Abstract
Alpha and beta interferons (IFN-α and IFN-β) are multifunctional cytokines that exhibit differential activities through a common receptor composed of the subunits IFNAR1 and IFNAR2. Here we combined biophysical and functional studies to explore the mechanism that allows the alpha and beta IFNs to act differentially. For this purpose, we have engineered an IFN-α2 triple mutant termed the HEQ mutant that mimics the biological properties of IFN-β. Compared to wild-type (wt) IFN-α2, the HEQ mutant confers a 30-fold higher binding affinity towards IFNAR1, comparable to that measured for IFN-β, resulting in a much higher stability of the ternary complex as measured on model membranes. The HEQ mutant, like IFN-β, promotes a differentially higher antiproliferative effect than antiviral activity. Both bring on a down-regulation of the IFNAR2 receptor upon induction, confirming an increased ternary complex stability of the plasma membrane. Oligonucleotide microarray experiments showed similar gene transcription profiles induced by the HEQ mutant and IFN-β and higher levels of gene induction or repression than those for wt IFN-α2. Thus, we show that the differential activities of IFN-β are directly related to the binding affinity for IFNAR1. Conservation of the residues mutated in the HEQ mutant within IFN-α subtypes suggests that IFN-α has evolved to bind IFNAR1 weakly, apparently to sustain differential levels of biological activities compared to those induced by IFN-β.
Type I interferons (IFNs) form a family of multifunctional cytokines initially described for their direct antiviral effect but now also recognized as major elements of the immune response (19, 46). Differential activities of IFN subtypes have been reported (3) and used in the clinic for the treatment of various pathologies, including viral hepatitis (IFN-α2) and multiple sclerosis (IFN-β) (32). All type I IFNs are recognized by a single shared receptor composed of two transmembrane proteins, IFNAR1 and IFNAR2. Because of the much faster kon and much slower koff of IFN-α2 towards IFNAR2 than those measured for IFNAR1 (18, 34, 40), a two-step assembling mechanism was proposed for the interaction between IFN and the two receptors (Fig. 1B). After binding of IFN-α2 to IFNAR2 (ka1), IFNAR1 transiently associates in a second step to the complex (ka2) (18, 25). Owing to the short lifetime of the IFN-α2-IFNAR1 interaction, the complex dissociates (kd2) and reassociates (ka2) in a fast manner. Thus, depending on the receptor surface concentrations and ligand binding affinities, only part of the bound ligand is involved in the active ternary complex.
FIG. 1.
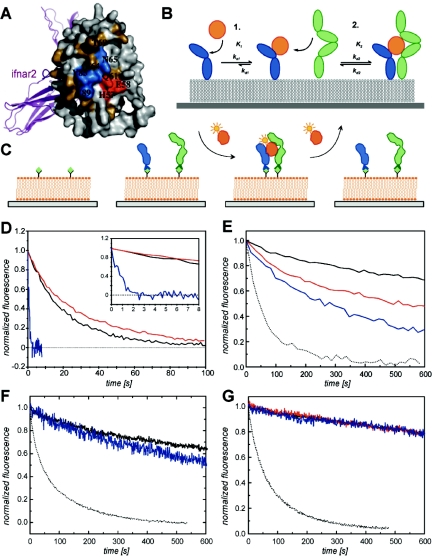
IFN binding to IFNAR1-EC and ternary complex formation, as studied by TIRFS. (A) Functional epitope for binding of IFNAR1 on IFN-α2, also showing the relative location of IFNAR2 (40, 41). The three residues constituting the HEQ mutations (residues 57, 58, and 61) are shown in red, mutations which reduce binding upon mutation to Ala are colored blue, and inert mutations are shown in brown. (B) Schematic representation of the two-step assembling mechanism. After IFN binding to IFNAR2 (blue) in the first step (K1), recruitment of IFNAR1 (green) to the ternary complex in the second step is determined by the affinity value K2 and the surface concentration of IFNAR1. (C) Schematic representation of ternary complex formation experiments with IFNAR2-EC and IFNAR1-EC tethered to solidly supported lipid bilayers through C-terminal histidine tags. Ternary complex formation was probed by measuring ligand dissociation kinetics at different receptor surface concentrations. (D) Comparison of dissociation kinetics of site-specifically fluorescently labeled wt IFN-α2 (blue), IFN-β (red), and the HEQ mutant (black) from IFNAR1-EC alone bound to the surface. The inset zooms into the first 8 s of the reaction. (E to G) Dissociation of wt IFN-α2 (E), IFN-β (F), and the HEQ mutant (G) from the ternary complex, with both IFNAR1-EC and IFNAR2-EC bound to the surface (IFNAR2-EC I47A was used for IFN-β), at IFNAR1-EC receptor surface concentrations of 4 fmol/mm2 (black), 2 fmol/mm2 (red), and 1 fmol/mm2 (blue). The dissociation from IFNAR2-EC alone is shown as a dotted line.
After formation of the ternary complex, the interferon signal is transduced through the receptor-associated JAK kinases, with the STAT transcription factors as their main targets (3). Typically recognized by the immunoglobulin G (IgG)-like folds of their extracellular domains, IFNAR2 and IFNAR1 are regarded as a binding protein and an accessory transducing factor, respectively (48). A difference in the ligand dissociation constants of the two chains is implicit in the definition (25, 33). Still, both contribute to the creation of high-affinity binding sites (6, 9, 18). The combination of a “common” beta chain with different recognition chains is a feature of heteromeric receptors that respond differentially to different ligands. The ability to interact with different alpha chains establishes potential network connections for differential receptor expression (22). In cases where the alpha chains, such as IFNAR1, possess the capacity of interacting with elements of different signaling pathways, they may establish connections for differential gene expression (37).
The human type I IFNs comprise 13 distinct nonallelic alpha subtypes, one beta subtype, and one omega subtype (2). As expected for a family with marked sequence and structural homology and a common receptor, the activities of the type I IFNs largely overlap (31). Nevertheless, numerous instances of relative differences in activity have been noted (8, 15, 27, 29, 36, 39, 44). The emerging picture is that functional differences appear only in specific physiological contexts. It was noted that a possible functional difference between the IFNs may reside in their potential to bind tightly to IFNAR1 (25, 45).
For the present study, we aimed to understand the molecular difference between IFN-β and IFN-α. By engineering an IFN-α2 mutant with similar binding properties for IFNAR1, we were able to reproduce key characteristics of differential IFN-β activity. These include a markedly increased antiproliferative but not antiviral activity, differential gene expression as monitored using gene arrays, and specific down-regulation of the IFNAR2 receptor. Our results support the notion that the type I interferons are not functionally redundant, despite sharing a common receptor and prominent activities.
MATERIALS AND METHODS
Site-directed mutagenesis.
Site-directed mutagenesis was carried out by PCR amplification of the expression plasmid pT72Cα2 with 18- to 21-nucleotide primers containing the mutated codon, using the high-fidelity pwo (Boehringer Mannheim) and Pfu (Stratagene) polymerases as described previously (36).
Protein expression, purification, and labeling.
IFN-α2 and IFNAR2-EC (extracellular domain) were expressed in Escherichia coli and purified by ion-exchange and size exclusion chromatography (35). IFNAR1-EC was expressed in Sf9 insect cells and purified as described previously (25). Both IFNAR2-EC and IFNAR1-EC carried a C-terminal decahistidine tag for oriented tethering on membranes. Protein purity was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis under nonreducing conditions. The concentration of active IFN-α2 protein was determined for all mutants by analytical gel filtration with IFNAR2-EC (36). For ligand binding studies, IFN-α2 and IFN-β were site-specifically labeled using maleimide chemistry. IFN-β was labeled directly at its free cysteine residue, C17, by incubation of a twofold molar excess of Oregon Green 488 maleimide (Molecular Probes) for 2 h in HEPES-buffered saline. IFN-α2 was labeled with Oregon Green 488 after incorporating an additional cysteine residue (S136C mutation), which has been shown not to affect the interaction with IFNAR1-EC or IFNAR2-EC (18).
