Abstract
Recoding of UGA from a stop codon to selenocysteine poses a dilemma for the protein translation machinery. In eukaryotes, two factors that are crucial to this recoding process are the mRNA binding protein of the Sec insertion sequence, SBP2, and the specialized elongation factor, EFsec. We sought to determine the subcellular localization of these selenoprotein synthesis factors in mammalian cells and thus gain insight into how selenoprotein mRNAs might circumvent nonsense-mediated decay. Intriguingly, both EFsec and SBP2 localization differed depending on the cell line but significant colocalization of the two proteins was observed in cells where SBP2 levels were detectable. We identify functional nuclear localization and export signals in both proteins, demonstrate that SBP2 undergoes nucleocytoplasmic shuttling, and provide evidence that SBP2 levels and localization may influence EFsec localization. Our results suggest a mechanism for the nuclear assembly of the selenocysteine incorporation machinery that could allow selenoprotein mRNAs to circumvent nonsense-mediated decay, thus providing new insights into the mechanism of selenoprotein translation.
Selenocysteine, the 21st amino acid, is the defining component of selenoproteins, a family whose members exhibit a wide range of functions, including roles in cellular oxidative status, male fertility, and thyroid function. Selenoprotein mRNAs are unique in that they recode UGA, which typically signals termination to the protein synthesis machinery, to instead specify cotranslational insertion of selenocysteine. In eukaryotes, this recoding process involves the assembly of complexes, termed Sec insertion sequence (SECIS) elements, at specific secondary structures in the 3′ untranslated regions of these mRNAs (5). The complexes include a SECIS binding protein (SBP2) (13) and a dedicated elongation factor (EFsec) (19, 35) complexed with selenocysteyl-tRNA[Ser]Sec (for a review, see references 4 and 17). Disrupting the functions of the factors involved in selenocysteine incorporation or limiting selenium availability results in premature termination of translation at UGA codons (6, 7, 13, 26, 30, 39). Under these circumstances, selenoprotein mRNAs are susceptible to degradation through a process termed nonsense-mediated decay (NMD) (for a review, see references 23 and 31), with different mRNAs exhibiting differential sensitivities to the NMD pathway (33, 38).
NMD or mRNA surveillance targets mRNAs containing premature termination codons for degradation, ensuring that they do not produce prematurely terminated polypeptides. In higher eukaryotes, a nonsense codon is usually recognized as premature if it is located more than 50 to 55 nucleotides upstream of the last intron in the pre-mRNA. Sensitivity to NMD is conferred by the deposition of an exon-junction complex upstream of exon-exon boundaries during mRNA splicing and export (23). Immunity to NMD is thought to be acquired upon removal of these proteins during the first round of translation. However, if an mRNA escapes NMD during the first round of translation, it becomes immune to subsequent decay via this pathway. While the protein synthesis machinery is thought to function primarily in the cytoplasm, mammalian NMD has been shown, in most cases, to occur on mRNAs that cofractionate or copurify with nuclei (10); this cofractionation or copurification is likely due to the association of mRNAs with the nuclear pore complex during export and concurrent initiation of the first round of translation.
For selenoprotein mRNAs to avoid NMD, it follows that the complexes necessary for decoding should assemble on selenoprotein mRNAs in the nucleus or during export. In this sense, shuttling events that determine subcellular localization of the components of the decoding complex may be as important for selenoprotein synthesis as the presence of selenium or the expression of these essential components.
To begin to investigate this possibility, we have undertaken a study of the subcellular localization of the selenoprotein synthesis factors in several cell lines. We have identified putative nuclear localization signals (NLSs) and nuclear export signals (NESs) in EFsec and putative NESs, NLSs, and a potential nucleolar localization signal (NoLS) in SBP2 and have shown that these signals are functional. We also demonstrate that the minimal functional domain of SBP2 undergoes nucleocytoplasmic shuttling and provide evidence that nuclear SBP2 may contribute to nuclear retention of EFsec. The implications of these findings for translational efficiency and NMD of selenoprotein mRNAs are discussed.
MATERIALS AND METHODS
Constructs, antibodies, and other reagents.
Full-length, truncated, and mutated EFsec cDNA with an N-terminal FLAG tag in pUHD10-3 has been described previously (35, 39). Inverse PCR was used to make mutations to the putative NLS in the truncated EFsec as previously described (17). Full-length SBP2 cDNA in pCR3.1 was a generous gift of Paul Copeland and Donna Driscoll (Cleveland Clinic Foundation). SBP2 was subcloned into the pUHD10-3 vector containing N-terminal hemagglutinin (HA) tag. SBP2 was also subcloned into a pDONR221 vector and recombined with a pcDNA-DEST40 vector with a C-terminal V5 tag using Gateway Technology (Invitrogen, Carlsbad, CA). The Xenopus tRNA[Ser]Sec gene in pGEM3 has been described previously (7). Putative NLS sequences were identified with the PSORTII algorithm (24), and putative NES sequences were identified manually as ΦX1-4ΦX2-3LXΦ, where Φ represents any hydrophobic residue and X represents any residue (16).
Primary antibodies included mouse monoclonal or rabbit polyclonal anti-FLAG (Sigma-Aldrich, St. Louis, MO), rabbit polyclonal anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-V5 (Invitrogen), and rabbit polyclonal anti-SBP2 (generous gift of Paul Copeland). All were titrated against isotype-control antibody or rabbit preimmune sera, found to be optimal in sensitivity and specificity at a final concentration of 1:500, and used at this dilution in subsequent experiments. Secondary anti-mouse or anti-rabbit antibodies included rhodamine, Texas Red, fluorescein isothiocyanate (FITC), and Alexa 488- and Alexa 586-conjugated (Molecular Probes, Eugene, OR) antibodies used at a final concentration of 1:500. In some experiments, cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (Molecular Probes) at 1:1,500. For some immunofluorescence assays, rhodamine-phalloidin (Molecular Probes) was used to visualize filamentous actin.
Cell culture, transfections, and subcellular fractionation.
HEK-293 human embryonic kidney, HepG2 human hepatoma, and NIH 3T3 murine fibroblast cells were cultured in Dulbecco's modified Eagle's medium (GIBCO-Invitrogen)-high glucose medium with 10% fetal bovine serum (Sigma or Invitrogen) at 37°C and 5% CO2. MSTO-211H human mesothelioma cells were cultured in RPMI 1640 (GIBCO-Invitrogen) supplemented with 10% fetal bovine serum and incubated at 37°C and 5% CO2. Cells were plated on glass coverslips, and at 18 to 24 h, transfections were performed using either a standard calcium phosphate method (8) or commercially available FuGene 6 (Roche) or TransIT-LT1 reagent (Mirus Bio Corporation, Madison, WI) according to the manufacturer's instructions. All transfections involving pUHD10-3 also included pUHD15 as transcriptional activator as previously described (22). Medium was supplemented with 100 nM sodium selenite 24 h after transfection. Subcellular fractionation was carried out by lysing cell pellets in 50 mM HEPES, pH 7.5, 75 mM NaCl, 40 mM NaF, 10 mM iodoacetamide, 0.25% Triton X-100, and 0.5 mM phenylmethylsulfonyl fluoride, pelleting and washing nuclei, and lysing nuclei in the same buffer containing 350 mM NaCl. This was followed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting with the indicated antibodies. Western blot analyses with antibodies to cytoplasmic (GRB2) and nuclear (histone H1) markers were carried out to assess the cross-contamination of subcellular fractions. Labeling of endogenous selenoproteins with sodium [75Se]selenite was carried out as described previously (35).
