Abstract
Cryptochromes (CRYs) are composed of a core domain with structural similarity to photolyase and a distinguishing C-terminal extension. While plant and fly CRYs act as circadian photoreceptors, using the C terminus for light signaling, mammalian CRY1 and CRY2 are integral components of the circadian oscillator. However, the function of their C terminus remains to be resolved. Here, we show that the C-terminal extension of mCRY1 harbors a nuclear localization signal and a putative coiled-coil domain that drive nuclear localization via two independent mechanisms and shift the equilibrium of shuttling mammalian CRY1 (mCRY1)/mammalian PER2 (mPER2) complexes towards the nucleus. Importantly, deletion of the complete C terminus prevents mCRY1 from repressing CLOCK/BMAL1-mediated transcription, whereas a plant photolyase gains this key clock function upon fusion to the last 100 amino acids of the mCRY1 core and its C terminus. Thus, the acquirement of different (species-specific) C termini during evolution not only functionally separated cryptochromes from photolyase but also caused diversity within the cryptochrome family.
Circadian rhythms in physiology, metabolism, and behavior are generated by a genetically determined clock with an intrinsic periodicity of approximately 24 h. In mammals, the master clock resides in the neurons of the suprachiasmatic nucleus (SCN) in the ventral hypothalamus. To keep pace with the light-dark cycle, the SCN clock is daily entrained by light perceived via the retina and transmitted to the SCN via the retinohypothalamic tract (27, 31). Subsequently, this master clock synchronizes peripheral oscillators via neuronal and humoral signaling (1, 19, 24, 46). Peripheral oscillators are thought to optimize organ performance by adjusting metabolic and physiological functions to the requirement at specific times of the day. SCN neurons, peripheral tissues, and in vitro-cultured fibroblasts generate circadian rhythms by means of a self-sustaining molecular oscillator that drives gene expression through interconnected positive and negative transcription/translation feedback loops (28, 47). In the positive limb of the circadian oscillator, transcription of the Period (mPer1, mPer2, and mPer3), Cryptochrome1 (mCry1), and Rev-Erbα clock genes is activated by a heterodimer of the two helix-loop-helix /PAS domain transcription factors CLOCK and BMAL1, which act via CACGTG E-box enhancer elements (3, 8, 26, 28, 29, 47). The mammalian CRY1 (mCRY1) and mCRY2 proteins are central components in the negative limb of this circuit, as they strongly inhibit CLOCK/BMAL1-mediated transcription and, as a consequence, shutdown their own expression (9, 15, 22). Cyclic expression of Bmal1 was found to occur through transcriptional activation by the orphan nuclear receptor RORα (36) and inhibition by REV-ERBα (5, 26).
Immunohistochemical analysis of the SCN has revealed synchronous circadian patterns of abundance and nuclear localization of mCRY and mammalian PER (mPER) proteins (6, 15). Moreover, as shown for mPER2, nuclear accumulation does not merely involve nuclear import but instead encompasses a delicate interplay of nuclear import signals (nuclear localization signals [NLSs]) and nuclear export signals (NESs) allowing the protein to shuttle between the cytoplasm and nucleus (15, 44). In addition, mPER proteins CLOCK and BMAL1 undergo circadian changes in protein phosphorylation, involving CK1ɛ (and presumably other kinases) and, as shown for mPER2, affecting protein stability (16). Protein stability also appears to be determined by ubiquitylation; mCRY proteins reduce the ubiquitylation status of mPER2 in vitro and are redundantly necessary for the stability of mPER2 in vivo (44). These findings strongly point to posttranslational modifications, nuclear translocation, and protein turnover of clock factors as critical events in shaping the approximately 6-h delay in mRNA and protein rhythms necessary to establish a near-24-h periodicity of the clock.
Coimmunoprecipitation studies with transiently expressed proteins, as well as yeast two-hybrid experiments, have uncovered direct interactions between mCRY proteins and multiple core clock components: mCRY proteins bind the C terminus of mPER2 and mPER1 (20, 44) as well as CLOCK, BMAL1, and TIMELESS (TIM) (9, 15, 37). Despite extensive studies, the underlying molecular mechanism for the synchronous nuclear accumulation of mCRY and mPER proteins has not been fully clarified (15, 20, 44). In addition, little is known about the mechanism of CRY-mediated inhibition of CLOCK/BMAL1. This is to a large extent due to the lack of information on mCRY domains involved in these processes.
Mammalian CRY proteins belong to the photolyase/cryptochrome protein family and were initially identified as homologs of the DNA repair protein photolyase, an enzyme that removes UV light-induced DNA damage using visible light as an energy source (reviewed in reference 34). Although animal cryptochromes share a high degree of homology with photolyases, they lack the NLS-containing N-terminal extension characteristic of eukaryotic photolyases and instead contain a C-terminal extension as also observed in plant cryptochromes (39, 41). Analysis of the amino acid sequences of mCRY1 and mCRY2 reveals over 80% amino acid identity in the core domain (the ∼500-amino-acid [aa] region shared by photolyases and cryptochromes), whereas their C-terminal tails are unique and distinct from those of plant and Drosophila melanogaster cryptochromes (14, 39). Since mCry1 and mCry2 single-knockout animals display short and long period of wheel-running behavior, respectively (40, 43), we sought to determine whether the C termini might orchestrate the function of mCRY proteins.
MATERIALS AND METHODS
Plasmids.