Ligand binding studies.
The binding of fluorescently labeled IFN to IFNAR1-EC and IFNAR2-EC and the formation of the ternary complex on solidly supported lipid bilayers were studied by simultaneous total internal reflection fluorescence (TIRFS) and reflectance interference (RIf) detection (18). Association and dissociation rate constants were determined from binding curves measured by TIRFS. Receptor surface concentrations were quantified from the RIf signal, which provides an absolute, mass-sensitive signal. Binding assays on solidly supported membranes were carried out with unilaminar vesicles containing chelator lipids fused on the surface of the RIf transducer (18, 25). IFNAR1-EC and IFNAR2-EC were tethered to these membranes through their C-terminal His tags, thus mimicking membrane anchoring of the receptor subunits. For this study, ternary complex stability was monitored as the rate of ligand dissociation and was studied at different receptor surface concentrations after saturating the IFNAR2 binding sites by injection of 100 nM fluorescently labeled IFN.
Antiviral and antiproliferative assays.
Antiviral activities of wild-type and mutant IFN-α2 were assayed as the inhibition of the cytopathic effect of vesicular stomatitis virus on human WISH cells (42).
The antiproliferative assay with WISH cells was conducted by adding IFN at serial dilutions to the growth medium in flat-bottomed microtiter plates and monitoring the cell density after 72 h by crystal violet staining.
The 50% activity concentrations (c50) as well as the sensitivities of cells to increasing amounts of interferon were deduced from an IFN dose-response curve (Kaleidagraph; Synergy Software), using the following equation (equation 1): y = A0 + A/[1 + (c/c50)s], where y is the absorbance and reflects the relative number of cells, A0 is the offset, A is the amplitude, c is the IFN concentration, and s is the slope.
The experimental error (σ) for both assays was 35%. Therefore, a 2σ confidence level would suggest that differences smaller than twofold between interferons are within the experimental error.
Fluorescence-activated cell sorting (FACS) analysis of cell surface receptors.
293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. They were treated with 1 nM IFN for 2 h, detached with 5 mM EDTA in phosphate-buffered saline, and resuspended in phosphate-buffered saline containing 3% fetal bovine serum and 0.5 mM EDTA. The mouse monoclonal antibodies (MAbs) AA3-AB2 and D5, specific for IFNAR1 or IFNAR2, were used at a concentration of 3 μg/ml. The signal was amplified with biotinylated rat anti-mouse IgG (Jackson Immunochemicals) and streptavidin-allophycocyanin (BD Biosciences). Samples were analyzed with a FACSCalibur instrument in the presence of propidium iodide to exclude dead cells.
IFN-induced luciferase activity.
The HL116 cell clone was derived from the HT1080 cell line after stable transfection of a plasmid carrying the luciferase gene controlled by the IFN-inducible 6-16 promoter. Cells were treated for 6 h with increasing doses of IFN and were lysed. The luciferase activity was quantified in a luminometer (EG&E Berthold) as described previously (27). The experiment was performed in replicates for each concentration.
Spotted oligonucleotide microarray experiments.
Poly-l-lysine-coated glass microarrays containing >18,000 different probes (human oligonucleotide set; Compugen) were purchased from the Center for Applied Genomics, New Jersey. The microarrays were probed with a mixture consisting of cyanine 3 (Cy3)- or Cy5-labeled cDNAs, representing IFN treatment versus no treatment (control). RNAs (100 μg) extracted from WISH cells (RNeasy Midi kit; QIAGEN) following IFN treatment or no treatment as a control were subjected to reverse transcription (Moloney murine leukemia virus H-minus point mutant reverse transcriptase enzyme; Promega) with aminoallyl-modified dUTP nucleotide (aa-dUTP; Ambion) at a 4:1 aa-dUTP-to-dTTP ratio. The cDNAs were labeled with an N-hydroxysuccinimide-activated Cy3 or Cy5 fluorescent probe (Amersham) through aa-dUTP. Labeled cDNAs from treatment and control cells were mixed with equivalent amounts of fluorescent dye (100 pmol each) in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.08% SDS, 6 μl of blocking solution (Amersham), and water to 100 μl. This target mixture was denatured at 95°C for 3 min, chilled, and applied between a raised coverslip (LifterSlip; Erie Scientific Company) and the array. The slide was then sealed in a microarray hybridization chamber and submerged in a darkened water bath set at 55°C for hybridization. After 12 h, the slide was washed for 5 minutes in 2× SSC-0.5% SDS at 55°C, 5 minutes in 0.5× SSC at room temperature, and 5 minutes in 0.05× SSC at room temperature. It was then quickly dried by spinning for 3 min at 1,000 rpm and stored in the dark until scanned. Each condition (IFN treatment) was represented by two dye-swap microarray replicates.
Microarray image and data analysis.
Scanning of the hybridized microarrays was performed with a DNA microarray scanner (Agilent Technologies), which excites the Cy3 and Cy5 fluorophores at 532 nm and 635 nm, respectively, with a spatial resolution of 10 μm. Automatic spot detection and green and red fluorescent signal quantitation for each spot were done with SpotReader software (Niles Scientific). Automatic background subtraction, data normalization, processing (e.g., log transformation of ratios), filtering, and cluster analysis were done with GeneSpring software (Silicon Genetics). To correct for possible differences in input RNA concentrations and in the Cy3 and Cy5 labeling efficiencies, the treatment signal was normalized to that of the control channel, using the ratio of the total signal from both channels (Loess normalization) per subarray (12-by-4 separated blocks in the array) to also account for spotting pin variations. The ratio between the normalized and control signals was taken as the level of gene induction. Data were filtered using SpotReader output flags, an indication of the quality of the spot, and by the distance between replicates for each gene.
The starting list of IFN-modulated genes was composed of genes whose expression was beyond a 1.7-fold threshold under at least two conditions, tolerating only one absent (A) flag (GeneSpring translation of SportReader flagging), indicative of a “noisy” spot, under all conditions per given gene. This criterion was chosen after the realization that genuine up- or down-regulation was always observed for more than one IFN treatment, so choosing two conditions was a minimum. A low-stringency threshold was necessary to include IFN-stimulated genes (ISGs) modified at relatively low levels. This gene subset was exported into an Excel worksheet, together with the mean treatment-to-control ratio value, the individual replicate values, the t test P value, which is an estimate of the microarray technical error based on the distance between replicates, and the flag codes. Genes were then screened for significance by comparing the distances between replicates and between conditions and also considering P values and flags. This manual analysis was necessary because statistical selection by analysis of variance left out clearly IFN-modified genes, i.e., gave false-negative results, because of the small number of replicates per condition. Mean values beyond the 1.7-fold threshold but having one replicate below this threshold, with no other conditions with similar and significant levels, were considered products of technical noise. Genes with no significant up-/down-regulation under any condition were excluded. Cluster analysis was done on the final list after importing it back into GeneSpring.