Immunofluorescence microscopy.
Immunofluorescence assays were performed 48 to 72 h after transfection as described previously (1), with modifications. Cells on slides were mounted in mounting medium with or without DAPI (Vector Laboratories, Burlingame, CA). Epifluorescence was performed using an Olympus IX71 microscope with an Olympus U-CMAD3 camera and MicroFire 1.0 software (Optronics, Goleta, CA). Confocal microscopy was performed in a Bio-Rad MRC-1024/2P system interfaced with a Zeiss Axiovert microscope.
Heterokaryon experiments.
Heterokaryon experiments were carried out as described previously (29), with modifications. Briefly, human MSTO-211H cells were plated and transfected as described above. After 48 h, murine NIH-3T3 cells were added at a ratio of approximately 2:1 and allowed to adhere to MSTO-211H cells for 3 h at 37°C in the presence of 50 μg/ml cycloheximide (Sigma-Aldrich), followed by 100 μg/ml for 30 min to inhibit protein synthesis. Cell fusion was then carried out by removing the medium and adding 50% (wt/vol) polyethylene glycol for 5 min at 37°C. The fused cells were then washed five times with phosphate-buffered saline, covered in medium containing 100 μg/ml cycloheximide, and incubated at 37°C for 3 h. Cells were then washed, fixed, permeabilized, and stained as described above.
Selenoprotein mRNA quantitation.
For comparison of the selenoprotein mRNA levels in HepG and HEK-293 cells, two 60-nucleotide-long oligonucleotide probes (37 to 60% GC) were designed for each gene, with positioning as close as possible to the 3′ end. Sequences were analyzed for the absence of secondary structure and cross-hybridization elsewhere in the genome. Probes were spotted onto GeneScreen Plus nylon membranes by using a V&P Scientific (San Diego, CA) 1,536-pin replicator and immobilized by alkali treatment. Total RNA (2 μg) was labeled via oligoT-directed first-strand cDNA synthesis using 400 U of murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA) and alpha-33P-dCTP (40 μCi). cDNA was purified using QIAquick PCR columns (QIAGEN, Valencia, CA), heat denatured, and hybridized in triplicate to arrays in MicroHyb buffer (Research Genetics, Huntsville, AL) overnight at 60°C. Arrays were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.5% SDS at 50°C, followed by 1 to 2× SSC and 0.5% SDS at 65°C, and then exposed to phosphor storage screens and signals quantified using a Molecular Dynamics PhosphorImager (Sunnyvale, CA). Signal readings were taken for each spot, and background readings were taken at empty spots. Raw data was further automatically processed using Microsoft Excel. Spot readings that failed to exceed the average background value by more than 3 standard deviations were excluded from the analysis. The remaining readings were scaled by the average signal in selected steadily expressed genes and then averaged among triplicate measurements. For comparison of GPX1 and selenoprotein P mRNAs in the presence versus the absence of SBP2 transfection, quantitative real-time PCR was performed on a LightCycler 2 (Roche) by using cDNA prepared from HEK-293 cells transfected with an either full-length or truncated version of SBP2 or empty pcDNA DEST40 vector. Student's unpaired t test was used to compare means of groups by using GraphPad Prism4 (San Diego, CA).
RESULTS
EFsec and SBP2 contain putative nuclear localization and export sequences.
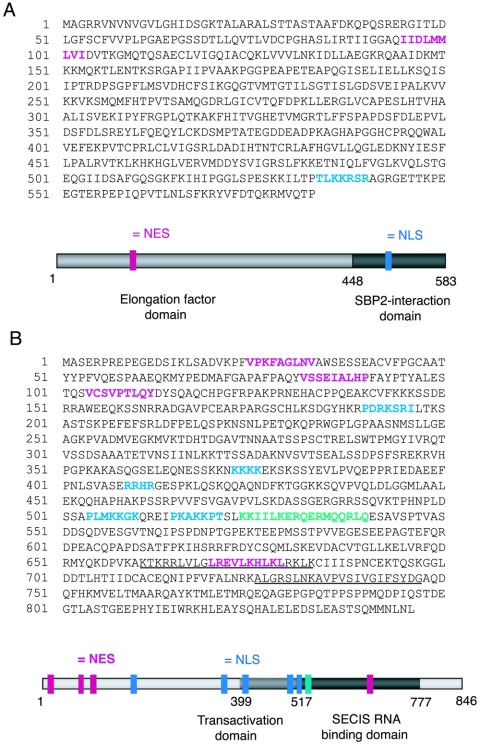
Analyses of the amino acid sequences of mouse EFsec and rat SBP2 (GenBank accession numbers AF268871 and NM024002, respectively) revealed the presence of putative nuclear localization and export signals in both proteins. The amino acid sequences of mouse EFsec and rat SBP2 with putative localization signals are shown in Fig. 1. Putative NLS sequences were identified with the PSORTII algorithm (24), and putative NES sequences were identified manually as ΦX1-4ΦX2-3LXΦ, where Φ represents any hydrophobic residue and X represents any residue (16). A putative NES was identified in the amino-terminal elongation factor domain of the murine EFsec sequence and was found to be conserved in the ortholog sequences from Homo sapiens, Drosophila melanogaster, Anopheles gambiae, and Ciona intestinalis but not from Caenorhabditis elegans or Archaea. A putative NLS was found in the carboxy-terminal region of the murine EFsec sequence, overlapping the region we previously identified as the SBP2 interaction domain (39). This NLS was also found to be conserved among the human, A. gambiae, and C. intestinalis ortholog sequences but not among the D. melanogaster, C. elegans, or Archaea sequences.
FIG. 1.
Amino acid sequence and primary structure model of EFsec and SPB2 with predicted NLS (blue), NES (pink), and NoLS (green) domains highlighted. (A) Mouse EFsec sequence and model, with the elongation factor domain in light gray and the SBP2-interaction domain in dark gray. (B) Rat SBP2 sequence and model, with transactivation domain in light gray and SECIS RNA binding domain in dark gray. Two RRM motifs previously identified in SBP2 are underlined. Gray bars in the cartoon schematics indicate the functional domains mapped previously (for a review, see reference 39 for EFsec and reference 14 for SBP2).