Hemagglutinin (HA)-tagged mCry1 (HA-mCry1) cloned into either pcDNA3 (Invitrogen) or pBluescript KS(−) (Stratagene) served as the starting construct for the generation of the various mutant mCRY1 expression constructs by using the QuikChange mutagenesis kit (Stratagene). The following forward primers were used: 5′-CAG CAG CTT TCC CGG TAC TGA GGG CGA ATT CTT CTC GCC TCG GTC CC-3′ (HA-CRY1Δtail), 5′-GGA GTT AAT TAC CCC AAA CCG ATG TGA AGA ATT CTG AGG CAA GCA GAC TG-3′ (HA-CRY1ΔCCtail), 5′-GGA GTT AAT TAC CCC AAA CCG ATG GTG GGA GGT GGA GGG CTA GGT CTT CTC GCC TCG GTC CC-3′ (HA-CRY1ΔCC), 5′-CAC CGG CCT CAG CAG TGG GGC GGC GCC TAG TCA GGA AGA GGA TGC-3′ (HA-CRY1mutNLSc), and 5′-CTA ACA GAT CTC TAC GCA GCG GTA AAG AAG AAT AGT TCC CCT CCC C-3′ (HA-CRY1mutNLSn) (EcoRI restriction sites are underlined). The reverse primers contained the reverse complementary sequence of the forward primers. To generate HA-CRY1ΔCCmutNLSc, we used a SacII site upstream of the NLS in the C-terminal end (NLSc) of mCry1 and a SacII site present in the vector to replace the last ∼100 bp of HA-CRY1ΔCC with those of HA-CRY1mutNLSc. mCRY1-EGFP was generated by PCR to create an AgeI site at the stop codon and subsequently cloned in frame in an enhanced green fluorescent protein (GFP) (EGFP) vector (Clontech). Primers used to create the AgeI site (underlined) are as follows: 5′-GTA CCG GTC CGT TAC TGC TCT G-3′ (forward) and 5′-GTA CCG GTC CGT TAC TGC TC-3′ (reverse). To generate the chimeric constructs between (6-4 photoproduct)photolyase [(6-4PP)PhL] and mCRY1, we used a PCR-based approach. The starting constructs were (6-4PP)PhL cloned into pcDNA3(EcoRI) and HA-CRY1 cloned in pcDNA3(EcoRI). (6-4PP)PhL-CT was made by creating an AflII site at the stop codon of (6-4PP)PhL using the QuikChange method (forward primer, 5′-CAG GAA CCA ACG ACC AAA ACT TAA GTA GGT AGA GGT ACC CCA GCG-3′ [the reverse primer was the reverse complement]) and an AflII site at aa 471 of mCRY1 (forward primer, 5′-GAG AAC TTA AGA ACC ATG CTG AGG-3′; reverse primer, 5′-GCG AAT TCT CAG TTA CTG CTC TGC-3′) (mutations are shown in boldface type, and restriction sites are underlined [AflII in the forward primers and EcoRI in the reverse primer]). The (6-4PP)PhL (aa 1 to 537) EcoRI/AflII fragment and mCRY1 (aa 471 to 606) AflII/EcoRI fragment were cloned into pcDNA3-linearized EcoRI. To generate (6-4PP)PhL-extCT, we created a PvuII site at aa 378 by introducing a silent mutation using the QuickChange method (forward primer, 5′-GTT TTC TTA CTC GTG GGG ATC TGT TCA TCA GCT GGG AAC AAG GGC GTG ATG-3′ [the reverse primer was the reverse complement]) and made use of a naturally occurring PvuII site at aa 371 of mCRY1. The (6-4PP)PhL (aa 1 to 378) EcoRI/PvuII fragment and an mCRY1 (aa 371 to 606) PvuII/NotI fragment were cloned into pcDNA3 EcoRI/NotI. EGFP-extCT was made by cloning the mCRY1 (aa 371 to 606) PvuII/NotI fragment into pEFGP (Clontech). The generation of the plasmids for FLAG-BMAL1 and EGFP-BMAL1 will be presented elsewhere (E. Jacobs, unpublished data).
Cell culture and MDFs.
COS7 cells and wild-type, Per1−/−, Per2Brdm1, and Per1−/− Per2Brdm1 mouse dermal fibroblasts (MDFs) were cultured in Dulbecco's modified Eagle's medium-F10-P/S-10% fetal calf serum and transfected with Fugene (Boehringer) according to the manufacturer's instructions. To generate MDFs, mice were killed by cervical dislocation, and a small piece of back skin of the mouse was removed and cut into pieces with a razor blade. Skin pieces were washed in ethanol, rinsed in phosphate-buffered saline, and incubated for 3 to 4 h at 37°C in TCH mix (0.125% trypsin, 1 mg/ml collagenase type V [Sigma], and 0.3 mg/ml hyaluronidase in phosphate-buffered saline). After the addition of 5 ml of Dulbecco's modified Eagle's medium-F10 with 20% fetal calf serum, cells were spun down for 5 min at 1,000 rpm, resuspended in similar medium, and seeded onto a 3.5-cm dish. MDFs were cultured in a low-oxygen incubator (5% CO2, 3% O2), and after a few days, cells were split. Leptomycin B (LMB) (Sigma) dissolved in methanol was added to the medium at a final concentration of 10 ng/ml.
Immunofluorescence and immunoprecipitation.
Immunofluorescence and immunoprecipitation techniques were performed as described previously (44). To detect the various proteins in single, double, and triple immunofluorescence studies, we employed rat anti-HA (1:1,000; Roche), mouse anti-HA (monoclonal 12CA5, 1:500), rabbit anti-HA (1:10,000; ECL Oregon), mouse anti-Flag (1:500; Sigma), rabbit anti-CLOCK (1:500, a generous gift from U. Schibler), and rabbit anti-(6-4PP)photolyase (13). The fluorescent secondary antibodies were Texas red-conjugated anti-rat immunoglobulin G (IgG) antibody (1:500; Jackson Immuno Research Laboratories), anti-mouse antibody-Alexa 350 (1:250; Molecular Probes), anti-mouse antibody-Alexa 488 (1:1,000), and anti-rabbit antibody-Alexa 594 (1:1,000). The subcellular localization of the different mCRY mutants and mPER2 was analyzed 24 h after transfection, and it was scored using an unbiased procedure by three investigators in at least two independent experiments. Approximately 200 cells were counted every time, and the data are presented as a percentage of the total amount of cells. The localization was defined as fully nuclear (black), nuclear and cytoplasmic (gray) and fully cytoplasmic (red). The cellular localizations of the proteins did not vary between 24 h and 48 h after transfection. Small changes in the localizations between experiments depended on the status of the cells in each experiment. Similar patterns of subcellular localizations were obtained with other cell lines tested (MRC5, NIH 3T3, and HEK293) (data not shown). We used anti-HA (Roche) and anti-GFP (Clontech) antibodies for the immunoprecipitation step and immunoblot analysis (1:2,000 dilution). As secondary antibodies, we used horseradish peroxidase-conjugated anti-rabbit IgG (Biosource Int.) and anti-rat IgG (DAKO) at a 1:1,000 dilution, visualized using the ECL system (Pharmacia Biotech).
CC prediction.
To predict the presence of coiled-coil (CC) domains, we used the program Coils (http://www.ch.embnet.org/software/COILS_form.html), which calculates the probability of an amino acid sequence adopting a coiled-coil conformation (18). We analyzed the sequences of mCRY1 (GenBank accession no. NM_007771) and mCRY2 (accession no. NM_009963) in this way. Other coiled-coil programs failed to predict the CC discussed in this study (data not shown), and modeling of the amino acid sequence of mCRY1 on the resolved structure of Escherichia coli photolyase has revealed that the N-terminal domain is an α-helix rather than a coil (10). In HA-CRY1ΔCC, the deletion of the CC was not complete at the sequence level (aa 471 to 485) (see Results); however, the program Coils confirmed the removal of the CC in HA-CRY1ΔCC (data not shown). Conversely, in HA-CRY1Δtail, this deletion included part of the CC (aa 485 to the end); however, the program still predicts the presence of the CC in this mutant (data not shown).
Luciferase reporter assay.
The luciferase transcription assay was performed as previously described (45). Cells were transfected with 750 ng of pcDNA3-hBMAL1, 750 ng of pcDNA3-mCLOCK, 5 ng of the pGL3 promoter containing the two E boxes from the mouse Dbp promoter, 0.1 ng of internal control plasmid pRL-CMV, and 10 to 100 ng of the plasmid containing wild-type or mutant HA-CRY1 by using Fugene (Roche) according to the manufacturer's instructions. The total amount of 2 μg DNA per transfection was adjusted by adding pcDNA3 vector. After 48 h, bioluminescence was measured using the Dual-Luciferase Reporter Assay system (Promega). For statistical analysis, a two-sample t test was applied.
RESULTS
Two routes for mCRY1 nuclear entry converge on its C terminus.
