galactose medium to induce GFPUfo1, and those with ts phenotypes and their isogenic wild types were transferred to 37°C for 2 h before addition of 3% glucose for promoter shutoff. Incubation continued at 37°C until the end of the experiment. Aliquots were collected at the times stated above each lane for anti-GFP immunoprecipitation and Western blotting.
These experiments show some degradation of Ufo1 in cdc34 and cdc53 mutants, and we tried to identify functions of an additional degradation pathway. Given the presence of the UIMs in proteins involved in intracellular trafficking and the finding that mammalian Hgs1 is ubiquitylated by the E3 Nedd4 (yeast Rsp5), we tested whether this may involve Rsp5 and the vacuole. We did not find a requirement for Rsp5 for degradation of Ufo1 and also no requirement for the autophagy pathway which is inhibited in atg7 mutants (33) (Fig. 5D). Nor was Ufo1 stabilized by phenylmethylsulfonyl fluoride, an inhibitor of certain lysosomal proteases (24) (Fig. 5E).
The Ufo1 UIMs are required for degradation of Ufo1.
The experiments described above suggest that the cell cycle arrest may be due to the Ufo1ΔUIM protein sequestering the endogenous Skp1-Cdc53 subunits of the SCF and acting as a dominant negative, preventing assembly of other SCF complexes. In this model, the UIMs of Ufo1 are critical for its degradation. To explore directly whether Ufo1 degradation is dependent on its UIMs, we collected aliquots of cells overexpressing pGAL-GFP-UFO1 and pGAL-GFP-UFO1ΔUIM over a 3-h period after transfer from galactose to glucose medium to obtain promoter shutoff. Ufo1 was present at the 1-h time point but had disappeared by 2 h. In contrast, Ufo1ΔUIM was markedly stabilized and could still be found at the 3 h time point. Thus, in the absence of its UIMs, the half-life of Ufo1 is extended, and this could explain inactivation of SCF function when it is overexpressed; it could also explain the lethality induced by deletion of the genomic UFO1 UIMs. Ufo1ΔUIMK134A,K141A in which the cell cycle arrest is suppressed, has a half-life comparable to that of Ufo1, further strengthening the correlation between extended half-life and cell cycle arrest (Fig. 6A).
FIG. 6.
The Ufo1 UIMs and Ddi1 are required for turnover of SCFUfo1 complexes. (A) Half-life of Ufo1, Ufo1ΔUIM, and Ufo1ΔUIMK134,K141 (Ufo1ΔUIM**) in wild-type cells as determined by anti-GFP immunoprecipitation and Western blotting. (B) Half-life of Ufo1 in wild-type (w.t.) and Δddi1, Δdsk2, and Δrad23 mutant cells determined as described in the text.
The UbL-UbA protein, Ddi1, is required for Ufo1 turnover.
Our results indicate that truncation of the Ufo1 UIMs did not affect the ability of Ufo1ΔUIM to form active SCF complexes as evidenced by normal degradation of Ho endonuclease (Fig. 2). However, Ufo1ΔUIM had a considerably extended half-life (Fig. 6A). Therefore, deletion of the Ufo1 UIMs may be interfering with a function downstream of ubiquitylation. We recently found that the ubiquitin domain protein Ddi1 is required for the final stages of degradation of Ho and that, in the absence of Ddi1, Ho accumulates in a stable ubiquitylated form (23). Ddi1 belongs to a family of UbL-UbA proteins proposed to act as proteasome receptors for ubiquitylated substrates (11, 35). We therefore tested whether any of the UbL-UbA proteins—Rad23, Dsk2, or Ddi1—is required for Ufo1 degradation. We induced pGFP-UFO1 in galactose medium overnight in rad23, dsk2, and ddi1 deletion mutants and collected aliquots of cells at 30-min intervals after transfer to glucose for promoter shutoff. Ufo1 is highly stabilized in Δddi1 mutants, whereas it was degraded with a half-life similar to that observed in wild-type cells in rad23 and dsk2 mutants (Fig. 6B). These results imply that Ddi1 has a role in Ufo1 degradation and thus in the turnover of SCFUfo1 complexes.