Five putative NLSs were identified in the rat SBP2 sequence, all of these being conserved in the mouse sequence. Three putative NLSs were identified in the human and A. gambiae sequences, and two were identified in the D. melanogaster sequence. The rat and mouse SBP2 proteins were both predicted to exhibit 74% nuclear localization, while the human, D. melanogaster, and A. gambiae protein were predicted to exhibit 65, 65, and 52% nuclear localization, respectively. In the rat and mouse sequences, two and three NLSs, respectively, were identified in the amino-terminal half of the protein, a region previously shown to be dispensable for selenocysteine incorporation in vitro (14). Another two NLS motifs were identified in the transactivation (putative EFsec interaction) domain, and the last was identified in the SECIS binding domain. Four NESs were predicted in the rat and mouse SBP2 sequences, three in the amino-terminal and one in the carboxy-terminal region. Of these, three were found to be conserved in the human sequence, whereas only the C-terminal putative NES was found to be present in the D. melanogaster and A. gambiae sequences.
Subcellular localization of EFsec and SBP2.
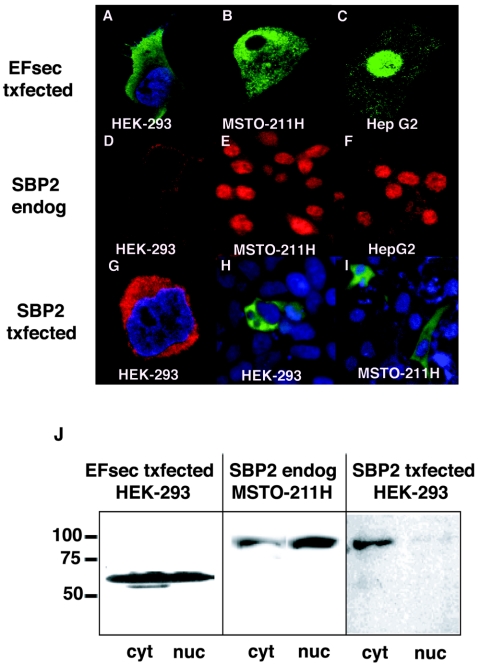
To determine the subcellular localization of full-length EFsec, an epitope tag was introduced at the amino terminus of the coding region and localization was assessed by immunofluorescence, following transient transfection. Intriguingly, EFsec localization varied depending on the cell line, being predominantly cytoplasmic with faint nuclear staining in HEK-293 cells (Fig. 2A), cytoplasmic and nuclear in MSTO-211H cells (Fig. 2B), and predominantly nuclear in HepG2 cells (Fig. 2C). The expression and subcellular localization of endogenous SBP2 were assessed in the same three cell lines by using antisera prepared against bacterially expressed SBP2. Strikingly, SBP2 exhibited patterns of localization similar to those of EFsec in MSTO-211H and HepG2 cells, i.e., nuclear and cytoplasmic staining in the former (Fig. 2E) and predominantly nuclear staining in the latter (Fig. 2F). As confirmation of the specificity of the anti-SBP2 antisera, staining was found to be completely blocked by preincubation of the antisera with bacterially expressed, purified SBP2 but not following incubation with an unrelated bacterially expressed, purified protein (data not shown). In contrast to the patterns in MSTO-211 and HepG2 cells, endogenous SBP2 was barely detectable in HEK-293 cells (Fig. 2D). Previous studies have shown that endogenous expression levels of SBP2 mRNA are low in most tissues (13) and in several cell lines (unpublished results). Localization of the factors by subcellular fractionation and Western blotting revealed a pattern similar to that observed with immunofluorescence for SBP2 in MSTO-211 cells (Fig. 2J, center panel), but a significant fraction of EFsec was detected in the nucleus in HEK-293 cells (Fig. 2J, left panel), suggesting differential sensitivity of the two methods. Western blotting of subcellular fractions with antibodies to cytoplasmic (GRB2) and nuclear (histone H1) markers showed no detectable cross-contamination of subcellular fractions.
FIG. 2.
Subcellular localization of EFsec and SBP2. Confocal microscopy after immunofluorescent localization of transiently expressed EFsec (EFsec txfected), endogenous SBP2 (SBP2 endog), and transiently expressed SBP2 (SBP2 txfected) was performed for HEK-293 (A, D, G, and H), MSTO-211H (B, E, and I), and HepG2 (C and F) cells. (A to C) EFsec (green) was visualized with 1:500 anti-FLAG/1:500 anti-mouse antibody-FITC after transfection with a FLAG-tagged expression construct. (D to F) Endogenous SBP2 (red) was visualized with 1:1,000 SBP2 antisera/1:500 anti-rabbit antibody-rhodamine or Texas Red. (G to I) Transiently expressed SBP2 was visualized with 1:500 anti-HA or 1:500 anti-V5 following transfection with epitope-tagged expression constructs. (G) N-terminal HA-tagged SBP2 (red) in HEK-293. (H and I) C-terminal V5-tagged SBP2 (green) visualized with 1:500 anti-V5/1:500 anti-mouse antibody-Alexa Fluor 488 in HEK-293 and MSTO-211H cells. Cell lines are indicated in white text on the images. In some panels, 1:1,500 DAPI staining (blue) indicates the nucleus of the cell. (J) Subcellular fractionation and Western blotting analysis with FLAG antibody (left panel) and SBP2 antibody (center and right panels) were carried out as described in Materials and Methods. Numbers on the left side of the panel indicate molecular masses in kilodaltons. cyt, cytoplasmic extract; nuc, nuclear lysate.
Localization of EFsec and SBP2 and expression levels of the latter in the three cell lines revealed two interesting findings. First, EFsec localization correlated with both the endogenous levels and localization of SBP2, being predominantly cytoplasmic in cells with undetectable levels of SBP2 (HEK-293), but colocalizing with SBP2 when levels of the latter are substantial (MSTO-211H and HepG2). Second, expression levels of SBP2 in the three cell lines correlated with previously reported selenoprotein expression levels. HEK-293 cells express low levels of the glutathione peroxidases (GPXs) and thioredoxin reductases and undetectable levels of the iodothyronine deiodinases and selenoprotein P (20, 36; unpublished results). MSTO-211H cells, a human mesothelioma tumor-derived cell line, express high levels of the mRNA for type 2 iodothyronine deiodinase (15), and HepG2 cells endogenously express iodothyronine deiodinases, selenoprotein P, and the GPXs and thioredoxin reductases (unpublished results). These results suggest that the levels of SBP2 may be the primary determinant for both EFsec localization and selenoprotein synthesis efficiency.
Overexpression of SBP2 results in predominantly cytoplasmic localization.