RESULTS
Alanine-scanning mutagenesis of the IFNAR1 binding interface on IFN-α2 suggested that this weak protein-protein interaction (KD = 1.5 to 5 μM) (24, 40) lacks binding hotspots. However, three mutations were found to increase binding two- to fourfold (40). For this study, these three IFN-α2 mutations (H57A, E58A, and Q61A) were combined to create a triple IFN-α2 mutant protein, hereafter termed the HEQ mutant (Fig. 1A, red patch). The mutated residues are spatially adjacent and are located next to the six residues found to impair IFNAR1 binding upon mutation (Fig. 1A, blue patch). The HEQ mutant residues are located opposite the modeled binding site for IFNAR2.
Ternary complex stability is enhanced by the HEQ mutant compared to that induced by IFN-α2.
First, we characterized the biophysical properties of the HEQ mutant. The binding of IFNs to IFNAR1-EC or the formation of the ternary complex including IFNAR1-EC and IFNAR2-EC was studied by TIRFS. IFNAR1-EC and/or IFNAR2-EC was tethered to solidly supported lipid bilayers (Fig. 1C). The dissociation kinetics of the binary complexes of wild-type (wt) IFN-α2, the HEQ mutant, and IFN-β from IFNAR1-EC alone are compared in Fig. 1D. A 30-fold increase in complex lifetime (kd) was observed for the HEQ mutant and IFN-β compared to IFN-α2 (Table 1). In contrast, very similar association rate constants (ka) were obtained for all three species. From these data, an equilibrium dissociation constant of 150 nM was determined according to the equation KD = kd/ka for the HEQ mutant/IFNAR1-EC complex. This can be compared to the KD of 5 μM for wt IFN-α2 and 100 nM for IFN-β. The binding affinity of the HEQ mutant for IFNAR2-EC is similar to that measured for wt IFN-α2 (5 nM), which is about 10-fold weaker than that of IFN-β (Table 1). Thus, only the affinity of the HEQ mutant towards IFNAR1, and not that towards IFNAR2, is similar to that measured for IFN-β.
TABLE 1.
Rate constants and affinities of interactions of wt IFN-α2, IFN-β and the HEQ mutant with IFNAR1-EC and IFNAR2-ECd
| IFN subtype | Parameter with IFNAR1-EC
|
Parameter with IFNAR2-EC
|
||||
|---|---|---|---|---|---|---|
| ka (M−1 s−1) | kd (s−1) | KD (nM) | ka (M−1 s−1) | kd (s−1) | KD (nM) | |
| IFN-α2a | ∼2 × 105 | 1.0 ± 0.3 | ∼5,000 | 3 ± 1 × 106 | 0.015 ± 0.003 | 5 ± 2 |
| 0.3 ± 0.1c | 100 ± 40c | |||||
| IFN-βb | 3 × 105 ± 1 × 105 | 0.030 ± 0.006 | 100 ± 40 | >5 × 106c | 0.002 ± 0.001 | <0.4 |
| 0.016 ± 0.005c | <3c | |||||
| HEQ mutanta | 3 × 105 ± 1 × 105 | 0.045 ± 0.005 | 150 ± 50 | 3 × 106 ± 1 × 106 | 0.015 ± 0.005 | 5 ± 2 |
| 0.3 ± 0.1c | 100 ± 40c | |||||
S136C mutant site-specifically labeled with Oregon Green 488 maleimide.
Site-specifically labeled with Oregon Green 488 maleimide at the free C17 residue.
Interaction with IFNAR2-EC I47A mutant. This mutant was used to probe IFN-β-induced ternary complex formation to put the IFNAR2 dissociation rate constant on par with that of IFN-α2.
As measured using TIRFS (see Fig. 1). Values are means ± standard deviations.
IFN induces biological activity by the formation of a ternary complex (IFNAR1-IFN-IFNAR2) in a 1:1:1 stoichiometry (3). Because of cooperative binding to the two surface-bound receptor subunits, the apparent ligand binding affinity for the ternary complex is higher than binding to either receptor alone and depends on the relative and absolute receptor surface concentrations (18, 25). The increased stability of the ternary complex was probed by measuring the IFN dissociation kinetics. The dissociation of IFN-α2, the HEQ mutant, and IFN-β was measured at different concentrations of IFNAR1-EC and IFNAR2-EC at stoichiometric ratios (Fig. 1E to G). Since we intended to compare the effects of the affinities of IFNs towards IFNAR1 on binding to the ternary complex, we used the I47A mutant of IFNAR2 for IFN-β measurements. The dissociation rate constant of IFN-β towards I47A IFNAR2 is exactly that of IFN-α2 towards wt IFNAR2 (Table 1). While for IFN-α2 the dissociation kinetics strongly depends on the receptor surface concentration (Fig. 1E), overlaying slow dissociation curves were observed for IFN-β and the HEQ mutant (Fig. 1F and G), demonstrating the higher efficiency of IFNAR1-EC recruitment to the ternary complex by IFN-β and the HEQ mutant. The concentration-dependent dissociation of wt IFN-α2 suggests that at sufficiently high receptor concentrations, it would dissociate as slowly as IFN-β or the HEQ mutant. Thus, the efficiency of ternary complex formation is a combined function of surface receptor concentrations and binding affinities towards the individual receptors.
Down-regulation of IFNAR receptors upon IFN induction.
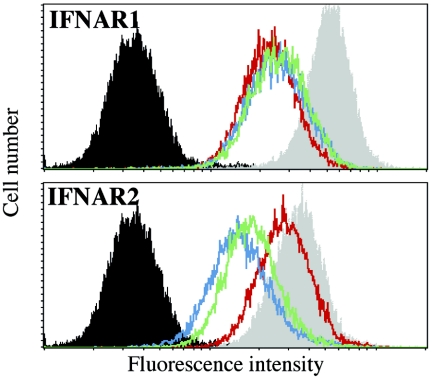
It was shown previously that the level of surface expression of the IFNAR1 receptor is down-regulated upon the addition of IFN-α2 (16, 23) and that the receptor complex formed by IFN-β is more stable than that formed by IFN-α (45). For this study, we measured the relative levels of expression of both the IFNAR1 and IFNAR2 receptor components upon treatment of 293T cells with 1 nM wt IFN-α2, HEQ mutant IFN, or IFN-β for 2 h. The level of IFNAR1 surface expression was reduced approximately twofold under these conditions for all three IFNs (Fig. 2, top panel). In contrast, IFNAR2 levels were reduced only by the addition of IFN-β or the HEQ mutant but not by wt IFN-α2 (Fig. 2, bottom panel). This result suggests that down-regulation of IFNAR2 is due to the increased stability of the ternary complex.
FIG. 2.
IFN-induced down-regulation of IFNAR surface receptors. 293T cells were treated with 1 nM of wt IFN-α2, the HEQ mutant, or IFN-β for 2 h. Cell surface levels of IFNAR1 and IFNAR2 were quantified by FACS analysis using AA3 and D5 MAbs, respectively. Black filled area, isotypic control; gray filled area, untreated cells; red line, wt IFN-α2; green line, HEQ mutant; blue line, IFN-β.
Antiviral and antiproliferative potencies.