Ufo1 binds Ddi1 in two-hybrid and co-IP experiments.
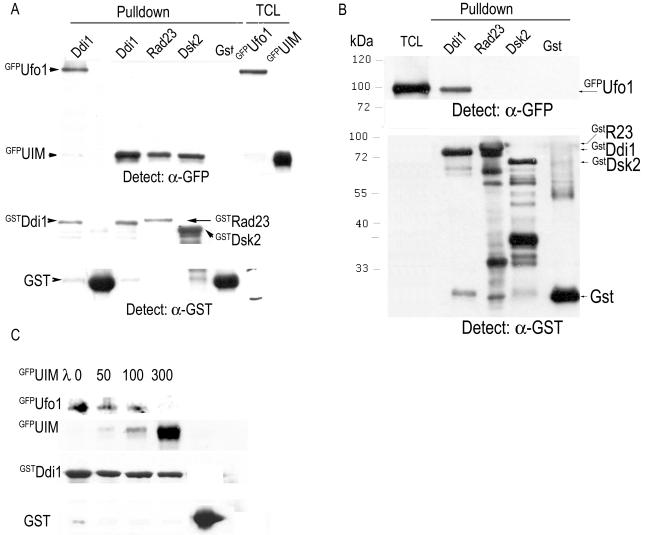
In accordance with this, when we tested for complex formation between Ufo1 and the UbL-UbA proteins Ddi1, Rad23, and Dsk2, we found that Ufo1 interacts only with Ddi1 and not with either Rad23 or Dsk2 in the two-hybrid protein interaction trap (Fig. 7A). Deletion of the UIMs of Ufo1 abrogated its interaction with Ddi1 in a co-IP experiment (Fig. 7B). An analysis of the domains of Ddi1 involved in interaction with Ufo1 showed that deletion of the UbL of Ddi1 abrogated binding to Ufo1 and that deletion of the UbA of Ddi1 reduced the interaction considerably (Fig. 7C). These results suggest that Ddi1 may have a role in the turnover of Ufo1 and, by implication, of SCFUfo1 complexes. This leads to the prediction that expressing full-length GFP-UFO1 in Δddi1 cells would phenocopy expression of GFP-UFO1ΔUIM in DDI1 cells. Indeed, we found that, whereas full-length UFO1 overexpression in wild-type cells did not affect cell cycle progression and it was only truncation of the Ufo1 UIM that led to arrest, in Δddi1 cells a very similar level of cell cycle arrest was obtained irrespective of the presence or absence of the Ufo1 UIMs (Fig. 7D). Ddi1 itself is very stable, with little sign of degradation over 3 h; its turnover was not affected by deletion of Ufo1 (Fig. 7E).
FIG. 7.