To begin to assess the function of the predicted NLS and NES sequences, we introduced epitope tags into full-length SBP2 expression constructs and carried out expression by transient transfection, followed by immunofluorescence staining. In either HEK-293 or MSTO-211H cells transfected with an SBP2 expression vector, SBP2 staining was predominantly cytoplasmic, which was in agreement with recent results in SBP2-transfected rat hepatoma cells (32). This pattern was observed with untagged SBP2 (not shown), SBP2 bearing an N-terminal HA tag (Fig. 2G), SBP2 bearing a C-terminal V5 tag (Fig. 2H and I), or SBP2 fused at the C terminus to green fluorescent protein (GFP) (data not shown), indicating that localization was not influenced by the identity of the epitope tag or its location in the protein. The difference in the localization of endogenous versus transiently expressed SBP2 does suggest, however, that localization may be affected by expression levels, possibly due to changes in stoichiometry relative to other cellular factors (see below). The localization pattern in transfected HEK-293 was also confirmed by subcellular fractionation and Western blotting, but in this case, a faint SBP2 band was also detected in the nuclear fraction (Fig. 2J, right panel).
EFsec contains functional nuclear export and nuclear localization signals.
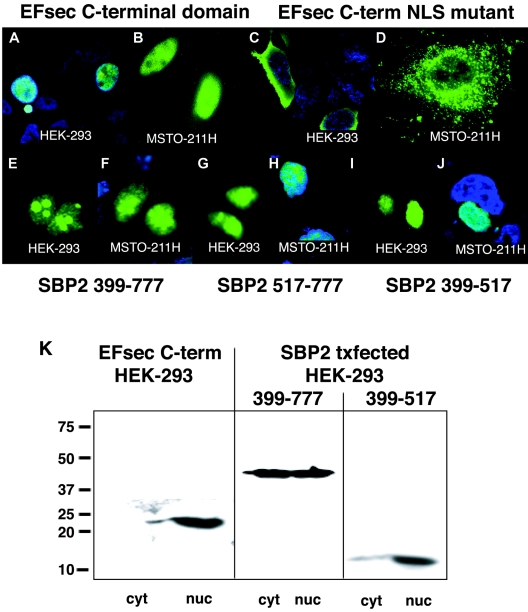
We next examined the localization of truncated versions of EFsec to delineate the functions of putative localization signals. The deletion of the N-terminal elongation factor domain, removing the putative NES (amino acids 95 to 103), resulted in the nearly exclusive nuclear localization of the C-terminal SBP2-interaction domain of EFsec in HEK-293 and MSTO-211H cells (Fig. 3A, B, and K) and the elimination of the faint cytoplasmic staining in HepG2 cells observed for full-length EFsec (data not shown). Thus, the deletion of the putative NES and the retention of the putative NLS in EFsec resulted in nuclear localization. NLS function was further confirmed by the mutation of two amino acids in the putative NLS, amino acids K536→E and K537→E, resulting in a reversion to cytoplasmic localization in HEK-293 (Fig. 3C) and MSTO-211H cells (Fig. 3D). Intriguingly, these two amino acids were previously shown to be essential for interaction of EFsec with SBP2 (39). The fact that the NLS and SBP2 interaction domains of EFsec overlap suggests that their functions may be mutually exclusive. In this view, the NLS would be masked when EFsec and SBP2 are associated but available when EFsec was free of SBP2 (see Discussion).
FIG. 3.
EFsec and SBP2 contain functional NES and NLS signals. (A and B) Localization of transiently expressed FLAG-tagged EFsec C-terminal domain in HEK-293 (A) and MSTO-211H (B) visualized by confocal microscopy using 1:500 anti-FLAG/1:500 anti-mouse antibody-FITC. (C and D) EFsec C-terminal domain NLS mutant (K536E, K537E) in HEK-293 (C) and MSTO-211H (D) cells stained as described for panel A. (E to J) Localization of mutant SBP2 constructs in HEK-293 (E, G, and I) and MSTO-211H (F, H, and J) cells visualized by immunostaining with 1:500 anti-V5/1:500 anti-mouse antibody-Alexa Fluor 488. Panels E and F indicate SBP2 399-777-V5 (minimal functional domain). Panels G and H indicate SBP2 517-777-V5 (transactivation domain). Panels I and J indicate SBP2 399-517-V5 (SECIS-RNA binding domain). In some panels, blue staining with 1:1,500 DAPI indicates nucleus of the cell. Cyan areas indicate the merging of blue and green fluorescence. (K) Subcellular fractionation and localization of EFsec C-terminal domain, SBP2 399-777, and SBP2 399-517 was carried out as described in Materials and Methods. Numbers on the left side of the panel indicate molecular masses in kilodaltons. Txfected indicates transfected. cyt, cytoplasmic extract; nuc, nuclear lysate.
NESs and NLSs of SBP2 are functional.
The presence of multiple putative NESs and NLSs in SBP2, the strong predicted nuclear localization, and the previous demonstration that the N-terminal domain containing three putative NESs and two putative NLSs is dispensable for in vitro function (14) all emphasized the importance of demonstrating the functionality of the putative localization signals. To this end, we assessed the localization of three truncated versions of SBP2: (i) amino acids 399 to 777 of the SBP2 region (SBP2 399-777), consisting of the minimal functional domain with three putative NLSs and one putative NES; (ii) amino acids 399 to 517 of the SBP2 region (SBP2 399-517), consisting of the transactivation domain with two putative NLSs; and (iii) amino acids 517 to 777 of the SBP2 region (SBP2 517-777), consisting of the SECIS RNA binding domain with one putative NLS and one putative NES. In contrast to the strong cytoplasmic localization of transiently expressed full-length SBP2 demonstrated in Fig. 2G to J, all three truncated proteins exhibited strong nuclear localization in both HEK-293 and MSTO-211H cells (Fig. 3E to J). The same patterns were observed for each of the truncated SBP2 proteins bearing C-terminal GFP fusions (not shown). This indicated that one or more of the N-terminal NESs are functional in nucleocytoplasmic export of the protein and that at least one NLS in the region of amino acids 399 to 517 as well as the NLS in the region of amino acids 517 to 777 is functional in import. Subcellular fractionation of HEK-293 cells following transfection with the same SBP2 expression constructs showed agreement in the localization of full-length and truncated SBP2, with the full-length protein being predominantly cytoplasmic and the truncated proteins exhibiting strong nuclear localization (Fig. 2J and 3K).
SBP2 contains a potential nucleolar localization signal.
Strikingly, SBP2 399-777 also exhibited nucleolar localization (Fig. 3E and F), as did SBP2 517-777 to a lesser extent (Fig. 3G and H), whereas SBP2 399-517 did not (Fig. 3I and J). This suggested the presence of a functional NoLS within the region of amino acids 517 to 777 of SBP2. These signals are not well defined but typically consist of runs of basic residues, predominantly arginine, lysine, and glutamine. A motif fitting this pattern is found at amino acids 524 to 540 (Fig. 1B). A role for ribosomal protein L30 in selenoprotein synthesis has recently been described (9), providing a potential explanation for nucleolar localization of SBP2, where recruitment of and/or interaction with L30 might take place. Intriguingly, a patient homozygous for a point mutation in this putative NoLS has been identified and found to express decreased levels of several selenoproteins, providing support for the functional importance of this region (18).