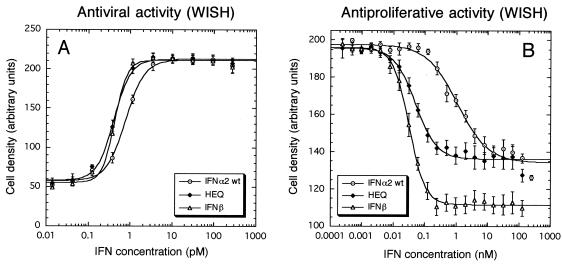
Antiviral and antiproliferative activities of interferons were measured with WISH cells (Fig. 3). Again, the HEQ mutant activity profile was comparable to that of IFN-β. Despite the large increase in binding affinity of the HEQ mutant towards IFNAR1, its antiviral potency was only twofold higher than that of wild-type IFN-α2. In contrast, the HEQ mutant's antiproliferative activity was increased ∼25-fold, which was comparable to the 42-fold increase for IFN-β (Table 2). The increased antiproliferative activities correlated with the increased affinities of these two IFNs towards IFNAR1. In addition, we found that the slope (s) in equation 1 was significantly steeper for IFN-β than for either IFN-α2 or the HEQ mutant (Fig. 3B and Table 2). Moreover, the recorded cell density using IFN-β was much lower than that with IFN-α2 or the HEQ mutant, even at the highest concentrations (Fig. 3B; note the difference in minimal absorbance values). The differences in slope and cell density relate to different rates of activation of the antiproliferative response, which is the only aspect in which the HEQ mutant behaves like wt IFN-α2 and is distinguished from IFN-β.
FIG. 3.
Antiviral and antiproliferative responses of WISH cells to IFN treatment. Each dot represents the mean value of six independent antiviral (A) or antiproliferative (B) experiments. The curves were fitted using equation 1.
TABLE 2.
Relative biological activities of wild-type IFN-α2, HEQ mutant, and IFN-β
| IFN subtype | Antiviral activity
|
Antiproliferative activity
|
||||||
|---|---|---|---|---|---|---|---|---|
| 50% (pM) | Ratioa | Maximumb ratio | Slopeb ratio | 50% (nM) | Ratioa | Maximumb ratio | Slopeb ratio | |
| IFN-α2 | 0.84 | 1 | 1 | 1 | 1.23 | 1 | 1 | 1 |
| HEQ mutant | 0.41 | 2.0 | 1 | 0.88 | 0.05 | 25 | 1.01 | 0.73 |
| IFN-β | 0.43 | 2.0 | 1 | 0.72 | 0.03 | 43 | 0.83 | 0.59 |
| P valuec | 0.00014 | 0.00014 | NS | 0.019 | <0.0001 | <0.0001 | 0.00038 | 0.012 |
Ratios are relative to wild-type IFN-α2 activities.
The maximal activity (A) and slope (s) were obtained from the dose-response curve fit (equation 1).
By analysis of variance. NS, not significant.
Expression regulation of 6-16 promoter.
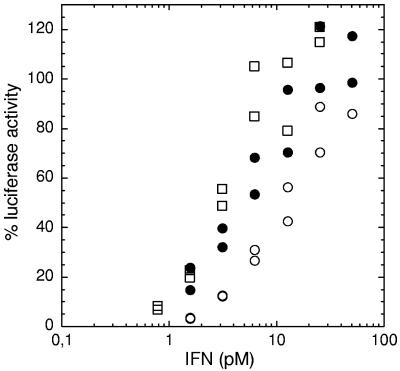
One of the more prominent IFN-stimulated response element-driven inducible genes is the abundantly expressed 6-16 mRNA, which has been classically used as a marker for STAT1 and STAT2 activation by type I IFNs, which, together with IRF9, will form the ISGF3 transcription factor (30). For this study, we treated HL116 cells (HT1080 cells expressing the luciferase gene controlled by the 6-16 promoter) with wt IFN-α2, the HEQ mutant, or IFN-β for 6 h and then measured the luciferase activity. The level of activity was expressed relative to that in untreated cells (0%) or cells treated with an excess of IFN (100%). Fifty percent activity was achieved by adding a 4 pM concentration of either the HEQ mutant or IFN-β or 10 pM wt IFN-α2 (Fig. 4). This difference in the midpoint of activation of the 6-16 promoter was in line with the difference in antiviral potencies between these IFNs but much below the difference in antiproliferative activities.
FIG. 4.
Expression regulation of the 6-16 promoter upon IFN induction. HL116 cells (HT1080 cell line expressing the luciferase gene controlled by the 6-16 promoter) were treated with wt IFN-α2 (open circles), the HEQ mutant (filled circles), or IFN-β (open squares) for 6 h. The luciferase activities are expressed relative to that in untreated cells (0%) or cells treated with an excess of IFN (100%).
The HEQ mutant induces an ISG expression profile similar to that of IFN-β.
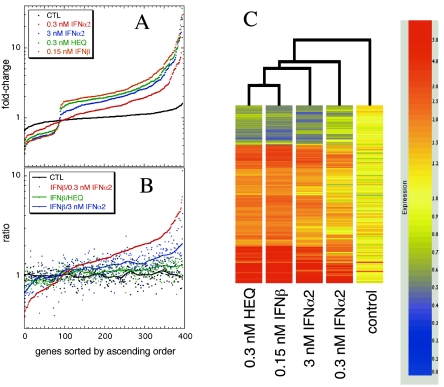
Gene microarray technology has been used for monitoring ISG expression (11, 12, 21, 26). For this study, the effects of IFNs on gene expression were monitored by spotted oligonucleotide microarray experiments. WISH cells were incubated for 16 h with 0.3 nM IFN-α2 (1,000 antiviral units [IU]/ml), 3 nM IFN-α2, 0.3 nM HEQ mutant, or 0.15 nM IFN-β (1,000 IU/ml). The last three promoted >90% antiproliferative responses in WISH cells, while for 0.3 nM wt IFN-α2 the antiproliferative response was below 10% (Fig. 3B), allowing a correlation between biological response and gene activation profiles. Each condition was represented by two dye-swap experiments of IFN treatment versus no treatment. In addition, four replicates of microarray experiments consisting of no treatment versus no treatment were used as controls. The ratio between the normalized and control signals was taken as the level of gene induction. The modified gene subset, defined by a 1.7-fold threshold in at least two different IFN treatments (for details, see Materials and Methods), was composed of 395 expression-modulated genes (data available on request). In Fig. 5A, the IFN-induced expression levels of these genes were plotted in ascending order according to their relative expression. The black dots represent the ratios between the two channels on the control chip, which provide an estimate of the random fluctuation; the low levels of random fluctuation in the control experiment (0.65- to 1.6-fold) provide a high degree of confidence in the quality of the data.
FIG. 5.
Effects of IFN on gene expression, as monitored by spotted oligonucleotide microarray experiments. We compared the expression profiles of four conditions consisting of WISH cells incubated for 16 h with 0.3 nM wt IFN-α2, 3 nM wt IFN-α2, 0.3 nM HEQ mutant, and 0.15 nM IFN-β. Each profile represents an experiment with IFN treatment versus no treatment with dye-swap microarray duplicates. In addition, a control expression profile was obtained from the ratio of both channels for untreated cells. (A) Changes in expression levels of IFN-stimulated 395-gene subset (data available on request), plotted in ascending order separately for each condition. (B) Expression levels relative to gene expression upon adding 0.15 nM IFN-β. Genes from the three treatment experiments and the control were sorted according to ascending order of IFN-β/IFN-α2 (0.3 nM) ratios. The trend lines were created with a 15% data smoothing function in Kaleidagraph (Synergy Software). (C) Cluster analysis, with distance correlation between genes and Spearman correlation between conditions (GeneSpring software).