Ufo1 interacts with Ddi1. (A) Two-hybrid interaction between three independent cotransformants of Ufo1 fused to the Gal4 activating domain (AD) and Ddi1 fused to the Gal4 DNA-binding domain (BD). Activation of the HIS3 and ADE2 reporter genes from the GAL promoter promotes growth on selective plates. Parallel cotransformants of Ufo1-AD with Rad23- or Dsk2-BD fusion proteins did not grow on the selective plates (not shown). (B) Interaction of Ufo1 with Ddi1 requires the Ufo1 UIMs. Extracts from cells producing full-length GFPUfo1 or GFPUfo1ΔUIM were incubated with extracts from wild-type (DDI1) or Δddi1 cells and immunoprecipitated with anti-Ddi1 antiserum. The Western blots were analyzed with anti-GFP antiserum. Control anti-GFP immunoprecipitations on 3% input indicate the presence of both full-length GFPUfo1 (U) and GFPUfo1ΔUIM (Δ). The Western blot was then incubated with anti-Ddi1 antiserum to detect the presence of Ddi1, which runs very close to the immunoglobulin G chains. (C) The UbL of Ddi1 is essential for interaction with Ufo1; the UbA domain is also very important. Extract from cells producing full-length GFPUfo1 was incubated with extracts from wild-type (DDI1), Δddi1, or Δddi1 cells expressing Ddi1ΔUbL or Ddi1ΔUbA as a fusion protein to the LexA activating domain. The mixtures were immunoprecipitated with anti-Ddi1 antiserum, and the Western blots were analyzed with anti-GFP antiserum. Control anti-GFP immunoprecipitations on 3% input indicate the presence of GFPUfo1; the presence of the different Ddi1 proteins was detected by Western blotting with anti-Ddi1 antiserum. (D) Percentage of arrested wild-type and Δddi1 cells expressing UFO1 or UFO1ΔUIM. Long buds were observed in cultures of Δddi1 cells expressing full-length UFO1. Compare these results with those for wild-type cells expressing UFO1 in Fig. 3C. (E) Ddi1 is not a degradation substrate of Ufo1. The half-life of Ddi1 was determined by pulse-chase and immunoprecipitation experiments in wild-type and Δufo1 cells.
The role of the Ufo1 UIMs in binding Ddi1 was pursued further in a series of pull-down experiments: a yeast subclone comprising the Ufo1 UIMs fused to GFP was found to interact with all three UbL-UbA proteins—Ddi1, Rad23, and Dsk2—made as GST fusion proteins in bacteria (Fig. 8A). However, when full-length Ufo1 was tested under the same conditions, it bound solely to Ddi1 (Fig. 8B). When increasing amounts of GFPUIMs were added to a reaction comprising GSTDdi1 on beads and yeast lysate with GFPUfo1, the presence of the UIMs inhibited complex formation between Ufo1 and Ddi1 in a dose-dependent manner (Fig. 8C).
FIG. 8.
The Ufo1UIMs interact with all three UbL-UbA proteins, whereas full-length Ufo1 interacts only with Ddi1. (A) The Ufo1 UIMs produced in yeast as a GFP fusion were incubated with immobilized GstDdi1, GstRad23, GstDsk2, or GST proteins purified from bacteria. TCL, total cell lysates. (B) Full-length GFPUfo1 was incubated with the same immobilized proteins. (C) An equal amount of GFPUfo1 yeast lysate was incubated with immobilized GstDdi1 in the presence of increasing amounts of yeast lysate with GFPUIMs.
DISCUSSION
Our results demonstrate that the turnover of the F-box protein, Ufo1, is essential for proper functioning of the SCF ubiquitylation pathway. SCF activity appears to be finely tuned, and malfunction in turnover of a subset of SCF complexes rapidly leads to cell cycle arrest. Most F-box proteins are inherently unstable (43); in the case of Ufo1, its unique UIMs are the determinants for its degradation and for SCFUfo1 turnover. The marked effect on SCF activity when Ufo1ΔUIM is overexpressed and even moreso when the UIMs of the genomic UFO1 gene are deleted is an indication that the levels of the core SCF components are stringently regulated and that there is not a large pool of free subunits. Skp1 participates in different complexes, the centromeric complex (6), and in a complex with the Golgi complex-related F-box protein, Rcy1 (10), that do not involve Cdc53. Our data imply that its partitioning must be very tightly regulated since it is not rapidly mobilized from either of these complexes. Transformants in which the UIMs of genomic UFO1 are deleted reach the stage of microcolonies and then die, whereas after about 20 h in liquid culture, cells expressing ectopic UFO1ΔUIM resume growth, and this could be due to upregulation of biosynthesis of the core SCF elements.