SBP2 minimal functional domain undergoes nucleocytoplasmic shuttling.
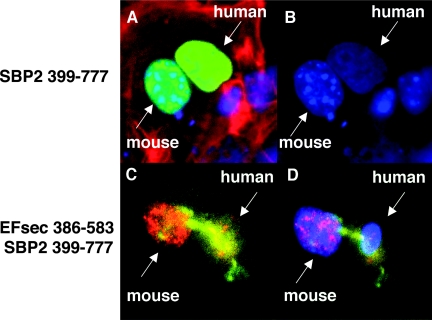
The presence of multiple NESs and NLSs in SBP2, its role in cytoplasmic selenocysteine incorporation and its potential role in circumventing nuclear-associated NMD suggest that this protein might undergo nucleocytoplasmic shuttling. To assess potential shuttling, we carried out a heterokaryon assay in which the SBP2 minimal functional domain was transfected into human MSTO-211H cells, followed by fusion with mouse NIH-3T3 cells. Following 3 h of fusion, SBP2 shuttling from the human to mouse nuclei was clearly apparent (Fig. 4A and B).
FIG. 4.
Nuclear colocalization and nucleocytoplasmic shuttling of SBP2 and EFsec. SBP2 minimal functional domain undergoes nucleocytoplasmic shuttling, as visualized in a heterokaryon assay using SBP 399-777-V5 transiently expressed in human MSTO-211H cells, followed by fusion with mouse NIH 3T3 cells. (A) V5 antibody staining in nuclei from both cell lines (green) merged with DAPI staining and filamentous actin localization with rhodamine-phalloidin (red). (B) DAPI staining used to distinguish human (diffuse) from mouse (punctuate) nuclei. EFsec 386-583 shuttles in the presence of SBP2 399-777. (C) Heterokaryon assay of EFsec 386-583 stained with anti-FLAG/1:500 followed by anti-rabbit antibody-Alexa Fluor 546 (red) and SBP2 399-777 stained with 1:200 anti-v5/1:500 followed by anti-mouse antibody-Alexa Fluor 488 (green) expressed in human MSTO-211H cells followed by fusion with mouse NIH-3T3 cells. (D) Blue fluorescence represents cell nucleus stained with DAPI. Yellow areas indicate the colocalization of green and red fluorescence, pink areas indicate the colocalization of blue and red fluorescence, and cyan areas indicate colocalization of blue and green fluorescence.
SBP2 coexpression results in shuttling of the SBP2-interaction domain of EFsec.
To assess the potential influence of EFsec-SBP2 interaction on localization, heterokaryon assays were performed with the SBP2-interaction domain of EFsec (amino acids 386 to 583 [EFsec 386-583]) in the presence or absence of the SBP2 minimal functional domain (SBP2 399-777). Consistent with the presence of an NLS but not an NES in the EFsec C-terminal region, nucleocytoplasmic shuttling of EFsec was not detected in the absence of SBP2 399-777. However, when SBP2 399-777 was included in the transfection, shuttling of both factors was observed (Fig. 4C and D), suggesting that EFsec may shuttle via its interaction with SBP2.
Coexpression of SBP2 and EFsec results in partial nuclear retention of EFsec.
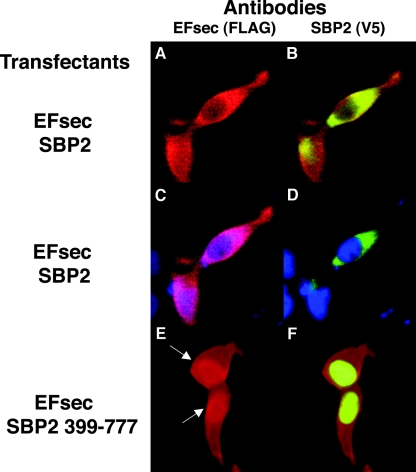
To further test the possibility that EFsec localization is influenced by its interaction with SBP2, we carried out cotransfections of EFsec plus tRNA[Ser]Sec with either the full-length SBP2 (predominantly cytoplasmic) or the SBP2 minimal functional domain (predominantly nuclear) in HEK-293 cells. The tRNA[Ser]Sec plasmid was included, as we have recently shown that its presence greatly enhances EFsec-SBP2 interaction (39). Cotransfection of full-length EFsec and SBP2 in the presence of tRNA[Ser]Sec resulted in the cytoplasmic localization of both factors (Fig. 5A to D), as seen above when either factor was expressed alone. However, upon the cotransfection of full-length EFsec with the minimal functional domain of SBP2 in the presence of tRNA[Ser]Sec, a significant fraction of EFsec was seen colocalizing with SBP2 in the nucleus (Fig. 5E and F). Thus, the localization of SBP2 to the nucleus apparently results in the redistribution of EFsec, possibly due to increased nuclear retention in a complex with SBP2 and/or other factors.
FIG. 5.
Coexpression of EFsec and SBP2 minimal functional domain results in partial nuclear localization of EFsec in HEK-293 cells. Localization was performed following cotransfection of EFsec and SBP2 (A to D) or EFsec and SBP2 399-777 (E and F). Panel A indicates EFsec (FLAG antibody, red). Panel B indicates SBP2 (V5 antibody, green) plus EFsec (FLAG antibody, red). (C and D) Antibody plus DAPI staining (blue) as for panels A and B, respectively. Panel E indicates EFsec (FLAG antibody). Panel F indicates SBP2 399-777 (V5 antibody, green) plus EFsec (FLAG antibody, red). Immunostaining conditions used were as described in the legend for Fig. 2; arrows indicate increased nuclear localization of EFsec in cells coexpressing SBP2 399-777.
EFsec is found in a complex in the nucleus but is predominantly free in the cytoplasm.
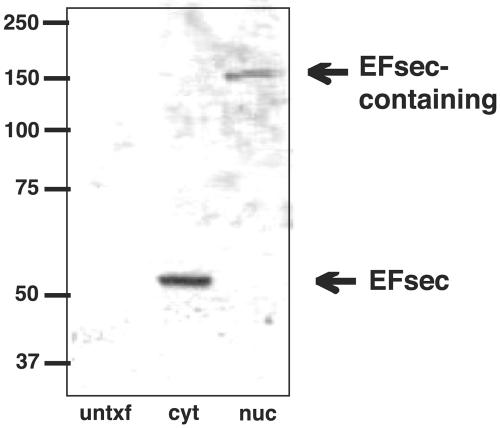
To further investigate the possible link between EFsec localization and association with SBP2, cytoplasmic and nuclear extracts from cells transfected with EFsec were analyzed on nondenaturing gels to preserve noncovalent interactions, followed by Western blotting. EFsec was found to exist predominantly as a monomer in the cytoplasmic extract (Fig. 6, middle lane) but migrated at ∼150 kDa in the nuclear lysate (Fig. 6, right lane), a position consistent with its existence in a heterodimer with SBP2. Gradient fractionation of cytoplasmic extracts revealed EFsec in both free mRNP and polysome fractions (33a), but complexes in association with ribosomes would be excluded from entering the nondenaturing gel.