In general, the gene expression levels for IFN-β and the HEQ mutant were similar (Fig. 5A), while those for wt IFN-α2 (0.3 nM) were significantly lower. Applying 3 nM wt IFN-α2 gave intermediate levels of gene expression. Figure 5B shows the levels of induction of all IFN-induced genes plotted relative to the levels of induction upon treatment with IFN-β. The genes were sorted in ascending order according to the ratios of expression induced by IFN-β to that induced by 0.3 nM IFN-α2 (IFN-β/0.3 nM IFN-α2 ratios). The figure shows that on a per-gene basis, the variation in expression levels between IFN-β and the HEQ mutant was similar to the random noise, as determined from the control (0.65- to 1.6-fold change). Also, comparing the relative expression levels induced by 0.3 and 3 nM wt IFN-α2 showed that while the levels were higher for the latter, the same set of differentially induced genes for 3 nM IFN-α2 was also differentially induced by 0.3 nM IFN-α2, albeit to a lesser extent. Cluster analysis of the gene activation profiles was done with GeneSpring analysis software. Figure 5C shows that the gene expression profiles for IFN-β and the HEQ mutant are the closest, followed by that for treatment with 3 nM wt IFN-α2. Again, the gene expression profile for 0.3 nM wt IFN-α2 is farther away. The gene expression profiling results are in excellent agreement with the differential biological activities observed for IFN-β and IFN-α2, also demonstrating that the HEQ mutant is an IFN-α2 with characteristics of IFN-β.
Table 3 lists a group of genes selected from the total data (data available on request) according to their differential levels of activation or functional impact. The functional classification in Table 3 results from the manual integration of gene ontology (data available on request) gene functions found at the GeneCards database (http://www.genecards.org/index.shtml) and through NCBI. The expression of most known ISGs was affected by all IFN treatments. However, 9-27 (IFITM1), a well-defined ISG, was not induced by any of the IFN conditions. This can be explained by a cell-type-specific IFN responsiveness, rather than technical error, resulting in a low signal-to-noise ratio in all eight IFN microarray experiments.
TABLE 3.
Gene expression levels from microarray experiments
| Functional group and GenBank accession no. | HUGOa name | Gene induction (fold)
|
||||
|---|---|---|---|---|---|---|
| IFN-β | HEQ mutant | IFN-α2 (0.3 nM) | IFN-α2 (3 nM) | Control (no IFN) | ||
| Innate immunity | ||||||
| NM_002462 | MX1 | 68.3 | 56.8 | 6.1 | 16.3 | 1.0 |
| NM_001548 | IFIT1 | 29.4 | 27.5 | 14.0 | 25.6 | 1.2 |
| NM_006820 | IFI44L | 27.1 | 27.8 | 5.8 | 16.0 | 0.7 |
| M30818 | MX2 | 25.5 | 18.5 | 7.1 | 20.2 | 0.9 |
| NM_002535 | OAS2 | 25.1 | 36.9 | 14.0 | 25.7 | 1.1 |
| NM_014314 | RIG-I | 21.1 | 14.1 | 5.1 | 12.4 | 0.9 |
| NM_016816 | OAS1 | 13.5 | 10.1 | 10.0 | 15.8 | 1.0 |
| AF026941 | RSAD2 | 9.9 | 9.5 | 2.1 | 6.2 | 1.6 |
| NM_004510 | IFI75 | 9.2 | 7.3 | 3.8 | 6.4 | 1.0 |
| NM_003265 | TLR3 | 7.7 | 6.1 | 1.3 | 2.5 | 1.3 |
| NM_002198 | IRF1 | 4.8 | 3.6 | 1.9 | 2.6 | 1.1 |
| NM_002038 | G1P3 | 4.3 | 3.2 | 3.1 | 4.7 | 1.0 |
| NM_002468 | MYD88 | 4.2 | 3.2 | 2.0 | 3.5 | 1.0 |
| NM_001549 | IFIT4 | 3.7 | 3.3 | 5.5 | 6.0 | 1.1 |
| U72882 | IFI35 | 3.6 | 3.1 | 7.4 | 11.8 | 1.2 |
| NM_004031 | IRF7 | 3.5 | 3.8 | 1.8 | 2.5 | 0.8 |
| NM_002759 | PRKR | 3.0 | 2.9 | 2.1 | 2.8 | 1.1 |
| NM_002053 | GBP1 | 2.9 | 2.0 | 1.2 | 2.1 | 1.0 |
| Acquired immunity | ||||||
| NM_018950 | HLA-F | 18.5 | 12.5 | 4.2 | 16.7 | 1.0 |
| NM_000592 | C4B | 6.4 | 4.7 | 2.0 | 2.6 | 1.1 |
| NM_004048 | B2M | 4.9 | 3.7 | 1.4 | 2.6 | 1.0 |
| M14584 | IL6 | 4.7 | 3.3 | 0.9 | 3.1 | 0.8 |
| JAK-STAT pathway | ||||||
| NM_007315 | STAT1 | 11.1 | 9.4 | 5.5 | 9.8 | 1.1 |
| NM_005419 | STAT2 | 2.8 | 2.3 | 1.2 | 2.1 | 0.8 |
| NM_006084 | ISGF3G | 2.0 | 2.0 | 2.2 | 3.7 | 1.1 |
| Cell growth inhibition | ||||||
| AF095844 | MDA5 | 15.0 | 10.7 | 5.0 | 8.7 | 1.0 |
| NM_005531 | IFI16 | 9.1 | 7.6 | 5.0 | 5.4 | 1.0 |
| NM_004585 | RARRES3 | 4.7 | 3.7 | 1.9 | 3.5 | 0.8 |
| NM_002539 | ODC1 | 0.51 | 0.51 | 0.58 | 0.55 | 0.90 |
| NM_005192 | CDKN3 | 0.47 | 0.53 | 0.57 | 0.46 | 1.00 |
| NM_004616 | TM4SF3 | 0.45 | 0.53 | 0.80 | 0.56 | 0.98 |
| Apoptosis | ||||||
| NM_003810 | TNFSF10 | 4.3 | 3.3 | 2.5 | 4.4 | 1.5 |
| NM_001225 | CASP4 | 3.6 | 2.7 | 1.8 | 2.6 | 0.7 |
| NM_001227 | CASP7 | 3.4 | 2.5 | 1.3 | 1.8 | 1.2 |
| NM_004760 | DRAK1 | 2.5 | 2.1 | 1.3 | 2.2 | 0.8 |
| NM_001230 | CASP10 | 2.1 | 1.9 | 1.1 | 1.4 | 1.1 |
| NM_006792 | MORF4 | 0.54 | 0.58 | 0.65 | 0.48 | 0.88 |
| NM_001344 | DAD1 | 0.48 | 0.59 | 0.62 | 0.52 | 1.00 |
| ISGylation and protein degradation | ||||||
| NM_017414 | USP18 | 15.8 | 14.4 | 6.9 | 13.5 | 1.0 |
| NM_002800 | PSMB9 | 14.1 | 12.7 | 4.2 | 11.4 | 0.9 |
| NM_002427 | MMP13 | 13.4 | 9.4 | 2.2 | 5.7 | 0.9 |
| NM_001223 | CASP1 | 8.6 | 6.9 | 3.0 | 5.7 | 1.0 |
| NM_004159 | PSMB8 | 5.2 | 5.0 | 2.8 | 4.4 | 1.2 |
| NM_005101 | ISG15 | 5.0 | 6.9 | 5.9 | 12.3 | 0.9 |
| NM_003335 | UBE1L | 4.6 | 4.0 | 2.5 | 3.8 | 0.9 |
| NM_002818 | PSME2 | 3.4 | 3.3 | 1.9 | 2.5 | 0.9 |
| NM_002801 | PSMB10 | 3.3 | 2.8 | 1.6 | 2.7 | 1.0 |
| Protein synthesis inhibition | ||||||
| NM_001959 | EEF1B2 | 0.47 | 0.55 | 0.58 | 0.52 | 0.99 |
| NM_001417 | EIF4B | 0.32 | 0.35 | 0.52 | 0.47 | 0.96 |
Naming convention by HUGO (http://www.gene.ucl.ac.uk/nomenclature).