To understand the mechanism of function of mCRY1, we initially studied how its nuclear localization is regulated in mammalian cells. Previously, Hirayama and coworkers reported the presence of a monopartite NLS at amino acid positions 274 to 278 of mCRY1 (10), hereafter referred to as NLSn (Fig. 1A). In addition to this conserved NLSn, mCRY2 contains a bipartite NLS in the C-terminal extension (KRKX13KRAR; boldface indicates conserved amino acids) (Fig. 1A), which appeared absent in mCRY1 (32). Nevertheless, a close inspection of the C terminus of mCRY1 uncovered a less-well-defined nucleoplasmin-like bipartite NLS (KRPX11KVQR, aa 585 to 602 [designated NLSc]) (Fig. 1A) (30). Inactivation of NLSc by a substitution of essential lysine and arginine residues (HA-CRY1mutNLSc) negatively affected nuclear accumulation of transiently expressed protein in transfected COS7 cells (Fig. 1B to D), as does, although to a significant lesser extent, the mutagenesis of NLSn (10). Yet the majority of cells (>80%) still display nuclear or nucleocytoplasmic distribution of HA-CRY1mutNLSn or HA-CRY1mutNLSc, which could suggest redundancy in NLSn and NLSc function. Importantly, the combined inactivation of NLSn and NLSc did not abolish nuclear translocation, as evident from the nucleocytoplasmic distribution of HA-CRY1mutNLSn+c in 75% of the transfected cells (Fig. 1B to D). Since we could not identify other NLS domains in the sequence of mCRY1, this finding points to the involvement of an additional mechanism in the nuclear localization of mCRY1.
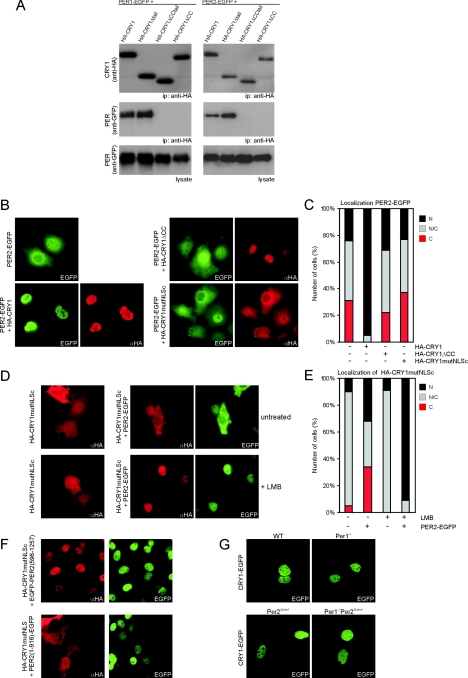
FIG.1.
Functional characterization of the C terminus of mCRY1 in nuclear import. (A) Protein sequence alignment of NLSn and NLSc of mCRY1, human CRY1 (hCRY1), and mCRY2 (amino acid numbers are indicated above the sequence; stop codons are represented by a star). For comparison, the bipartite NLS of nucleoplasmin is aligned with NLSc. Conserved basic amino acids in the NLSs are in boldface type and underlined. Amino acid substitutions, as present in HA-CRY1mutNLSc, HA-CRY1mutNLSn, and HA-CRY1mutNLSn+c, are also indicated. (B) Schematic representation of HA-CRY1, HA-CRY1mutNLSc, HA-CRY1mutNLSn, and HA-CRY1mutNLSn+c. Mutations in NLSc and NLSn are indicated by a star. The photolyase-like domain is shown in dark blue, and the C terminus is shown in light blue. (C) Immunofluorescence pictures of COS7 cells transfected with the constructs shown in B. (D) Quantification of the cellular localization of the above-described proteins. Nuclear (N) signal is black, nuclear/cytoplasmic (N/C) signal is gray, and cytoplasmic (C) signal is red. (E) Schematic representation of HA-CRY1ΔCC, HA-CRY1ΔCCmutNLSc, HA-CRY1ΔCCtail, and HA-CRY1Δtail. The tail is indicated in light blue, and the CC domain is shown in yellow. The left and right red bars represent NLSn and NLSc, respectively, and the star is the mutated NLSc. (F) Immunofluorescence pictures of COS7 cells transfected with the constructs shown in E. αHA, anti-HA. (G) Quantification of the cellular localization of the transiently expressed proteins.
“Coiled-coil output” analysis revealed potential coiled-coil domains at the N terminus (aa 53 to 78) and at the start of the C-terminal extension (aa 471 to 493) of mCRY1 (see Fig. S1 in the supplemental material) and at similar positions in mCRY2 (data not shown). Since we focus here on the C terminus of mCRY1, we further analyzed the C-terminal CC domain. For simplicity, the remaining part of the C-terminal extension of mCRY1 (and mCRY2) is termed “tail.” Deletion of the CC (HA-CRY1ΔCC) did not significantly change the subcellular localization of transiently expressed mCRY1 (Fig. 1E to G). Interestingly, deletion of both the CC and tail (HA-CRY1ΔCCtail) rendered the protein completely cytoplasmic, while deletion of the tail (HA-CRY1Δtail) resembles the nucleocytoplasmic distribution of HA-CRY1mutNLSc (Fig. 1E to G). This indicates that the NLSc in HA-CRY1ΔCC could obscure a functional role of the CC in nuclear accumulation. Indeed, HA-CRY1ΔCCmutNLSc (lacking the CC and containing the tail with a mutant NLSc) localizes exclusively in the cytoplasm. This finding not only shows that the CC contributes to the nuclear localization of mCRY1 but also points to a minor role of NLSn.
In conclusion, we identified an essential role of the C terminus of mammalian CRY1 in nuclear localization of the protein, which is achieved through two distinct domains, NLSc and CC. While the NLSc is the classic target for the importin α/β heterodimer and is dominant over the CC in this cellular assay, the CC modulates the localization of mCRY1 (and likely mCRY2), likely through an association with an unknown endogenous factor(s).
The C terminus of mCRY1 regulates the subcellular localization of the mCRY1/mPER2 complex.
mPER proteins play a critical role in the synchronous nuclear localization of mCRY/mPER complexes in vivo (16), but the contribution of mCRY proteins to this process is unclear due to the enhanced degradation of mPER2 and the constitutive expression levels of mPER1 in the absence of mCRY proteins (37). Nonetheless, overexpression studies in mammalian cells strongly indicate an mCRY1- as well as an mPER2-dependent mechanism for the nuclear translocation of this protein complex (15, 20).
To gain further insight in the mechanism that controls subcellular localization of the mCRY1/mPER2 complex, we characterized the mPER binding domain in mCRY1. In coimmunoprecipitation assays of COS7 cell lysates (following coexpression of C-terminally EGFP-tagged mPER1 or mPER2 and HA-tagged wild-type or mutant CRY proteins), we could only pull down mPER proteins with full-length HA-CRY1 and HA-CRY1Δtail (Fig. 2A, middle panels) and HA-CRY1mutNLSc (data not shown). Comparable results were obtained in reverse immunoprecipitation experiments (data not shown). From the inability of HA-CRY1ΔCC and HA-CRY1ΔCCtail to pull down mPER1 or mPER2, we conclude that the CC is involved in mPER association.
FIG.2.