FIG. 6.
EFsec is predominantly monomeric in the cytoplasm but exists in a complex in the nucleus. Cytoplasmic and nuclear extracts from cells transfected with an EFsec expression plasmid were electrophoresed on nondenaturing gels to preserve noncovalent interactions, followed by Western blotting for EFsec. EFsec was found to exist predominantly as a monomer in the cytoplasmic extract (middle lane) but migrated at ∼150 kDa in the nuclear lysate (right lane). untxtf, untransfected; cyt, cytoplasmic extract; nuc, nuclear lysate.
SBP2 levels influence the levels of selenoprotein mRNAs and proteins.
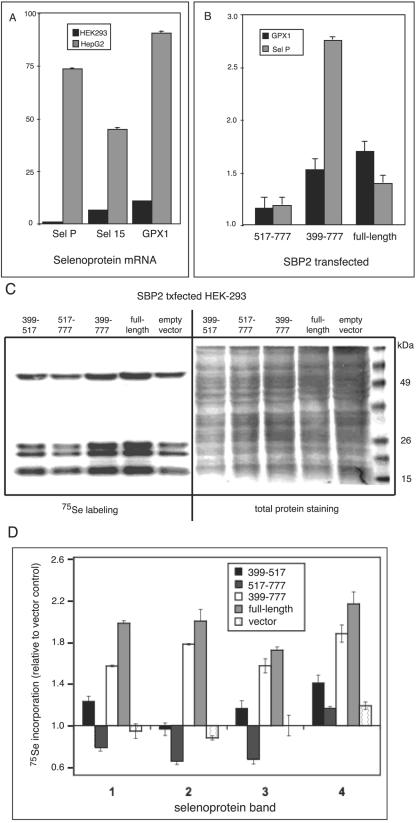
Quantitation of three representative selenoprotein mRNAs in cell lines with low or high endogenous SBP2 revealed expression at levels ∼7- to 90-fold higher in HepG2 (high endogenous SBP2) cells than that in HEK-293 cells (low endogenous SBP2) when normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (Fig. 7A). Transfection of either full-length SBP2 or the minimal functional domain (amino acids 399 to 777) into HEK-293 cells increased the levels of endogenous selenoprotein mRNAs encoding GPX1 and selP by ∼1.4- to 2.8-fold (Fig. 7B). Similar but less pronounced effects were seen with selenoprotein S and GPX4 mRNAs (not shown). In contrast, the effect of transfecting the SECIS binding domain alone (amino acids 517 to 777) on selenoprotein mRNA levels was not statistically different from the effect of transfecting empty vector. Thus, either full-length SBP2 or the truncated, nuclearly localized SBP2 399-777 fragment is capable of increasing the steady-state levels of multiple, distinct selenoprotein mRNAs.
FIG. 7.
SBP2 expression increases selenoprotein mRNA and protein levels. (A) mRNA levels for the 15-kDa selenoprotein (sel 15), GPX1, and selenoprotein P (sel P) in HepG2 and HEK-293 cells were assessed by spotted array hybridization and PhosphorImager quantitation. All mRNA levels were normalized to GAPDH mRNA, and hybridizations were performed at least in triplicate. (B) mRNA levels for GPX1 and selenoprotein P (sel P) were assessed in HEK-293 cells by quantitative real-time reverse transcriptase PCR following transient transfection of the indicated SBP2 expression constructs. Note that transient transfection is not highly efficient; thus, increases in mRNA levels in the presence of SBP2 likely reflect a fraction of the total cell population. Means were compared using Student's t test; P was <0.01 for all samples. (C) Selenoproteins were labeled with sodium [75Se]selenite following transfection of the indicated SBP2 constructs and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. (D) Densitometric quantitation of the four prominent selenium-labeled bands shown in panel C, numbered from top to bottom of the gel. Means from two labeling experiments that were derived from independent transfections are given.
As previous studies have implicated the nonsense-mediated decay pathway in degradation of selenoprotein mRNAs and a hallmark of this pathway is intron dependence, the effect of SBP2 on the expression of mRNA from an intronless GPX1 cDNA was assessed. Upon transfection of either the full-length SBP2 or the minimal functional domain, the level of GPX1 mRNA that was derived from the intronless cDNA did not increase but, in fact, decreased. To ascertain the reason for this decrease, we carried out subcellular fractionation prior to reverse transcription and real-time PCR. This revealed greatly increased retention of mRNA from the intronless cDNA in the nuclear fraction in response to SBP2 transfection. Nuclear retention fully accounted for the decrease in cytoplasmic mRNA. While the nuclear retention of intronless mRNAs is consistent with previous reports, the reasons for the increase with SBP2 transfection will require further investigation.
We also assessed the effects of expressing the SBP2 full-length and truncated fragments on selenoprotein synthesis. 75Se labeling of endogenous selenoproteins increased ∼1.5- to 2.2-fold following transfection of full-length SBP2 or the SBP2 399-777 region. Incorporation of label was minimally affected by transfection with the SBP2 399-517 region and was decreased upon transfection of amino acids 517 to 777 of the SECIS binding domain (Fig. 7C and D).