To date, research on substrate degradation by the proteasome has focused on how substrates are marked for recruitment by F-box proteins. An extreme case is the CKI, Sic1, that requires a minimum of six phosphorylations to form a stable complex with Cdc4. The actual residues phosphorylated are less important than their total number, and this is interpreted to be a mechanism that links nutritional availability with entry into the cell cycle (32). The underlying assumption is that once a stable complex between phosphorylated Sic1 and Cdc4 is formed, substrate degradation followed by SCF turnover are assured. In contrast, we find that extraction of Ufo1 from SCFUfo1 complexes is a highly regulated process and depends specifically on its unique UIMs.
The SCF alternates between an active and inactive state regulated by neddylation of Cdc53 (43). In Schizosaccharomyces pombe and higher eukaryotes the signalosome (CSN) is responsible for inactivating SCF function by deneddylation. In conditions when SCF activity is limiting, the deneddylation by the CSN and the deubiquitylating activity of its associated DUB are critical for survival. This is due to their stabilization of the F-box protein, which would otherwise be rapidly eliminated in an autocatalytic reaction on binding active SCF core elements (42). Our experiments show that Ufo1 degradation requires the SCF functions Cdc53 and its associated E2, Cdc34. Stabilization of Ufo1 deleted for its F-box domain indicates the importance of incorporation of Ufo1 into SCF complexes for its degradation. Deletion of the Ufo1 UIMs leads to extension of the half-life of Ufo1. Ufo1ΔUIM resembles canonical F-box proteins that do not have UIMs, and its extended half-life may perhaps be attributed to the UIMs superseding internal degron sequences similar to those identified in Cdc4 and Met30 (9, 31, 46). The S. pombe ortholog of Ufo1, Pof10, is also very stable, and in this species overexpression of the full-length protein leads to loss of viability (16). The finding that Ufo1ΔUIMK134A,K141A does not lead to cell cycle arrest despite the absence of UIMs suggests that interactions of Ufo1ΔUIM with other protein(s) are important. These mutations are in the vicinity of the F-box domain and may affect the strength of binding between the Ufo1 F-box do- main and Skp1 and/or result in a different positioning of Ufo1ΔUIMK134A,K141A within the SCFUfo1 complex. For example, in S. pombe a defined point mutation in the F-box binding region of Skp1 differentially affects its binding affinity for different F-box proteins (28). Alternatively, F-box proteins may have a unique pathway of degradation: the proteasome receptor, Cic1, that binds an α subunit of the 20S CP mediates degradation of the F-box proteins Cdc4 and Grr1. Cic1 is not required for degradation of Ufo1 (not shown); however, the existence of a different 20S CP-interacting proteasome receptor cannot be excluded (18). Mutant Ufo1ΔUIMK134A,K141A may be treated as an SCF ubiquitylated substrate rather than as a component of the SCF complex and may be degraded via a different postubiquitylation pathway.
There is growing evidence for regulation of additional stages in the assembly of active SCF complexes. Entry of Skp1 into SCF complexes is inhibited by Cadmium: in the presence of Cadmium the F-box protein, Met30, binds its substrate Met4 but no longer binds Skp1 (2, 44). A further downstream regulated interaction is demonstrated between the Skp1-like BTB-adaptor, Keap1, that binds its substrate, the transcription factor Nrf2, but no longer binds the Cul3 scaffold under stress conditions. This abrogates ubiquitylation of Nrf2 and leads to its stabilization (45). These experiments show that in addition to F-box protein recruitment of a substrate that is marked for degradation by signal transduction pathways, assembly of the substrate-F-box protein complex into active SCF complexes is a highly regulated process.