The effects of IFN-β and the HEQ mutant on the expression of most of the genes were much stronger than the effects induced by IFN-α2 at equivalent antiviral units. This trend was also observed at other time points (unpublished data). For a few interesting exceptions, namely, IFIT4 and IFI35, upregulation by 0.3 nM IFN-α2 was consistently lower than that by IFN-β and the HEQ mutant. The stronger IFN-β effect on cell growth can be correlated with the earlier induction/repression of many genes involved in proliferation or apoptosis (e.g., repression of the apoptosis inhibitor DAD1 and ornithine decarboxylase [ODC1], a gene involved in cell growth and proliferation control whose overexpression was linked to tumor progression). In addition, nine histone genes were up-regulated, and two were down-regulated (unpublished data), which might correlate with changes in genomic DNA structure during cell growth inhibition or apoptosis. Still, not a single gene was found which was strongly up- or down-regulated by either the HEQ mutant or IFN-β but not regulated by wt IFN-α2. For example, the MX1 gene was up-regulated 60-fold by IFN-β and the HEQ mutant, but only 6- and 16-fold by 0.3 nM and 3 nM wt IFN-α2. The TLR3 gene was up-regulated 7.7-fold by IFN-β and 6.1-fold by the HEQ mutant, but only 1.3- and 2.5-fold by 0.3 nM and 3 nM wt IFN-α2.
DISCUSSION
The question of whether IFNs have differential activities has been asked repeatedly since the different IFN genes have been discovered. Most of the attention has gone toward investigating whether the response to IFN-β is different from that induced by IFN-α (1, 7, 11, 15, 17). At the level of receptor recruitment, a prominent feature of IFN-β compared to IFN-α2 is an ∼50-fold higher affinity towards IFNAR1 (18). For this study, we have engineered an IFN-α2 triple mutant (with H57A, E58A, and Q61A mutations) that binds IFNAR1 30-fold tighter than the wild-type protein. Thus, the binding affinity of the HEQ mutant for IFNAR1 is about equal to that of IFN-β, although its binding affinity for IFNAR2 is 10-fold lower. Strikingly, the HEQ mutant exhibits several functional characteristics of IFN-β, including (i) substantially increased antiproliferative but only slightly changed antiviral activity and ISGF3 formation; (ii) ligand-induced down-regulation of IFNAR2 cell surface expression; and (iii) similar patterns of gene activation for the HEQ mutant and IFN-β. Our study clearly demonstrates that important functional differences between IFN-β and wt IFN-α2 are due to their different affinities for IFNAR1. A higher affinity for IFNAR1 increases the apparent binding affinity of the cellular receptor, but furthermore, it has the following consequences on ternary complex stability: (i) more efficient recruitment of IFNAR1 and (ii) a longer lifetime of the ternary complex, as seen in our biophysical studies (Fig. 1). The fact that differential signal activation of IFNs apparently depends on receptor cell surface concentrations and can be compensated for by the ligand concentration suggests that the affinity towards IFNAR1 can limit ternary complex formation, which leads to higher potencies of IFN-β and the HEQ mutant than that of IFN-α2. Interestingly, differential recruitment of IFNAR1 affects the antiproliferative response much more strongly than the antiviral response, leading to differential response patterns at low ligand concentrations. Thus, the signal transduction pathways required for antiproliferative activity make use of a stable ternary complex, while ISGF3 formation requires only a transient receptor complex. Studying effector recruitment in vivo will be required for elucidating the molecular basis of these differences, and the HEQ mutant will be a powerful tool for such studies. Furthermore, our results confirm previous reports that the differential activity of IFN-β (and also the HEQ mutant) is not qualitative but can only be observed at specific ligand concentrations. This was observed for both biological activities and gene induction patterns.
The HEQ mutant boosts binding affinity and biological activity, similar to IFN-β. In a previous study, we analyzed separately the three IFN-α2 mutations that comprise the mutations in the HEQ mutant (40). The individual mutations increased binding to IFNAR1 and biological activity two- to fivefold. For the H57A and Q61A mutants, which had twofold increased binding, a similar increase in biological activity was observed. However, for the E58A mutant, which had a fivefold increase in affinity for IFNAR1, some differential increase in antiproliferative over antiviral activity was observed (3.3- versus 5.4-fold). Taken together with the HEQ mutant data presented here, this suggests a clear maximum for increased antiviral activity stemming from increased IFNAR1 binding.
Two additional observations are important for understanding the mechanisms of IFN activation. One is that in our experimental system, some IFN-induced activities are not differentiated between IFN-α2 and IFN-β, while others are. The second is that different IFN concentrations activate the antiviral versus the antiproliferative response in WISH cells (picomolar versus nanomolar concentrations), albeit for IFN-α2 the difference is 1,000-fold, whereas for IFN-β or the HEQ mutant the difference is only 50-fold. In Daudi cells, low concentrations of IFNs (pM) already cause growth arrest, which may be linked to higher levels of the interferon receptors on the surfaces of Daudi cells than on WISH cells (6). A recent report has suggested that tumor cells have a reduced level of interferon receptors, making them resistant to the antiproliferative activity of IFNs (49). Thus, it seems that interferon receptor concentrations affect the biological activities of IFNs. This model would nicely explain the IFN-β-specific activity in a Tyk2-deficient background which results in a reduced number of IFNAR1 receptors on the cell surface (38). Figure 1 clearly shows that at low IFNAR1 concentrations, the ternary complex is formed only with the HEQ mutant and IFN-β, which bind IFNAR1 tightly, and not with IFN-α2. Indeed, the native, low receptor surface concentrations have been suggested to limit ternary complex formation (50).
Surprisingly, the three mutated residues of the HEQ mutant are fully conserved between all IFN-α subtypes. Since these mutations cause an increase in antiproliferative activity but only a small change in antiviral potency, one may conclude that IFN-α subtypes are not optimized to promote strong antiproliferative activity. This conclusion is supported by the similar binding affinities and biological activities found for IFN-α2 and four other alpha interferon subtypes (alpha1, alpha 8, alpha16, and alpha21) (J. Piehler and G. Uze, unpublished data). This should not be surprising, as the foremost activity of IFN-α is to induce an antiviral state, whereas IFN-β, which is specifically induced in response to nonviral inducers such as Toll-like receptor stimulation (4), is most likely to be involved in subtle regulations of cellular differentiation of the monocyte lineage (5, 46) that could be reflected by the antiproliferative activity on cell lines. Of course, for a drug against a disease such as cancer, an increased antiproliferative activity of IFN would be desired. Our data suggest that by further increasing the affinity of IFN for IFNAR1, such an increased activity could be achieved, even in cancer cells that have lost their responsiveness to IFNs by receptor down-regulation (49).
Our proposed mechanistic model for the establishment of differential IFN effects might be similar to other signaling systems. For example, a viral mimic of interleukin-10 (vIL-10) has been identified, which confers only a subset of activities found for IL-10 (28). In this case, differential signal activation by vIL-10 was not due to the structure of the complex, which is similar, but was ascribed to the ∼200-fold lower affinity toward IL-10 receptor 1 (IL-10R1) (51). Indeed, a single mutation of vIL-10 increasing the affinity toward IL-10R1 was able to recover IL-10-like activities (13). Strikingly, the activity of vIL-10 also critically depends on the cell surface concentration of IL-10R1 (14), confirming the key role of binding affinities for receptor recruitment to the plasma membrane. A second example of differential activation would be the activities of insulin and insulin-like growth factor. It was shown that, depending on their concentrations, insulin and insulin-like growth factor can bind to either their homologous or heterologous receptors and trigger different biological responses, which also depend on the surface receptor concentration (10, 47).