DISCUSSION
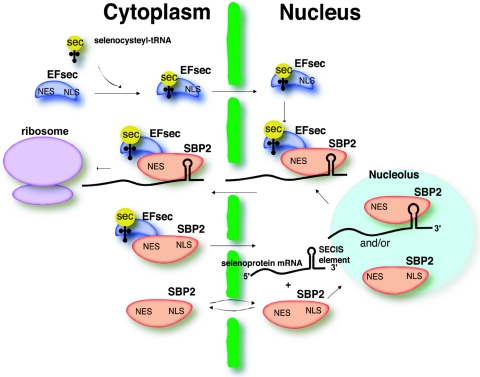
The role of the selenoprotein synthesis factors in facilitating efficient selenocysteine incorporation, thus circumventing NMD, presents an intriguing problem which is clearly dependent on the early and efficient recruitment of these factors to the selenoprotein mRNA. Import of SBP2 to the nucleus would allow binding to the SECIS element either during or immediately after its transcription. However, for selenocysteine incorporation to occur, EFsec and selenocysteyl-tRNA[Ser]Sec would also need to be recruited to the mRNA. Full-length EFsec in HEK-293 exhibits both cytoplasmic and nuclear localization, but nuclear localization is more pronounced upon coexpression of SBP2. Further, coimmunoprecipitation studies identify an EFsec-SBP2 complex in the nucleus of cells transfected with both plasmids (33a). submitted for publication). The overlap of the EFsec NLS with the SBP2-interaction domain and likewise of the EFsec NES with the elongation factor domain invites speculation about the relationship between the interaction of these factors and their localization. Interaction between selenocysteyl-tRNA[Ser]Sec-EFsec and SBP2 in the cytoplasm is necessary for selenocysteine incorporation. However, some fraction of this complex may also be imported into the nucleus via the NLSs of SBP2. Noncomplexed SBP2 (e.g., newly synthesized) would be predicted to undergo more rapid or efficient import than SBP2 associated with EFsec, as three of the predicted NLSs in SBP2 lie within the transactivation domain and thus might be masked by interaction with the elongation factor. In addition, one of the putative NESs overlaps the RNA binding domain of SBP2 (13); thus, SECIS binding may result in masking of this signal. EFsec or the selenocysteyl-tRNA[Ser]Sec-EFsec complex may also undergo import in the absence of SBP2 via the EFsec NLS, followed by interaction with SBP2 in the nucleus (see Fig. 6). Nuclear retention of the complex may occur via association with a SECIS element until the selenoprotein mRNA and associated complex are exported. Furthermore, upon import of EFsec, export via its NES may also be masked by selenocysteyl-tRNA[Ser]Sec binding, allowing sufficient time for interaction with SBP2. Thus, the import of these factors into the nucleus and the assembly of selenocysteine incorporation complexes at the SECIS element early in the life of selenoprotein mRNAs could allow these UGA-containing mRNAs to circumvent NMD during the first round of translation. Import and export of EFsec and SBP2, either individually or complexed with each other and their respective RNA ligands, is depicted in the model in Fig. 8.
FIG. 8.
Schematic model for localization and interaction of EFsec and SBP2. Selenocysteyl-tRNA (yellow) binds to the N-terminal portion of EFsec (blue) in the cytoplasm, and the complex of sec-tRNASec-EFsec is imported into the nucleus via the C-terminal NLS of EFsec. SBP2 (red) shuttles between the cytoplasm and nucleus via its NLSs and NESs. SBP2 binds to SECIS elements of selenoprotein mRNAs in the nucleus, either prior to or after shuttling in and out of the nucleolus. SBP2 binds the selenocysteyl-tRNA[Ser]Sec-EFsec complex in either the cytoplasm or nucleus. If this occurs in the cytoplasm, the NLSs of SBP2 are still exposed for import of the selenocysteyl-tRNA[Ser]Sec-EFsec-SBP2 complex into the nucleus and binding of selenoprotein mRNA. Alternatively, SBP2 may interact with selenocysteyl-tRNA[Ser]Sec-EFsec in the nucleus. Finally, either the mRNA export pathway or the NES of SBP2 would ultimately be responsible for the export of the complex into the cytoplasm for binding to ribosomes and translation of selenoproteins. This model would explain not only the localization data presented herein but also the observation that levels of SBP2 determine levels of selenoprotein expression.
Several lines of evidence support a role for SBP2 as a key player in circumventing NMD in higher eukaryotes. As mentioned in Results, selenoprotein expression levels differ significantly in the cell lines under investigation, with GPX1, selenoprotein P, and 15-kDa selenoprotein mRNA levels being up to 2 orders of magnitude higher in HepG2 versus those in HEK-293 cells, when normalized to β-actin and GAPDH mRNA. Transfection of SBP2 into HEK-293 cells increased the levels of GPX1 and selenoprotein P mRNAs, suggesting that SBP2 may stabilize these mRNAs.
Numerous studies have reported on the differential sensitivity of different selenoprotein mRNAs or even the same mRNAs in different tissues to degradation upon selenium depletion (12, 25, 27). For example, classical cytoplasmic GPX1 mRNA levels fall to ∼5 to 10% in liver following selenium depletion in rats (11, 33, 38). The decline in GPX1 mRNA upon selenium depletion has been shown to exhibit all the hallmarks of NMD (33), resulting in the down-regulation of the levels of this mRNA when selenium is not abundant. In contrast, phospholipid hydroperoxide glutathione peroxidase is highly resistant to the effects of selenium depletion (3, 27, 34, 37). The selenoprotein mRNA and tissue-specific hierarchy have long been speculated to be results of the differential retention of selenium in different organs (2). However, the studies reported herein suggest that other factors, including SBP2 levels and subcellular localization, SBP2-SECIS interactions, and sensitivity to NMD, may also play a significant role in the hierarchy of selenoprotein synthesis.
Comparative analysis of the EFsec and SBP2 sequences in higher and lower eukaryotes as well as archaea sheds additional light on the importance of the localization signals in selenoprotein synthesis and NMD. Localization signals were not detected in EFsec of C. elegans, and the only homology to SBP2 we could identify in this organism was found in a protein comprising only the SECIS binding domain. In accord with this, the sole selenoprotein in C. elegans, a thioredoxin reductase, is predicted to be insensitive to NMD, as the selenocysteine and termination codons are both located in the last exon. The NLS in EFsec was also not conserved in D. melanogaster. Intriguingly, recent studies have indicated that the identification of termination codons as premature in this organism does not occur via exon junction complexes as is the case in vertebrates (21), suggesting that NMD may not occur in association with the nuclear fraction in insects. Localization signals are also lacking in the archaeal homolog of EFsec, which is not surprising, given the absence of nuclei. No SBP2-like factor has been identified in archaea to date, but a recent report indicates that this function may be served by the C-terminal extension of archaeal EFsec (28), suggesting that recruitment of the archaeal elongation factor may occur in a manner that combines features of prokaryotes and eukaryotes.
Further studies of the selenocysteine incorporation process and the roles, localization, and levels of the factors involved will provide exciting new insights into the cellular mechanisms involved when an mRNA faces the “UGA, stop or selenocysteine” molecular dilemma.
Acknowledgments
We thank Michelle Lowe for technical assistance at the Brigham and Women's Hospital Confocal Facility, Tina Carvalho and Qingping He for technical support at the Pacific Biomedical Research Center Confocal Facility at University of Hawaii at Manoa, and Paul Copeland for the generous gift of SBP2 antisera.
This study was supported by NIH grants DK47320, DK52963, and RR16467.