The interaction of Ufo1 with Ddi1 is important for the degradation of Ufo1 and for the turnover of SCFUfo1 complexes. This may be by recycling the SCF core components Skp1 and Cdc53 into alternative SCF complexes. Ddi1 is a very stable protein and is not a degradation substrate of Ufo1. The Ufo1 UIMs bind all yeast UbL-UbA proteins indiscriminately; however, full-length Ufo1 only binds Ddi1. The UIMs are required for its interaction with Ddi1. The importance of the interaction between Ufo1 and Ddi1 is shown most clearly by the extended half-life of Ufo1 in Δddi1 mutants and by the cell cycle arrest of Δddi1 cells induced by overexpression of full-length UFO1. These genetic data are supported by experimental data obtained with the two-hybrid system and in co-IP experiments that demonstrate that both the UbL and UbA domains of Ddi1 are required for interaction with Ufo1. Regulation of the SCF pathway is a novel function for an UbL-UbA protein. The UbL domain of Ddi1 is required for interaction with the 19S RP of the proteasome, and one model for activation of an UbL-UbA protein could be that binding of a K48-linked ubiquitin chain to the UbA domain frees the UbL domain to bind the proteasome. This model is suggested by our finding that Ddi1 can bind ubiquitylated Ho in the absence of its UbL domain (23). Furthermore, the UbL of Rad23 interacts with its UbA domain, and this may represent an inactive form of the protein (37). The UbA domain of Ddi1 is a cis-stabilization element (13), and we have shown that it interacts with ubiquitin-conjugated Ho (23). Here, too, we find that it is important for the interaction of Ddi1 with Ufo1. Furthermore, it has been shown that the UbL-UbA KPC2 subunit is required to stabilize the KPC1 RING subunit of the heterodimeric E3 (12). This mechanism regulates function of the E3 for p27, a cyclin-dependent kinase inhibitor in mammalian cells. It will be interesting to decipher how these competing interactions for the Ddi1 UbL are regulated. Our experiments identify further regulatory steps that determine whether the ubiquitylated substrate will be processed further by functions of the pathway that leads to its degradation. In addition, Ddi1, alias Vsm1, has a role in intracellular membrane transport (29, 30), and it is not clear whether this role involves its association with Ufo1.
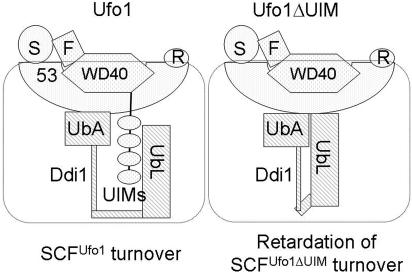
We propose a model in which Ufo1 and Ufo1ΔUIM are incorporated into SCF complexes and can support substrate ubiquitylation, but after that their fates differ. Interaction between the Ufo1 UIMs and the UbL of Ddi1 could be required for activation of Ddi1 for destabilization of the SCFUfo1 complex. In this model the UbL and UbA domains would play a regulatory role and gate access to the Ddi1 core. (In this scenario one possible explanation for the marked reduction [ca. 90%] in the binding of Ufo1 to Ddi1 lacking its UbA domain may reflect a similar lack of accessibility to the Ddi1 core.) The interaction between the Ufo1 UIMs and Ddi1 is not required for substrate extraction but is specifically required for turnover of SCFUfo1 complexes. This may involve repositioning of Ufo1 by Ddi1 within the SCF complex so that it now becomes accessible to the E2 and/or on the proteasome so that it can be deubiquitylated, unfolded, and degraded. In the absence of the Ufo1 UIMs or of Ddi1 the half-life of Ufo1 is extended, and this leads to inhibition of the SCF pathway of substrate degradation (Fig. 9).
FIG. 9.
Model for interaction between Ufo1 and Ddi1. Ufo1 and Ufo1ΔUIM are shown in a complex with Skp1 (S) and Cdc53 (53) via contacts of both their F-box (F) and WD40 domains. (R is Rbx1). Interaction between the Ufo1 UIMs and the UbL domain of Ddi1 promotes degradation of Ufo1 and recycling of SCFUfo1 complexes. The interaction between Ddi1 and Ufo1 also involves its UbA domain that may need to be released from interaction with the UbL domain to make contact with the ubiquitin chains on Ufo1.