Subcellular localization equilibrium of the mCRY1-mPER2 complex. (A) Western blot of an immunoprecipitation (ip) of COS7 cells transfected with HA-CRY1 (wild type or mutant) and either PER1-EGFP (left) or PER2-EGFP (right). Proteins were precipitated from the lysate with anti-HA antibodies and then analyzed on Western blots using anti-HA (top panels) and anti-GFP (middle panels) antibodies. The bottom panels show Western blots of the total lysate using anti-GFP antibody. (B) Immunofluorescence pictures of COS7 cells transfected with PER2-EGFP alone or with PER2-EGFP and HA-CRY1, HA-CRY1ΔCC, or HA-CRY1mutNLSc. αHA, anti-HA. (C) Quantification of the cellular localization of PER2-EGFP shown in panel B. (D) Immunofluorescence pictures of COS7 cells transfected with HA-CRY1mutNLSc alone (top panels) or with HA-CRY1mutNLSc and PER2-EGFP (bottom panels). Treatment of cells with LMB is indicated (+ LMB). (E) Quantification of the cellular localization of HA-CRY1mutNLSc, as shown in panel D. (F) Immunofluorescence pictures of COS7 cells transfected with HA-CRY1mutNLSc and EGFP-PER2(596-1257) (top) or PER2(1-916)-EGFP (bottom). (G) Fluorescence microscopy pictures of wild-type (WT), Per1−/−, Per2Brdm1/Brdm1, and Per1−/− Per2Brdm1/Brdm1 MDFs transiently expressing mCRY1-EGFP.
Overexpressed mPER2 uses NLSs and NESs to continuously shuttle between the nucleus and cytoplasm, and as result of this activity, its localization is either cytoplasmic, nuclear, or nucleocytoplasmic (42, 44). Coexpression with mCRY1 (or mCRY2) has been shown to cause complete nuclear localization of mPER2 (15) (Fig. 2B). In contrast, coexpression of HA-CRY1ΔCC, HA-CRY1mutNLSc (Fig. 2B and C), or HA-CRY1Δtail (data not shown) did not affect the nucleocytoplasmic localization pattern of mPER2. These data reveal an important function of the NLSc in determining the nuclear localization of the mCRY1/mPER2 complex and further support the involvement of the CC in mPER association.
Interestingly, when the cellular distribution of HA-CRY1mutNLSc in the presence of mPER2 was analyzed, we noticed an unexpected change: the pattern of HA-CRY1mutNLSc now closely resembled the distribution of mPER2, as evident from an increase in the percentage of cells with exclusively nuclear, or cytoplasmic, localization (compare Fig. 2C and E). Similar results were obtained when HA-CRY1mutNLSc was replaced by HA-CRY1Δtail, when mPER2 was replaced by mPER1, or when the above-described experiments were performed with NIH 3T3 cells (see Fig. S2 in the supplemental material). To further investigate the supportive role of mPER2 in the nuclear localization of mCRY1, we analyzed the effect of LMB, an inhibitor of CRM1-mediated nuclear export (7). LMB promotes mCRY1-independent nuclear accumulation of mPER2 by inhibiting nuclear export of the latter protein (44). Strikingly, when cells were grown in the presence of LMB, coexpressed mPER2-EGFP causes near-exclusive nuclear accumulation of HA-CRY1mutNLSc (Fig. 2D and E) or HA-CRY1Δtail (see Fig. S2 in the supplemental material). In the absence of PER2-EGFP, LMB did not detectably affect the subcellular localization of HA-CRY1mutNLSc (Fig. 2D and E). These results suggest that the HA-CRY1mutNLSc/mPER2 complex is shuttling between the nucleus and cytoplasm. This finding is further supported by the observation that nuclear EGFP-PER2(596-1257), lacking the N-terminal and middle NES domains but containing the NLS and the CRY binding domain (44), is capable of fully relocalizing HA-CRY1mutNLSc to the nucleus in the absence of LMB, while nuclear EGFP-PER2(1-916), containing the NLS but lacking the C-terminal NES and mCRY binding domain (44), is not (Fig. 2F).
The present data, together with those reported previously by Miyazaki et al. (20), indicate that neither the NLSc of mCRY1 nor the NLS of mPER2 is by itself sufficient to counteract the nuclear export (i.e., cytoplasmic localization) mediated by the three NESs present in mPER2. To shift the cellular localization equilibrium of the mCRY1/mPER2 complex from the cytoplasm to the nucleus, the combined activities of the two NLSs are necessary. Finally, the nuclear localization of HA-CRY1ΔCC (Fig. 1F and G) as well as wild-type mCRY1-EGFP in Per1−/− Per2Brdm1 mouse dermal fibroblasts (Fig. 2G) indicates that the association with mPER proteins is not an absolute requirement for the nuclear localization of mCRY1.
The C-terminal extension of mCRY1 is required for inhibition of CLOCK/BMAL1-mediated transcription.
mCRY proteins redundantly inhibit transcription activation of E-box-containing promoters by the CLOCK/BMAL1 heterodimer (15). To investigate whether the tail and/or CC of mCRY1 is required for this function, and after having shown that COS7 cells express mutant CRY1 proteins at wild-type levels (see Fig. S3 in the supplemental material), we performed a Dbp promoter-driven luciferase reporter assay with COS7 cells. Nuclear accumulation of mCRY1 is an important aspect of this assay. HA-CRY1ΔCCtail does not significantly (P > 0.1) inhibit CLOCK/BMAL1 (Fig. 3A), which could be explained by the cytoplasmic localization of the mutant CRY protein (Fig. 1F to G). Somewhat surprisingly, as these mutant proteins show less pronounced nuclear localization (nuclear and nucleocytoplasmic in 10% and ∼75% of the cells respectively) (Fig. 1D and H), HA-CRY1Δ tail and HA-CRY1mutNLSc inhibit CLOCK/BMAL1 as efficiently as wild-type mCRY1 (Fig. 3A) (see Fig. S3 in the supplemental material).
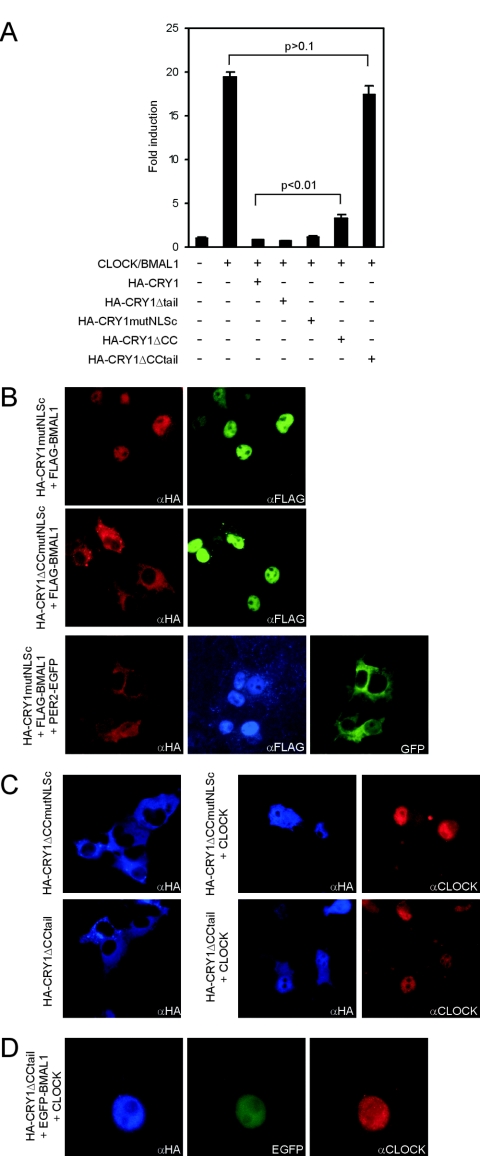
FIG. 3.