REFERENCES
- 1.Baqui, M. M., B. Gereben, J. W. Harney, P. R. Larsen, and A. C. Bianco. 2000. Distinct subcellular localization of transiently expressed types 1 and 2 iodothyronine deiodinases as determined by immunofluorescence confocal microscopy. Endocrinology 141:4309-4312. [DOI] [PubMed] [Google Scholar]
- 2.Behne, D., H. Hilmert, S. Scheid, H. Gessner, and W. Elger. 1988. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim. Biophys. Acta 966:12-21. [DOI] [PubMed] [Google Scholar]
- 3.Bermano, G., J. R. Arthur, and J. E. Hesketh. 1996. Role of the 3′ untranslated region in the regulation of cytosolic glutathione peroxidase and phospholipid-hydroperoxide glutathione peroxidase gene expression by selenium supply. Biochem. J. 320:891-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry, M. J. 2000. Recoding UGA as selenocysteine, p. 763-783. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Berry, M. J., L. Banu, Y. Y. Chen, S. J. Mandel, J. D. Kieffer, J. W. Harney, and P. R. Larsen. 1991. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353:273-276. [DOI] [PubMed] [Google Scholar]
- 6.Berry, M. J., L. Banu, J. W. Harney, and P. R. Larsen. 1993. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 12:3315-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, M. J., J. W. Harney, T. Ohama, and D. L. Hatfield. 1994. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res. 22:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brent, G. A., J. W. Harney, Y. Chen, R. L. Warne, D. D. Moore, and P. R. Larsen. 1989. Mutations of the rat growth hormone promoter which increase and decrease response to thyroid hormone define a consensus thyroid hormone response element. Mol. Endocrinol. 3:1996-2004. [DOI] [PubMed] [Google Scholar]
- 9.Chavatte, L., B. A. Brown, and D. M. Driscoll. 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12:408-416. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, J., and L. E. Maquat. 1993. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol. Cell. Biol. 13:1892-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, M. J., and K. W. Burgener. 1992. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J. Nutr. 122:1620-1626. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, M. J., P. M. Cammack, and C. D. Wray. 1995. Tissue specificity of selenoprotein gene expression in rats. J. Nutr. Biochem. 6:367-372. [DOI] [PubMed] [Google Scholar]
- 13.Copeland, P. R., J. E. Fletcher, B. A. Carlson, D. L. Hatfield, and D. M. Driscoll. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Copeland, P. R., V. A. Stepanik, and D. M. Driscoll. 2001. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 21:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curcio, C., M. M. Baqui, D. Salvatore, B. H. Rihn, S. Mohr, J. W. Harney, P. R. Larsen, and A. C. Bianco. 2001. The human type 2 iodothyronine deiodinase is a selenoprotein highly expressed in a mesothelioma cell line. J. Biol. Chem. 276:30183-30187. [DOI] [PubMed] [Google Scholar]
- 16.Dostie, J., M. Ferraiuolo, A. Pause, S. A. Adam, and N. Sonenberg. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 19:3142-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driscoll, D. M., and P. R. Copeland. 2003. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 23:17-40. [DOI] [PubMed] [Google Scholar]
- 18.Dumitrescu, A. M., X. H. Liao, M. S. Abdullah, J. Lado-Abeal, F. A. Majed, L. C. Moeller, G. Boran, L. Schomburg, R. E. Weiss, and S. Refetoff. 2005. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat. Genet. 37:1247-1252. [DOI] [PubMed] [Google Scholar]
- 19.Fagegaltier, D., N. Hubert, K. Yamada, T. Mizutani, P. Carbon, and A. Krol. 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasdaska, J. R., J. W. Harney, P. Y. Gasdaska, G. Powis, and M. J. Berry. 1999. Regulation of human thioredoxin reductase expression and activity by 3′-untranslated region selenocysteine insertion sequence and mRNA instability elements. J. Biol. Chem. 274:25379-25385. [DOI] [PubMed] [Google Scholar]
- 21.Gatfield, D., L. Unterholzner, F. D. Ciccarelli, P. Bork, and E. Izaurralde. 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hentze, M. W., and A. E. Kulozik. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307-310. [DOI] [PubMed] [Google Scholar]
- 24.Hicks, G. R., and N. V. Raikhel. 1995. Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 11:155-188. [DOI] [PubMed] [Google Scholar]
- 25.Hill, K. E., P. R. Lyons, and R. F. Burk. 1992. Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem. Biophys. Res. Commun. 185:260-263. [DOI] [PubMed] [Google Scholar]
- 26.Himeno, S., H. S. Chittum, and R. F. Burk. 1996. Isoforms of selenoprotein P in rat plasma. Evidence for a full-length form and another form that terminates at the second UGA in the open reading frame. J. Biol. Chem. 271:15769-15775. [DOI] [PubMed] [Google Scholar]
- 27.Lei, X. G., J. K. Evenson, K. M. Thompson, and R. A. Sunde. 1995. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 125:1438-1446. [DOI] [PubMed] [Google Scholar]
- 28.Leibundgut, M., C. Frick, M. Thanbichler, A. Bock, and N. Ban. 2005. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 24:11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lykke-Andersen, J., M. D. Shu, and J. A. Steitz. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103:1121-1131. [DOI] [PubMed] [Google Scholar]
- 30.Ma, S., K. E. Hill, R. M. Caprioli, and R. F. Burk. 2002. Mass spectrometric characterization of full-length rat selenoprotein P and three isoforms shortened at the C terminus. Evidence that three UGA codons in the mRNA open reading frame have alternative functions of specifying selenocysteine insertion or translation termination. J. Biol. Chem. 277:12749-12754. [DOI] [PubMed] [Google Scholar]
- 31.Maquat, L. E. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5:89-99. [DOI] [PubMed] [Google Scholar]
- 32.Mehta, A., C. M. Rebsch, S. A. Kinzy, J. E. Fletcher, and P. R. Copeland. 2004. Efficiency of mammalian selenocysteine incorporation. J. Biol. Chem. 279:37852-37859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriarty, P. M., C. C. Reddy, and L. E. Maquat. 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 18:2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Small-Howard, A., N. Morozova, Z. Stoytcheva, E. P. Forry, J. B. Mansell, J. W. Harney, B. A. Carlson, X. M. Xu, D. L. Hatfield, and M. J. Berry. Supramolecular complexes mediate selenoystein incorporation in vivo. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 34.Sun, X., X. Li, P. M. Moriarty, T. Henics, J. P. LaDuca, and L. E. Maquat. 2001. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol. Biol. Cell 12:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tujebajeva, R. M., P. R. Copeland, X. M. Xu, B. A. Carlson, J. W. Harney, D. M. Driscoll, D. L. Hatfield, and M. J. Berry. 2000. Decoding apparatus for eukaryotic selenocysteine incorporation. EMBO Rep. 2:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tujebajeva, R. M., J. W. Harney, and M. J. Berry. 2000. Selenoprotein P expression, purification, and immunochemical characterization. J. Biol. Chem. 275:6288-6294. [DOI] [PubMed] [Google Scholar]
- 37.Weiss Sachdev, S., and R. A. Sunde. 2001. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem. J. 357:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss, S. L., and R. A. Sunde. 1998. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA 4:816-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zavacki, A. M., J. B. Mansell, M. Chung, B. Klimovitsky, J. W. Harney, and M. J. Berry. 2003. Coupled tRNASec dependent assembly of the selenocysteine decoding apparatus. Mol. Cell 11:773-781. [DOI] [PubMed] [Google Scholar]