Acknowledgments
We thank Dorota Skowyra for many stimulating discussions and extremely useful advice. We thank Kay Hofmann for pointing out to us that Ufo1 has UIMs.
These experiments were supported by the Israel Cancer Research Fund, by the Association for International Cancer Research, by the German-Israel Foundation for Scientific Research and Development, by the Israel Cancer Association, and by FP5 EU contract HPRN-CT-2002-00238.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Barbey, R., P. Baudouin-Cornu, T. A. Lee, A. Rouillon, P. Zarzov, M. Tyers, and D. Thomas. 2005. Inducible dissociation of SCF(Met30) ubiquitin ligase mediates a rapid transcriptional response to cadmium. EMBO J. 24:521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. [DOI] [PubMed] [Google Scholar]
- 4.Cagney, G., P. Uetz, and S. Fields. 2001. Two-hybrid analysis of the Saccharomyces cerevisiae 26S proteasome. Physiol. Genomics 7:27-34. [DOI] [PubMed] [Google Scholar]
- 5.Cardozo, T., and M. Pagano. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell. Biol. 5:739-751. [DOI] [PubMed] [Google Scholar]
- 6.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deffenbaugh, A. E., K. M. Scaglione, L. Zhang, J. M. Moore, T. Buranda, L. A. Sklar, and D. Skowyra. 2003. Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114:611-622. [DOI] [PubMed] [Google Scholar]
- 8.Elsasser, S., R. R. Gali, M. Schwickart, C. N. Larsen, D. S. Leggett, B. Muller, M. T. Feng, F. Tubing, G. A. Dittmar, and D. Finley. 2002. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4:725-730. [DOI] [PubMed] [Google Scholar]
- 9.Galan, J. M., and M. Peter. 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. USA 96:9124-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galan, J. M., A. Wiederkehr, J. H. Seol, R. Haguenauer-Tsapis, R. J. Deshaies, H. Riezman, and M. Peter. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21:3105-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glickman, M. H., and D. Raveh. 2005. Proteasome plasticity. FEBS Lett. 579:3214-3223. [DOI] [PubMed] [Google Scholar]
- 12.Hara, T., T. Kamura, S. Kotoshiba, H. Takahashi, K. Fujiwara, I. Onoyama, M. Shirakawa, N. Mizushima, and K. I. Nakayama. 2005. Role of the UBL-UBA protein KPC2 in degradation of p27 at G1 phase of the cell cycle. Mol. Cell. Biol. 25:9292-9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heessen, S., M. G. Masucci, and N. P. Dantuma. 2005. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol. Cell 18:225-235. [DOI] [PubMed] [Google Scholar]
- 14.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, K., and L. Falquet. 2001. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26:347-350. [DOI] [PubMed] [Google Scholar]
- 16.Ikebe, C., K. Kominami, T. Toda, and K. Nakayama. 2002. Isolation and characterization of a novel F-box protein Pof10 in fission yeast. Biochem. Biophys. Res. Commun. 290:1399-1407. [DOI] [PubMed] [Google Scholar]
- 17.Ito, T., K. Tashiro, S. Muta, R. Ozawa, T. Chiba, M. Nishizawa, K. Yamamoto, S. Kuhara, and Y. Sakaki. 2000. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 97:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jager, S., J. Strayle, W. Heinemeyer, and D. H. Wolf. 2001. Cic1, an adaptor protein specifically linking the 26S proteasome to its substrate, the SCF component Cdc4. EMBO J. 20:4423-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jelinsky, S. A., P. Estep, G. M. Church, and L. D. Samson. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20:8157-8167.11027285 [Google Scholar]
- 21.Kaplun, L., Y. Ivantsiv, A. Bakhrat, and D. Raveh. 2003. DNA damage response-mediated degradation of Ho endonuclease via the ubiquitin system involves its nuclear export. J. Biol. Chem. 278:48727-48734. [DOI] [PubMed] [Google Scholar]