Role of the C terminus of mCRY1 in the inhibition of CLOCK/BMAL1-driven transcription. (A) CLOCK/BMAL1 transcription assay using a Dbp E-box promoter-luciferase reporter construct. Luminescence, shown as a severalfold induction from the basal level, is indicated on the y axis. pcDNA3, pRL-CMV, and the pGL3 promoter (Dbp) were added in all conditions. The presence or absence of the other expression plasmids is indicated. We tested HA-CRY1, HA-CRY1Δtail, HA-CRY1mutNLSc, HA-CRY1ΔCC, and HA-CRY1ΔCCtail (100 ng plasmid/assay). Error bars represent the standard deviations. (B) Immunofluorescence pictures of COS7 cells expressing FLAG-BMAL1 and HA-CRY1mutNLSc (top panels), FLAG-BMAL1 and HA-CRY1ΔCCmutNLSc (middle panels), or FLAG-BMAL1, PER2-EGFP, and HA-CRY1mutNLSc (bottom panels). The HA-CRY1 mutants were detected with rabbit anti-HA (αHA) antibodies. (C) Immunofluorescence pictures of COS7 cells expressing HA-CRY1mutNLSc or HA-CRY1ΔCCtail either alone (left panels) or together with CLOCK (right panels). (D) Immunofluorescence pictures of COS7 cells expressing HA-CRY1ΔCCtail, EGFP-BMAL1, and CLOCK (note the higher magnification used).
Previously, it has been shown that transiently expressed BMAL1 localizes in the nucleus, whereas CLOCK is nucleocytoplasmic and becomes completely nuclear and more stable after coexpression with BMAL1 (see Fig. S3 in the supplemental material). We therefore investigated whether BMAL1 can promote nuclear accumulation of mCRY1. Indeed, coexpression of BMAL1 relocalizes HA-CRY1mutNLSc to the nucleus in 100% of the cells, whereas deletion of the CC (as in HA-CRY1Δ CCmutNLSc) abolishes this effect (Fig. 3B). Similar results were obtained with HA-CRY1Δtail (data not shown). These findings strongly suggest that the transcriptional repressing activities of HA-CRY1mutNLSc and HA-CRY1Δtail are due to BMAL1-mediated enhancement of their nuclear localizations and that BMAL1 binds mCRY1 through the CC domain of the latter protein. Moreover, in cells coexpressing BMAL1, mPER2, and HA-CRY1mutNLSc, the mutant CRY1 protein colocalizes exclusively with mPER2 (indicative for shuttling mPER2/mCRY1 complexes), whereas BMAL1 localizes exclusively in the nucleus (Fig. 3B, lower panel). Apparently, mPER2 and BMAL1 compete for association with the CC of mCRY1 and do so with different affinities (mPER2 > BMAL1).
In addition, we noticed that nuclear HA-CRY1ΔCC inhibits CLOCK/BMAL1, although with significantly lower (P < 0.01) efficiency (Fig. 3A; see Fig. S3 in the supplemental material). Since the previous experiment suggests that HA-CRY1ΔCC is unlikely to bind BMAL1, interaction of mCRY1 with CLOCK is sufficient to inhibit CLOCK/BMAL1 transcription activation. However, interaction with both CLOCK and BMAL1 is required for complete inhibition.
We next set out to investigate the interactions between CLOCK and mCRY1 in more detail. The CLOCK binding domain of zebrafish CRY1a has been mapped at amino acids 196 to 263 of the (photolyase-like) core domain (10). To analyze whether the core domain of mCRY1 associates with CLOCK, we coexpressed HA-CRY1ΔCCmutNLSc or HA-CRY1ΔCCtail (both exclusively cytoplasmic) (Fig. 1G) with CLOCK in COS7 cells. As shown in Fig. 3C, the mCRY mutant proteins colocalize with nucleocytoplasmic CLOCK, indicating that the latter protein binds to the core domain of mCRY1 and promotes its nuclear accumulation. Importantly, when HA-CRY1ΔCCtail is coexpressed with both CLOCK and BMAL1, the protein could only be detected in the nucleus (Fig. 3D), which suggests that the exclusive nuclear localization of HA-CRY1ΔCCtail is the net result of CLOCK binding to the mCRY core domain and subsequent BMAL1-mediated relocalization of CLOCK (and thus mCRY1) to the nucleus.
Importantly, the nuclear localization of HA-CRY1ΔCCtail in the presence of CLOCK and BMAL1 prompted us to reinterpret the observed lack of inhibition of CLOCK/BMAL1-driven transcription by this mutant CRY1 protein (Fig. 3A), which we initially attributed to cytoplasmic localization. Apparently, the C-terminal extension of mCRY1 (CC and tail), in addition to its role in the regulation of subcellular localization of clock protein complexes, is required for the inhibition of CLOCK/BMAL1-mediated transcription. However, no clear domains (CC, NLSc, or tail) could be directly assigned to the latter function, although the CC mediates BMAL1 association.
Functional conservation between the photolyase-like domain of mCRY1 and Arabidopsis (6-4PP)PhL.
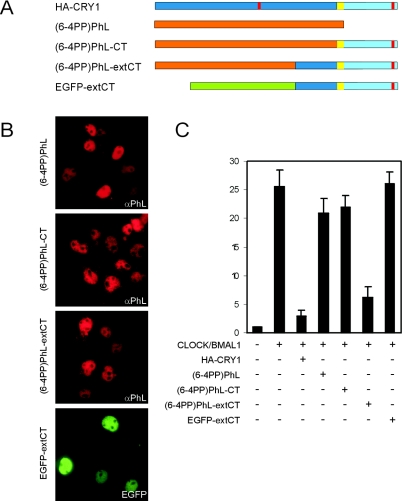
Given the involvement of the C-terminal extension in CLOCK/BMAL inhibition, and the strong homology between the core domains of cryptochromes and photolyases, we next asked whether the addition of the mCRY1 C-terminal extension could convert a photolyase into a cryptochrome. To this end, we created chimeric proteins in which the photolyase-like domain of mCRY1 was replaced by Arabidopsis thaliana (6-4PP)PhL (Fig. 4A). In (6-4PP)PhL-CT, full-length (6-4PP)PhL (aa 1 to 537) was fused to the C terminus of mCRY1 (aa 471 to 606, containing CC and tail). In (6-4PP)PhL-extCT, aa 1 to 378 of (6-4PP)PhL were hooked up to an N-terminally extended version of the mCRY1 C terminus (aa 371 to 606, referred to as extCT) containing the last 100 amino acids of the core domain, CC, and tail. In addition, we fused the extCT sequence to the C terminus of EGFP. Analysis of the subcellular localization of (6-4PP)PhL, (6-4PP)PhL-CT, (6-4PP)PhL-extCT, and EGFP-extCT in transfected COS7 cells reveals exclusive nuclear localization of these proteins (Fig. 4B). While (6-4PP)PhL and (6-4PP)PhL-CT have no effect on CLOCK/BMAL1 activity in the transcription inhibition assay, (6-4PP)PhL-extCT efficiently suppresses E-box promoter transcription (Fig. 4C). Thus, the C-terminal part of the mCRY1 core domain, together with the C-terminal extension not shared with photolyases (i.e., CC and tail), can confer CLOCK/BMAL1 transcription-inhibitory capacity to a photolyase. In marked contrast, EGFP-extCT does not inhibit transcription of the E-box promoter, indicating that the extCT sequence is necessary but not sufficient for the transcription-inhibitory capacity of mCRY1. Apparently, as will be discussed below, the remainder of the photolyase/cryptochrome core domain is vital for this activity.