A more unified model of signal transduction promoted by IFN emerges from our results: we suggest that differential activation is a function of surface receptor concentrations and of binding affinities towards individual receptors. The assumption made in previous reports that the uniqueness of IFN-β is a consequence of structural differences in the receptor complex or that IFN-β binds additional factors (15, 27) does not fit with the results obtained with the HEQ mutant. In light of our findings, it was interesting to reanalyze the work of Runkel et al. (44) describing the loss of IFN-β-specific activities for mutants with mutations at positions 86 and 92. These mutations, which are located at the IFNAR1 binding site on interferon (20, 40, 43), apparently reduce the binding affinity for IFNAR1, and hence it is not surprising that the mutants now have the characteristics of IFN-α. Our data also demonstrate that the binding affinity towards IFNAR1 is directly related to the antiproliferative activity of IFN. The gene activation pattern also suggests a linear response to the IFN concentration and binding affinity. All genes upregulated by the HEQ mutant or IFN-β are also upregulated by wt IFN-α2, although at higher ligand concentrations. All of these observations support a relatively simple mechanism of induction of differential responses by differential recruitment of IFNAR1, which is a limiting factor in ternary complex formation. The next stage of study is to elucidate which signaling pathways are sensitive to differential IFNAR1 recruitment and how this is converted into complex biological responses.
Acknowledgments
We thank Olga Davydov (WIS) and Ron Ophir (WIS) for their guidance in setting up and analyzing the microarray experiments, Renne Abramovich (WIS) for help with protein purification, and Laura Runkel (Biogen Inc.) for the gift of anti-IFNAR1 and two MAbs.
This work was supported by Human Frontier Science Program grant RGP 60/2002.
REFERENCES
- 1.Abramovich, C., J. Chebath, and M. Revel. 1994. The human interferon alpha-receptor protein confers differential responses to human interferon-beta versus interferon-alpha subtypes in mouse and hamster cell transfectants. Cytokine 6:414-424. [DOI] [PubMed] [Google Scholar]
- 2.Allen, G., and M. O. Diaz. 1996. Nomenclature of the human interferon proteins. J. Interferon Cytokine Res. 16:181-184. [DOI] [PubMed] [Google Scholar]
- 3.Brierley, M. M., and E. N. Fish. 2002. Review: IFN-alpha/beta receptor interactions to biologic outcomes: understanding the circuitry. J. Interferon Cytokine Res. 22:835-845. [DOI] [PubMed] [Google Scholar]
- 4.Coccia, E. M., M. Severa, E. Giacomini, D. Monneron, M. E. Remoli, I. Julkunen, M. Cella, R. Lande, and G. Uze. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34:796-805. [DOI] [PubMed] [Google Scholar]
- 5.Coelho, L. F., G. M. de Freitas Almeida, F. J. Mennechet, A. Blangy, and G. Uze. 2005. Interferon-α and -β differentially regulate osteoclastogenesis: role of differential induction of chemokine CXCL11 expression. Proc. Natl. Acad. Sci. USA 102:11917-11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, B., D. Novick, S. Barak, and M. Rubinstein. 1995. Ligand-induced association of the type I interferon receptor components. Mol. Cell. Biol. 15:4208-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook, J. R., C. M. Cleary, T. M. Mariano, L. Izotova, and S. Pestka. 1996. Differential responsiveness of a splice variant of the human type I interferon receptor to interferons. J. Biol. Chem. 271:13448-13453. [DOI] [PubMed] [Google Scholar]
- 8.Croze, E., D. Russell-Harde, T. C. Wagner, H. Pu, L. M. Pfeffer, and H. D. Perez. 1996. The human type I interferon receptor. Identification of the interferon beta-specific receptor-associated phosphoprotein. J. Biol. Chem. 271:33165-33168. [DOI] [PubMed] [Google Scholar]
- 9.Cutrone, E. C., and J. A. Langer. 1997. Contributions of cloned type I interferon receptor subunits to differential ligand binding. FEBS Lett. 404:197-202. [DOI] [PubMed] [Google Scholar]
- 10.De Meyts, P. 2004. Insulin and its receptor: structure, function and evolution. Bioessays 26:1351-1362. [DOI] [PubMed] [Google Scholar]
- 11.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 13.Ding, Y., L. Qin, S. V. Kotenko, S. Pestka, and J. S. Bromberg. 2000. A single amino acid determines the immunostimulatory activity of interleukin 10. J. Exp. Med. 191:213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding, Y., L. Qin, D. Zamarin, S. V. Kotenko, S. Pestka, K. W. Moore, and J. S. Bromberg. 2001. Differential IL-10R1 expression plays a critical role in IL-10-mediated immune regulation. J. Immunol. 167:6884-6892. [DOI] [PubMed] [Google Scholar]
- 15.Domanski, P., O. W. Nadeau, L. C. Platanias, E. Fish, M. Kellum, P. Pitha, and O. R. Colamonici. 1998. Differential use of the betaL subunit of the type I interferon (IFN) receptor determines signaling specificity for IFNalpha2 and IFNbeta. J. Biol. Chem. 273:3144-3147. [DOI] [PubMed] [Google Scholar]
- 16.Dupont, S. A., S. Goelz, J. Goyal, and M. Green. 2002. Mechanisms for regulation of cellular responsiveness to human IFN-beta1a. J. Interferon Cytokine Res. 22:491-501. [DOI] [PubMed] [Google Scholar]
- 17.Fleischmann, C. M., and W. R. J. Fleischmann. 1988. Differential antiproliferative activities of IFNs alpha, beta and gamma: kinetics of establishment of their antiproliferative effects and the rapid development of resistance to IFNs alpha and beta. J. Biol. Regul. Homeost. Agents 2:173-185. [PubMed] [Google Scholar]
- 18.Gavutis, M., S. Lata, P. Lamken, P. Muller, and J. Piehler. 2005. Lateral ligand-receptor interactions on membranes probed by simultaneous fluorescence-interference detection. Biophys. J. 88:4289-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gresser, I., and F. Belardelli. 2002. Endogenous type I interferons as a defense against tumors. Cytokine Growth Factor Rev. 13:111-118. [DOI] [PubMed] [Google Scholar]
- 20.Hu, R., J. Bekisz, H. Schmeisser, P. McPhie, and K. Zoon. 2001. Human IFN-alpha protein engineering: the amino acid residues at positions 86 and 90 are important for antiproliferative activity. J. Immunol. 167:1482-1489. [DOI] [PubMed] [Google Scholar]
- 21.Khabar, K. S., L. Al-Haj, F. Al-Zoghaibi, M. Marie, M. Dhalla, S. J. Polyak, and B. R. Williams. 2004. Expressed gene clusters associated with cellular sensitivity and resistance towards anti-viral and anti-proliferative actions of interferon. J. Mol. Biol. 342:833-846. [DOI] [PubMed] [Google Scholar]