- 22.Kaplun, L., Y. Ivantsiv, D. Kornitzer, and D. Raveh. 2000. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. USA 97:10077-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplun, L., R. Tzirkin, A. Bakhrat, N. Shabek, Y. Ivantsiv, and D. Raveh. 2005. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol. Cell. Biol. 25:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katunuma, N., E. Kominami, S. Hashida, and N. Wakamatsu. 1982. Modification of rat liver fructose biphosphate aldolase by lysosomal proteinases. Adv. Enzyme Regul. 20:337-350. [DOI] [PubMed] [Google Scholar]
- 25.Katz, M., K. Shtiegman, P. Tal-Or, L. Yakir, Y. Mosesson, D. Harari, Y. Machluf, H. Asao, T. Jovin, K. Sugamura, and Y. Yarden. 2002. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 3:740-751. [DOI] [PubMed] [Google Scholar]
- 26.Kitagawa, K., D. Skowyra, S. J. Elledge, J. W. Harper, and P. Hieter. 1999. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4:21-33. [DOI] [PubMed] [Google Scholar]
- 27.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann, A., S. Katayama, C. Harrison, S. Dhut, K. Kitamura, N. McDonald, and T. Toda. 2004. Molecular interactions of fission yeast Skp1 and its role in the DNA damage checkpoint. Genes Cells 9:367-382. [DOI] [PubMed] [Google Scholar]
- 29.Lustgarten, V., and J. E. Gerst. 1999. Yeast VSM1 encodes a v-SNARE binding protein that may act as a negative regulator of constitutive exocytosis. Mol. Cell. Biol. 19:4480-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marash, M., and J. E. Gerst. 2001. t-SNARE dephosphorylation promotes SNARE assembly and exocytosis in yeast. EMBO J. 20:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathias, N., S. Johnson, B. Byers, and M. Goebl. 1999. The abundance of cell cycle regulatory protein Cdc4p is controlled by interactions between its F box and Skp1p. Mol. Cell. Biol. 19:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 33.Ohsumi, Y. 2001. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell. Biol. 2:211-216. [DOI] [PubMed] [Google Scholar]
- 34.Oldham, C. E., R. P. Mohney, S. L. Miller, R. N. Hanes, and J. P. O'Bryan. 2002. The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr. Biol. 12:1112-1116. [DOI] [PubMed] [Google Scholar]
- 35.Pickart, C. M., and D. Fushman. 2004. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8:610-616. [DOI] [PubMed] [Google Scholar]
- 36.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 37.Ryu, K. S., K. J. Lee, S. H. Bae, B. K. Kim, K. A. Kim, and B. S. Choi. 2003. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J. Biol. Chem. 278:36621-36627. [DOI] [PubMed] [Google Scholar]
- 38.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 39.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 40.Smith, S., J. Y. Hwang, S. Banerjee, A. Majeed, A. Gupta, and K. Myung. 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:9039-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solsbacher, J., P. Maurer, F. R. Bischoff, and G. Schlenstedt. 1998. Cse1p is involved in export of yeast importin alpha from the nucleus. Mol. Cell. Biol. 18:6805-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wee, S., R. K. Geyer, T. Toda, and D. A. Wolf. 2005. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 7:387-391. [DOI] [PubMed] [Google Scholar]
- 43.Wolf, D. A., C. Zhou, and S. Wee. 2003. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5:1029-1033. [DOI] [PubMed] [Google Scholar]
- 44.Yen, J. L., N. Y. Su, and P. Kaiser. 2005. The yeast ubiquitin ligase SCFMet30 regulates heavy metal response. Mol. Biol. Cell 16:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, P., and P. M. Howley. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2:571-580. [DOI] [PubMed] [Google Scholar]