FIG. 4.
Cross talk between the photolyase-like domain of mCRY1 and its C terminus. (A) Schematic representation of HA-CRY1, (6-4PP)PhL, and the fusion proteins (6-4PP)PhL-CT, (6-4PP)PhL-extCT, and EGFP-extCT. (B) Immunofluorescence pictures of transfected COS7 cells showing the subcellular localization of the aforementioned proteins. αPhL, anti-PhL. (C) Graphic representation of the CLOCK/BMAL1-inhibitory capacity of the proteins in the Dbp E-box promoter-luciferase reporter assay (using 100 ng of plasmid). Error bars represent the standard deviations. The difference between HA-CRY1 and (6-4PP)Phl-extCT is not statistically significant (t test P value is 0.07).
DISCUSSION
All members of the photolyase/cryptochrome protein family share a 500-amino-acid core domain (containing two chromophore binding sites) but differ in the presence of N- or C-terminal extensions (39, 41). The large variation in length and amino acid composition of the C-terminal extension of CRY proteins suggests that this region constitutes the major structural determinant in shaping its function as either circadian core oscillator protein and/or (circadian) photoreceptor protein. In the present study, we have performed a detailed structure/function analysis of the C terminus of the mammalian circadian core oscillator protein CRY1.
By expressing a set of mutant CRY1 proteins in COS7 cells, we successfully identified domains that control the subcellular localization of the protein. We first uncovered a functional bipartite NLSc of the mouse CRY1 protein, which resembles the previously identified N-terminal monopartite NLSn (10) in that it is evolutionary conserved in all vertebrate CRY homologs (48). However, combined inactivation of the NLSc and the NLSn (by in vitro mutagenesis) could not prevent mCRY1 from entering the nucleus, suggesting the presence of an additional nuclear import mechanism, such as nuclear cotranslocation via interaction with another protein (piggyback transport). In the search for potential protein binding domains, we identified a putative CC domain at the beginning of the C-terminal extension of CRY1. Deletion of this CC in combination with inactivation of the NLSc resulted in complete cytoplasmic localization of mCRY1 (Fig. 1G). Proof of piggyback transport was obtained by our observation that mPER2 and BMAL1 can competitively bind to the CC of mCRY1 and facilitate nuclear localization of mCRY1mutNLSc (Fig. 3B). However, as HA-CRY1mutNLSn+c can still enter the nucleus in the absence of other overexpressed proteins, and endogenous mPER proteins and the CLOCK/BMAL1 heterodimer are expressed at very low levels in COS7 cells (data not shown), the endogenous factor mediating the CC-dependent nuclear import of mCRY1 remains to be identified. We consider TIMELESS an excellent candidate, as it is expressed robustly in these cells, and overexpressed TIM also associates with mCRY1, causing nuclear translocation of the latter protein (F. Tamanini, unpublished data). In conclusion, the C-terminal extension of mCRY1 is involved in a bimodal mechanism for nuclear import; (i) the binding of nuclear import factors to NLSc serves classical nuclear translocation, and (ii) the association of other (core clock) proteins to the CC can mediate nuclear cotranslocation. Yet, as cotranslocation has only been visualized with mutant mCRY1 proteins in a cellular transfection assay, the physiological relevance of the latter mechanism remains to be resolved.
Previously, we have shown that mPER2 shuttles between the nucleus and cytoplasm and accumulates in the nucleus after association with mCRY1, which led us to propose that signal-mediated regulation of subcellular localization of mCRY/mPER complexes controls their stability (and therefore activity) and as such can contribute to the phase delay in mRNA and protein rhythms (15, 44). In the present study, we have shown that mPER2 (and mPER1) binds to the CC of mCRY1 to form mCRY/mPER complexes and that the NLSc of mCRY1 is necessary to shift the subcellular equilibrium of the mCRY1/mPER2 complex from the cytoplasm to the nucleus. Oppositely, Miyazaki and coworkers provided evidence that the NLS of mPER2 is required for nuclear translocation of mCRY1 (20). We have shown that the NLSc of mCRY1 and NLS of mPER2 are both required for nuclear accumulation of the complex (Fig. 2C and E). Importantly, inactivation of mPER2 NES domains or treatment of cells with the nuclear export inhibitor LMB promotes nuclear accumulation of mCRY1 (as visualized using mutant proteins with a defective NLSc). These findings suggest that mCRY and mPER shuttle as a complex (requiring the concerted action of NLS and NES sequences in the complex) and are in agreement with a recent report in which microinjected mCRY1 was shown to leave the Xenopus laevis oocyte nucleus via association with mPER1 (17). In conclusion, our data provide a molecular basis for the synchronous nuclear accumulation of mCRY and mPER proteins observed in vivo (15, 16) and define the C terminus of mCRY1 as a central domain in this process.
Despite the lack of amino acid sequence homology between the C-terminal extensions of mCRY1 and mCRY2, the latter protein also contains a putative CC and a bipartite NLSc (32). Interestingly, serine 557 (underlined), which is in close proximity to the first basic cluster of the NLS (SPKRK) of mCRY2, can be phosphorylated by mitogen-activated protein kinase (33). Phosphorylation has been identified as a posttranslational event for the regulation of the activity of NLS domains, including that of mPER1 (12, 38). The opposite circadian phenotype of mCry1−/− and mCry2−/− mice, carrying fast- and slow-ticking circadian oscillators, respectively (40, 43), suggests that nuclear accumulation of mCRY2/mPER complexes runs slightly ahead of mCRY1/mPER complexes. A difference in kinetics could be achieved by phosphorylation-mediated control over the relative strength of NLS (and NES) sequences that together determine the equilibrium between nuclear and cytoplasmically localized mCRY/mPER complexes.
The prime function of mCRY proteins is to inhibit CLOCK/BMAL1-mediated transcription activation of E-box genes in the negative limb of the mammalian circadian core oscillator. Neither the CC, nor the tail, nor the NLSc of mCRY1, when deleted individually, affects the CLOCK/BMAL1-inhibitory capacity of mCRY1, which suggests that these domains are dispensable. Xenopus CRY1 and CRY2 (xCRY) resemble mammalian CRY proteins in carrying a CC and NLS-containing tail in their C-terminal extension (48). Any truncation of the tail of xCRY caused CLOCK/BMAL1 inhibition in the transcription assay. However, this was only achieved after compensation with an exogenous NLS, which implies that tail-deficient xCRY proteins, unlike their mammalian counterparts, do not reach the nucleus by cotranslocation with CLOCK and/or BMAL1. Although a comparison between the Xenopus and mammalian systems is difficult, taken together, these observations suggest differences between mCRY1 and xCRY1 in their cross talk with the CLOCK/BMAL1 heterodimer. Furthermore, we have established the first mutant CRYPTOCHROME with a deficiency in CLOCK/BMAL1-inhibitory capacity; HA-CRY1ΔCCtail (nuclear as a result of cotranslocation with BMAL1 and CLOCK) could no longer inhibit E-box gene expression. Thus, the presence of at least part of the C-terminal extension (CC or tail) of mCRY1 is mandatory for CLOCK/BMAL1 inhibition activity.