- 22.Kotenko, S. V., and J. A. Langer. 2004. Full house: 12 receptors for 27 cytokines. Int. Immunopharmacol. 4:593-608. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, K. G., W. Tang, A. K. Ravindranath, W. A. Clark, E. Croze, and S. Y. Fuchs. 2003. SCF(HOS) ubiquitin ligase mediates the ligand-induced down-regulation of the interferon-alpha receptor. EMBO J. 22:5480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamken, P., M. Gavutis, I. Peters, J. Van der Heyden, G. Uze, and J. Piehler. 2005. Functional cartography of the ectodomain of the type I interferon receptor subunit ifnar1. J. Mol. Biol. 350:476-488. [DOI] [PubMed] [Google Scholar]
- 25.Lamken, P., S. Lata, M. Gavutis, and J. Piehler. 2004. Ligand-induced assembling of the type I interferon receptor on supported lipid bilayers. J. Mol. Biol. 341:303-318. [DOI] [PubMed] [Google Scholar]
- 26.Leaman, D. W., M. Chawla-Sarkar, B. Jacobs, K. Vyas, Y. Sun, A. Ozdemir, T. Yi, B. R. Williams, and E. C. Borden. 2003. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J. Interferon Cytokine Res. 23:745-756. [DOI] [PubMed] [Google Scholar]
- 27.Lewerenz, M., K. E. Mogensen, and G. Uzè. 1998. Shared receptor components but distinct complexes for alpha and beta interferons. J. Mol. Biol. 282:585-599. [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., R. de Waal Malefyt, F. Briere, C. Parham, J. M. Bridon, J. Banchereau, K. W. Moore, and J. Xu. 1997. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J. Immunol. 158:604-613. [PubMed] [Google Scholar]
- 29.Mogensen, K. E., M. Lewerenz, J. Reboul, G. Lutfalla, and G. Uzè. 1999. The type I interferon receptor: structure, function and evolution of a family business. J. Interferon Cytokine Res. 19:1069-1098. [DOI] [PubMed] [Google Scholar]
- 30.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestka, S. 2000. The human interferon alpha species and receptors. Biopolymers 55:254-287. [DOI] [PubMed] [Google Scholar]
- 32.Pfeffer, L. M., C. A. Dinarello, R. B. Herberman, B. R. Williams, E. C. Borden, R. Bordens, M. R. Walter, T. L. Nagabhushan, P. P. Trotta, and S. Pestka. 1998. Biological properties of recombinant alpha-interferons: 40th anniversary of the discovery of interferons. Cancer Res. 58:2489-2499. [PubMed] [Google Scholar]
- 33.Philo, J. S., K. H. Aoki, T. Arakawa, L. O. Narhi, and J. Wen. 1996. Dimerization of the extracellular domain of the erythropoietin (EPO) receptor by EPO: one high-affinity and one low-affinity interaction. Biochemistry 35:1681-1691. [DOI] [PubMed] [Google Scholar]
- 34.Piehler, J., L. C. Roisman, and G. Schreiber. 2000. New structural and functional aspects of the IFN-receptor interaction revealed by comprehensive mutational analysis of the binding interface. J. Biol. Chem. 275:40425-40433. [DOI] [PubMed] [Google Scholar]
- 35.Piehler, J., and G. Schreiber. 1999. Biophysical analysis of the interaction of human Ifnar2 expressed in E. coli with IFNa2. J. Mol. Biol. 289:57-67. [DOI] [PubMed] [Google Scholar]
- 36.Piehler, J., and G. Schreiber. 1999. Mutational and structural analysis of the binding interface between type I interferons and their receptor ifnar2. J. Mol. Biol. 294:223-237. [DOI] [PubMed] [Google Scholar]
- 37.Platanias, L. C., and E. N. Fish. 1999. Signaling pathways activated by interferons. Exp. Hematol. 27:1583-1592. [DOI] [PubMed] [Google Scholar]
- 38.Ragimbeau, J., E. Dondi, A. Alcover, P. Eid, G. Uze, and S. Pellegrini. 2003. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J. 22:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rani, M. R., C. Gauzzi, S. Pellegrini, E. N. Fish, T. Wei, and R. M. Ransohoff. 1999. Induction of beta-R1/I-TAC by interferon-beta requires catalytically active TYK2. J. Biol. Chem. 274:1891-1897. [DOI] [PubMed] [Google Scholar]
- 40.Roisman, L. C., D. A. Jaitin, D. P. Baker, and G. Schreiber. 2005. Mutational analysis of the IFNAR1 binding site on IFNalpha2 reveals the architecture of a weak ligand-receptor binding-site. J. Mol. Biol. 353:271-281. [DOI] [PubMed] [Google Scholar]
- 41.Roisman, L. C., J. Piehler, J. Y. Trosset, H. A. Scheraga, and G. Schreiber. 2001. Structure of the interferon-receptor complex determined by distance constraints from double-mutant cycles and flexible docking. Proc. Natl. Acad. Sci. USA 98:13231-13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubinstein, S., P. C. Familletti, and S. Pestka. 1981. Convenient assay for interferons. J. Virol. 37:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Runkel, L., C. deDios, M. Karpusas, M. Betzenhauser, C. Muldowney, M. Zafari, C. D. Benjamin, S. Miller, P. S. Hochman, and A. Whitty. 2000. Systematic mutational mapping of sites on human interferon-b-1a that are important for receptor binding and functional activity. Biochemistry 39:2538-2551. [DOI] [PubMed] [Google Scholar]
- 44.Runkel, L., L. Pheffers, M. Lewerenz, D. Monneron, C. H. Yang, A. Murti, S. Pellegrini, S. Goelz, G. Uzè, and K. Mogensen. 1998. Differences in activity between a and b interferons explored by mutational analysis. J. Biol. Chem. 273:8003-8008. [DOI] [PubMed] [Google Scholar]
- 45.Russel-Harde, D., C. T. Wagner, D. H. Perez, and E. Croze. 1999. Formation of a uniquely stable type I interferon receptor complex by interferon b is dependent upon particular interactions between interferon b and its receptor and independent of tyrosine phosphorylation. Biochem. Biophys. Res. Commun. 255:539-544. [DOI] [PubMed] [Google Scholar]
- 46.Santini, S. M., P. T. Di, C. Lapenta, S. Parlato, M. Logozzi, and F. Belardelli. 2002. The natural alliance between type I interferon and dendritic cells and its role in linking innate and adaptive immunity. J. Interferon Cytokine Res. 22:1071-1080. [DOI] [PubMed] [Google Scholar]
- 47.Shymko, R. M., P. De Meyts, and R. Thomas. 1997. Logical analysis of timing-dependent receptor signalling specificity: application to the insulin receptor metabolic and mitogenic signalling pathways. Biochem. J. 326:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stahl, N., and G. D. Yancopoulos. 1993. The alphas, betas, and kinases of cytokine receptor complexes. Cell 74:587-590. [DOI] [PubMed] [Google Scholar]
- 49.Wagner, T. C., S. Velichko, S. K. Chesney, S. Biroc, D. Harde, D. Vogel, and E. Croze. 2004. Interferon receptor expression regulates the antiproliferative effects of interferons on cancer cells and solid tumors. Int. J. Cancer 111:32-42. [DOI] [PubMed] [Google Scholar]
- 50.Whitty, A., N. Raskin, D. L. Olson, C. W. Borysenko, C. M. Ambrose, C. D. Benjamin, and L. C. Burkly. 1998. Interaction affinity between cytokine receptor components on the cell surface. Proc. Natl. Acad. Sci. USA 95:13165-13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon, S. I., B. C. Jones, N. J. Logsdon, and M. R. Walter. 2005. Same structure, different function: crystal structure of the Epstein-Barr virus IL-10 bound to the soluble IL-10R1 chain. Structure (Cambridge) 13:551-564. [DOI] [PubMed] [Google Scholar]