It has been suggested that the inhibition of CLOCK/BMAL-mediated transcription requires the interaction of mCRY proteins with either CLOCK alone or both CLOCK and BMAL1 (37). We provide mechanistic evidence in support of the first hypothesis by showing that the association of mCRY1 with only CLOCK is necessary (e.g., HA-CRY1ΔCC), yet not sufficient (e.g., HA-CRY1ΔCCtail), to mediate this function. Given the slightly reduced CLOCK/BMAL1-inhibitory capacity of HA-CRY1ΔCC, CRY1/BMAL1 interactions could serve to stabilize the binding of mCRY1 to the promoter complex during the negative phase of the circadian loop.
Finally, we identified amino acids 371 to 470 from the core domain as being very important for the transcription-inhibitory activity of mCRY1. An extended C terminus (aa 371 to 606), but not the C-terminus itself (aa 471 to 606), when fused to Arabidopsis thaliana (6-4PP)-specific photolyase, is able to confer CLOCK/BMAL1 transcription-inhibitory activity to the resulting chimeric protein. However, the extended C terminus requires the remainder of the core region (either mCRY1 or photolyase derived) to fully exert its function, possibly by mediating the association with CLOCK. These observations suggest that mCRY1 requires a complex network of interactions and intrinsic structural requirements for proper transcription inhibition, which involve the core domain and the C terminus. This concept is further illustrated by our observation that from a panel of full-length mCRY1 proteins with random insertions of 5 amino acids, mutant proteins that are not able to repress CLOCK/BMAL1-mediated transcription are most likely unfolded (F. Tamanini, unpublished data). Interestingly, it has recently been shown that whereas the C termini of animal and plant CRY proteins are intrinsically unstructured, they acquire a stable tertiary structure upon intermolecular interaction with the core domain (25). Given the critical roles of amino acids 371 to 470 and amino acids 471 to 606, it is tempting to speculate that these two regions could be involved in the proposed interaction. This intermolecular binding can cause a structural change within the mCRY1 molecule, which in turn would activate the C terminus. While CLOCK and BMAL1 keep mCRY1 in the proximity of the E-box promoter, the activated C-terminal domains may recruit multiple transcriptional corepressor complexes (histone deacetylases and Sin3A) to silence CLOCK/BMAL1-driven transcription. This model (Fig. 5) takes into account the fact that transcriptional corepressors have been recently shown to associate and possibly regulate the function of mCRY1 (21) and that the binding of CLOCK/BMAL1 at the promoter is not affected by the coexpression of mCRY proteins (11). In addition, the C terminus would be engaged in posttranslational processes (e.g., stability of mPER proteins, competitive association with clock factors, and nuclear import) that are essential for the robustness of the circadian mechanism.
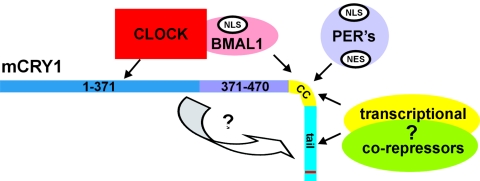
FIG. 5.
Model for the mechanism of action of mCRY1 within the mammalian core oscillator. The C terminus of mCRY1 is involved in association with mPER1 and mPER2 proteins and therefore regulates the stability and cellular localization of the latter proteins. The formation of the mCRY/mPER complex occurs possibly through CC interactions, as a predicted CC is also present in the mCRY binding region of mPER proteins (data not shown). To achieve full nuclear localization, this protein complex requires both the mCRY1 NLSc and the mPER2 NLS to counteract the NES-mediated nuclear export of mPER2. This molecular mechanism may explain the synchrony in nuclear localization of these proteins observed in vivo. Moreover, the photolyase-like core domain of mCRY1 interacts with CLOCK, while the CC is involved in association with BMAL1. Our data also suggest that there is competition between mPER2 and BMAL1 for binding with mCRY1, which supports the concept that the periodicity of the oscillator depends on temporal abundance and the strength of the interaction between the partners as well as on the transcription-inhibitory capacity of each complex (23). The inhibitory action of mCRY1 is likely achieved through an intermolecular interaction, which may impose a structural change in the C terminus, thereby allowing this domain to recruit transcriptional corepressor complexes.
Functional cross talk between the photolyase-like domain and the C-terminal extension of CRY proteins is not unprecedented. Homodimerization of the Arabidopsis CRY photoreceptor through the photolyase-like domain has been shown to be essential for light-mediated activation of the C terminus (35). In the case of Drosophila CRY, the photolyase-like domain is essential and sufficient for light detection, while the C terminus is involved in the degradation of Drosophila TIM in response to light (2, 4). Our study on mammalian CRY1 suggests that communication between the core domain and the C-terminal extension may be a common feature of cryptochromes. Yet we propose that, in terms of protein function, the acquirement of different species-specific C-terminal extensions during evolution not only functionally separated cryptochromes from photolyases but also caused diversity within the cryptochrome protein family, making cryptochromes act like photoreceptors (as in Arabidopsis), both photoreceptors and core oscillators proteins (as in Drosophila and zebra fish), or pure core oscillator proteins (as in Xenopus and mammals).
Supplementary Material
Acknowledgments
We thank Xavier Bonnefont and André Eker for stimulating discussions and Shun Yamaguchi for help with the luciferase assay.
This work was supported in part by grants from The Netherlands Organization for Scientific Research (ZonMW Vici 918.36.619 and NWO-CW 700.51.304) and the European Community (BrainTime QLG3-CT-2002-01829) to G.T.J.V.D.H.
REFERENCES
- 1.Balsalobre, A., F. Damiola, and U. Schibler. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929-937. [DOI] [PubMed] [Google Scholar]
- 2.Busza, A., M. Emery-Le, M. Rosbash, and P. Emery. 2004. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 304:1503-1506. [DOI] [PubMed] [Google Scholar]
- 3.Cermakian, N., and P. Sassone-Corsi. 2000. Multilevel regulation of the circadian clock. Nat. Rev. Mol. Cell Biol. 1:59-67. [DOI] [PubMed] [Google Scholar]
- 4.Dissel, S., V. Codd, R. Fedic, K. J. Garner, R. Costa, C. P. Kyriacou, and E. Rosato. 2004. A constitutively active cryptochrome in Drosophila melanogaster. Nat. Neurosci. 7:834-840. [DOI] [PubMed] [Google Scholar]
- 5.Etchegaray, J. P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421:177-182. [DOI] [PubMed] [Google Scholar]
- 6.Field, M. D., E. S. Maywood, J. A. O'Brien, D. R. Weaver, S. M. Reppert, and M. H. Hastings. 2000. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron 25:437-447. [DOI] [PubMed] [Google Scholar]
- 7.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 8.Gekakis, N., D. Staknis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher, D. P. King, J. S. Takahashi, and C. J. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564-1569. [DOI] [PubMed] [Google Scholar]
- 9.Griffin, E. A., Jr., D. Staknis, and C. J. Weitz. 1999. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286:768-771. [DOI] [PubMed] [Google Scholar]
- 10.Hirayama, J., H. Nakamura, T. Ishikawa, Y. Kobayashi, and T. Todo. 2003. Functional and structural analyses of cryptochrome. Vertebrate CRY regions responsible for interaction with the CLOCK:BMAL1 heterodimer and its nuclear localization. J. Biol. Chem. 278:35620-35628. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa, T., J. Hirayama, Y. Kobayashi, and T. Todo. 2002. Zebrafish CRY represses transcription mediated by CLOCK-BMAL heterodimer without inhibiting its binding to DNA. Genes Cells 7:1073-1086. [DOI] [PubMed] [Google Scholar]
- 12.Jans, D. A., M. J. Ackermann, J. R. Bischoff, D. H. Beach, and R. Peters. 1991. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J. Cell Biol. 115:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jans, J., W. Schul, Y. G. Sert, Y. Rijksen, H. Rebel, A. P. Eker, S. Nakajima, H. van Steeg, F. R. de Gruijl, A. Yasui, J. H. Hoeijmakers, and G. T. van der Horst. 2005. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 15:105-115. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi, K., S. Kanno, B. Smit, G. T. van der Horst, M. Takao, and A. Yasui. 1998. Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucleic Acids Res. 26:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 16.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867. [DOI] [PubMed] [Google Scholar]
- 17.Loop, S., M. Katzer, and T. Pieler. 2005. mPER1-mediated nuclear export of mCRY1/2 is an important element in establishing circadian rhythm. EMBO Rep. 6:341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 19.McNamara, P., S. P. Seo, R. D. Rudic, A. Sehgal, D. Chakravarti, and G. A. FitzGerald. 2001. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell 105:877-889. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki, K., M. Mesaki, and N. Ishida. 2001. Nuclear entry mechanism of rat PER2 (rPER2): role of rPER2 in nuclear localization of CRY protein. Mol. Cell. Biol. 21:6651-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naruse, Y., K. Oh-hashi, N. Iijima, M. Naruse, H. Yoshioka, and M. Tanaka. 2004. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 24:6278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamura, H., S. Miyake, Y. Sumi, S. Yamaguchi, A. Yasui, M. Muijtjens, J. H. Hoeijmakers, and G. T. van der Horst. 1999. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286:2531-2534. [DOI] [PubMed] [Google Scholar]
- 23.Oster, H., S. Baeriswyl, G. T. Van Der Horst, and U. Albrecht. 2003. Loss of circadian rhythmicity in aging mPer1−/−mCry2−/− mutant mice. Genes Dev. 17:1366-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pando, M. P., D. Morse, N. Cermakian, and P. Sassone-Corsi. 2002. Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110:107-117. [DOI] [PubMed] [Google Scholar]
- 25.Partch, C. L., M. W. Clarkson, S. Ozgur, A. L. Lee, and A. Sancar. 2005. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44:3795-3805. [DOI] [PubMed] [Google Scholar]
- 26.Preitner, N., F. Damiola, L. Lopez-Molina, J. Zakany, D. Duboule, U. Albrecht, and U. Schibler. 2002. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251-260. [DOI] [PubMed] [Google Scholar]
- 27.Ralph, M. R., and M. Menaker. 1989. GABA regulation of circadian responses to light. I. Involvement of GABAA-benzodiazepine and GABAB receptors. J. Neurosci. 9:2858-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reppert, S. M., and D. R. Weaver. 2001. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63:647-676. [DOI] [PubMed] [Google Scholar]
- 29.Ripperger, J. A., L. P. Shearman, S. M. Reppert, and U. Schibler. 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14:679-689. [PMC free article] [PubMed] [Google Scholar]
- 30.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 31.Rusak, B., J. H. Meijer, and M. E. Harrington. 1989. Hamster circadian rhythms are phase-shifted by electrical stimulation of the geniculo-hypothalamic tract. Brain Res. 493:283-291. [DOI] [PubMed] [Google Scholar]
- 32.Sakakida, Y., Y. Miyamoto, E. Nagoshi, M. Akashi, T. J. Nakamura, T. Mamine, M. Kasahara, Y. Minami, Y. Yoneda, and T. Takumi. 2005. Importin alpha/beta mediates nuclear transport of a mammalian circadian clock component, mCRY2, together with mPER2, through a bipartite nuclear localization signal. J. Biol. Chem. 280:13272-13278. [DOI] [PubMed] [Google Scholar]
- 33.Sanada, K., Y. Harada, M. Sakai, T. Todo, and Y. Fukada. 2004. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cells 9:697-708. [DOI] [PubMed] [Google Scholar]
- 34.Sancar, A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103:2203-2237. [DOI] [PubMed] [Google Scholar]
- 35.Sang, Y., Q. H. Li, V. Rubio, Y. C. Zhang, J. Mao, X. W. Deng, and H. Q. Yang. 2005. N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato, T. K., S. Panda, L. J. Miraglia, T. M. Reyes, R. D. Rudic, P. McNamara, K. A. Naik, G. A. FitzGerald, S. A. Kay, and J. B. Hogenesch. 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43:527-537. [DOI] [PubMed] [Google Scholar]
- 37.Shearman, L. P., S. Sriram, D. R. Weaver, E. S. Maywood, I. Chaves, B. Zheng, K. Kume, C. C. Lee, G. T. van der Horst, M. H. Hastings, and S. M. Reppert. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288:1013-1019. [DOI] [PubMed] [Google Scholar]
- 38.Takano, A., Y. Isojima, and K. Nagai. 2004. Identification of mPer1 phosphorylation sites responsible for the nuclear entry. J. Biol. Chem. 279:32578-32585. [DOI] [PubMed] [Google Scholar]
- 39.Todo, T., H. Ryo, K. Yamamoto, H. Toh, T. Inui, H. Ayaki, T. Nomura, and M. Ikenaga. 1996. Similarity among the Drosophila (6-4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science 272:109-112. [DOI] [PubMed] [Google Scholar]
- 40.van der Horst, G. T., M. Muijtjens, K. Kobayashi, R. Takano, S. Kanno, M. Takao, J. de Wit, A. Verkerk, A. P. Eker, D. van Leenen, R. Buijs, D. Bootsma, J. H. Hoeijmakers, and A. Yasui. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630. [DOI] [PubMed] [Google Scholar]
- 41.van der Spek, P. J., K. Kobayashi, D. Bootsma, M. Takao, A. P. Eker, and A. Yasui. 1996. Cloning, tissue expression, and mapping of a human photolyase homolog with similarity to plant blue-light receptors. Genomics 37:177-182. [DOI] [PubMed] [Google Scholar]
- 42.Vielhaber, E. L., D. Duricka, K. S. Ullman, and D. M. Virshup. 2001. Nuclear export of mammalian PERIOD proteins. J. Biol. Chem. 276:45921-45927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vitaterna, M. H., C. P. Selby, T. Todo, H. Niwa, C. Thompson, E. M. Fruechte, K. Hitomi, R. J. Thresher, T. Ishikawa, J. Miyazaki, J. S. Takahashi, and A. Sancar. 1999. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. USA 96:12114-12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yagita, K., F. Tamanini, M. Yasuda, J. H. Hoeijmakers, G. T. van der Horst, and H. Okamura. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi, S., S. Mitsui, L. Yan, K. Yagita, S. Miyake, and H. Okamura. 2000. Role of DBP in the circadian oscillatory mechanism. Mol. Cell. Biol. 20:4773-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo, S. H., S. Yamazaki, P. L. Lowrey, K. Shimomura, C. H. Ko, E. D. Buhr, S. M. Siepka, H. K. Hong, W. J. Oh, O. J. Yoo, M. Menaker, and J. S. Takahashi. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 101:5339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, M. W., and S. A. Kay. 2001. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2:702-715. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, H., F. Conte, and C. B. Green. 2003. Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr. Biol. 13:1653-1658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.